Abstract
Inflammatory breast cancer (IBC) is a rare and aggressive presentation of invasive breast cancer with a 62% to 68% 5-year survival rate. It is the most lethal form of breast cancer, and early recognition and treatment is important for patient survival. Like non-inflammatory breast cancer, IBC comprises multiple subtypes, with the triple-negative subtype being overrepresented. Although the current multimodality treatment regime of anthracycline- and taxane-based neoadjuvant therapy, surgery, and radiotherapy has improved the outcome of patients with triple-negative IBC, overall survival continues to be worse than in patients with non-inflammatory locally advanced breast cancer. Translation of new therapies into the clinics to successfully treat IBC has been poor, in part because of the lack of in vitro preclinical models that can accurately predict the response of the original tumor to therapy. We report the generation of a preclinical IBC patient-derived xenograft (PDX)-derived ex vivo (PDXEx) model and show that it closely replicates the tissue architecture of the original PDX tumor harvested from mice. The gene expression profile of our IBC PDXEx model had a high degree of correlation to that of the original tumor. This suggests that the process of generating the PDXEx model did not significantly alter the molecular signature of the original tumor. We demonstrate a high degree of similarity in drug response profile between a PDX mouse model and our PDXEx model generated from the same original PDX tumor tissue and treated with the same panel of drugs, indicating that our PDXEx model had high predictive value in identifying effective tumor-specific therapies. Finally, we used our PDXEx model as a platform for a robotic-based high-throughput drug screen of a 386-drug anti-cancer compound library. The top candidates identified from this drug screen all demonstrated greater therapeutic efficacy than the standard-of-care drugs used in the clinic to treat triple-negative IBC, doxorubicin and paclitaxel. Our PDXEx model is simple, and we are confident that it can be incorporated into a PDX mouse system for use as a first-pass screening platform. This will permit the identification of effective tumor-specific therapies with high predictive value in a resource-, time-, and cost-efficient manner.
Introduction
Inflammatory breast cancer (IBC) is a rare, clinically and pathologically unique breast cancer subtype [1] that accounts for 2% to 6% of all breast cancers diagnosed in the United States [2–6] but 7% to 10% of all breast-cancer-related deaths [6–10]. It is the most aggressive subtype of breast cancer and associated with a poor prognosis [6,11–13], due in part to the high frequency of misdiagnosis at the onset of IBC [14–17] and its propensity to rapidly metastasize [18]. Some patients with IBC present without an underlying palpable mass [17,19–20]. The 5-year overall survival rate for patients with primary IBC is typically 62% to 68% [2, 21]. In an attempt to improve this unfavorable outcome, the International Expert Panel on IBC proposed clinical guidelines important for the diagnosis of this disease [17,19, 22]. These guidelines state that in order for IBC to be diagnosed, the clinical presentation should include erythema, edema, peau d’orange, and/or warm breast with or without an underlying palpable mass and a duration of no more than 6 months with erythema occupying at least one-third of the breast. Although patients with primary IBC present with characteristic clinical signs and symptoms [2], the molecular mechanisms modulating these presentations are poorly understood.
Van Laere et al [23] showed that IBC is a heterogeneous disease comprising luminal, HER2-positive, and triple-negative subtypes. The triple-negative subtype of IBC is overrepresented [21, 23], accounting for 20% to 40% of all IBC cases. Although improvements in IBC patient survival have been noted with the introduction of trastuzumab-based systemic therapy to treat patients with the HER2-expressing subtype [21, 24–29], this improved outcome is not seen in the triple-negative IBC group, who are excluded from hormonal therapy and HER2 targeting as treatment options [30–31]. In the absence of any known druggable target, treatment of patients with triple-negative IBC follows the standard-of-care treatment prescribed to patients with triple-negative non-inflammatory breast cancer [1], which is a combination of taxane- and anthracycline-based neoadjuvant chemotherapy followed by modified radical mastectomy including axillary clearance and postoperative chest wall and/or nodal radiotherapy [1, 14, 24]. The reasons for the poor outcomes of treatment in patients with triple-negative IBC are the limited treatment options [23] and onset of resistance to the standard of care therapy [5, 21, 32–34]. There is thus an urgent need to identify new and effective therapies to prolong the disease-free survival of patients with triple-negative IBC.
One of the reasons for the lack of success to date in identifying effective therapies for triple-negative IBC is the use of simplified in vitro cell culture models that fail to recapitulate the complex 3-dimensional (3D) tumor microenvironment as screening platforms to identify new drugs [35–37]. Another reason for the lack of success to date is that the cell lines used in these models may be genetically distinct from tumor cells in patients as a result of their adaptation to growth outside a natural tumor microenvironment [38]. Although complex 3D multicellular tumor models that recapitulate important aspects of the 3D microenvironment have shown promise in the development of new anticancer therapies [35, 37, 39–45], these 3D models fail to replicate tumor heterogeneity [46]. Currently, the most effective preclinical models are patient-derived xenograft (PDX) models [38]. PDX models recapitulate the heterogeneity of tumors in patients and demonstrate genetic stability that closely and stably replicates the human tumor microenvironment [38, 47]. PDX models respond to the standard-of-care therapies in a manner similar to that observed in patients and have played a major part in furthering our understanding of cancer and our ability to develop new effective therapies against cancer. The generation of PDX models, however, requires immense resources and is costly and time-consuming, and thus PDX models have limited utility as platforms for high-throughput drug screens [38, 48]. Ex vivo culture models [39] and organoid models [49–50] are cheaper alternatives that are currently gaining popularity as clinically relevant preclinical models for the development of new and effective anticancer drugs. As these models recapitulate most of the characteristics of the original tumor, they are useful for studying tumor biology and developing tumor-specific therapies.
Herein, we chronicle the development of an IBC PDX-derived ex vivo (PDXEx) model generated from the cellular milieu released from a PDX tumor harvested from a mouse. Our PDXEx model closely replicates the tissue architecture of the original PDX tumor. We show a highly significant correlation between the gene expression signatures of the ex vivo tumor tissue model and the original PDX tumor, suggesting that our PDXEx model retains certain complex intra- and extra-cellular communication networks, cell signaling pathways, and differentiated cell types characteristic of the in vivo tumor and thus will have physiological responses similar to those observed in the in vivo model. We confirm the value of our model in terms of predicting effective tumor-specific therapies and demonstrate its potential application as a drug screening platform to identify novel and effective IBC tumor-specific therapies. Generating the PDXEx model was time, resource, and cost effective, and we predict that incorporating this PDXEx model as a first-pass screening platform within a PDX system will permit the cost- and time-efficient identification of therapies with a high probability of efficacy against the original tumor. Combining the PDXEx model with a PDX system has the potential to identify new therapies that would result in benefit if incorporated into an individual patient’s adjuvant therapy regimen.
Materials and methods
Culture medium
Advanced DMEM (Gibco Life Technologies) was supplemented with Pen/Strep (50 units/ml), antibiotic/antimycotic (1x), glutamine (4 mM), epidermal growth factor (20 ng/ml), and 0.5% fetal calf serum.
Generation of tumors in mice
Animal experiments were conducted in full compliance with MDACC Institutional Animal Care and Utilization Committee (IACUC) policies and procedures (protocol ID: 00001305-RN00). Mice were housed under pathogen-free conditions and treated in accordance with NIH guidelines and standard rodent chow and water were available ad libitum. Tumors were implanted into NSG mice under Ketamine/Xylazine light anesthesia and Ketoprofen was used as an analgesic. At time of tumor harvest, mice were euthanized through CO2 inhalation followed by secondary exsanguination Previously archived, frozen IBC PDX tissue fragments (~100 mg) of the Bcx087 IBC PDX model were thawed and rinsed in PBS. Pieces of tumor tissue were mechanically fragmented with scissors prior to digestion in 3 mL of collagenase A (1 mg/mL solution in PBS) for 15 minutes at 37°C. Digested tissue samples were filtered through a 70-μm pore strainer, and the collagenase A was inactivated by addition of an equal amount of DMEM supplemented with 10% fetal calf serum. Samples were pelleted by centrifugation (400 x g, 2 minutes, room temperature) and resuspended in 1 mL of red cell lysis buffer (Roche) for 2 minutes. Samples were centrifuged, and the cell pellet was resuspended in a 50:50 PBS:Matrigel solution to a final concentration of 2.0x107 cells/mL. Next, 100 μL of the PDX cell suspension was implanted into the mammary fat pad of 6- to 8-week old nude mice (Nu/Nu mice) following the protocol approved by the Institutional Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center. Tumor growth in the mice was monitored once per week until tumors reached a volume of 500 mm3 to 800 mm3. Mice were monitored for signs of morbidity: failure to reach food and water; failure to thrive; inappropriate behavior such as lack of grooming, circling, etc.; a hunched posture, lateral recumbency, or distended abdomen; dyspnea; or weight loss >20%. Moribund animals were euthanized, and tumors and other tissues were removed for weighing and molecular marker studies.
Harvest of tumors
Prior to tumor harvest, mice were euthanized in a chamber containing CO2 from a cylinder as recommended by the American Veterinary Panel on Euthanasia [51]. Every effort was made to minimize the pain and discomfort of all animals used in the proposed work.
Generation of PDXEx models
Freshly harvested PDX tumors were introduced into 10 mL of room-temperature PBS in a sterile 50-mL centrifuge tube and delivered to the laboratory within 30 minutes. Tumors were photographed, weighed, and placed in a 35-mm sterile tissue culture dish containing culture medium. Tumors were mechanically chopped using a pair of sterile scalpels to release the cellular milieu. The cell suspension was passed through a 100-μM filter to separate the homogeneous cell suspension from tissue aggregates, fat, and necrotic tissue, and the concentration of the cell suspension was determined. Ex vivo tumor tissue was generated by magnetic levitation using the Bio-AssemblerTM (Nan03D Biosciences,Inc.) as previously described [52]. Briefly, cells were incubated with nanoshuttleTM (nanoparticle assembly of iron oxide and iron nanoparticles cross-linked with poly-L-lysine) for about 18 to 24 h at a ratio of 1 μL of nanoshuttleTM to 150,000 cells to permit the binding of the iron nanoparticles to the surface of the cells. The nanoshuttle-coated cells were then placed in the Bio-Assembler system to bring the cells into a levitating aggregate (Fig 1A). Cells were maintained within the Bio-Assembler system for 10 to 14 days at 37°C in a humidified environment with medium changed every 3 to 4 days. The best results were obtained from cells released from PDX tumors that were vascular with a fat content between 20% and 30% of the total tumor weight. Tumors weighing 0.8 gram to 1 gram generally yielded the best results.
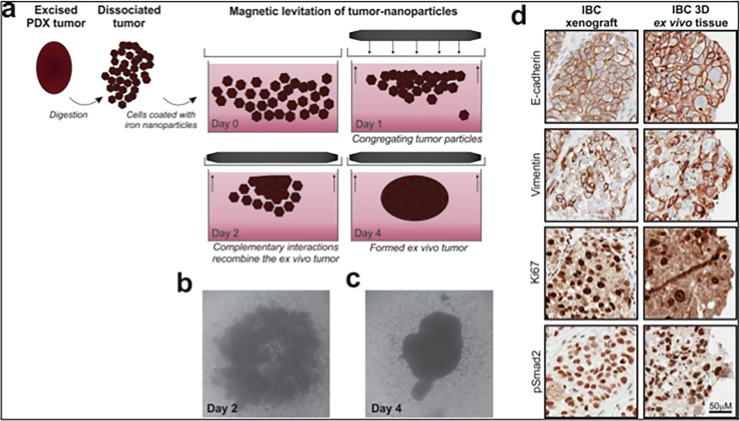
Fig 1. Generation of preclinical PDX-derived ex-vivo model.
(A) A freshly harvested PDX tumor from a mouse was finely chopped to release all its cellular content. The released cells are filtered to separated them form fat and necrotic tissue and tagged with a nanoparticle assembly of iron oxide and iron nanoparticles cross-linked with poly-L-lysine (NanoshuttleTM) by an overnight incubation prior to been placed under a magnetic field. (Bio-AssemblerTM) (n3D Biosciences Inc.). (B) The levitating mass of cells developed into a loose unstructured mass by day 2 of incubation and (C) into a more structured compact mass by day 4 of incubation. (D) Immunohistochemistry analysis of PDX tissue and PDXEx tissue revealed a similar tissue architecture and staining for E-cadherin, Vimentin, Ki67 and pSMAD2.
Magnetic bio-printing of PDXEx to generate an in vitro screening platform
Cells harvested from IBC PDX tumors were incubated overnight with nanoshuttleTM as described in the preceding section. The iron nanoparticle–coated cells were then dispensed into a 96-well tissue culture plate placed on a 96 well magnetic drive (Fig 2A) at a final concentration of 75,000 cells per well and incubated at 37°C in a humidified environment.
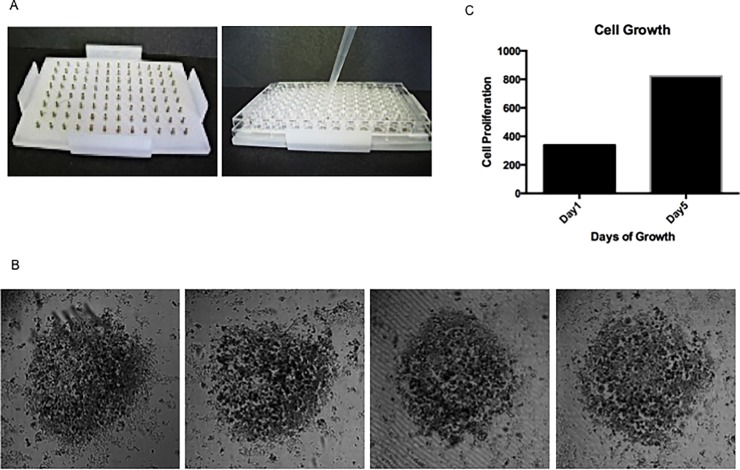
Fig 2. Tissue characteristics of the ex-vivo tumor model.
(A) Bio-printing cells–Tumor cells tagged with a nanoparticle assembly of iron oxide and iron nanoparticles cross-linked with poly-L-lysine (NanoshuttleTM) were dispensed into an ultralow attachment 96 well tissue culture plate placed on a 96 well magnetic drive (n3D Biosciences, Inc). (B) Morphology (4x magnification) of the ex-vivo tumor tissue prints after 5 days of growth at 37°C revealing uniformly sized structures. (C) Proliferative capacity of the PDXEx tissue following 5 days of culture at 37°C.
Purification of total RNA
Total RNA isolated from the PDX tumors as well as from the ex vivo tumor tissue after 5 days of growth in culture was purified with the Trizol method (Invitrogen) according to the manufacturer’s instructions.
Whole transcriptome expression profiling
Whole transcriptome expression profiling was performed using an Affymetrix GeneChip human Clariom D array (PN 902923, Life Technologies). Total RNA sample labeling and processing, GeneChip hybridization, and scanning were performed according to instructions in the Affymetrix user manual of GeneChip WT PLUS Reagent Kit–“Target Preparation for GeneChip Whole Transcript Expression Arrays (P/N 703174)”. Briefly, the first-strand cDNA was reverse synthesized from 100 ng of total RNA spiked in controls using a whole transcript plus kit, using primers containing a T7 promoter sequences (Genset, La Jolla, CA). cRNAs were generated and amplified by in vitro transcription from double-strand cDNA templates containing T7 promoter. Fifteen micrograms of purified cRNA was reverse transcribed into second-cycle single-strand sense cDNA using second-cycle primers and dNTP mix with fixed dUTP/dTTP ratio in reaction mix. The single sense strand cDNA in second cycle was released from RNA/DNA hybrids by RNase H digestion. Then 5.5 μg of purified sense-strand cDNA was fragmented by uracil-DNA glycosylase and apurinic/apyrimidinic endonuclease 1 at the unnatural dUTP residues to break the DNA strand. The fragmented cDNA was labeled by terminal deoxynucleotidyl transferase using the Affymetrix proprietary DNA labeling reagent, which is covalently linked to biotin. Five micrograms of biotin-labeled sense-strand cDNA fragments in 200 μL of hybridization mix was loaded onto a human Clariom D array and hybridized to probes on the array in a GeneChip Hybridization Oven 645 (Affymetrix) for 16 h at 45°C with 60 rpm rotation. After hybridization, each array was washed and stained with streptavidin–phycoerythrin (Invitrogen), and the signal was further amplified with biotinylated anti-streptavidin antibody (Vector Laboratories) on the GeneChip Fluidics Station 450 (Affymetrix). Arrays were scanned with the GeneArray G7 scanner (Affymetrix) and quantified into CEL files with image and signal intensities.
Bioinformatic analysis
The CEL files generated were analyzed through Transcriptome Analysis Console 4.0, which normalizes (and applies the log2 function to) array signals using a robust multiarray averaging algorithm. The Spearman's rank-order correlation test was applied to measure the strength of the association between gene expression in different arrays. The graphical representation of data and statistical analysis were performed in R (version 3.0.1) (http:///www.r-project.org/), and statistical significance was defined as p less than 0.05.
Immunohistochemistry
Excised tumor tissue was fixed in formalin and archived in paraffin. For analysis of proteins by immunohistochemistry, 5-μm slides were cut from archived paraffin blocks and subsequently adhered to glass slides. The sections were deparaffinized, rehydrated, and subjected to citrate buffer antigen retrieval. Staining was performed on an automated immunohistochemical autostainer (Lab Vision Corp.). Briefly, the slides were blocked for nonspecific binding and incubated with relevant antibodies for 1 hour at room temperature. The primary antibody was detected with the labeled streptavidin biotin plus peroxidase kit (DAKO). Slides were developed with DAB chromogen to reveal immunoreactivity, counterstained with hematoxylin, and covered with a glass coverslip. Antibodies used for immunohistochemistry were as follows: anti-E-Cadherin rabbit mAb (1:200) (Cell Signaling Technology, clone 24E10, #3195), anti-Vimentin XP rabbit mAb (1:200) (Cell Signaling Technology, clone D21H3, #5741), anti-Ki67 rabbit mAb (1:200) (GeneTex, GTX16667), and anti-pSmad2 (Ser465/467) rabbit mAb (1:200) (EMD Millipore, clone A5S, #04–953).
Statistical analysis of the high-throughput anticancer drug screen
Median scores of viability across 3 replicates were normalized relative to the plate-level controls (i.e., the median viability was divided by the median viability of the controls). The resulting values were then ranked from lowest to highest. Drugs ranking highest exhibited the strongest reduction in viability relative to the control. The statistical analysis for the in-vivo data described in the section “Value of the PDXEx model for predicting tumor response to drugs” is present in the paper by KURT et al. [53].
High-throughput anticancer drug screen
Using robotics, 10 μL of the small molecules comprising the Anti-Cancer 386 Compound Library (Selleckchem) were dispensed into each well of a 96-well bio-printed PDXEx. Each well in the array contained 100 μL of medium. The small molecules were dispensed into 3 sets of plates for final concentrations of 1 μM, 0.5 μM, and 0.25 μM. Each drug concentration set was performed in triplicate. The bio-printed PDXEx was incubated with the drug concentrations for 5 days. Viability of the ex vivo tumor tissue was determined on day 5 using the Cell Titer Glo Luminescent cell viability assay (Promega).
Results and discussion
Generation of the PDXEx model
Using previously described methodology [52], the cellular milieu released from a Bcx087 (triple-negative IBC) PDX tumor was tagged with a nanoparticle assembly of iron oxide and iron nanoparticles cross-linked with poly-L-lysine (NanoshuttleTM) and placed under a magnetic field (Bio-AssemblerTM), resulting in the cells’ been drawn together into a levitating mass (Fig 1A). Prolonged incubation of the levitating cells at 37°C resulted in a morphological change of this cellular mass. A loose, poorly organized structure was observed after 2 days of incubation (Fig 1B), and this structure gradually transformed into a more structured compact mass by day 4 (Fig 1C). To determine the similarity between the tissue architecture of the original PDX tumor and the PDXEx model, tissue sections were generated from the respective samples and analyzed by immunohistochemistry. We observed a high degree of similarity between the tissue architecture of the 2 sets of tissue (Fig 1D). In addition, subpopulations of IBC cells within tumor xenografts and our PDXEx tissues were found to be Ki67 positive, which is indicative of active proliferation in both tissues. Positive staining was also for E-cadherin, Vimentin, and pSMAD (Fig 1D). E-cadherin is highly expressed in IBC [54]. The similarities between the 2 sets of tissue samples with respect to tissue architecture and expression levels of these biomarkers suggested that the PDXEx model closely recapitulates the tumor microenvironment of the original PDX tumor.
Comparison of molecular signatures of the original IBC PDX tumor and the IBC PDXEx model
Having demonstrated that our IBC PDXEx model retained both the tissue architecture and expression profile of key biomarkers of the original PDX tumor, it was important to confirm that our model also retained the molecular signature of the original PDX tumor, which would allow our model to serve as a relevant preclinical platform for high-throughput drug screens. To compare the molecular signatures, we utilized magnetic 3D bio-printing (n3D BiosciencesTM, Inc.), which has been shown to have a wide range of applications in tissue engineering [55–60]. Briefly, viable cells released from the PDX tumor and incubated with poly-L-lysine–tagged magnetic nanoparticle assembly for 24hr were harvested and dispensed into a 96-well ultra-low attachment tissue culture plate at a concentration of 50,000 cells per well, and the plate was placed on a bed of magnets (Fig 2A). Well-formed and equally sized PDXEx “bio-prints” were observed at 5 days of incubation, a time corresponding to the duration of the planned high-throughput drug screens (Fig 2B).
We predict that using the cellular milieu of an entire tumor to generate the PDXEx bio-prints for use as a screening platform will results in the majority of the cellular components of the heterogeneous tumor being distributed throughout the PDXEx bio-print. At present, we typically harvest 60 million to 90 million cells from a single PDX tumor weighing approximately 0.8 to 1.2 grams. This yield of cells permits the bio-printing of approximately twelve 96-well-plate arrays. This number is sufficient to perform a high-throughput drug screen in triplicate utilizing a 380-compound drug library. We are currently working towards developing a 384-well tumor tissue array to screen larger drug libraries.
For our preclinical PDXEx model to serve as a clinically relevant screening platform, (i) the cells in the model must retain the capacity to proliferate in culture to enable relatively rapid assessment of the anti-proliferative activity of novel therapeutic agents, and (ii) the method used to produce the model must produce robust and reproducible results with respect to similarity between the genetic signature of the original PDX tumor and the genetic signature of the PDXEx model. To assess the proliferative capacity of our PDXEx model, the viability of the bio-printed tissue was determined at 24 hours and 5 days after printing using the Cell Titer Glo Luminescent cell viability assay. The rationale for determining cell viability after 5 days of culture is that when the bio-printed PDXEx platform is used for drug screens, the bio-printed PDXEx will be grown in the presence of the drugs for 5 days before viability is measured. Our data clearly showed tissue proliferation during this 5-day period (Fig 2C).
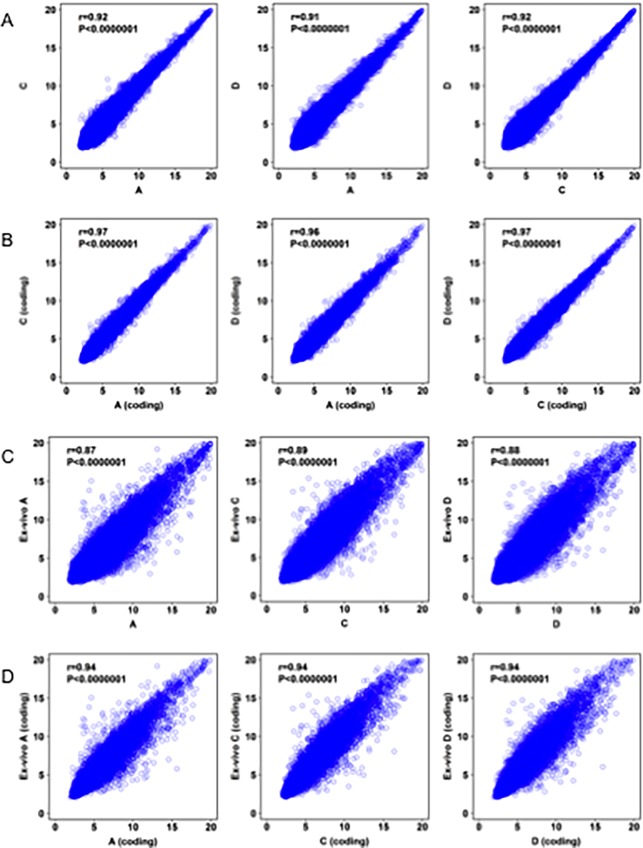
To evaluate the robustness and reproducibility of the method used to generate the PDXEx model, we performed the method 3 times using PDX tumors harvested from 3 host mice, all of which had been implanted with the same original PDX tumor tissue. To ensure that the harvested PDX tumors had similar genetic signatures, we isolated purified RNA from these tumors and analyzed it using Clarion D human Affymetrix arrays comprising more than 6,765,500 probes derived from >134,700 characterized genes of both poly(A) and non-poly(A) mRNA, to provide complete and unbiased coverage of the transcriptome and profile differentially expressed genes. This complete gene set comprises coding genes, noncoding genes, microRNA, precursor micro RNA, pseudogenes, small RNA, and unassigned genes. The Spearman rank correlation was used to test the strength of the association in gene expression profiles between the 3 PDX tumors. We observed a highly significant genetic correlation (>90%) between the PDX tumors harvested from the 3 mice (Fig 3A). A higher degree of correlation (>96%) was observed when only the coding genes were selected (Fig 3B). These data indicated that the host mice exerted a minimal effect on the genetic signature of the original PDX tumor.
Fig 3. Correlation of gene expression profiles between three PDX tumors harvested from mice implanted with tissue from the same original PDX tumor.
(A) Highly significant correlation between the entire gene set between tumor A and C, A and D and D and C (B) Selecting the coding genes in this analysis increased the significance of correlation. (C) Highly significant correlation between the entire gene set between each PDX tumor and its corresponding PDXEx tumor model (D). Selecting the coding genes in this analysis increased the significance of correlation.
Next, in a similar manner, we determined the significance of the association in gene expression profiles between each PDX tumor and the resulting PDXEx model. Here too, a highly significant correlation (>87%) was observed between the gene expression profiles within each set (Fig 3C), and a higher correlation (>90%) was observed when only the coding genes were selected (Fig 3D).
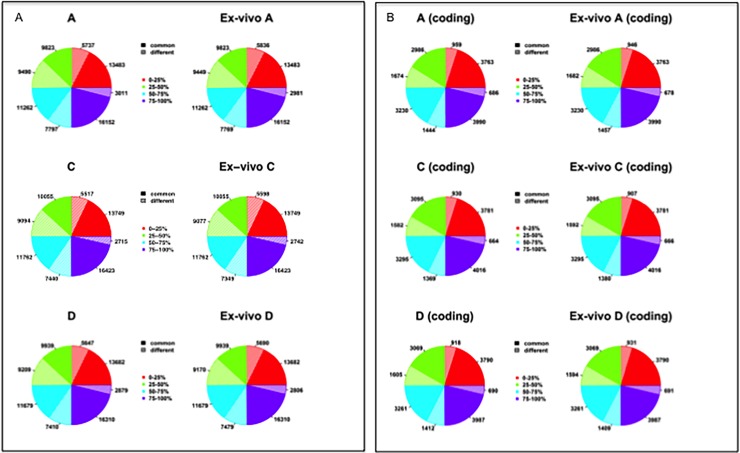
We next determined the proportions of genes that were similarly expressed and differentially expressed between each PDX tumor and the resulting PDXEx model. We first performed the analysis with the entire gene set (Fig 4A) and then performed the analysis with only the coding genes (Fig 4B). For the sake of completeness, the genes were divided into 4 quantiles according to their expression levels. For original PDX tumor A, patterns were as follows: for genes in the top quantile (76%-100%), 59% of all genes and 69% of coding genes had similar expression levels between the PDX tumor and the resulting PDXEx model; for genes in the third quantile (56%-75%), 84% of all genes and 85% of coding genes had similar expression levels; for genes in the second quantile (26%-50%), 51% of all genes and 64% of coding genes had similar expression levels; and for genes in the first quantile (0–25%), 70% of all genes and 80% of coding genes had similar expression levels. Comparisons of gene expression between the original PDX tumor and PDXEx model for original PDX tumors B and C showed similar results (Fig 4A and 4B). We speculate that the differences between quadrants in the ratios of similar and differential gene expression observed in each PDX/PDXEx tumor model set are associated with the heterogeneity of each tumor and PDXEx model and not a result of a genetic drift resulting from the generation of the PDXEx model nor the biocompatibility of the nanoparticles. Studies previously performed to address the biocompatibility of the nanoparticles [52, 57, 60–66] confirmed that the nanoparticles are bio-inert as they do not affect cell viability, oxidative stress, protein expression, inflammatory response, or stem cell differentiation. Our finding that the majority of genes and especially the majority of coding genes had similar expression in PDX tumors and their PDXEx models suggests that our model recapitulates important properties of the tumor microenvironment of the original tumor. This possibility needs further investigation, which is currently under way.
Fig 4. Comparison of genes either similarly or differentially expressed.
(A) Entire gene set divided into 4 quantiles based on their level of expression to enable determination percentage of similarly expressed genes and differentially expressed genes. (B) Coding gene set divided into 4 quantiles based on their level of expression to enable determination percentage of similarly expressed genes and differentially expressed genes.
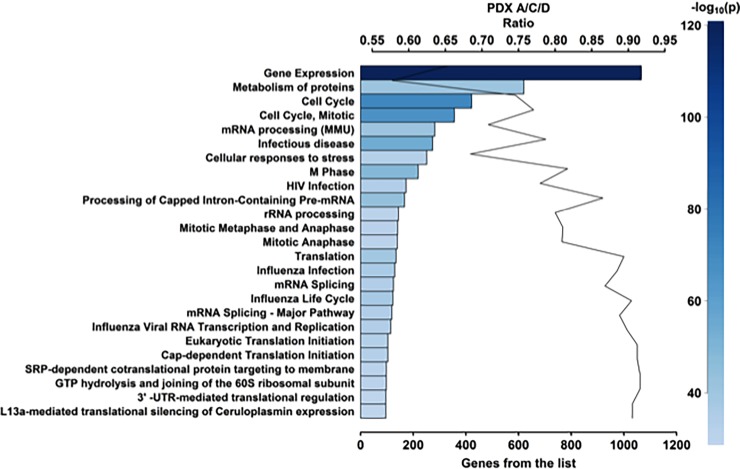
The lack of clearly defined molecular drivers for IBC contributes to the difficulty of identifying effective therapies to treat this disease. We too were unable to identify unique molecular drivers of the Bcx087 IBC PDXEx model. In Table 1, we present the top 50 genes based on their expression levels. Twenty-two of these genes are members of the family of ribosomal proteins. Two potential targets within this group are the redox regulator thioredoxin-1 (Txn-1) and aspartate beta-hydroxylase (ASPH). Thioredoxin-1 [67–69] and aspartate beta-hydroxylase [70] are highly expressed in many cancers and contribute to the aggressive nature of these cancers. We are currently investigating the role of these molecules in the biology of IBC and determining if targeting these molecules will improve our ability to treat this very aggressive subtype of breast cancer. Applying gene ontology to our derived expression data, we identified several distinct biological pathways that are associated with Bcx087 tumors (Fig 5) (Table 2). As IBC is a rapidly growing tumor, it is not surprising that the pathways with the highest numbers of highly expressed genes included those regulating the cell cycle, metabolism, and protein metabolism.
Table 1. Gene expression levels of the top 50 ranked genes.
| Gene Symbol | Fold Expression | Gene Symbol | Fold Expression |
|---|---|---|---|
| RPS7 | 19.928 | PPIA | 18.838 |
| RPL30 | 19.928 | RPS6 | 18.837 |
| TXN | 19.928 | HSPE1 | 18.81 |
| RPS21 | 19.725 | MTRNR2L8 | 18.8 |
| RPD20 | 19.723 | CANX | 18.763 |
| ASPH | 19.578 | RPL38 | 18.762 |
| LAPTM4A | 19.545 | CRABP2 | 18.637 |
| RPL41 | 19.51 | TCEB1 | 18.617 |
| RPS15A | 19.508 | SNRPE | 18.613 |
| HSP90AB1 | 19.502 | RPS24 | 18.59 |
| RPL35A | 19.453 | RPL9 | 18.553 |
| HSPA8 | 19.445 | YWHAQ | 18.515 |
| RPS23 | 19.438 | SOD1 | 18.29 |
| RPL37A | 19.323 | RPN1 | 18.242 |
| THEM123 | 19.272 | RPL39 | 18.215 |
| COX6C | 19.2 | RPL31 | 18.155 |
| TMSB10 | 19.168 | RPL7 | 18.145 |
| COX7C | 19.167 | NDUFA1 | 18.122 |
| NDUFS5 | 19.065 | VKORC1 | 18.115 |
| H3F3A | 19.054 | RPS12 | 18.102 |
| RPS8 | 19.028 | TPT1 | 10.062 |
| ITGB1 | 18.97 | RPL27 | 18.023 |
| EEF1A1 | 18.917 | ATP6VOE1 | 18.081 |
| UBB | 18.89 | ILF2 | 18.015 |
| RPL34 | 8.882 | RPL5 | 17.94 |
Fig 5. Layout of biological pathways common to all 3 PDX tumors.
The Top 25 out of 1116 pathways are presented.
Table 2. Top 25 pathways among 1116 identified to be activated in the Bcx087 IBC model.
| Pathway | Number | Total | pValue |
|---|---|---|---|
| Gene expression | 1066 | 1631 | 3.10E-121 |
| Cell Cycle | 422 | 566 | 3.40E-72 |
| Cell Cycle, Mitotic | 356 | 462 | 2.50E-67 |
| Infectious disease | 274 | 348 | 2.26E-55 |
| M Phase | 219 | 268 | 1.66E-49 |
| Processing of capped intron-containing pre mRNA | 167 | 193 | 9.64E-45 |
| Metabolism of Proteins | 620 | 1074 | 1.16E-41 |
| mRNA processing (MMU) | 282 | 398 | 3.17E-41 |
| Translation | 135 | 151 | 6.03E-40 |
| Influenza life cycle | 123 | 136 | 9.86E-38 |
| Influenza Infection | 130 | 147 | 2.49E-37 |
| Influenza viral RNA Transcription and Replication | 115 | 128 | 1.09E-34 |
| mRNA Splicing–Major Pathway | 119 | 134 | 1.10E-34 |
| HIV Infection | 173 | 222 | 2.63E-34 |
| mRNA Splicing | 125 | 144 | 4.25E-34 |
| Cap-dependent Translation initiation | 104 | 114 | 6.70E-33 |
| Eukaryotic Translation initiation | 104 | 114 | 6.70E-33 |
| Cellular response to stress | 251 | 367 | 8.55E-33 |
| Mitotic Metaphase and Anaphase | 141 | 174 | 1.55E-33 |
| GTP hydrolysis and joining of the | 98 | 107 | 1.99E-31 |
| SRP-dependent co-translational protein targeting to membrane | 98 | 107 | 1.99E-31 |
| Mitotic Anaphase | 140 | 173 | 3.36E-31 |
| rRNA processing | 144 | 180 | 5.05E-31 |
| L13a-mediated translational silencing of Ceruloplasmin expression | 96 | 106 | 7.84E-30 |
| 3’-UTR-mediated translational regulation | 96 | 106 | 7.84E-30 |
Value of the PDXEx model for predicting tumor response to drugs
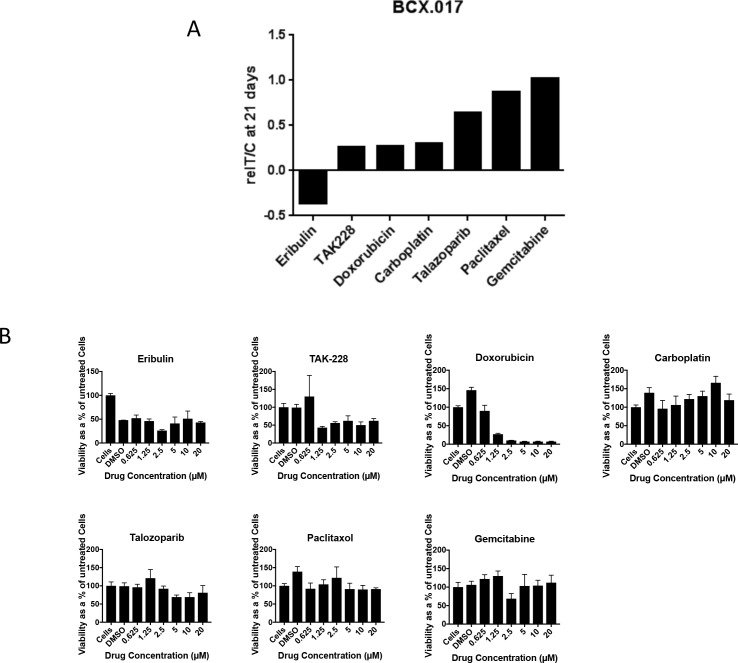
To determine the value of our PDXEx model for predicting tumor response to drugs, we performed a pilot drug screen utilizing this platform and determined the similarity between the drug response profile identified in the in vitro screen and the drug response profile identified in a previously described PDX mouse system [53]. Both systems were generated from the same original PDX tumor tissue and treated with the same panel of chemotherapeutic agents: eribulin, TAK228, doxorubicin, carboplatin, talazoparib, paclitaxel, and gemcitabine. The Bcx017 triple-negative IBC PDX model was used in this comparative study. Drug doses ranged from 0.625 μM to 20 μM (Fig 6). The drug response profile identified using our PDXEx model (Fig 6B) mirrored that identified in the corresponding animal study (Fig 5A) except in the case of carboplatin, which showed an anti-tumor effect in the in vivo PDX mouse study but not in the PDXEx model. These findings confirmed the value of the PDXEx model in identifying effective tumor-specific therapies. The outcome of this study strengthens our belief that incorporating this PDXEx model within a PDX system to serve as a high-throughput screening platform will permit the identification of effective tumor-specific therapies in a time- and cost-efficient manner.
Fig 6. Comparative drug response profile between IBC PDX Bcx017 mouse model and Bcx017 PDXEx tissue model.
A) Actively growing (~200–350 mm3) implants in mice were grouped and treated with the indicated drugs for 21days. The graph summarizes data from a previously published in vivo study [69]. B) Tumor cells tagged with a nanoparticle assembly of iron oxide and iron nanoparticles cross-linked with poly-L-lysine (NanoshuttleTM) were bio-printed into ultra low attachment plates. The PDXEx bio-print was treated with the indicated drugs at the indicated dose range for 5 days.
Proof-of-principle drug screen utilizing the PDXEx platform
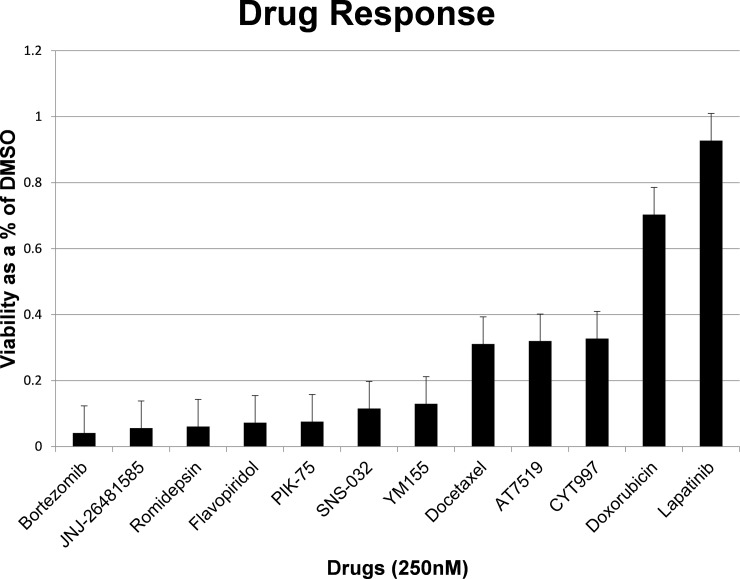
Utilizing the Bcx087 bio-printed triple-negative IBC PDXEx platform, we performed a high-throughput small molecule screen with the Anti-Cancer 386 Compound Library (Selleckchem). The drug screen was performed in triplicate, and drug sensitivity as a percentage of the sensitivity of the solvent (DMSO)-treated cells was determined by measuring cellular proliferation using the Cell Titer Glo Luminescent cell viability assay (Promega). The drug screen was performed at drug concentrations of 1 μM, 0.5 μM, and 0.25 μM, and the platform was incubated with the drugs at 37°C for 5 days. Included in this screen were similar concentrations of the standard-of-care drugs doxorubicin and docetaxel [24], which served as positive controls. As the Bcx087 PDX model is a triple-negative IBC model, lapatinib, which targets HER2, was selected as the negative control. Following statistical analysis, we identified the following drugs as our lead candidates with activity superior to that of the standard-of-care drugs at a concentration of 0.25 μM: bortezomib (potent 20s proteasome inhibitor), quisinostat (JNJ-26481585) (second-generation histone deacetylase [HDAC] inhibitor), romidepsin (potent HDAC 1 and 2 inhibitor), flavopiridol (CDK 1, 2, 4, and 6 inhibitor), PIK-75 (p110a inhibitor), SNS-032 (selective inhibitor for CDK2, also inhibits CDK 1, 4, 7, and 9), and YM155 (potent inhibitor of survivin) (Fig 7). In, addition, AT7519 (CDK inhibitor) and CYT997 (microtubule associated) had activity similar to that of the standard-of-care agent docetaxel (Fig 7). Among the 9 top drug candidates, 3 were CDK inhibitors and 2 were HDAC inhibitors. Analyzing the list of genes whose expression levels placed them within the top 25% of the coding genes, we observed that 13 CDK family members were present in this cohort of genes, including CDK1, CDK2, CDK4, and CDK7 and the 9 targets of the CDK inhibitors identified in our screen. Members of the Histone families 1, 2, and 3 were also well represented in this cohort of genes and could have been targets of the identified HDAC inhibitors.
Fig 7. Lead candidates identified from a high throughput drug screen utilizing the PDXEx screening platform.
A high throughput small molecule screen was performed utilizing the Bcx087 triple negative PDXEx screening platform and the small molecule Anti-Cancer 386 Compound Library (Selleckchem). The ex vivo tumor tissue array was incubate with the drugs at 37oC for 5 days. The top hits were identified as agents having a superior anticancer effect to that of docetaxel and doxorubicin at 250nM.
Anticancer drugs developed utilizing 2-dimensional cell culture model systems have had minimal success when translated to the clinic [37]. This failure is often due to the fact that primary tumor cells grown on 2-dimensional monolayer cultures do not accurately mimic cells in the tumor microenvironment [37]. Our group [71] and others [35, 37, 72–73] have identified clinically relevant targets for drug development utilizing 3D cell culture models. However, even 3D cell culture models have limitations as they can still fail to capture the heterogeneity associated with the tumor’s microenvironment. Moreover, often utilized immortalized cancer cell lines, because of their long period of growth outside their natural environment, have acquired unique genetic changes not typically seen in the corresponding tumors in patients [38]. Currently, the most clinically relevant models for drug development are PDX models [38]. Although PDX models are furthering our understanding of cancer biology and our ability to develop more effective cancer therapies, establishing these models is time consuming, cost prohibitive, and resource intensive [38, 48]. In addition, it is not logistically feasible to use PDX model systems for high-throughput screening. One of the approaches to circumvent the need for PDX model systems is the generation of ex vivo cultures of primary human tissue. These are preclinical models generated from tumor tissue slices and have been shown to recapitulate the tissue architecture of the original tumor and key features of the tumor microenvironment, including tumor heterogeneity [74]. The use of ex vivo model systems in a preclinical setting enables robust quantitative evaluation of clinically relevant anticancer therapies, drug resistance, and biomarkers and thus contributes significantly to identifying successful tumor-specific therapies and improving patients’ quality of life. Another benefit of ex vivo models is that by permitting recapitulation of inter-patient heterogeneity, they can be used to differentiate responders from non-responders to a treatment and thus aid in the selection of patients for clinical trials [74]. All previously published organoid models [75–79], including our PDXEx model, are promising models developed for identifying patient-specific therapies. These models have phenotypic and genomic profiles highly similar to those of the original PDX tumor. However, the lack of an immune system and angiogenesis system represents a limitation of the culture system, especially when the aim is to study tumor vasculature and immune interactions. In this regard, we deem it necessary to validate all therapeutic outcomes determined from our PDXEx model system in patient-derived xenografts (PDXs) generated from the same original tumor. PDX models have proved to be useful models for human cancer, because they retain tumor histopathology, including tumor-infiltrating immune cells and generation of tumor vasculature. The importance of validation in PDXs was recently highlighted in a study by Vlachogainnis et al [75]. They tested the ability of their organoid model to recapitulate response to regorafenib, a multiple tyrosine kinase inhibitor blocking oncogenic and angiogenic signaling pathways. No response to regorafenib was observed in their organoid screening assays. However, strong inhibition of tumor growth was observed in an orthotopic human tumor xenograft model. The authors concluded that regorafenib is mainly driven by its antiangiogenic effect.
The IBC PDXEx model that we describe appears to have potential as a solution to the current lack of suitable models for high-throughput identification of effective tumor-specific therapies for IBC. In this study, we clearly showed that i) the tissue architecture of our PDXEx model was very similar to that of the original tumor (Fig 1D), ii) there was a highly significant correlation between the gene expression profiles of the PDXEx model and the original PDX tumor (Fig 3B, 3C and 3D), and iii) our PDXEx model demonstrated a significant predictive power in identifying tumor-specific therapies (Fig 6A and 6B).
Our PDXEx model is simple in design, taking only 11 days to reveal a tissue architecture similar to that of the original PDX tumor (Fig 1). In comparison, it takes about 2 months to grow IBC PDX tumors in mice. At present, we have generated our PDXEx model from PDX tumors and thus have to consider the time taken for the generation of the PDX system. Our future goal is to generate our model directly from human tumor tissue and use it as a screening platform to identify patient-specific therapies. Success in this endeavor will enable us bypass the need for PDX systems and significantly shorten the time it would take to identify patient-specific therapies. Our bio-printed PDXEx screening platform will permit screening of a large number of drugs in just a few days. Incorporating our system as a first-past screening platform to identify the most promising lead agents will cut down on the number of agents that ultimately need to be tested in mice. Our PDXEx screening platform closely replicates the cellular milieu of a tumor microenvironment. To determine the degree to which the tumor microenvironment of the PDX tumor was represented in the PDXEx model derived from this PDX tumor, we analyzed our microarray data from three independent PDX/PDXEx sets for the expression profiles of genes representing the stroma and ECM. The stoma comprises fibroblasts, vasculature (endothelial cells), tumor-associated inflammatory cells and immune cells. Cancer-associated fibroblasts (CAF) are the predominant cell type in the stromal component of the tumor environment [80–81] and are identified by a gene signature comprising ã-smooth muscle actin (ã -SMA), fibroblast activating protein (FAP), tenascin-C (TNC), CD36, IL6 and platelet-derived growth factor ã and ß (PDGFR ã / ß). Our microarray data revealed the levels of expression of fibroblast activating protein (FAP), tenascin-C (TNC), CD36, IL6 and platelet-derived growth factor ã and ß (PDGFR ã / ß). The expression levels of CD34 (endothelial cells), CD24 (B-lymphocytes) and CD3E (T-lymphocytes) were also determined. The ECM is a complex network of macromolecules and is built up of a large collection of biochemically distinct components, including proteins, glycoproteins, proteoglycans, and polysaccharides [82–83]. An acceptable ECM gene signature comprises components of the collagen chain, matrix metalloproteinases (MMP2, MMP11 and MMP14) and ECM regulation and modeling molecules such as SPARC, LoxL1, ADAM12, Plod2 and PALUR [82–83]. Comparing the expression levels of this panel of genes, we observed a high degree of correlation of this panel of genes between each PDX and its corresponding PDXEx. The high correlation of gene expression within this panel of genes suggests a high probability that the our PDXEx model was able to generate a tumor microenvironment similar to that of the original PDX tumor.
Reproducing the tumor microenvironment with a high degree of fidelity is a requirement for any cell culture system used for drug development because the tumor microenvironment not only influences the development of cancer but also has a strong influence on the tumor response to radiation and chemotherapy [84]. The high degree of correlation in gene expression profiles between the PDX tumor and PDXEx model for all 3 of the original PDX tumors shows that the methodology used in the generation of the PDXEx model was both robust and reproducible. Furthermore, the fact that all 3 of the PDXEx models had gene expression similar to that in the original PDX tumor clearly indicates the suitability of our PDXEx model for high-throughput drug screens to identify new and effective therapies and for other studies to further our understanding of IBC.
The diagnosis of IBC relies largely on its clinical presentation; despite a characteristic phenotype, the molecular mechanisms underlying IBC are poorly understood [2]. Analysis of the gene expression profile of our IBC PDXEx model (Table 1) and pathway analysis (Fig 5) (Table 2) also failed to demonstrate a key underlying pathway or driver gene or genes responsible for IBC. The lack of targeted molecular mechanism in triple-negative IBC together with the evidence that patients with triple-negative IBC derive limited benefit from standard therapy [85] highlights the importance of a PDXEx screening platform to identify new and effective therapies for IBC. We predict that by identifying potential therapeutic agents, we could work backwards to further our understanding of the molecular mechanisms mediating the onset and progression of IBC and perhaps identify better therapeutic targets for IBC. We performed a 386-drug Anti-Cancer Compound Library screen utilizing our PDXEx platform, and at a concentration of 250 nM, we identified 7 drugs with a stronger growth inhibitory effect than the standard-of-care drugs and 2 drugs with a growth-inhibitory effect similar to that of the positive control docetaxel (Fig 7). The 9 top candidates were 1 proteosome inhibitor, 2 HDAC inhibitors, 3 CDK inhibitors, 1 PI3K inhibitor, 1 potent inhibitor of survivin, and 1 microtubule inhibitor. HDAC inhibitors [86] have shown favorable outcomes in the treatment of IBC. PI3K/AKT signaling has been suggested to be important for the progression of IBC, and thus PI3K inhibitors might produce favorable outcomes in patients with IBC. CDK inhibitors have been shown to be important for the treatment of hormone-responsive breast cancer [87] and, according to our data, might also be important for the treatment of IBC. The fact that 6 of the 9 top drug candidates identified in our study have published evidence suggesting that the agent might produce favorable outcomes in IBC supports the potential of our preclinical model to identify effective tumor-specific therapies. Our findings also suggest that inhibiting survivin might lead to clinical benefits for patients with triple-negative IBC. Survivin, the smallest member of the inhibitor of apoptosis protein family, is overexpressed in cells of almost all cancers but not in most normal tissues in adults. Survivin expression is required for cancer cell survival, and knocking down its expression or inhibiting its function using molecular approaches results in spontaneous apoptosis. Thus, survivin is an attractive and perhaps ideal target for cancer drug discovery [88]. Studies are currently under way to further our understanding of the role of survivin in IBC and the clinical benefits of inhibitors of survivin as single agents or in combination with other potential therapeutic agents for the treatment of IBC and triple-negative IBC. We are currently planning an in vivo study of the Bcx087 PDX model to validate the top drug candidates identified in our bio-printed PDXEx platform.
Although our PDXEx model has potential clinical benefits, it also has one minor drawback. Despite many attempts, we were unable to prolong viability of our ex vivo tumor tissue beyond 2 weeks. We do not consider this outcome a major impediment to use of our model since most ex vivo models derived from slices of tumor maintain cell viability over culture periods of at least 1 week [74]. Similar to what is true for other ex vivo models, use of our PDXEx model as a screening platform requires good availability of biopsy tissue, which could be a limitation in patients with IBC. However, the use of PDX tumor tissue harvested from mice will overcome this limitation, affording us the ability to identify agents with a high likelihood of clinical efficacy before clinical trials or even animal studies. This outcome would undoubtedly improve the efficiency of translating preclinical research to clinical practice. Incorporating ex vivo culture of IBC tumors into the development pipeline could help us to achieve this outcome, and this approach has been increasingly used to investigate potential prostate cancer therapeutics [74] using tissue pieces. Ex vivo models such as organoid models have been generated from specific cells within tumors with characteristics similar to those of the original tumor [79]. The advantage of these systems is that they can be maintained for long periods of time and used to generate tissue biobanks. Generating such models from IBC tumors is limited by the nature of the disease. At present, no unique structure has been identified in IBC that could be used for the generation of organoid structures; thus, our model might be the best for IBC.
Conclusions
Further work is needed to determine if our preclinical PDXEx platform will serve as a clinically relevant predictor of tumor response to drugs and improve understanding of the biology of IBC. We have shown in the work described here the importance that ex vivo cultures can enable quantitative evaluation of multiple drugs. We postulate that if a drug cannot elicit an anti-proliferative or proapoptotic response in a high proportion of patient tumors in an ex vivo system, that drug is unlikely to be effective in the clinics. Here, we show the importance of ex vivo cultures enabling the evaluation of drug efficacy using a high-throughput strategy. We are confident that our preclinical ex vivo tumor tissue array platform will ultimately be confirmed as a clinically relevant predictor of tumor response to drugs and improve understanding of the biology of IBC.
Acknowledgments
We thank Stephanie Deming for her editing assistance and Sandra Bishnoi, an IBC patient advocate, who through her courage battling IBC is an inspiration to us all.
Data Availability
Array data has been submitted to the GEO database (accession number GSE106685).
Funding Statement
This work was supported by an Avon Foundation grant 02-2015-067 (https://www.avonfoundation.org/programs/breast-cancer/) to Geoffrey A. Bartholomeusz, the Morgan Welch Inflammatory Breast Cancer Research Program and Clinic (https://www.mdanderson.org/research/departments-labs-institutes/programs-centers/inflammatory-breast-cancer-research-program.html) to Geoffrey A. Bartholomeusz, Cancer Prevention and Research Institute of Texas grants RP110532 and RP150578 (www.cprit.state.tx.us) to Arvind Rao, an American Cancer Society grant RSG-16-005-01 (Cancer.org) to Arvind Rao, and a Cancer Center Support Grant P30CA016672 from the National Cancer Institute, which supports the Bioinformatics Shared Resource to Arvind Rao. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dawood S. Biology and management of inflammatory breast cancer. Expert Rev Anticancer Ther. 2010;10(2):209–20. doi: 10.1586/era.09.90 . [DOI] [PubMed] [Google Scholar]
- 2.Costa R, Santa-Maria CA, Rossi G, Carneiro BA, Chae YK, Gradishar WJ, et al. Developmental therapeutics for inflammatory breast cancer: Biology and translational directions. Oncotarget. 2017;8(7):12417–32. doi: 10.18632/oncotarget.13778 ; PubMed Central PMCID: PMCPMC5355355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson FM, Bondy M, Yang W, Yamauchi H, Wiggins S, Kamrudin S, et al. Inflammatory breast cancer: the disease, the biology, the treatment. CA: a cancer journal for clinicians. 2010;60(6):351–75. doi: 10.3322/caac.20082 . [DOI] [PubMed] [Google Scholar]
- 4.Cristofanilli M, Buzdar AU, Hortobagyi GN. Update on the management of inflammatory breast cancer. The oncologist. 2003;8(2):141–8. . [DOI] [PubMed] [Google Scholar]
- 5.Dawood S, Cristofanilli M. Inflammatory breast cancer: what progress have we made? Oncology. 2011; 25(3):264–70,73. . [PubMed] [Google Scholar]
- 6.Anderson WF, Schairer C, Chen BE, Hance KW, Levine PH. Epidemiology of inflammatory breast cancer (IBC). Breast disease. 2005;22:9–23. ; PubMed Central PMCID: PMC2852616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine PH, Veneroso C. The epidemiology of inflammatory breast cancer. Seminars in oncology. 2008;35(1):11–6. doi: 10.1053/j.seminoncol.2007.11.018 . [DOI] [PubMed] [Google Scholar]
- 8.Fouad TM, Kogawa T, Liu DD, Shen Y, Masuda H, El-Zein R, et al. Overall survival differences between patients with inflammatory and noninflammatory breast cancer presenting with distant metastasis at diagnosis. Breast cancer research and treatment. 2015;152(2):407–16. doi: 10.1007/s10549-015-3436-x ; PubMed Central PMCID: PMCPMC4492876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matro JM, Li T, Cristofanilli M, Hughes ME, Ottesen RA, Weeks JC, et al. Inflammatory breast cancer management in the national comprehensive cancer network: the disease, recurrence pattern, and outcome. Clin Breast Cancer. 2014;15(1):1–7. doi: 10.1016/j.clbc.2014.05.005 ; PubMed Central PMCID: PMCPMC4422394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamm CA, Moran D, Rao K, Trusk PB, Pry K, Sausen M, et al. Genomic and Immunological Tumor Profiling Identifies Targetable Pathways and Extensive CD8+/PDL1+ Immune Infiltration in Inflammatory Breast Cancer Tumors. Molecular cancer therapeutics. 2016;15(7):1746–56. doi: 10.1158/1535-7163.MCT-15-0353 . [DOI] [PubMed] [Google Scholar]
- 11.van Uden DJ, van Laarhoven HW, Westenberg AH, de Wilt JH, Blanken-Peeters CF. Inflammatory breast cancer: An overview. Critical reviews in oncology/hematology. 2015;93(2):116–26. doi: 10.1016/j.critrevonc.2014.09.003 . [DOI] [PubMed] [Google Scholar]
- 12.Fouad TM, Kogawa T, Reuben JM, Ueno NT. The role of inflammation in inflammatory breast cancer. Advances in experimental medicine and biology. 2014;816:53–73. doi: 10.1007/978-3-0348-0837-8_3 . [DOI] [PubMed] [Google Scholar]
- 13.Cariati M, Bennett-Britton TM, Pinder SE, Purushotham AD. "Inflammatory" breast cancer”. Surgical oncology. 2005;14(3):133–43. doi: 10.1016/j.suronc.2005.07.004 . [DOI] [PubMed] [Google Scholar]
- 14.Iniesta MD, Mooney CJ, Merajver SD. Inflammatory breast cancer: what are the treatment options? Expert Opin Pharmacother. 2009;10(18):298797 doi: 10.1517/14656560903401638 . [DOI] [PubMed] [Google Scholar]
- 15.Hance KW, Anderson WF, Devesa SS, Young HA, Levine PH. Trends in inflammatory breast carcinoma incidence and survival: the surveillance, epidemiology, and end results program at the National Cancer Institute. J Natl Cancer Inst. 2005;97(13):966–75. doi: 10.1093/jnci/dji172 ; PubMed Central PMCID: PMC2844937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cristofanilli M, Valero V, Buzdar AU, Kau SW, Broglio KR, Gonzalez-Angulo AM, et al. Inflammatory breast cancer (IBC) and patterns of recurrence: understanding the biology of a unique disease. Cancer. 2007;110(7):1436–44. doi: 10.1002/cncr.22927 . [DOI] [PubMed] [Google Scholar]
- 17.Abeywardhana DY, Nascimento VC, Dissanayake D, Taylor D, Metcalf C, Saunders C, et al. Review of ultrasound appearance in inflammatory breast cancer: A pictorial essay. Journal of medical imaging and radiation oncology. 2016;60(1):83–7. doi: 10.1111/1754-9485.12418 . [DOI] [PubMed] [Google Scholar]
- 18.Molckovsky A, Fitzgerald B, Freedman O, Heisey R, Clemons M. Approach to inflammatory breast cancer. Canadian family physician Medecin de famille canadien. 2009;55(1):25–31. ; PubMed Central PMCID: PMC2628841. [PMC free article] [PubMed] [Google Scholar]
- 19.Dawood S, Merajver SD, Viens P, Vermeulen PB, Swain SM, Buchholz TA, et al. International expert panel on inflammatory breast cancer: consensus statement for standardized diagnosis and treatment. Annals of oncology: official journal of the European Society for Medical Oncology. 2011;22(3):515–23. doi: 10.1093/annonc/mdq345 ; PubMed Central PMCID: PMCPMC3105293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee KW, Chung SY, Yang I, Kim HD, Shin SJ, Kim JE, et al. Inflammatory breast cancer: imaging findings. Clin Imaging. 2005;29(1):22–5. . [PubMed] [Google Scholar]
- 21.Masuda H, Brewer TM, Liu DD, Iwamoto T, Shen Y, Hsu L, et al. Long-term treatment efficacy in primary inflammatory breast cancer by hormonal receptor- and HER2-defined subtypes. Annals of oncology: official journal of the European Society for Medical Oncology. 2014;25(2):384–91. doi: 10.1093/annonc/mdt525 ; PubMed Central PMCID: PMC3905780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang WT, Le-Petross HT, Macapinlac H, Carkaci S, Gonzalez-Angulo AM, Dawood S, et al. Inflammatory breast cancer: PET/CT, MRI, mammography, and sonography findings. Breast cancer research and treatment. 2008;109(3):417–26. doi: 10.1007/s10549-007-9671-z . [DOI] [PubMed] [Google Scholar]
- 23.Van Laere SJ, Ueno NT, Finetti P, Vermeulen P, Lucci A, Robertson FM, et al. Uncovering the molecular secrets of inflammatory breast cancer biology: an integrated analysis of three distinct affymetrix gene expression datasets. Clin Cancer Res. 2013;19(17):5685–96. doi: 10.1158/1078-0432.CCR-12-2549 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natori A, Hayashi N, Soejima K, Deshpande GA, Takahashi O, Cristofanilli M, et al. A comparison of epidemiology, biology, and prognosis of inflammatory breast cancer in Japanese and US populations. Clin Breast Cancer. 2013;13(6):460–4. doi: 10.1016/j.clbc.2013.08.004 . [DOI] [PubMed] [Google Scholar]
- 25.Suarez-Arroyo IJ, Feliz-Mosquea YR, Perez-Laspiur J, Arju R, Giashuddin S, Maldonado-Martinez G, et al. The proteome signature of the inflammatory breast cancer plasma membrane identifies novel molecular markers of disease. American journal of cancer research. 2016;6(8):1720–40. ;PubMed Central PMCID: PMC5004075. [PMC free article] [PubMed] [Google Scholar]
- 26.Yagi N, Manabe I, Tottori T, Ishihara A, Ogata F, Kim JH, et al. A nanoparticle system specifically designed to deliver short interfering RNA inhibits tumor growth in vivo. Cancer Res. 2009;69(16):6531–8. doi: 0008-5472.CAN-08-3945 [pii] doi: 10.1158/0008-5472.CAN-08-3945 . [DOI] [PubMed] [Google Scholar]
- 27.Tsai CJ, Li J, Gonzalez-Angulo AM, Allen PK, Woodward WA, Ueno NT, et al. Outcomes After Multidisciplinary Treatment of Inflammatory Breast Cancer in the Era of Neoadjuvant HER2-directed Therapy. Am J Clin Oncol. 2015;38:3242–7. doi: 10.1097/COC.0b013e3182937921 ; PubMed Central PMCID: PMCPMC3880388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Limentani SA, Brufsky AM, Erban JK, Jahanzeb M, Lewis D. Phase II study of neoadjuvant docetaxel, vinorelbine, and trastuzumab followed by surgery and adjuvant doxorubicin plus cyclophosphamide in women with human epidermal growth factor receptor 2-overexpressing locally advanced breast cancer. J Clin Oncol. 2007;25(10):1232–8. doi: 10.1200/JCO.2005.05.3306 . [DOI] [PubMed] [Google Scholar]
- 29.Hurley J, Doliny P, Reis I, Silva O, Gomez-Fernandez C, Velez P, et al. Docetaxel, cisplatin, and trastuzumab as primary systemic therapy for human epidermal growth factor receptor 2-positive locally advanced breast cancer. J Clin Oncol. 2006;24(12):1831–8. doi: 10.1200/JCO.2005.02.8886 . [DOI] [PubMed] [Google Scholar]
- 30.Li J, Gonzalez-Angulo AM, Allen PK, Yu TK, Woodward WA, Ueno NT, et al. Triple-negative subtype predicts poor overall survival and high locoregional relapse in inflammatory breast cancer. The oncologist. 2011;16(12):1675–83. doi: 10.1634/theoncologist.2011-0196 ; PubMed Central PMCID: PMC3248766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawood S, Ueno NT, Valero V, Woodward WA, Buchholz TA, Hortobagyi GN, et al. Differences in survival among women with stage III inflammatory and noninflammatory locally advanced breast cancer appear early: a large population-based study. Cancer. 2011;117(9):1819–26. doi: 10.1002/cncr.25682 . [DOI] [PubMed] [Google Scholar]
- 32.Yamasaki F, Johansen MJ, Zhang D, Krishnamurthy S, Felix E, Bartholomeusz C, et al. Acquired resistance to erlotinib in A-431 epidermoid cancer cells requires down-regulation of MMAC1/PTEN and up-regulation of phosphorylated Akt. Cancer Res. 2007;67(12):5779–88. doi: 10.1158/0008-5472.CAN-06-3020 . [DOI] [PubMed] [Google Scholar]
- 33.Cabioglu N, Gong Y, Islam R, Broglio KR, Sneige N, Sahin A, et al. Expression of growth factor and chemokine receptors: new insights in the biology of inflammatory breast cancer. Annals of oncology: official journal of the European Society for Medical Oncology. 2007;18(6):1021–9. doi: 10.1093/annonc/mdm060 . [DOI] [PubMed] [Google Scholar]
- 34.ACS. Cancer Facts and Figures 2007. 2007.
- 35.Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nature reviews Molecular cell biology. 2007;8(10):839–45. Epub 2007/08/09. doi: nrm2236 [pii] doi: 10.1038/nrm2236 . [DOI] [PubMed] [Google Scholar]
- 36.Lee J, Cuddihy MJ, Kotov NA. Three-dimensional cell culture matrices: state of the art. Tissue Eng Part B Rev. 2008;14(1):61–86. Epub 2008/05/06. doi: 10.1089/teb.2007.0150 . [DOI] [PubMed] [Google Scholar]
- 37.Hess MW, Pfaller K, Ebner HL, Beer B, Hekl D, Seppi T. 3D versus 2D cell culture implications for electron microscopy. Methods in cell biology. 2010;96:649–70. Epub 2010/09/28. doi: S0091- 679X(10)96027-5 [pii] doi: 10.1016/S0091-679X(10)96027-5 . [DOI] [PubMed] [Google Scholar]
- 38.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumor xenografts as models for oncology drug development. Nature Reviews Clinical Oncology. 2012;9:338–50. doi: 10.1038/nrclinonc.2012.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranga A, Gjorevski N, Lutolf MP. Drug discovery through stem cell-based organoid models. Advanced drug delivery reviews. 2014;69–70:19–28. [DOI] [PubMed] [Google Scholar]
- 40.Breslin S, O'Driscoll. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today. 2012;18(5–6):240–9. doi: 10.1016/j.drudis.2012.10.003 . [DOI] [PubMed] [Google Scholar]
- 41.Bartholomesuz G, Rao A. A Three Dimensional Spheroid Cell Culture Model for Robust High Throughput RNA Interference Screens In: Trip RA, Karpilow J, editors. Frontiers in RNAi. 1: Bentham Science Publishers; 2014. p. 215–31. [Google Scholar]
- 42.Lamhamedi-Cherradi SE, Santoro M, Ramammoorthy V, Menegaz BA, Bartholomeusz G, Iles LR, et al. 3D tissue-engineered model of Ewing's sarcoma. Advanced drug delivery reviews. 2014;79–80:155–71. doi: 10.1016/j.addr.2014.07.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desoize B. Contribution of three-dimensional culture to cancer research. Critical reviews in oncology/hematology. 2000;36(2–3):59–60. Epub 2000/10/18. doi: S1040842800000767 [pii]. . [DOI] [PubMed] [Google Scholar]
- 44.Friedrich J, Ebner R, Kunz-Schughart LA. Experimental anti-tumor therapy in 3-D: spheroids—old hat or new challenge? International journal of radiation biology. 2007;83(11–12):849–71. doi: 10.1080/09553000701727531 . [DOI] [PubMed] [Google Scholar]
- 45.Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. Spheroid-based drug screen: considerations and practical approach. Nature protocols. 2009;4(3):309–24. Epub 2009/02/14. doi: nprot.2008.226 [pii] doi: 10.1038/nprot.2008.226 . [DOI] [PubMed] [Google Scholar]
- 46.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92. doi: 10.1056/NEJMoa1113205 ; PubMed Central PMCID: PMCPMC4878653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alpaugh ML, Tomlinson JS, Shao ZM, Barsky SH. A novel human xenograft model of inflammatory breast cancer. Cancer Res. 1999;59(20):5079–84. . [PubMed] [Google Scholar]
- 48.Moro M, Bertolini G, Tortoreto M, Pastorino U, Sozzi G, Roz L. Patient-derived xenografts of non small cell lung cancer: resurgence of an old model for investigation of modern concepts of tailored therapy and cancer stem cells. J Biomed Biotechnol. 2012;2012:568567 doi: 10.1155/2012/568567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Astashkina A, Grainger DW. Critical analysis of 3-D organoid in vitro cell culture models for high- throughput drug candidate toxicity assessments. Advanced drug delivery reviews. 2014;69–70:1–18. doi: 10.1016/j.addr.2014.02.008 . [DOI] [PubMed] [Google Scholar]
- 50.Hynds RE, Giangreco A. Concise review: the relevance of human stem cell-derived organoid models for epithelial translational medicine. Stem Cells. 2013;31(3):417–22. doi: 10.1002/stem.1290 ; PubMed Central PMCID: PMCPMC4171682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.1993 Report of the AVMA Panel on Euthanasia. Journal of the American Veterinary Medical Association. 1993;202(2):229–49. [PubMed] [Google Scholar]
- 52.Haisler WL, Timm DM, Gage JA, Tseng H, Killian TC, Souza GR. Three-dimensional cell culturing by magnetic levitation. Nature protocols. 2013;8(10):1940–9. Epub 2013/09/14. doi: 10.1038/nprot.2013.125 . [DOI] [PubMed] [Google Scholar]
- 53.Evans KW, Yuca E, Akcakanat A, Scott SM, Arango NP, Zheng X, et al. A Population of Heterogeneous Breast Cancer Patient-Derived Xenografts Demonstrate Broad Activity of PARP Inhibitor in BRCA1/2 Wild-Type Tumors. Clin Cancer Res. 2017;23(21):6468–77. doi: 10.1158/1078-0432.CCR-17-0615 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kleer CG, van Golen KL, Braun T, Merajver SD. Persistent E-cadherin expression in inflammatory breast cancer. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2001;14(5):458–64. doi: 10.1038/modpathol.3880334 . [DOI] [PubMed] [Google Scholar]
- 55.Desai A, Webb B, Gerson SL. CD133+ cells contribute to radioresistance via altered regulation of DNA repair genes in human lung cancer cells. Radiotherapy and oncology: journal of the European Society for Therapeutic Radiology and Oncology. 2014. doi: 10.1016/j.radonc.2013.10.040 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Souza GR, Tseng H, Gage JA, Mani A, Desai P, Leonard F, et al. Magnetically Bioprinted Human Myometrial 3D Cell Rings as A Model for Uterine Contractility. International journal of molecular sciences. 2017;18(4). doi: 10.3390/ijms18040683 ; PubMed Central PMCID: PMCPMC5412269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tseng H, Gage JA, Shen T, Haisler WL, Neeley SK, Shiao S, et al. A spheroid toxicity assay using magnetic 3Dbioprinting and real-time mobile device-based imaging. Scientific reports. 2015;5:13987. doi: PMC4568483. doi: 10.1038/srep13987 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Timm DM, Chen J, Sing D, Gage JA, Haisler WL, Neeley SK, et al. A high-throughput three-dimensional cell migration assay for toxicity screening with mobile device-based macroscopic image analysis. Scientific reports. 2013;3:3000 doi: 10.1038/srep03000 ; PubMed Central PMCID:PMCPMC3801146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tseng H, Gage JA, Haisler WL, Neeley SK, Shen T, Hebel C, et al. A high-throughput in vitro ring assay for vasoactivity using magnetic 3D bioprinting. Scientific reports. 2016;6:30640 doi: 10.1038/srep30640 ; PubMed Central PMCID: PMCPMC4967891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desai PK, Tseng H, Souza GR. Assembly of Hepatocyte Spheroids Using Magnetic 3D Cell Culture for CYP450 Inhibition/Induction. International journal of molecular sciences. 2017;18(5). Epub 2017/05/20. doi: 10.3390/ijms18051085 ; PubMed Central PMCID: PMCPMC5454994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Souza GR, Molina JR, Raphael RM, Ozawa MG, Stark DJ, Levin CS, et al. Three-dimensional tissue culture based on magnetic cell levitation. Nat Nanotechnol. 2010;5(4):291–6. Epub 2010/03/17. doi: 10.1038/nnano.2010.23 ; PubMed Central PMCID: PMCPMC4487889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Becker JL, Souza GR. Using space-based investigations to inform cancer research on Earth. Nat Rev Cancer. 2013;13(5):315–27. Epub 2013/04/16. doi: 10.1038/nrc3507 . [DOI] [PubMed] [Google Scholar]
- 63.Tseng H, Balaoing LR, Grigoryan B, Raphael RM, Killian TC, Souza GR, et al. A three-dimensional co- culture model of the aortic valve using magnetic levitation. Acta Biomater. 2014;10(1):173–82. Epub 2013/09/17. doi: 10.1016/j.actbio.2013.09.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tseng H, Gage JA, Raphael RM, Moore RH, Killian TC, Grande-Allen KJ, et al. Assembly of a three- dimensional multitype bronchiole coculture model using magnetic levitation. Tissue Eng Part C Methods. 2013;19(9):665–75. Epub 2013/01/11. doi: 10.1089/ten.TEC.2012.0157 . [DOI] [PubMed] [Google Scholar]
- 65.Jaganathan H, Gage J, Leonard F, Srinivasan S, Souza GR, Dave B, et al. Three-dimensional in vitro co- culture model of breast tumor using magnetic levitation. Scientific reports. 2014;4:6468 Epub 2014/10/02. doi: 10.1038/srep06468 ; PubMed Central PMCID: PMCPMC4180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin H, Dhanani N, Tseng H, Souza GR, Wang G, Cao Y, et al. Nanoparticle Improved Stem Cell Therapy for Erectile Dysfunction in a Rat Model of Cavernous Nerve Injury. J Urol. 2016;195(3):788–95. Epub 2015/11/01. doi: 10.1016/j.juro.2015.10.129 . [DOI] [PubMed] [Google Scholar]
- 67.Lincoln DT, Ali Emadi EM, Tonissen KF, Clarke FM. The thioredoxin-thioredoxin reductase system: over- expression in human cancer. Anticancer Res. 2003;23(3B):2425–33. . [PubMed] [Google Scholar]
- 68.Kim SJ, Miyoshi Y, Taguchi T, Tamaki Y, Nakamura H, Yodoi J, et al. High thioredoxin expression is associated with resistance to docetaxel in primary breast cancer. Clin Cancer Res. 2005;11(23):8425–30. doi: 11/23/8425 [pii] doi: 10.1158/1078-0432.CCR-05-0449 . [DOI] [PubMed] [Google Scholar]
- 69.Li C, Thompson MA, Tamayo AT, Zuo Z, Lee J, Vega F, et al. Over-expression of Thioredoxin-1 mediates growth, survival, and chemoresistance and is a druggable target in diffuse large B-cell lymphoma. Oncotarget. 2012;3(3):314–26. doi: 10.18632/oncotarget.463 ; PubMed Central PMCID: PMCPMC3359887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong X, Lin Q, Aihara A, Li Y, Huang CK, Chung W, et al. Aspartate beta-Hydroxylase expression promotes a malignant pancreatic cellular phenotype. Oncotarget. 2015;6(2):1231–48. doi: 10.18632/oncotarget.2840 ; PubMed Central PMCID: PMCPMC4359229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J, Galloway R, Grandjean G, Jacob J, Humphries J, Bartholomeusz C, et al. Comprehensive Two- and Three-Dimensional RNAi Screening Identifies PI3K Inhibition as a Complement to MEK Inhibitor AS703026 for Combination Treatment of Triple-Negative Breast Cancer. Journal of Cancer. 2015;6(12):1306–19. doi: 10.7150/jca.13266 ; PubMed Central PMCID: PMC4643087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bissell MJ, Rizki A, Mian IS. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol. 2003;15(6):753–62. Epub 2003/12/04. doi: S0955067403001431 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang Y, Pjesivac-Grbovic J, Cantrell C, Freyer JP. A multiscale model for avascular tumor growth. Biophys J. 2005;89(6):3884–94. Epub 2005/10/04. doi: S0006-3495(05)73031[pii] doi: 10.1529/biophysj.105.060640 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM. Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol. 2013;10(8):483–7. doi: 10.1038/nrurol.2013.126 . [DOI] [PubMed] [Google Scholar]
- 75.Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920–6. doi: 10.1126/science.aao2774 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23(12):1242–435. doi: 10.1038/nm.4438 ; PubMed Central PMCID: PMCPMC5722201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie BY, Wu AW. Organoid Culture of Isolated Cells from Patient-derived Tissues with Colorectal Cancer. Chin Med J (Engl). 2016;129(20):2469–75. doi: 10.4103/0366-6999.191782 ; PubMed Central PMCID: PMCPMC5072260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1–2):328–38. doi: 10.1016/j.cell.2014.12.021 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–45. doi: 10.1016/j.cell.2015.03.053 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buchsbaum RJ, Oh SY. Breast Cancer-Associated Fibroblasts: Where We Are and Where We Need to Go. Cancers (Basel). 2016;8(2). doi: 10.3390/cancers8020019 ; PubMed Central PMCID: PMCPMC4773742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shiga K, Hara M, Nagasaki T, Sato T, Takahashi H, Takeyama H. Cancer-Associated Fibroblasts: Their Characteristics and Their Roles in Tumor Growth. Cancers (Basel). 2015;7(4):2443–58. doi: 10.3390/cancers7040902 ; PubMed Central PMCID: PMCPMC4695902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395–406. doi: 10.1083/jcb.201102147 ; PubMed Central PMCID: PMCPMC3283993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giussani M, Merlino G, Cappelletti V, Tagliabue E, Daidone MG. Tumor-extracellular matrix interactions: Identification of tools associated with breast cancer progression. Semin Cancer Biol. 2015;35(3–10). doi: 10.1016/j.semcancer.2015.09.012 . [DOI] [PubMed] [Google Scholar]
- 84.Miki Y, Ono K, Hata S, Suzuki T, Kumamoto H, Sasano H. The advantages of co-culture over mono cell culture in simulating in vivo environment. The Journal of steroid biochemistry and molecular biology. 2012;131(3–5):68–75. doi: 10.1016/j.jsbmb.2011.12.004 . [DOI] [PubMed] [Google Scholar]
- 85.Cristofanilli M. Inflammatory breast cancer: a new approach. The lancet oncology. 2016;17(5):544–6. doi: 10.1016/S1470-2045(16)00064-4 . [DOI] [PubMed] [Google Scholar]
- 86.Robertson FM, Chu K, Boley KM, Ye Z, Liu H, Wright MC, et al. The class I HDAC inhibitor Romidepsin targets inflammatory breast cancer tumor emboli and synergizes with paclitaxel to inhibit metastasis. J Exp Ther Oncol. 2013;10(3):219–33. . [PubMed] [Google Scholar]
- 87.Shah AN, Cristofanilli M. The Growing Role of CDK4/6 Inhibitors in Treating Hormone Receptor-Positive Advanced Breast Cancer. Curr Treat Options Oncol. 2017;18(1):6 doi: 10.1007/s11864-017-0443-7 . [DOI] [PubMed] [Google Scholar]
- 88.Peery RC, Liu JY, Zhang JT. Targeting survivin for therapeutic discovery: past, present, and future promises. Drug Discov Today. 2017. doi: 10.1016/j.drudis.2017.05.009 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Array data has been submitted to the GEO database (accession number GSE106685).