Abstract
Circular RNAs (circRNAs) are a class of long noncoding RNAs that are characterized by the presence of covalently linked ends, and have been found in all life kingdoms. Exciting studies in regulatory roles of circRNAs are emerging. Here we summarize classification, characteristics, biogenesis and regulatory functions of circRNAs. CircRNAs are found to be preferentially expressed along neural genes and in neural tissues. We thus highlight the association of circRNA dysregulation with neurodegenerative diseases such as Alzheimer’s disease. Investigation of regulatory role of circRNAs will shed novel light in gene expression mechanisms during development and under disease conditions, and may identify circRNAs as new biomarkers for aging and neurodegenerative disorders.
Keywords: circular RNA, back-splicing, regulatory RNA, microRNA sponge, neurodegeneration
Broad diversity of circRNAs
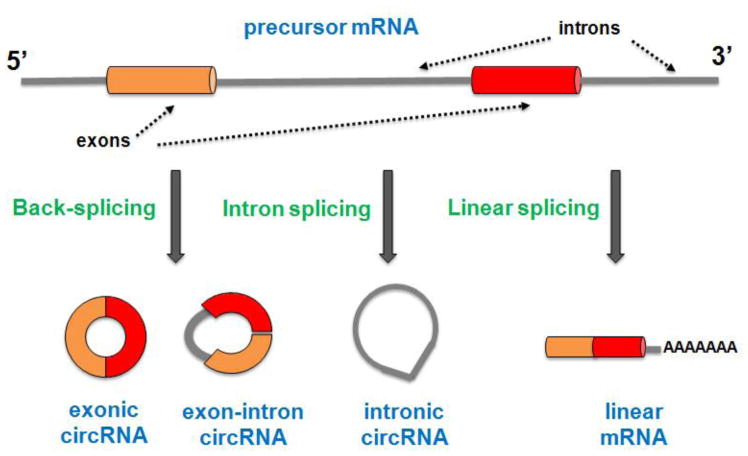
Circular RNAs (circRNAs) are a class of long noncoding RNAs (lncRNAs) characterized by the presence of covalently linked ends produced in a non-canonical splicing event called “back-splicing” [1–3]. CircRNAs can arise from coding and non-coding exons, introns (including intron lariats), or from both exons and introns [4–6]. They also can be derived from 3′ and 5′ untranslated regions (UTR), intergenic sequences, pseudogenes, and from long intergenic noncoding RNAs [4,7]. Here we summarize the most studied subclasses of circRNAs (Figure 1).
Figure 1. Three major subclasses of circRNAs.
Exonic circRNAs (ecircRNAs) consist of only exon(s) (usually less than five) and represent the most important group of circRNA class. EcircRNAs have cytoplasmatic location and may regulate microRNA and protein functions. Exon-intron circRNAs (EIciRNAs) are composed of at least two exons and one retained intron. EIciRNAs have nuclear localization and have been found to be able to regulate gene transcription in cis and probably also in trans. Intronic circRNAs (ciRNAs) are derived from intron lariats and are accumulated in the nucleus in which regulate gene transcription in cis.
Exonic circRNAs (ecircRNAs)
This category includes all circRNAs that are exclusively composed of exon(s) joined together with the classical 3′-5′ covalent carbon link (Figure 1). Currently ecircRNAs represent the largest class of circRNAs found in animals as well as in plants [4,8–11]. In a genome-wide analysis, at least 5923 of 7112 total circRNAs (83%) were found overlapping with protein-coding regions [4]. Most ecircRNAs span less than 5 exons, and the length of ecircRNAs can range from hundreds to thousands nucleotides (nt) with an average estimated length of about 547nt [4,11]. Several studies reveal cytoplasmic location of ecircRNAs and their high stability in cells [3]. Some ecircRNAs can interact with microRNAs (miRNAs) and/or RNA binding proteins (RBPs), and many of them encircle the second exon that contains the canonical translation start codon [3,13–15]. These findings suggest the potential regulatory role of ecircRNAs in gene expression regulation and translation.
Intronic circRNAs (ciRNAs)
This subclass includes all circRNAs that are derived only from introns (including intron lariats) (Figure 1). They represent a small fraction of circRNAs, of which only 19.2% found in humans [7] and a very low fraction in plants [10]. Unlike ecircRNAs, ciRNAs have 2′-5′ head-tail link joint and different features in terms of stability, subcellular localization, abundance, conservation and functions. CiRNAs display nuclear localization and less conserved sequence than ecircRNAs, and many ciRNA are exclusive in human cells [5]. Moreover, ciRNAs appear to be less regulated and their expression is positively correlated with the expression of their parental mRNAs [5]. ciRNAs can be associated with the RNA Polymerase II (Pol II) complex and can increase expression of their parental genes. These studies support the idea that ciRNAs are not simple byproducts of splicing and they may play different, but important roles compared to the ecircRNAs.
Exon-Intron circRNA (EIciRNAs)
Recently a new subclass of circRNAs, in which the intron is retained between exons, was identified (Figure 1) [6]. A study has shown that a fraction (~20%) of ecircRNAs can retain introns [4]. The intron retention between exons makes this subclass unique, even though they share some features with both ecircRNAs and ciRNAs. Similar to ecircRNAs, EIciRNAs have reverse complementary sequences in long flanking introns, suggesting the existence of a common mechanism for their biogenesis. Like ciRNAs, EIciRNAs are predominantly in the nucleus and have been shown to be associated with Pol II to promote the transcription of their parental genes in cis through interaction with U1 small nuclear ribonucleoprotein (snRNP) [6].
Characteristics of circRNAs
The most important feature of circRNAs is their characteristic circular structure, which is associated with their important functions in cells [12]. Firstly, the lack of free terminals allows circRNAs to avoid exonucleolytic degradation by RNase R or/and RNA exonuclease, making them stable in cells up to 48 hours [3]. Secondly, circRNAs are used as a template in an enzymatic process known as “rolling circle amplification (RCA)”, which is important for Hepatitis Delta Virus (HDV) and viroids replication models [16,17]. It has been recently demonstrated that exogenous circRNAs are efficiently translated into functional proteins by RCA mechanism in living human cells [18]. Finally, circularization provides a means for increasing RNA structural stability and gaining potential evolutionary advantage, for example in thermophilus organisms [19]. A recent work revealed that circularization is essential for restoring signal recognition particles (a universal ribonucleoprotein complex), a functionality in Thermoproteus Tenax [20].
CircRNAs are ancient and well conserved RNA isoforms found in all life kingdoms. Detection of circRNAs with similar structural features in protist, fungi and plants, traced back these molecules to more than one billion years ago, indicates their important and conserved function retained during evolution [19,21]. Moreover, several studies have shown that many circRNAs display sequence conservation across species. For example Rybak-Wolf et al. found that about 28% (4522 out of 15,849) of circRNAs in mouse are conserved in human [22]. Interestingly they also found conservation in the expression patterns of neural circRNAs [22]. Furthermore, homology analysis of sequences among three different Drosophila species revealed that circularization among evolutionary divergent species is broadly conserved, and some fly brain circRNAs also have been found expressed in mammalian brains [22,23].
CircRNAs are usually found expressed at low levels [2,4,24], and represent a low fraction of transcriptome (0,2–1%), suggesting that the majority of circRNAs could be inert transcriptome byproducts [4]. However, Jeck et al. found that at least 50 genes in which circular forms are likely more abundant than their linear analogies, and Rybak-Wolf et al. found that almost a hundred of circRNAs, detected in the human and mouse brain, are expressed much higher than their linear cognates [8,22]. Moreover, some circRNAs are found to be expressed in a tissue- and cell-specific pattern, and many factors such as aging, developmental stage, stress condition, synaptic activities, Alzheimer’s disease (AD), and tumors, can lead to significant changes in circRNA abundance, which is often uncoupled from the linear RNA counterparts [10,23,25–29]. These studies suggest that many circRNAs may have an important role in physiological and pathological conditions and support the hypothesis that circRNAs have their own specific function.
Biogenesis of circRNAs
The majority of circRNAs derived from a precursor mRNA (pre-mRNA) and the process of its formation is called “back-splicing”, also known as “head to tail junction”. Since the canonical GT-AG splicing sites are typically required for circularization, the spliceosome machinery has been implicated in circRNA biogenesis [15,30–32]. In addition, using the specific splicing inhibitor isoginkgetin, Starke et al. found a significant reduction of the circRNA level [32]. However, how exactly the spliceosome acts to produce circRNAs and how it may be able to discriminate between linear splicing and back-splicing is unclear.
CircRNAs biogenesis appears to be initiated when the two putative splicing sites are brought into close proximity and facilitate the back-splicing events. Complementary repeats in flanking introns, which are positively correlated with the biogenesis of many circRNAs, may work through base pairing for the juxtaposition of the putative splicing sites [33,34]. Likewise, RNA binding proteins (RBPs) involved in alternative splicing such as Quaking I and Muscleblind may bind specific sequence in flanking introns and promote the biogenesis of some circRNAs through this mechanism [15,28].
The question whether circRNAs are co- or post-transcriptional products remains controversial [15,35,36]. A recent study in Drosophila suggests that both mechanisms exist, depending on the length of flanking repeats in bracketing introns. In particular, long repeats promote co-transcriptional circularization (400nt), whilst short repeats (<40nt) promote post-transcriptional circularization [37]. Here, we summarize the biogenesis mechanisms proposed for different subclasses of circRNAs.
Formation of ecircRNAs and EIciRNAs
Studies have shown that exon circularization involves the 3′-tail of a downstream exon (known as splice acceptor) joining with the 5′-head of an upstream exon (known as splice donor). Long exons tend to be more susceptible for circularization, especially in the case that ecircRNAs contain a single exon [30,33,35]. Moreover, many ecircRNAs or EIciRNAs can arise from the same gene locus with a mechanism named “alternative circularization” [6,33]. Currently two mechanisms involved in back-splicing are proposed: “direct back-splicing” and “exon skipping” [38]. Direct back-splicing involves two pathways: “intron pairing-driven circularization”, and “RBP pairing driven circularization” that depends on sequence specific RNA-binding proteins [39]. Moreover, these two pathways may work in concert to ensure an appropriate production of circRNAs and fine-tune the ratio between linear and circular transcripts [37]. The second mechanism involves the exon skipping model and is known as “lariat-driven circularization” [40,41]. A recent study using computational analysis of a RNA-sequencing (RNA-seq) dataset demonstrates that exon circularization is well correlated with exon skipping [40], suggesting that both mechanisms might be equally important.
Direct back-splicing
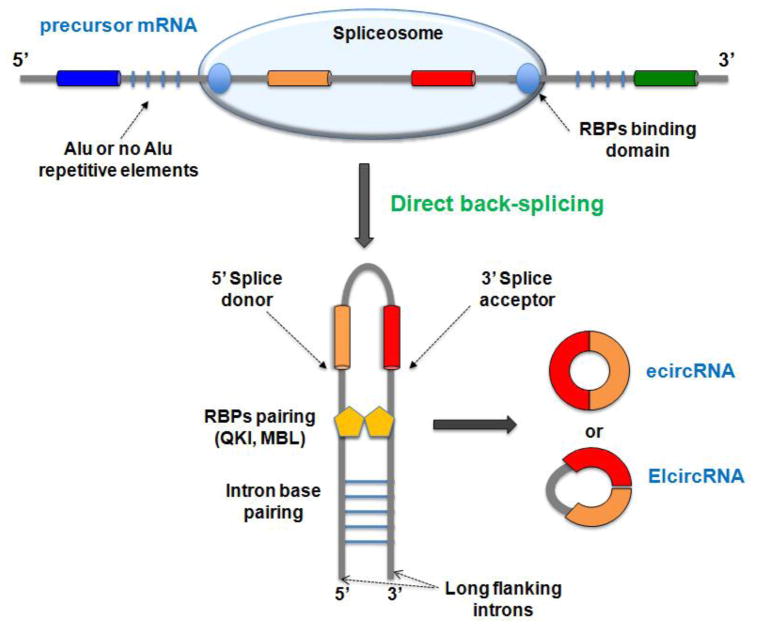
Direct back-splicing is independent on exon skipping, and both cis-acting factors and/or trans-acting factors are required in order to accost the exon splicing sites together and facilitate back-splicing. Cis-acting factors such as reverse complementary sequence in flanking introns (as Alu or not Alu elements), which form stable base pairs, are critical to enhance circRNA biogenesis [8,31,33–35] (Figure 2). Many circRNAs seem not to be able to form strong intron base pairing (62% and 91% in C elegans and humans, respectively) [34]. Moreover, Westholm et al. showed lack of nucleotide motif for intron paring in many gene loci that produce abundant circRNAs in a genome-wide analysis in Drosophila [23]. These studies suggest that intron pairing-driving circularization mechanism alone is unlikely a widespread mechanism for circularization. In addition, studies have shown that the length of flanking introns seems to be a critical factor for back-splicing [8,23,31,34]. However, Starke et al. were able to detect circRNAs in human cells using a minigene expression vector in which engineered flanking introns are considerably shortened and have no motif to form base pairing [32]. Taken together, it appears that reverse complementary sequence or long flanking exon are important factors for promoting back-splicing.
Figure 2. Direct back-splicing model of circRNA formation.
CircRNAs are derived from precursor mRNA, and the back-splicing reaction is catalyzed by the spliceosome. Reverse complementary sequence, such as Alu or not Alu elements, and RNA binding proteins (RBPs), such as Quaking I (QKI) and Muscleblind (MBL), might work in a combinatorial manner to bring the splicing sites into close proximity and facilitate back-splicing reaction. Long flanking introns might facilitate back splicing by inducing structural flexibility. Finally, the intervening intron is removed or retained to generate ecircRNA or EIciRNA.
The deposition of sequence specific RBPs are also required to circRNA biogenesis [15,28]. Both RBP Quaking I (QKI) and Muscleblind (MBL) have been found to be able to facilitate circRNA biogenesis, since the knockdown of QKI and the over expression of MBL can in turn decrease or increase circRNA production, respectively. This “sequence-specific” protein requires a specific (MBL/QKI) binding sites within flanking introns for promoting circularization. Thus, they may regulate circRNA biogenesis only in the case in which these motifs are present [15,28]. Conversely the double-strand RNA-editing enzyme ADAR1 (ADAR1) can negatively regulate circRNA biogenesis [22,34]. The role of ADAR1 may involve its editing activity, in which adenosine-to-inosine editing in flanking intron repeats could interfere with the strength of base-pair formation [34]. The circRNA biogenesis mediated by RBPs is in agreement with the idea that many circRNAs are regulated independently during development or in a cell-type specific manner [15,27,39].
Together, these studies suggest that circRNA biogenesis may depend on different factors that likely work in concert to regulate back-splicing outcomes. Moreover, different circRNAs can be regulated with different mechanisms and their production in cells seems to be more complicated than previously appreciated.
Exon skipping
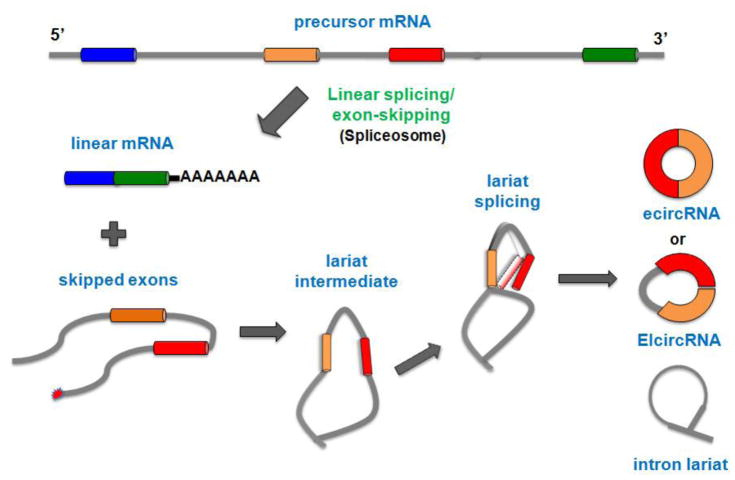
Exon skipping is a common type of alternative splicing with a well-established impact in mRNA formation [42]. However, a recent study suggest that exon skipping may also have a pivotal role in ecircRNA biogenesis [40,41]. During exon skipping, a big lariat containing the exon(s) is produced. Then, this lariat undergoes internal splicing in order to remove the intron and generate the ecircRNA or EIciRNA (Figure 3) [38]. The RNA-seq dataset analysis has shown that the largest part (if not all) of skipped exon(s) is able to produce ecircRNAs in human endothelial cells stimulated with tumor growth factor-β or tumor necrosis factor-α [40]. Moreover, a recent study has found that a lariat containing exon(s) production (probably from exon skipping) is a very common step in ecircRNA biogenesis of S Pombe [41]. More studies are required to test whether the circularization proceeds only by intrinsic features of the lariat-containing exon(s) or whether other factors such as RBPs may be involved.
Figure 3. Exon-skipping model of circRNA formation.
During exon skipping, a linear mRNA and a skipped exon(s) are formed. The skipped exon(s) then generate a big lariat intermediate, which undergoes internal splicing by inducing the juxtaposition of the putative splice sites and in turn produces ecircRNA or EIciRNA.
Formation of ciRNAs
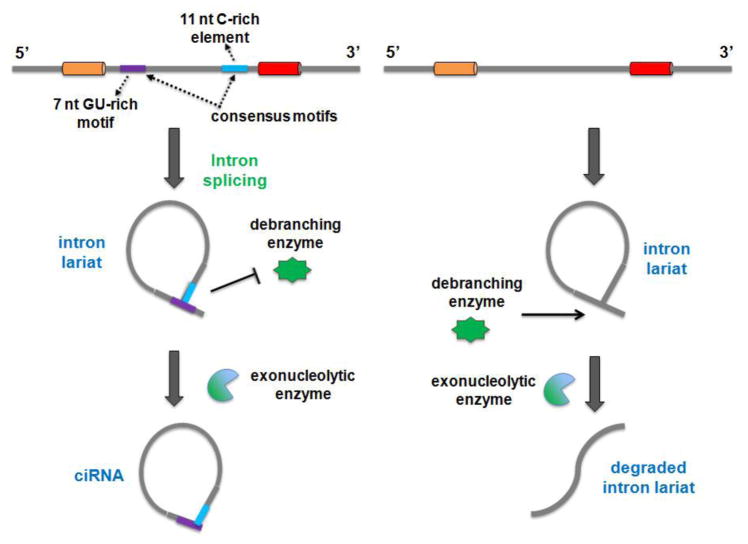
ciRNA and/or intron lariat can somehow escape debranching and accumulate in cells after eukaryotic spliceosomal activity. The process of ciRNA formation depends on a consensus motif near the 5′ splice site that contains 7nt GU-rich motifs and 11nt C-rich element near the branch point site. Moreover, their formation can be recapitulated with expression vectors [5]. The intronic circularization requires the release of the 3′exon by spliceosomal activity and then, the intron terminal group OH-2′ attacks the 5′ intron-exon junction to form the 2′-5′ ciRNA [38] (Figure 4). As discussed above, the consensus motif necessary for ciRNA processing could be essential to escape debranching [5]. However, how this key motif works to avoid debranching and whether other elements are involved in such process is still not clear.
Figure 4. Model of intronic circRNA (ciRNA) formation.
The process of ciRNA formation depends on a consensus motif near the 5′ splice site that contains 7nt GU-rich motifs and 11nt C-rich element near to the branch point site. Intron are excised during precursor mRNA processing. Intronic circularization requires the release of the 3′ exon, leaving the 2-OH terminal group free. The intron terminal group then attacks the 5′ intron-exon junction, resulting in 2′-5′ loop formation. The consensus motifs asisit escape of debranching induced by the debranching enzyme. The intron lariat is finally cleaved by the exonucleolytic enzyme to form ciRNA.
The regulatory role of circRNAs
CircRNAs have been perceived for decades as splicing errors and only recently their potential role as major gene regulators is becoming appreciated. Dozens of recent studies have revealed that circRNAs are involved in a wide range of life processes as well as in many human pathologies. It has been suggested that their regulatory roles pass through all steps of gene regulation, ranging from mRNA transcription and splicing to RNA decoy and translation. Although functions of a few circRNAs have been demonstrated, the full aspect of functions for these molecules is still unclear.
CircRNAs modulate miRNA function
miRNAs are small noncoding RNAs (20–22nt), which negatively modulate the stability and translation of many target mRNAs containing miRNA response elements (MREs) [43]. Although miRNAs are powerful negative regulators for most mRNAs, many of mRNAs are actively translated, suggesting the existence of mechanisms counteracting miRNA silencing regulation [44]. A genome wide analysis has revealed that thousands of mammalian circRNAs harbor MREs, suggesting a potential role as competitive endogenous RNAs (ceRNAs) [45]. Moreover, many circRNAs in flies contain conserved miRNA seed sites and some of them host more than one thousand miRNA binding elements [23].
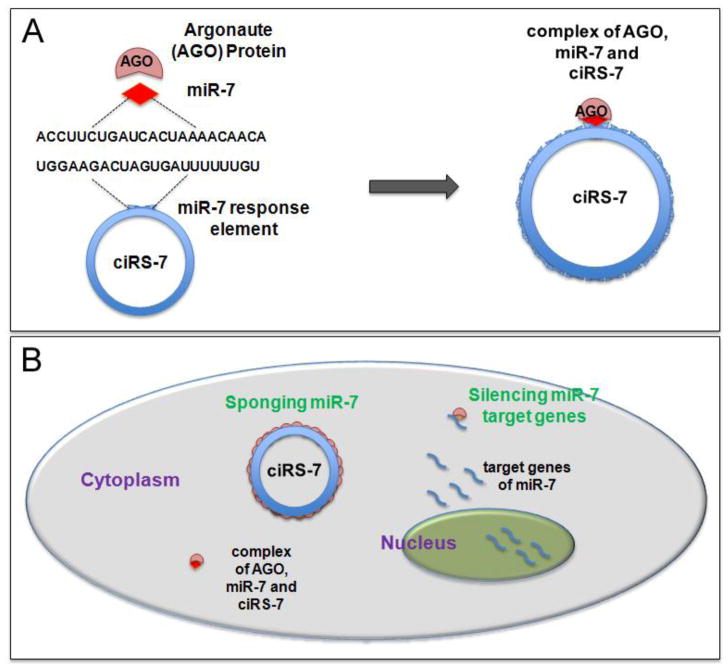
Recent two studies by Hansen et al. and Memczak et al. demonstrated that circular CDR1 antisense, also known as ciRS-7 (Circular RNA sponge for miRNA-7), can function as a powerful “miRNA sponges” [13,14]. In particular, ciRS-7 has been found contain more than 70 putative binding sites for miRNA-7 (miR-7) and can bind miR-7 without being degradated [13]. Moreover, its stability, abundance, and cytoplasmatic enrichment make it able to decrease the availability of miR-7 to inhibit its target mRNAs.
ciRS-7 is suggested to form a complex with Argonaute protein (AGO) (the catalytic core of RNA induce silence complex) in a miR-7 dependent manner in order to regulate miRNA (Figure 5). Hence, ciRS-7 can function as a platform for binding miR-7 and AGO, and allowing miR-7 degradation. Coherently, knockdown of ciRS-7 decreases the expression of miR-7 target genes, whilst its overexpression is able to prevent the down regulation of miR-7 targets [13].
Figure 5. Mechanism of the role of circular RNA sponge for miRNA-7 (ciRS-7).
(A) ciRS-7 contains over 70 miR-7 target sites and is capable to form a complex with Argonaute (AGO) protein in a miR-7 dependent manner. (B) Target genes of miR-7 is transcribed and silenced by miR-7. ciRS-7, which is localized in the cytoplasm, can function as miR-7 sponge to block miR-7 silencing activity and release miR-7 inhibition to its target genes.
Moreover, ectopic expression of ciRS-7 results in abnormal midbrain development in zebrafish, suggesting important function in the developmental brain [14]. In a post-mortem brain study, it has been shown a dysregulation in miR-7/ciRS-7 system in a sporadic form of AD [25]. Furthermore, as miR-7 regulates the expression of many oncogenes, miR-7/ciRS-7 system may have an important role in cancer formation and progression [46]. Recently, Xu et al. provided evidence for a role of ciRS-7 in pancreatic islet cells. They found that the reduction of insulin content and secretion in a diabetic mouse model in which miR-7 overexpression causes the pathology, can be reverted by overexpressing ciRS-7, suggesting a potential role of ciRS-7 as a therapeutic target in diabetes [47].
Besides ciRS-7, only a few circRNAs can function efficiently as miRNA sponges, for example circular SRY that harbors sixteen binding sites for miR-138 in mice but only one in human [13]. Moreover, it has been shown that circular ITCH harbors many binding sites for different miRNAs (including miR-7, miR-17, and miR-214) to titrate miRNAs and allow the transcription of ITCH linear cognate [48]. Circular HIPK3, derived from exon 2 of the HIPK3 gene, has 18 putative binding sites for 9 different miRNAs, and is able to regulate cell growth by functioning as a sponge for multiple miRNAs [49].
Taken together, these findings suggest that some mammalian circRNAs contain MREs, and can act as endogenous sponges for miRNAs.
CircRNAs regulate protein production
The evidence that circRNAs interact with Pol II and AGO proteins raise the possibility that they also can function as protein regulators and might work to localize, sort, and store RBPs [3]. It has been shown that the circular Mbl (circMbl) has conserved MBL binding sites and can bind MBL, while MBL can in turn regulate the formation of circMbl, suggesting an existence of a sophisticated auto-regulatory mechanism to fine-tune the production and availability of the protein [15]. Moreover, the circRNA circ-Foxo3 has been found to regulate the cell cycle progression by binding CDK2 and P21 proteins, resulting in the formation of a ternary complex that inhibits cell cycle progression and cell proliferation [50].
CircRNAs regulate gene transcription
Despite the fact that most circRNAs are cytoplasmatic, some circular isoforms can be detected also in the nucleus. Zhang et al. described a role for nuclear intronic circRNAs (ci-ankrd52, ci-mcm5 and ci-sirt7), which accumulate in the nucleus and do not exhibit enrichment of MREs [5]. Knockdown of the most abundant ciRNA ci-ankrd52 caused a significant down-regulation of the linear mRNA ankrd52, but had no effects on upstream or downstream genes, suggesting that ciRNAs act in cis and can only regulate the expression of their parental genes [5]. Finally they found an interaction between ciRNAs and the elongation complex of Pol II, suggesting a potential mechanism of these ciRNAs in regulating gene transcription [5].
Another mechanism of circRNA regulation of gene transcription involves a direct and stable interaction with the U1 snRNP [36]. In EIciRNAs, the retained intron has one putative U1 snRNP binding site. A multiple interaction among EIciRNAs, U1 snRNP, Pol II and the promoter of the host genes has been detected using the RNA-DNA double fluorescence in situ hybridization [6]. It appears that although many circRNAs may interfere and/or compete with their linear mRNA cognate formation [15], once EIciRNAs are produced, they can induce a positive feedback that enhances their own expression and the expression of their linear counterpart as well [6].
Other putative function of circRNAs
The fact that most circRNAs are derived from protein-coding sequences, carry open reading frames (ORF), and are located in the cytoplasm, raises the question that some of them might be able to translate into proteins. It has been suggested that translation can proceed on circRNAs that possess internal ribosome entry sites (IRES) [51]. An engineered circRNA containing IRES has been shown to produce functional proteins in transfected cells [31]. A recent study has shown that circRNAs can be translated into proteins even without IRES and other any particular sequence required for canonical translation (a cap structure and poly-A tail) [18]. However, even though exogenous circRNAs can undergo translation, there is still no evidence of translation from endogenous circRNAs [4,8,27,52].
Some ecircRNAs have been shown to have the translation start codon in their sequence [4]. Having the translation start codon in an ecircRNA may leave the linear transcript unable to produce protein, resulting in a down regulation of the final product. This regulation, named “mRNA traps”, suggests that the inclusion of the start codon into the circle is likely to function as traps to control translation rather than to produce canonical proteins from circRNAs [53]. It has been shown that 34% of the single circular exon contains the start codon in human fibroblasts, suggesting a widespread role of circRNAs as mRNA traps [3].
CircRNAs in the brain
The evidence that circRNAs are preferentially expressed from neural genes and are accumulated in neural tissues during aging has attracted attention in the neuroscience field [15,23]. Sequencing analyses of differentiated neuronal cell lines and dissected brain tissues revealed that thousands of circRNAs are highly expressed in the mammalian brain, developmentally regulated, conserved between rodents and humans, and enriched in synaptic fractions in neurons more than their linear counterparts [22,27]. Rybak-Wolf et al. found that a large part of circRNAs is upregulated during neuronal differentiation and many of them derived from host genes playing major roles in neuronal functions [22]. Moreover, You et al. found an enrichment of circRNAs with a prevalent fraction being derived from genes encoding for synaptic proteins in brain samples, and confirmed synaptic localization of circRNAs using a high resolution RNA in situ hybridization technique [27]. The presence of many circRNAs in synapses suggests that they might be selectively positioned to respond to synaptic activity, and might serve as keeper molecules of cellular memory, due to their long-lived nature.
Furthermore, Veno et al. provided a detailed profile of circRNA landscape during the course of porcine embryonic brain development and found that thousands of circRNAs display spatio-temporal expression pattern, with the maximal expression amount and complexity from embryonic day 48 to day 60, which corresponds to the period of major neurogenesis [26]. Szabo et al. found hundreds of genes in which circular isoforms exceed linear counterparts in human fetal tissues [30]. Functional studies of circRNAs expressed in the brain should provide new insights into their roles in brain development and neurogenesis.
CircRNAs and neurological disorders
CircRNAs have been found associated with many neurological diseases such as AD, Parkinson’s disease, multiple sclerosis, and schizophrenia [25,54]. For example, ciRS-7 that regulates miR-7 availability has been proposed to be associated with neurological disorders [55]. Studies have shown that miR-7 is highly expressed in cortical neuronal progenitors, and the silencing of miR-7 using miRNA sponges causes microcephaly-like brain defects [56]. Likewise, it has been shown that ectopic expression of human ciRS-7 in zebrafish impairs midbrain development in a similar manner to the effect induced by miR-7 knockdown [14]. Dysregulation of miR7-ciRS-7 interaction has been found in the hippocampus of AD patients [25]. Ubiquitin protein ligase A (UBE2A) is an autophagic, phagocytic protein that is essential for the clearance of AD-amyloid peptides, and also is a miR-7 target gene. UBE2A has been found down-regulated in AD, likely due to depletion of ciRS-7 sponge activity on miR-7 expression [25].
Moreover, a genome-wide association study has shown a link among circRNAs with single nucleotide polymorphisms and neurological diseases [54]. Alpha-synuclein, whose overexpression is associated with development of Parkinson’s disease, is a target gene of miR-7. miR-7-ciRS-7 interaction might be involved in Parkinson’s disease [54]. Szabo et al. showed that several circRNAs are derived from the gene loci that have genetic links to abnormal neurodevelopmental phenotypes, such as FBXW7, DOPEY2, and RMST [30].
In addition, a cytoplasmic accumulation of RBP TDP-43 has been found in some forms of sporadic amyotrophic lateral sclerosis (ALS). Armakola et al. found that cytoplasmatic increase of intron lariats, due to knockdown of debranching enzyme 1 activity, is effective for suppressing TDP-43 toxicity in the human neuronal cell line and primary rat neurons, suggesting possibility of using circRNAs as a potential therapeutic means for ALS [57]. Finally, some differentially expressed circRNAs were found in peripheral blood mononuclear cells of patients with major depressive disorder (MDD). hsa_circRNA_103636, which is downregulated in MDD patients compare to healthy controls, is significantly altered after 8 weeks of antidepressant regimens [58]. These results suggest that hsa_circRNA_103636 could be a novel potential biomarker for the diagnosis and treatment of MDD.
Perspectives
The fast advance in next-generation RNA sequencing, coupled with biochemical enrichment strategies, has allowed the discovery of thousands of circRNAs, and of a new layer of gene expression regulation. While studies in circRNAs are becoming a new frontier, there are still more questions than answers on these intriguing noncoding RNA molecules. In particular, little is known about how and when circRNAs are produced and regulated by cis regulatory elements and transacting factors in different tissues and under physiological/pathological conditions. Although roles of a few circRNAs have been described, understanding functions of thousands of circRNAs remains a challenge, for example the biological function of dynamic spatio-temporal expression and subcellular localization of circRNAs in the brain.
Moreover, how circRNAs are degraded in cells also is an important question to be addressed. The stability and specific expression of many circRNAs make them ideal candidates as biomarkers for aging and neurodegenerative diseases. Dysregulation of circRNAs may play a crucial role in many neurodegenerative diseases. Better understanding of molecular mechanisms of circRNAs in disease progression will help developing circRNA-based diagnostic tools and therapeutic strategies for abnormal aging and neurodegenerative diseases such as AD.
Acknowledgments
This work was supported by a grant from the National Science Foundation of China (81471152), the Hirschl/Weill-Caulier Trust (T.S.), and an R01-MH083680-07 grant from the NIH/NIMH (T.S.).
References
- 1.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Huang C, Bao C, Chen L, Lin L, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 7.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Huang C, Wang X, Shan G. Circular RNAs in Eukaryotic Cells. Curr Genomics. 2015;16:312–318. doi: 10.2174/1389202916666150707161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- 11.Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D, Luo J, Wang Y, Tian Q, Feng Q, Huang T, Han B. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 14.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 15.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Flores R, Grubb D, Elleuch A, Nohales MÁ, Delgado S, Gago S. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis δ virus: variations on a theme. RNA Biol. 2011;8:200–206. doi: 10.4161/rna.8.2.14238. [DOI] [PubMed] [Google Scholar]
- 17.Abe N, Hiroshima M, Maruyama H, Nakashima Y, Nakano Y, Matsuda A, Sako Y, Ito Y, Abe H. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem Int Ed Engl. 2013;52:7004–7008. doi: 10.1002/anie.201302044. doi:0.1002/anie.201302044. [DOI] [PubMed] [Google Scholar]
- 18.Abe N, Matsumoto K, Nishihara M, Nakano Y, A Shibata, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, Abe H. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plagens A, Daume M, Wiegel J, Randau L. Circularization restores signal recognition particle RNA functionality in Thermoproteus. Elife. 2015:4. doi: 10.7554/eLife.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen T, Han M, Wei G, Ni T. An intriguing RNA species--perspectives of circularized RNA. Protein Cell. 2015;6:871–880. doi: 10.1007/s13238-015-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Abdelmohsen K, Panda AC, De S, Grammatikakis I, Kim J, Ding J, Noh JH, Kim KM, Mattison JA, de Cabo R, Gorospe M. Circular RNAs in monkey muscle: age-dependent changes. Aging (Albany NY) 2015;7:903–910. doi: 10.18632/aging.100834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Wang Z. Efficient back splicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 35.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C, Shan G. What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription. 2015;6:61–64. doi: 10.1080/21541264.2015.1071301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Kelly S, Greenman C, Cook PR, Papantonis A. Exon Skipping Is Correlated with Exon Circularization. J Mol Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilusz JE. Repetitive elements regulate circular RNA biogenesis. Mob Genet Elements. 2015;5:1–7. doi: 10.1080/2159256X.2015.1045682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taulli R, Loretelli C, Pandolfi PP. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat Struct Mol Biol. 2013;20:541–543. doi: 10.1038/nsmb.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas LF, Saetrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30:2243–2246. doi: 10.1093/bioinformatics/btu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 47.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 52.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 53.Chao CW, Chan DC, Kuo A, Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med. 1998;4:614–628. [PMC free article] [PubMed] [Google Scholar]
- 54.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016 doi: 10.1136/jmedgenet-2016-103758. [DOI] [PubMed] [Google Scholar]
- 56.Pollock A, Bian S, Zhang C, Chen Z, Sun T. Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell Rep. 2014;7:1184–96. doi: 10.1016/j.celrep.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, Diaz Z, Fang X, Shorter J, Krogan NJ, Finkbeiner S, Farese RV, Jr, Gitler AD. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet. 2012;44:1302–1309. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui X, Niu W, Kong L, He M, Jiang K, Chen S, Zhong A, Li W, Lu J, Zhang L. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomark Med. 2016 Jul 12; doi: 10.2217/bmm-2016-0130. Epub ahead of print. [DOI] [PubMed] [Google Scholar]