Abstract
Purpose of review
In this review, we will discuss (i) how the recent advancements in digital technology and computational engineering are currently applied to nephropathology in the setting of clinical research, trials, and practice; (ii) the benefits of the new digital environment; (iii) how recognizing its challenges provides opportunities for transformation; and (iv) nephropathology in the upcoming era of kidney precision and predictive medicine.
Recent findings
Recent studies highlighted how new standardized protocols facilitate the harmonization of digital pathology database infrastructure and morphologic, morphometric, and computer-aided quantitative analyses. Digital pathology enables robust protocols for clinical trials and research, with the potential to identify previously underused or unrecognized clinically useful parameters. The integration of digital pathology with molecular signatures is leading the way to establishing clinically relevant morphoomic taxonomies of renal diseases.
Summary
The introduction of digital pathology in clinical research and trials, and the progressive implementation of the modern software ecosystem, opens opportunities for the development of new predictive diagnostic paradigms and computer-aided algorithms, transforming the practice of renal disease into a modern computational science.
Keywords: computational disease, convolutional neural network, deep learning, focal segmental glomerulosclerosis, morphometry, nephrotic syndrome, podocytes, structural feature extraction
INTRODUCTION
During the last decade, the rapid evolution of digital image technology has challenged the established light microscopy-based protocols. Virtual microscopy, encompassing telepathology and whole slide imaging (WSI), and digital pathology, a dynamic, image-based environment for the acquisition, management, and interpretation of disease information generated from WSI or other digitized images, are becoming the new standard of practice in the setting of clinical trials [1] and research [2▪▪,3,4▪]. Simultaneously, telepathology and WSI are consciously, but progressively, introduced to patient care [5–8]. In this review, we discuss how the modern software ecosystem is transforming clinical trials, research, and practice in nephropathology, opening opportunities for the development of new diagnostic paradigms, and computer-aided algorithms. The modernization of disease assessment, however, carries new challenges as well as new advances (Tables 1 and 2).
Table 1.
Advantages of digital pathology
| Digital pathology: advantages | |
|---|---|
| Work flow | Remote access by multiple users simultaneously |
| Application of multiple scoring systems in different studies | |
| Targeted adjudication process of annotated structures | |
| Limits multiple mailing of unreplaceable material | |
|
| |
| Long term investment | Permanent library of data/images available to multiple investigators, studies, and to test different approaches |
| Cyber space storage | |
| Abatement of costs in long term | |
|
| |
| Transparency | For intra and interworking groups/consortia collaborations in clinical research |
| For regulatory agencies in clinical trials | |
|
| |
| Standardization | Definition: ‘The process of implementing and developing technical standards can help to maximize compatibility, interoperability, repeatability and quality and to facilitate commoditization of formerly custom processes’ |
|
| |
| Globalization of the renal biopsy | Achieved by implementing, across different studies worldwide, sharable standardized digital platforms and protocols for disease material acquisition, uploading, organization in the digital pathology repositories, and analysis |
|
| |
| Accuracy | ‘Degree of closeness of measurements of a quantity to that quantity’s true value’. For example quantification of annotated (enumerated) structures |
|
| |
| Reproducibility | One of main principles of scientific methods – facilitated by remote access to same images by multiple observers assessing the same structures |
|
| |
| Quantitative disease | Term used for stereology and morphometry or when visual assessment is based on (digital) quantitative metrics. ‘Quantitative disease’ arises from a demand for quantitation, objectivity, awareness that parameters detectable with quantitative analysis would otherwise escape observation, consistency, reproducibility and standardization |
|
| |
| Modalities of assessment | Visual morphologic
|
|
| |
| Computational nephropathology | Quantitative disease generating biologically and clinically relevant information using mathematical models at the individual and population levels, leading to diagnostic and outcome predicting algorithms |
Goals:
| |
Table 2.
Challenges of digital pathology
| Digital pathology: challenges | |
|---|---|
| Build the infrastructure | Financial investment |
| Organizational – logistic module | |
| Participation of investigators | |
| Regulatory (i.e., IRBs) | |
| Training of all personnel involved | |
|
| |
| Technical standardization | Preanalytic
|
|
| |
| Standardization of protocols | Pathology material collection |
| Deidentification | |
| Organization of the digital pathology repository | |
| Disease assessment | |
| Recording of data | |
| Data management | |
|
| |
| Compliance with regulatory agenciesa | Food and Drug Administration
|
|
| |
| Data access and data sharing ethics | HIPAA (consent and anonymization) |
| Regulate intra and interconsortia collaboration | |
Code of conduct
| |
|
| |
| Overcome the pathologist’s fear | The microscope was used for >100 years |
| Perception that whole slide imaging is a disruptive technology | |
| Perception of loss of control over individual/local case set | |
| Fear of transparency/judgment | |
| Learning new skills | |
HIPAA, Health Insurance Portability and Accountability Act; IRB, Institutional Review Board.
United States only and limited to the use of digital pathology in clinical practice.
TRANSFORMING CHALLENGES INTO OPPORTUNITIES
Leveraging financial investments
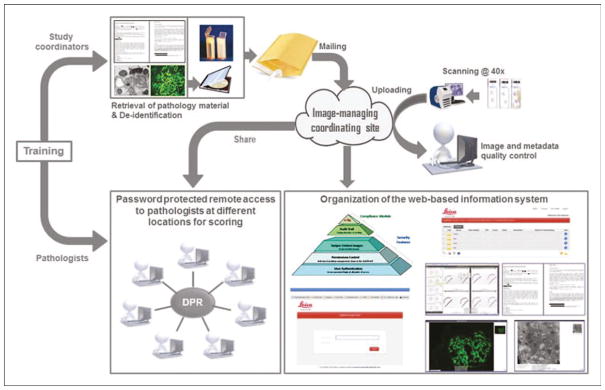
Digital pathology repositories (DPRs) exemplify a new cost-efficient form of systematic organization of resources (Fig. 1). Although the initial investment to establish a DPR may be significant, historical light microscopy-based protocols, requiring multiple mailing steps of disease material, interaction of numerous personnel, and repeated resubmissions across multiple studies, are reduced to a single mailing event. While glass slides may fade, break, be lost, and need to be warehoused, high-quality WSI are permanent and occupy ‘virtual’ space. Web-hosted image libraries can be accessed by multiple users, in underresourced locations, and for multiple purposes (e.g., to test different scoring systems simultaneously or consecutively), with abatement of costs with time and improved workflow [2▪▪,3,4▪].
FIGURE 1.
Overall workflow to establish a DPR.
Implementing new standards
The evolution of nephropathology from the ‘expert opinion’ model toward validated evidence-based models demands a robust pathology schema that can be carried across studies separated by distance in location and time. Digital pathology is a suitable platform for the implementation of the critical elements to achieve robustness: reproducibility, accuracy, standardization, objectivity of the observations, and comprehensiveness of the approach capturing the structural complexity of the disease studied under the form of quantifiable information.
One of the main challenges is standardization of protocols. Standardization of preanalytic and analytic phases is particularly critical in the setting of international large consortia and/or computational imaging applications, and assures that an adequate product (tissue properly handled, fixed, processed, cut, and stained) is generated by the histology laboratory, prior to scanning into WSI [2▪▪]. The standardization of these steps on large scale is often underestimated or not addressed because it is perceived as insurmountable. These challenges represent an opportunity for the nephropathology community to elaborate solutions applicable across laboratories.
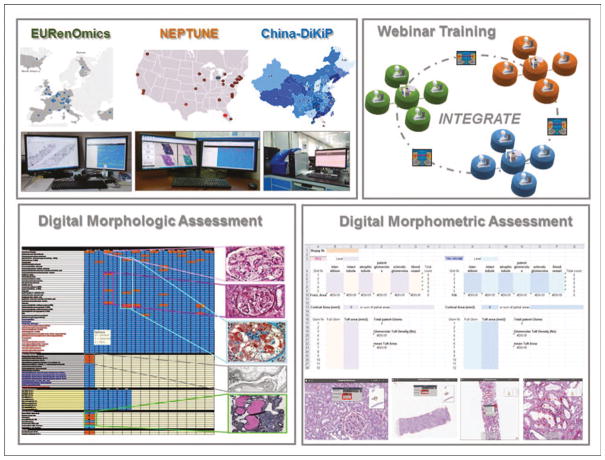
The availability of webinar-based meetings and WSIs accessible remotely by multiple investigators provides the opportunity for harmonization of repositories and quantitative assessment across institutions, regions, states, and continents [2▪▪]. Recently, several investigators focused on standardization and reproducibility of the postanalytic phase (disease analysis), recognizing that, while diagnostic scoring protocols and classification systems are continuously created, reproducibility of morphologic analysis remains problematic [2▪▪,3,9]. This led the NEPhrotic syndrome sTUdy NEtwork (NEPTUNE) investigators to exploit a DPR of renal biopsies to test different metrics and statistical approaches, with the goal of increasing accuracy [10▪] and reproducibility of data collection [11▪]. Utilizing NEP-TUNE as a model, the INTErnational diGital nephRopAThology nEtwork (INTEGRATE) investigators harmonized repositories in three different continents by sharing protocols and employed modern telecommunication, such as webinar-based meetings, to increase international reproducibility via consensus [2▪▪].
Regulating the new ecosystem
The introduction of new techniques and environments mandates that regulatory agencies and the medical community establish guidelines for approval of new devises, validation of new tests, and data access/sharing ethics.
Although the use of digital pathology in clinical research and trials is not under the control of the Food and Drug Administration (FDA), FDA approval is a requirement in the United States before a device such a scanning machine or a camera connected to a remote computer can be used in clinical practice. Recently, the first scanner was approved for primary diagnoses [12]. Furthermore, prior implementation of new clinical tests (i.e., the use of WSI or video images of glass slides), individual laboratories are also required to comply with the College of American Pathologists regulations to validate the performance of the test [8].
In the setting of large consortia for clinical research, data sharing, and analytic platforms that facilitate mining of different large-scale datasets are necessary [13▪▪]. Two glomerular diseases consortia, NEPTUNE and CureGN, adopted tranSMART, an open source software (transmartfoundation.org) originally developed by Johnson & Johnson (New Brunswick, New Jersey, USA) [14], as a platform for data mining and sharing.
Data sharing needs to be regulated. Data sharing ethics for genetic and genomic databases has been formalized, and could be transferred to other analytic platforms [15,16]. Conversely, intra and inter-consortia digital pathology-specific data sharing ethics and code of conduct have yet to be well defined (Table 2).
Training
Different factors modulate how a new technology is perceived; fortunately, almost invariably, the perception changes with time and gain of knowledge. Despite some initial resistance (see Table 2), overall pathologists’ acceptance of digital pathology as the new standard has increased [7], but investigators participation rate or enthusiasm needs continuous enticements, especially in low-budget circumstances. The establishment of any new digital pathology-based project also requires the implementation of new protocols for all categories of operators involved. With changes in the diagnostic ecosystem, the training curriculum for healthcare-associated personnel and pathologists must evolve accordingly. Future generations of pathologists will need to acquire new skills and languages to closely collaborate with computational engineers and statisticians, as their participation is integral to the creation of novel tools to extract actionable information from the large set of digital images generated and stored.
VIRTUAL MICROSCOPY IN CLINICAL PRACTICE
Both telepathology and WSI were recently implemented for patient care (Fig. 2). FDA approved telepathology systems, used for screening, primary diagnosis, second opinion, and education, can be designed as a static mode (images are captured, transmitted, and then viewed at distance), dynamic mode (live video images are transmitted and viewed at distance in real time), or dynamic robotic mode (when the viewer controls the microscope at the originating site) [17,18]. The practice of telepathology has been validated in surgical pathology (frozen section) and cytology [19,20▪▪,21–23], however, in renal pathology it is currently under testing. The two major indications that best fit the use of telepathology are assessment of adequacy of tissue procurement at the time of the renal biopsy (currently under validation at the University of Miami), and adequacy of renal parenchyma from frozen sections prior to transplantation.
FIGURE 2.

Virtual microscopy in clinical practice: telepathology (left panel) is used to evaluate frozen sections from cadaveric kidney transplant preimplantation. Frozen sections from a wedge biopsy is viewed with microscope connected to a camera for evaluation by a renal pathologist remotely located. Telepathology can also be used to evaluate adequacy of native and transplant kidney biopsies by placing the cores of fresh tissue on a glass slide. By using the microscope connected to a camera, the renal pathologist can remotely evaluate the percentage of cortex in the biopsy prior triaging and fixation. Whole slide images (right panel) are created from scanned glass slides, and stored in a web-accessible digital pathology repository to be shared with other renal pathologists for consultation, second opinion, or primary diagnosis (the later has still restricted use in the United States). An expert renal pathologist can access the repository remotely for visualization, assessment, and reporting of findings.
The routine use of WSI for clinical applications, already largely used in Europe [24▪▪], was tested and validated in selected centers in the United States [20▪▪,25]. Validation studies demonstrated that workflow is improved, with general concordance of diagnoses between digital pathology and light microscopy [6,26–28], and with good inter and intrareader concordance for diagnoses and scoring criteria [4▪,29,30], although some authors reported notable differences in concordance between diagnostic groups [31]. Pathologists recorded mixed experiences about using WSI over glass slides [26,32]
DIGITAL PATHOLOGY AND CLINICAL TRIALS
The introduction of digital pathology in clinical trials has been beneficial by: a) providing transparency to regulatory agencies; b) employing software for digital annotation; c) allowing different metrics for assessment; and alleviating workflow difficulties in respect of operator’s time and industry budget [1,33].
Digital pathology as standard of practice in drug development
Ongoing or recently completed digital pathology-based clinical trials include evaluation of a) globotriaosylceramide (GL) inclusions in Fabry’s disease in patients receiving a chaperone (NCT00214500) [1] or enzyme replacement therapy (NCT02795676), b) proliferative lupus nephritis in patients treated with anti-Tumor necrosis factor-like Weak Inducer of Apoptosis mAb (NCT01499355) [34], and c) the amount of transplant glomerulopathy after treatment with C1 esterase inhibitor (NCT02547220) [33]. Some of these trials used annotation (enumeration) of objects of interest, so that pathologists score exactly the same structures, and not only the same biopsy, resulting in increased accuracy and reproducibility. For example, when a quantitative scoring system to assess GL inclusions in 300 cortical peritubular capillaries per biopsy was applied to both glass slides and annotated WSI, interreader concordance was greater with the digital pathology protocol [1]. In a recent clinical trial for Fabry’s disease (NCT02795676), the establishment of a DPR consented to use the same set of annotated images to compare quantitative [1] and semiquantitative [35] metrics to test drug efficacy, and to streamline the adjudication process, limiting the adjudication to the individual annotated structures with discordant results, rather than of the entire annotated set.
DIGITAL PATHOLOGY IN CLINICAL RESEARCH
Our increasing understanding of molecular (genetic, epigenetic, transcriptomic, proteomic, metabolomics) [36], clinical, environmental, socioeconomic data, and of the renal disease pathophysiology [37] demand that novel quantitative approaches, suitable for dataset integration, are developed. Three different tissue interrogation approaches exploit digital pathology in clinical research: visual morphologic assessment; visual morphometric assessment; and computer-aided assessment (Table 1). This increasing comprehensiveness and sophistication of data collection in pathology opens opportunities for better integration of structural changes with the complexity and variety of other patient-related datasets.
Changing paradigms to establish clinically relevant patient categorization
Until now, pathologists applied conventional interpretative diagnoses by light microscopy and the one-size-fits-all approach, in which diagnoses and classification strategies are developed for the average, ‘classic’ presentation. Furthermore, pathologists typically first generated disease categories and then tested them against outcome. Novel quantitative digital visual protocols for morphologic profiling of native and transplant kidney biopsies were recently proposed and tested across the globe. [2▪▪,3,4▪,9,10▪,11▪,38] One example is provided by the NEPTUNE digital pathology protocol for pathology material acquisition and digital annotation, and the NEPTUNE/INTEGRATE scoring system for harmonization of language, comprehensiveness of the descriptor (observational data) used in the analysis, quantification of the parameters observed, and data collection and reporting (Fig. 3) [2▪▪]. Highly granular data collection allows for the identification of previously unutilized or underrecognized parameters to generate and test new disease categories [39], to change historical paradigms for disease categorization, and to explore new predictors of molecular mechanisms and outcome. For example, utilizing patient cluster analysis models based on frequency of descriptors, outcome can be used to establish which of these patient clusters are clinically significant and test whether common pathways and gene expression are activated [40▪]. Reproducible descriptors such as interstitial fibrosis and tubular atrophy reliably correlate with other features of chronicity (global sclerosis and arteriosclerosis) and inflammation, and predict outcome independently from disease category. Strong correlation between tubulointerstitial mRNA gene expression signatures and morphology validated the hypothesis that visual assessment of interstitial fibrosis reflects underlying transcriptional programs linked to intrarenal fibrosis and inflammation [41].
FIGURE 3.
Standardization of protocols for globalization of the renal biopsy morphologic and morphometric profiles. Standardize protocols for pathology material acquisition, uploading and organization into the digital pathology repository, data collection using an electronic scoring matrix are implemented across different consortia and continents. Cross-training of investigators to implement reproducibility is performed via international webinar meetings. Adapted with permission [2▪▪].
Changing paradigms for renal biopsy adequacy
The utility of digital pathology protocols-based discoveries and their translation into clinical practice can be exemplified by two sets of consecutive observations. First, two separate studies, one in North America (NEPTUNE) and the other one in Europe (EURenOMICS), independently demonstrated that a quantitative protocol for manual enumeration of glomeruli on WSI through all biopsy levels can improve accuracy in measuring the total number (denominators) or the number of affected (numerators) glomeruli per biopsy [10▪,38], and reported that denominators and numerators as documented in clinical practice are significantly underestimated compared to using annotated WSI [10▪,38]. A recent study highlighted the utility of glomerular annotation by demonstrating that the percentage of digitally annotated globally sclerotic glomeruli had prognostic value if exceeding age-appropriate levels. Interestingly, when these data were compared with those obtained by conventional counting by light microscopy, the ratio between total number of glomeruli and number of globally sclerotic glomeruli was similar between light microscopy and digital pathology [42▪]. These observations carry two important messages: first, parameters conventionally not incorporated in current classification systems may have significant prognostic value; second, such discovery derives from a very accurate, albeit time consuming, digital pathology-based methodology to gather reproducible data across different populations, thus it should be considered robust. Although annotation is impractical in clinical practice, assessment of global sclerosis in daily clinical practice using light microscopy was proved to provide useful prognostic data, even though the conventional light microscopy protocol for glomerular counting is less accurate [42▪].
While visual assessment on WSI is successfully leading to new discoveries, morphometry is still considered the gold standard for quantitative analysis. Morphometry on WSI provides standardized parameters (assessment of cortical interstitial volume, fractional interstitial area, average glomerular tuft volume, cortical density of glomeruli) that correlate with various aspects of renal function and may play a role in estimating the baseline risk of future loss of function [43]. Similar studies applying convolutional neural network demonstrated the value of cortical structural changes, such as interstitial fibrosis, in predicting estimated glomerular filtration rate decline [44]. Furthermore, utilizing visual morphologic and computer-aided morphometric protocols to measure fibrosis, novel, and useful information residing in the analysis of the medulla can be retrieved [4▪]. Overall, these studies point to an imminent adjustment of historical paradigms and parameters for renal biopsy adequacy, measured as number of glomeruli and cortex/medulla ratio.
Introducing human–computer interactive protocols
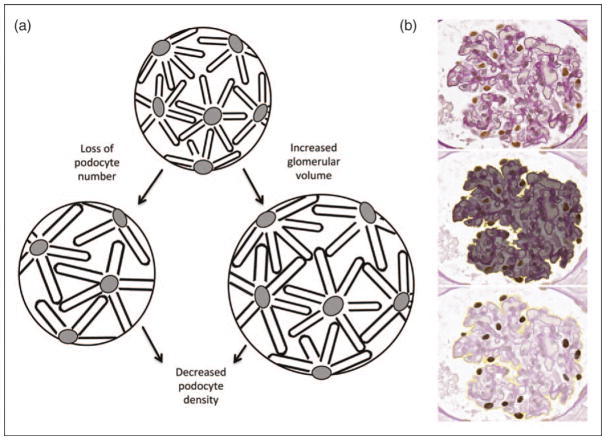
As machine training progress, we can envision future human–computer synergistic protocols for calculation of quantitative parameters or for extraction of subvisual features critical to outcome prediction [45]. Recently, pioneering studies using computational imaging software and algorithms demonstrated segmentation of glomeruli from murine and rat renal cortex [46▪]. The ability to segment glomeruli is the first step toward the analysis of quantitative glomerular features such as cellularity, capillary width, and tuft, mesangium and podocyte volumes. Several studies have reported reduced absolute or relative podocyte density associated with glomerular destabilization, glomerulosclerosis and proteinuria in diabetic glomerulopathy, immunoglobulin A nephropathy, aging, and in the setting of transplant kidneys [47–51]. Computer-aided estimation of podocyte density, named podometrics (Fig. 4), is an example of clinically relevant digital quantitative disease, and would allow an estimation of podocyte density that cannot be achieved with the human eye, with potential predictive power of risk of loss of renal function and/or treatment response. The application of such methodologies enables a semisupervised pipeline for digital pathology workflow, data collection, and feature extraction that could improve diagnostic efficiency in clinical research, trials, and practice.
FIGURE 4.
Podometrics. (a) The cartoon illustrates the events leading to a reduction in relative or absolute podocyte density, resulting in glomerulosclerosis. When podocytes are lost or the glomerular size increases, podocytes become hypertrophic in an attempt to maintain the filtration barrier, resulting in a decrease in podocyte density. When the threshold of hypertrophic stress and loss of adaptive capacity is reached, podocytes detach, resulting in progressive podocytopenia. (b) From a glomerulus (upper panel), measurements of tuft area (middle panel) and podocyte features (bottom panel) can be obtained from a single section stained with periodic acid–Schiff and immunohistochemistry for Wilm’s Tumor 1 by employing National Institute of Health ImageJ software. With this modified protocol based on the measurements are then used to calculate podocyte density per glomerular volume. (Histology images kindly provided by Chris O’Connor and Markus Bitzer at the University of Michigan, Division of Nephrology) [47].
Integration of pathology in kidney precision medicine
Recently, the Kidney Precision Medicine Project was launched by the National Institute of Health in an effort to refine biologically and clinically relevant categories of kidney diseases, through identification of molecular composition of kidney cell types and their structural relationship within the tissue (cell–cell and cell–matrix), new disease pathways, and targets for novel therapies [52]. Concurrently to the emerging precision medicine approach, seeking to comprehensively integrate large multiscale datasets at the patient level, powerful new computational disease approaches are being developed for analyzing large digital pathology datasets using mathematical models and algorithms [53–55]. However, the intelligent combination of divergent large-scale datasets for patient profiles with predictive power must be able to withstand disparate dimensionalities of the data [53]. One successful approach has employed machine learning and data fusion methods to computationally combine computer-derived morphometric features and protein expression data from prostate specimens that more accurately predict risk of biochemical recurrence than with either feature alone. [56] The heterogeneity of the normal renal parenchyma and the even more extreme heterogeneity of renal structural damage, however, makes the computational image analysis challenging.
In summary, digital pathology grants control of the research material simply not possible with standard glass slides. These improvements allow for much granular detail of structural information, broader profiling, and superior integration with other datasets.
CONCLUSION
In parallel with the effort of the nephrology community in establishing a multiscale understanding of renal diseases, nephropathologists will have to leave their comfort zone and embrace the challenges accompanying the modern software ecosystem. Confronting and addressing such imminent technological transformation creates opportunities to lay the foundation for integration of quantitative structural changes into an interdisciplinary computational science. This process shall result in better definitions of disease subcategories, identification of critical single cells, pathways and therapeutic targets, and the establishment of kidney tissue atlases and clinically relevant morphoomic taxonomies of kidney diseases.
KEY POINTS.
The introduction of telepathology and WSI in nephropathology is changing pathology practice, clinical trials, and research.
Digital pathology allows to apply multiple approaches to the same set of digital images, from visual morphologic and morphometric assessment, to deep learning and convolutional neural network for tissue interrogation and feature extraction.
Quantitative accurate and reproducible methodologies to capture the structural changes in the diseased kidney can be computationally integrated with omic science to create fused predictors of outcome and address questions in precision and predicting medicine.
Acknowledgments
The authors would like to thank Dr David Thomas for critical review of the manuscript and the NEPTUNE and International Digital Nephropathology Network investigators for the long-standing collaboration.
Financial support and sponsorship
The authors acknowledge financial support from the Nephrotic Syndrome Study Network Consortium (NEPTUNE), part of the National Center for Advancing Translational Sciences (NCATS), the Rare Disease Clinical Research Network (RDCRN), and supported through a collaboration between the Office of Rare Diseases Research (ORDR), NCATS, and the National Institute of Diabetes, Digestive, and Kidney Diseases. RDCRN is an initiative of ORDR and NCATS. Additional funding and/or programmatic support for this project has also been provided by the University of Michigan, NephCure Kidney International, and the Halperin Foundation.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1.Barisoni L, Jennette JC, Colvin R, et al. Novel quantitative method to evaluate globotriaosylceramide inclusions in renal peritubular capillaries by virtual microscopy in patients with fabry disease. Arch Pathol Lab Med. 2012;136:816–824. doi: 10.5858/arpa.2011-0350-OA. [DOI] [PubMed] [Google Scholar]
- 2▪▪.Barisoni L, Gimpel C, Kain R, et al. Digital pathology imaging as a novel platform for standardization and globalization of quantitative nephropathology. Clin Kidney J. 2017;10:176–187. doi: 10.1093/ckj/sfw129. The study illustrates how digital renal biopsies is facilitating standardization processes across multiple consortia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barisoni L, Nast CC, Jennette JC, et al. Digital pathology evaluation in the multicenter Nephrotic Syndrome Study Network (NEPTUNE) Clin J Am Soc Nephrol. 2013;8:1449–1459. doi: 10.2215/CJN.08370812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4▪.Liapis H, Gaut JP, Klein C, et al. Banff Working Group. Banff histopathological consensus criteria for preimplantation kidney biopsies. Am J Transplant. 2017;17:140–150. doi: 10.1111/ajt.13929. The study is an example of digital pathology applied to current classification system to test reproducibility of observations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Janabi S, Huisman A, Vink A, et al. Whole slide images for primary diagnostics of gastrointestinal tract pathology: a feasibility study. Hum Pathol. 2012;43:702–707. doi: 10.1016/j.humpath.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Al-Janabi S, Huisman A, Vink A, et al. Whole slide images for primary diagnostics in dermatopathology: a feasibility study. J Clin Pathol. 2012;65:152–158. doi: 10.1136/jclinpath-2011-200277. [DOI] [PubMed] [Google Scholar]
- 7.Chordia TD, Vikey A, Choudhary AB, et al. Current status and future trends in telepathology and digital pathology. J Oral Maxillofac Pathol. 2016;20:178–182. doi: 10.4103/0973-029X.185924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pantanowitz L, Sinard JH, Henricks WH, et al. College of American Pathologists Pathology and Laboratory Quality Center. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the college of American pathologists pathology and laboratory quality center. Arch Pathol Lab Med. 2013;137:1710–1722. doi: 10.5858/arpa.2013-0093-CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rijnink EC, Teng YK, Wilhelmus S, et al. Clinical and histopathologic characteristics associated with renal outcomes in lupus nephritis. Clin J Am Soc Nephrol. 2017;12:734–743. doi: 10.2215/CJN.10601016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10▪.Rosenberg AZ, Palmer M, Merlino L, et al. The Application of digital pathology to improve accuracy in glomerular enumeration in renal biopsies. PloS One. 2016;11:e0156441. doi: 10.1371/journal.pone.0156441. The study demonstrated that the use of ditial pathology and tools for annoatation increases accuracy of quantitative disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11▪.Barisoni L, Troost JP, Nast C, et al. Reproducibility of the NEPTUNE descriptor-based scoring system on whole-slide images and histologic and ultrastructural digital images. Mod Pathol. 2016;29:671–684. doi: 10.1038/modpathol.2016.58. The study explore how digital pathology can facilitate reproducibility studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. [Accessed 21 July 2017];2017 http://www.fdanews.com/articles/181386-philips-whole-slide-imaging-system-wins-fda-approval.
- 13▪▪.Mariani LH, Pendergraft WF, 3rd, Kretzler M. Defining glomerular disease in mechanistic terms: implementing an integrative biology approach in nephrology. Clin J Am Soc Nephrol. 2016;11:2054–2060. doi: 10.2215/CJN.13651215. This manuscript illustrates how individual descriptors, predict outcome independently from conventional classification systems and can reflect underlying transcriptional programs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheufele E, Aronzon D, Coopersmith R, et al. tranSMART: an open source knowledge management and high content data analytics platform. AMIA Jt Summits Transl Sci Proc. 2014;2014:96–101. [PMC free article] [PubMed] [Google Scholar]
- 15.Knoppers BM, Harris JR, Tasse AM, et al. Towards a data sharing code of conduct for international genomic research. Genome Med. 2011;3:46. doi: 10.1186/gm262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugano S Regulatory and Ethics Working Group, Global Alliance for Genomics & Health. International code of conduct for genomic and health-related data sharing. Hugo J. 2014;8:1. doi: 10.1186/1877-6566-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantanowitz L, Wiley CA, Demetris A, et al. Experience with multimodality telepathology at the university of Pittsburgh medical center. J Pathol Inform. 2012;3:45. doi: 10.4103/2153-3539.104907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh S, Badaya S. Tele-cytology: an innovative approach for cervical cancer screening in resource-poor settings. J Cancer Res Ther. 2016;12:481–485. doi: 10.4103/0973-1482.157343. [DOI] [PubMed] [Google Scholar]
- 19.Agarwal S, Zhao L, Zhang R, Hassell L. FaceTime validation study: low-cost streaming video for cytology adequacy assessment. Cancer Cytopathol. 2016;124:213–220. doi: 10.1002/cncy.21636. [DOI] [PubMed] [Google Scholar]
- 20▪▪.Farris AB, Cohen C, Rogers TE, Smith GH. Whole slide imaging for analytical anatomic pathology and telepathology: practical applications today, promises, and perils. Arch Pathol Lab Med. 2017;141:542–550. doi: 10.5858/arpa.2016-0265-SA. The study demonstrate the interrelateddness of the cortex and medulla measured using digital pathology and computer-aided imaging. [DOI] [PubMed] [Google Scholar]
- 21.Hanna MG, Monaco SE, Cuda J, et al. Comparison of glass slides and various digital-slide modalities for cytopathology screening and interpretation. Cancer. 2017 doi: 10.1002/cncy.21880. [DOI] [PubMed] [Google Scholar]
- 22.Pradhan D, Monaco SE, Parwani AV, et al. Evaluation of panoramic digital images using panoptiq for frozen section diagnosis. J Pathol Inform. 2016;7:26. doi: 10.4103/2153-3539.181770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vitkovski T, Bhuiya T, Esposito M. Utility of telepathology as a consultation tool between an off-site surgical pathology suite and affiliated hospitals in the frozen section diagnosis of lung neoplasms. J Pathol Inform. 2015;6:55. doi: 10.4103/2153-3539.168515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24▪▪.Eccher A, Neil D, Ciangherotti A, et al. Digital reporting of whole-slide images is safe and suitable for assessing organ quality in preimplantation renal biopsies. Hum Pathol. 2016;47:115–120. doi: 10.1016/j.humpath.2015.09.012. The study is an example of application of digital pathology in clinical practice. [DOI] [PubMed] [Google Scholar]
- 25.Thrall MJ, Wimmer JL, Schwartz MR. Validation of multiple whole slide imaging scanners based on the guideline from the college of American pathologists pathology and laboratory quality center. Arch Pathol Lab Med. 2015;139:656–664. doi: 10.5858/arpa.2014-0073-OA. [DOI] [PubMed] [Google Scholar]
- 26.Furness P. A randomized controlled trial of the diagnostic accuracy of internet-based telepathology compared with conventional microscopy. Histopathology. 2007;50:266–273. doi: 10.1111/j.1365-2559.2006.02581.x. [DOI] [PubMed] [Google Scholar]
- 27.Houghton JP, Ervine AJ, Kenny SL, et al. Concordance between digital pathology and light microscopy in general surgical pathology: a pilot study of 100 cases. J Clin Pathol. 2014;67:1052–1055. doi: 10.1136/jclinpath-2014-202491. [DOI] [PubMed] [Google Scholar]
- 28.Ozluk Y, Blanco PL, Mengel M, et al. Superiority of virtual microscopy versus light microscopy in transplantation pathology. Clin Transplant. 2012;26:336–344. doi: 10.1111/j.1399-0012.2011.01506.x. [DOI] [PubMed] [Google Scholar]
- 29.Jen KY, Olson JL, Brodsky S, et al. Reliability of whole slide images as a diagnostic modality for renal allograft biopsies. Hum Pathol. 2013;44:888–894. doi: 10.1016/j.humpath.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Sima CS, Beasley MB, et al. Classification of thymic epithelial neoplasms is still a challenge to thoracic pathologists: a reproducibility study using digital microscopy. Arch Pathol Lab Med. 2014;138:658–663. doi: 10.5858/arpa.2013-0028-OA. [DOI] [PubMed] [Google Scholar]
- 31.Shah KK, Lehman JS, Gibson LE, et al. Validation of diagnostic accuracy with whole-slide imaging compared with glass slide review in dermatopathology. J Am Acad Dermatol. 2016;75:1229–1237. doi: 10.1016/j.jaad.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 32.Guo H, Birsa J, Farahani N, et al. Digital pathology and anatomic pathology laboratory information system integration to support digital pathology sign-out. J Pathol Inform. 2016;7:23. doi: 10.4103/2153-3539.181767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mroz P, Parwani AV, Kulesza P. Central pathology review for phase III clinical trials: the enabling effect of virtual microscopy. Arch Pathol Lab Med. 2013;137:492–495. doi: 10.5858/arpa.2012-0093-RA. [DOI] [PubMed] [Google Scholar]
- 34.Furie R, Malvar A, Navarra SV, et al. Evaluation of the efficacy, safety, and tolerability of BIIB023 as an adjunct to standard of care in subjects with lupus nephritis [Abstract] Arthritis Rheumatol. 2016:68. http://acrabstracts.org/abstract/evaluation-of-the-efficacy-safety-and-tolerability-of-biib023-as-an-adjunct-to-standard-of-care-in-subjects-with-lupus-nephritis/
- 35.Thurberg BL, Rennke H, Colvin RB, et al. Globotriaosylceramide accumulation in the Fabry kidney is cleared from multiple cell types after enzyme replacement therapy. Kidney Int. 2002;62:1933–1946. doi: 10.1046/j.1523-1755.2002.00675.x. [DOI] [PubMed] [Google Scholar]
- 36.He JC, Chuang PY, Ma’ayan A, Iyengar R. Systems biology of kidney diseases. Kidney Int. 2012;81:22–39. doi: 10.1038/ki.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kretzler M, Sedor JR. Introduction: precision medicine for glomerular disease: the road forward. Semin Nephrol. 2015;35:209–211. doi: 10.1016/j.semnephrol.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gimpel C, Royal V, Simic I, et al. Standardized assessment of digital renal biopsies: accuracy and reproducibility studies. Nephrol Dial Transplant. 2016;31:i373. [Google Scholar]
- 39.Nast CC, Lemley KV, Hodgin JB, et al. Morphology in the digital age: integrating high-resolution description of structural alterations with phenotypes and genotypes. Semin Nephrol. 2015;35:266–278. doi: 10.1016/j.semnephrol.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40▪.Mariani LH, Zee J, Hodgin JB, et al. Minimal change disease and focal segmental glomerulosclerosis pateint subgroups using cluster analysis of morphologic descriptors: early findings from the nephrotic syndrome study network (NEPTUNE) [Abstract] J Am Soc Nephrol. 2016;27:511A. Digital pathology allows novel protocol for visual moprhologic analysis, resulting in change in paradigms for clinically relevant patient categorization. [Google Scholar]
- 41.Mariani LH, Martini S, Barisoni L, et al. Interstitial fibrosis scored on whole-slide digital imaging of kidney biopsies is a predictor of outcome in proteinuric glomerulopathies. Nephrol Dial Tansplant. 2017 doi: 10.1093/ndt/gfw443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42▪.Hommos M, Zeng C, Liu Z, et al. Abnormal global glomerulosclerosis in nephrotic syndrome. Am J Kidney Dis. 2017;69:A53. Standardization of digital pathology protocols and the descritor-based analysis allows the identification of clinically relevant parameters not currently incorporated in conventional classifications. [Google Scholar]
- 43.Lemley KV, Bagnasco SM, Nast CC, et al. Morphometry predicts early GFR change in primary proteinuric glomerulopathies: a longitudinal cohort study using generalized estimating equations. PloS One. 2016;11:e0157148. doi: 10.1371/journal.pone.0157148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ledbetter D, Ho LV, Lemley KV. [Accessed 21 July 2017];Prediction of Kidney Function from Biopsy Images Using Convolutional Neural Networks. 2017 :1–11. https://arxivorg/pdf/170201816pdf.
- 45.Antunes J, Viswanath S, Rusu M, et al. Radiomics analysis on FLT-PET/MRI for characterization of early treatment response in renal cell carcinoma: a proof-of-concept study. Transl Oncol. 2016;9:155–162. doi: 10.1016/j.tranon.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46▪.Ginley B, Tomaszewski JE, Yacoub R, et al. Unsupervised labeling of glomerular boundaries using Gabor filters and statistical testing in renal histology. J Med Imaging (Bellingham) 2017;4:021102. doi: 10.1117/1.JMI.4.2.021102. The conputer-aided algorithms for the identification of stuctural features and quantitative parameters in the renal parenchyma can be paired with the human effort incclinical reasearch and ultimately clinical practice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkatareddy M, Wang S, Yang Y, et al. Estimating podocyte number and density using a single histologic section. J Am Soc Nephrol. 2014;25:1118–1129. doi: 10.1681/ASN.2013080859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Hodgin JB, Afshinnia F, et al. The two kidney to one kidney transition and transplant glomerulopathy: a podocyte perspective. J Am Soc Nephrol. 2015;26:1450–1465. doi: 10.1681/ASN.2014030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pagtalunan ME, Miller PL, Jumping-Eagle S, et al. Podocyte loss and progressive glomerular injury in type II diabetes. J Clin Invest. 1997;99:342–348. doi: 10.1172/JCI119163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemley KV, Lafayette RA, Safai M, et al. Podocytopenia and disease severity in IgA nephropathy. Kidney Int. 2002;61:1475–1485. doi: 10.1046/j.1523-1755.2002.00269.x. [DOI] [PubMed] [Google Scholar]
- 51.Hodgin JB, Bitzer M, Wickman L, et al. Glomerular aging and focal global glomerulosclerosis: a podometric perspective. J Am Soc Nephrol. 2015;26:3162–3178. doi: 10.1681/ASN.2014080752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. [Accessed 21 July 2017]; http://www.niddk.nih.gov/research-funding/research-programs/kidney-precision-medicine-project-kpmp.
- 53.Bhargava R, Madabhushi A. Emerging themes in image informatics and molecular analysis for digital pathology. Annu Rev Biomed Eng. 2016;18:387–412. doi: 10.1146/annurev-bioeng-112415-114722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Louis DN, Feldman M, Carter AB, et al. Computational pathology: a path ahead. Arch Pathol Lab Med. 2016;140:41–50. doi: 10.5858/arpa.2015-0093-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Louis DN, Gerber GK, Baron JM, et al. Computational pathology: an emerging definition. Arch Pathol Lab Med. 2014;138:1133–1138. doi: 10.5858/arpa.2014-0034-ED. [DOI] [PubMed] [Google Scholar]
- 56.Lee G, Singanamalli A, Wang H, et al. Supervised multiview canonical correlation analysis (sMVCCA): integrating histologic and proteomic features for predicting recurrent prostate cancer. IEEE Trans Med Imaging. 2015;34:284–297. doi: 10.1109/TMI.2014.2355175. [DOI] [PubMed] [Google Scholar]