We report a new strategy uncovered for iron acquisition in globally important unicellular photosynthetic organisms.
Abstract
Phytoplankton growth is limited in vast oceanic regions by the low bioavailability of iron. Iron fertilization often results in diatom blooms, yet the physiological underpinnings for how diatoms survive in chronically iron-limited waters and outcompete other phytoplankton when iron becomes available are unresolved. We show that some diatoms can use siderophore-bound iron, and exhibit a species-specific recognition for siderophore types. In Phaeodactylum tricornutum, hydroxamate siderophores are taken up without previous reduction by a high-affinity mechanism that involves binding to the cell surface followed by endocytosis-mediated uptake and delivery to the chloroplast. The affinity recorded is the highest ever described for an iron transport system in any eukaryotic cell. Collectively, our observations suggest that there are likely a variety of iron uptake mechanisms in diatoms besides the well-established reductive mechanism. We show that iron starvation–induced protein 1 (ISIP1) plays an important role in the uptake of siderophores, and through bioinformatics analyses we deduce that this protein is largely diatom-specific. We quantify expression of ISIP1 in the global ocean by querying the Tara Oceans atlas of eukaryotic genes and show a link between the abundance and distribution of diatom-associated ISIP1 with ocean provinces defined by chronic iron starvation.
INTRODUCTION
Most living organisms have evolved to depend on iron (Fe) for survival and growth, but their individual cellular requirements vary depending on their functional capabilities and their Fe uptake and storage capacities. Fe is a cofactor for proteins involved in a range of essential life processes, which include photosynthetic and respiratory electron transport, reactions in the tricarboxylic acid cycle, nitrate and nitrite reduction, as well as chlorophyll synthesis. Unicellular photosynthetic species have particularly high Fe demands (1), as do plants, for which it has been estimated that approximately half of their intracellular iron is bound to photosynthesis proteins (2). It is considered that in large swathes of the ocean, rich in nutrients but low in chlorophyll, growth of primary producers is limited by the availability of Fe. This inferred iron limitation is confirmed by mesoscale ocean fertilization experiments, which stimulate iron-induced blooms visible from space [for example, see (3)]. Intriguingly, the blooms are often dominated by diatoms (4), which are cosmopolitan, ubiquitous stramenopile algae responsible for an estimated 20% of carbon fixation on Earth (5). This suggests that diatoms, without dispensing with the need for Fe, to which they are highly sensitive, have evolved mechanisms to cope with chronic Fe limitation. It also signals the need to better understand the diatom response to Fe, an element whose chemical speciation is affected by temperature and pH changes associated with ongoing ocean acidification due to anthropogenic climate change.
Iron stress in diatoms has been studied on a genetic level using a model pennate diatom, Phaeodactylum tricornutum, which has a sequenced genome and can be easily transformed (6). P. tricornutum has also been found to survive chronically low iron concentrations and to respond readily to iron stimulation, similarly to the “low–iron quota” diatoms found in iron-deprived regions of the global ocean (7, 8). A combined transcriptome and metabolome analysis of P. tricornutum found that iron deprivation leads to the up-regulation of genes encoding proteins designated as “iron starvation–induced proteins” (ISIPs), specifically ISIP1, ISIP2, and ISIP3 (9). Since their designation, studies have corroborated the importance of ISIPs in the diatom response to iron stress. Up-regulation of these genes during iron limitation has been demonstrated in laboratory cultures of Thalassiosira oceanica (another low–iron quota diatom), as well as in field populations of diatoms (10, 11). Although the role and function of ISIP1 and ISIP3 remain unknown, it has been demonstrated experimentally that ISIP2a is involved in the nonreductive uptake of ferric [Fe(III)] iron by Morrissey et al. (12). Because of its binding Fe(III) at the cell surface, this protein was suggested to correspond to the hypothetical protein “phytotransferrin” (12), proposed more than 30 years ago by Anderson and Morel working on Thalassiosira weissflogii (13). This suggestion has recently been confirmed by McQuaid et al. (14) who showed that ISIP2a and human transferrin are functional equivalents. Putative ISIP functions may include whole-cell regulation of iron stress responses, iron storage, or involvement in the uptake process of discrete Fe sources from the environment.
The chemical speciation of Fe in seawater is dynamic and controlled by processes that occur at different spatiotemporal scales. Although geological processes such as lithogenic input and sedimentation are better characterized, biological regulation of Fe cycling is elusive, particularly due to presumed microbial preferences for Fe species within the iron pool and flux between community members (15). Defining discrete functional groups of phytoplankton and their preferences for the various fractions of the Fe pool is paramount for a better understanding of the biological component of Fe biochemistry. To this purpose, ongoing oceanography research is investigating which Fe forms are more “bioavailable” to diatoms in natural populations. Although dissolved uncomplexed ferrous [Fe(II)] iron (Fe′) appears to be the form most readily taken up by all phytoplankton, it is a scarce micronutrient, present at concentrations that are insufficient to satisfy the requirements of diatom species in bloom (16). A possible supplement to Fe′ is the organic pool of complexed Fe [typically present as Fe(III)], which is estimated to comprise >99.9% of the dissolved Fe in the marine environment [(17) and references therein]. Experimental incubation of natural seawater has shown active production of Fe-binding ligands concomitant with diatom growth under Fe-limiting conditions (18, 19), suggesting that these ligands play an important role in the diatom iron stress response. The organic chelators of Fe that have been described include discrete ligands, such as porphyrins, exopolymers, and humic substances, as well as siderophores. Siderophores are the strongest chelators, with thermodynamic stability constants in the range of 1025 to 1050 M−1.
In unicellular eukaryotes, two main strategies of iron acquisition from bound Fe(III) are known (20). One is reductive uptake, in which extracellular ferric complexes are dissociated by reduction before uptake via ferrireductases at the plasma membrane (Fre proteins). In the second strategy, known as nonreductive uptake, ferric iron is taken up without previous dissociation via specific high-affinity receptors. Iron is then released (often by reduction) from the chelates once inside the cell. Nonreductive iron uptake from siderophores is particularly well developed in yeast and other fungal species, as well as in bacteria (21, 22). Not only do these species have receptors able to recognize specific siderophores: Many of them are also producers of the same siderophore molecules, a factor that enables them to compete for iron more efficiently than other organisms. Conversely, no eukaryotic algae are known to produce siderophores (early studies that identified siderophore-producing algae subsequently have been deemed unreliable because of the likely presence of bacterial contaminants in the laboratory cultures). In diatoms, although reductive iron uptake is well known (1), and the ability to concentrate ferric iron at the cell surface in an ISIP2a-dependent process has been described (12, 14), there is currently no evidence for the nonreductive uptake of ferric iron bound to siderophores. Although some reports suggest that certain species of diatoms are capable of relying on siderophores for growth (23–25), these processes are not thought to rely on the uptake of siderophore-complexed iron directly into the cell as in fungal and bacterial systems, but are based on indiscriminate reductive processes at the cell surface (26).
Identification of diatoms capable of using siderophore-bound iron nonreductively would have significant implications. First, it would provide further support for the importance of nonreductive processes of iron uptake in these organisms. Second, knowledge of the mechanism of cellular uptake and the extent of its phylogenetic distribution could lead to the identification of new marker genes for assessing iron homeostasis in different kinds of diatoms, which is currently beyond our reach. Furthermore, if some diatoms can recognize specific complexes of Fe, then it would have consequences for understanding their functional ecology. In particular, specific recognition is the hallmark of well-characterized siderophore-based exchanges in terrestrial microbial communities. In this case, siderophores are considered as a “public good,” capable of shaping competitive/cooperative dynamics, and species can be partitioned into producers, users, and cheaters who cannot synthesize siderophores, engaged in a survival race driven by the evolution of novel siderophore molecules and associated recognition systems (27). The ability to classify diatoms into such functional categories based on their relationship with a diverse range of iron sources, identified via molecular markers, would be a breakthrough for ocean models, because it would begin to open the “black box” of functional phytoplankton ecology in the context of global ocean biogeochemistry.
Here, we present the first evidence for direct intracellular uptake of siderophores by diatoms. We demonstrate that there is a species-specific preference for the form of siderophore recognized. This finding argues against the more general “nonspecific reduction at the cell surface” model proposed to explain the ability of diatoms to use chelated Fe (26). Furthermore, we use the model pennate diatom P. tricornutum to show that the siderophore uptake process involves endocytosis and delivery of siderophores to the chloroplast, where the bound Fe is finally released presumably by chloroplast-associated reductases. We show that the process of siderophore uptake is highly efficient, with an estimated affinity higher than for any other described eukaryote. Although the molecular machinery required for siderophore uptake remains unresolved, we link the ability to take up siderophores to the presence of the ISIP1 gene. Using knockdown ISIP1 lines, we show that the protein is necessary for the endocytic mechanism of uptake and exclude its role in reductive uptake of Fe. Finally, we use large sequencing data sets, from the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) initiative (28) and the Tara Oceans atlas of eukaryotic genes (29), to verify that ISIP1 is largely evolutionarily restricted to diatoms and is highly expressed in Fe-poor regions of the world’s ocean. We propose that siderophore uptake via an ISIP1-mediated mechanism is an adaptive strategy that has allowed some diatom lineages to flourish in iron-limited environments.
RESULTS
Siderophore uptake by marine diatoms is species-specific
To determine whether diatoms can exploit iron complexed with siderophores, we first measured the rates of iron uptake from hydroxamate and catecholate siderophores in three diatom species whose genomes have been sequenced—P. tricornutum, Thalassiosira pseudonana, and T. oceanica—and compared these rates to uptake of Fe′ (Fig. 1). We found that both P. tricornutum and T. oceanica could take up iron from siderophores, albeit with contrasting uptake patterns. P. tricornutum could only take up hydroxamate-type siderophores, such as ferrioxamine B (FOB) and ferrichrome (FCH), whereas T. oceanica preferentially took up catecholate-type siderophores, such as enterobactin (ENT). T. pseudonana could not take up any kind of siderophore. The uptake from Fe′ is denoted by the black line (ferric EDTA treatment) in Fig. 1. Further, we performed a series of experiments with P. tricornutum to parametrize the kinetics of uptake for the two hydroxamate siderophores (summarized in fig. S1). This allowed us to calculate their KM and Vmax to be 5 to 7 nM and 0.5 to 0.7 amol/hour per cell, respectively, the highest affinity ever described for any iron transport system in any eukaryotic cell. For comparison, in yeast, the KM values for FCH and FOB uptake are in the range of 1 to 2 μM (30). We also found that uptake of FCH could be inhibited by addition of FOB and by gallium analogs of FCH (desferrichrome bound to Ga, denoted GaDFCH in fig. S1C). This confirms that all three compounds are taken up by the same mechanism and therefore exhibit competitive inhibition, in accordance to the estimated Michaelis-Menten kinetics.
Fig. 1. Iron uptake from different iron sources by T. oceanica, P. tricornutum, and T. pseudonana.
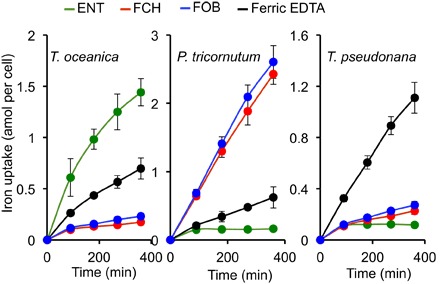
The figure shows specificity of siderophore uptake in three species of diatoms. The cells were precultured in iron-deficient medium, harvested at mid-exponential growth, and washed once with iron-free medium. The cells were then resuspended in growth medium containing one of the following 55Fe-labeled sources supplied at 1 μM: ferric EDTA (black), FCH (red), FOB (blue), or ENT (green). The cells were washed on filters at intervals with the washing buffer, and 55Fe associated to the washed cells was counted by scintillation. Data are means ± SD from four experiments (biological replicates).
Knocking down ISIP1 inhibits the ability of P. tricornutum to acquire Fe from siderophores
To locate putative genes that may be involved in the uptake of siderophores, we turned to the previously identified ISIPs. We were intrigued particularly by ISIP1, a putative membrane-associated protein with no recognizable functional domains, because it is the most highly expressed gene in P. tricornutum under iron-limiting conditions (9), and its presence/absence matches the ability of the tested diatoms to use siderophores—ISIP1 is found in T. oceanica and P. tricornutum but not in T. pseudonana (10). To test whether ISIP1 may be involved in siderophore uptake, we examined the effect of knocking down the gene in transgenic P. tricornutum cell lines (see fig. S2 for summary of the knockdown). We found that the silenced strain was unable to grow on hydroxamate siderophores as a sole source of Fe (Fig. 2A) and confirmed that there was a marked decrease in the rate of iron uptake from FOB (Fig. 2B) in this mutant. Through further experimentation, we were able to pinpoint this to the inability of the cells to import the siderophores across the cell membrane, whereas binding at the cell surface remained unaffected (fig. S3). As a control, we compared the behavior of the ISIP1 knockdown to that of the ISIP2a knockdown that was generated previously (12). In this previous work, we found that ISIP2a, the second most highly up-regulated gene under conditions of iron starvation, is involved in the nonreductive uptake of unbound Fe(III). As expected, the ISIP2a knockdown line demonstrated a decreased iron uptake from ferric EDTA, as previously reported, but showed no significant difference in its uptake of FOB compared to wild type (Fig. 2B).
Fig. 2. P. tricornutum ISIP1 knockdown cells are unable to grow on FOB as the sole source of iron.
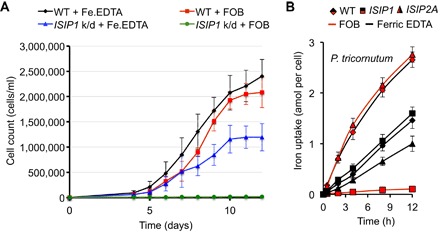
(A) P. tricornutum is able to grow on 15 pM Fe′ (black line) and 10 nM FOB (red line) as sole Fe sources in Aquil media. The ISIP1 knockdown (k/d) line shows reduced growth on Fe′ (blue line) and no growth on FOB (green). Error bars denote SDs from three biological replicates. Before the start of the experiment, cultures were depleted of Fe through a 10-day preculture on low-Fe Aquil. (B) Comparison of siderophore uptake by P. tricornutum wild-type (WT) cells (rhombus markers) versus ISIP1 (square) and ISIP2a (triangle) knockdown cell lines.
Further, we conducted an experiment that looked at siderophore uptake over a longer time course. We examined uptake of siderophores versus ferric EDTA in P. tricornutum cells maintained in exponential growth phase (wild-type and ISIP1 knockdown lines) in iron-rich and iron-deficient media over a period of 2 weeks. This approach allowed us to follow ferrireductase activity and compare the response of the cells to ferric EDTA versus FOB. We found that uptake of FOB was maximally induced in wild-type cells after 1 week of growth under iron-deficient conditions, whereas the FOB uptake capacity of ISIP1 knockdown lines remained low under all conditions (Fig. 3). The maximum FOB uptake capacity in wild-type cells did not correspond to the peak(s) of iron uptake capacity from ferric EDTA. Unlike FOB, iron uptake from ferric EDTA was not significantly different between wild-type and ISIP1 knockdown lines (Fig. 3A), although the inducible ferrireductase activity was higher in ISIP1 knockdown lines than in wild-type cells (Fig. 3C), indicating that the loss of FOB uptake activity in these cells was probably not related to reductase activity.
Fig. 3. Iron uptake from siderophores and from ferric EDTA is not regulated in the same way.
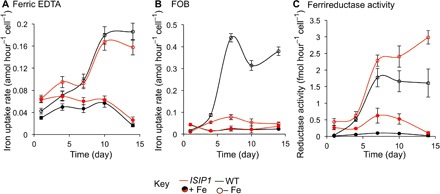
The figure shows rates of iron uptake from ferric EDTA (A) and FOB (B) as well as ferrireductase activity (C) as a function of time in wild-type (black) and ISIP1 knockdown cell lines (red) grown in iron-rich (closed symbols) or iron-deficient medium (open symbols). The cells were grown for 1 week in iron-rich medium (1 μM ferric citrate in Mf medium), washed twice with iron-free Mf medium, and re-inoculated at 2 × 106 cells/ml in iron-free (open symbols) or iron-rich medium (1 μM ferric citrate; closed symbols). The cultures were diluted every 2 days with the same media to maintain cell density at 2 × 106 cells/ml for 2 weeks. For ferrireductase activity, initial rates of uptake (first 30 min of kinetics) are plotted against the day since the start of the experiment. Data are means ± SD from four experiments (two biological replicates with two technical replicates). ISIP1 knockdown cell lines show a marked descrease in Fe uptake from FOB, but not from ferric EDTA, and a higher ferrireductase activity than wild-type cells. Maximal induction of FOB uptake activity under iron-deficient condition occurred before maximal induction of ferric EDTA uptake activity.
On the basis of these collective findings, we propose that the mechanism of iron uptake from siderophores does not involve a reductive step. We also infer that there are several iron uptake systems at play in P. tricornutum, whose individual impairment has consequences on whole-cell physiology. This hypothesis would explain our observation of reduced growth on ferric EDTA in ISIP1 knockdown lines (Fig. 2A), despite increased ferrireductase activity and unimpaired uptake of iron from ferric EDTA (Fig. 3C). It is also possible that the ISIP1 gene has further cell functions or is involved in the post-uptake processing of iron sourced from ferric EDTA.
Siderophores are taken up by endocytosis
To track the uptake and intracellular fate of siderophores in P. tricornutum, we used a conjugated FOB coupled to a fluorescent moiety, nitrobenz-2-oxa-1,3-diazole (FOB-NBD), which we previously synthesized as described by Ouchetto et al. (31). For the initial experiments, we used the gallium analog of FOB-NBD (Ga-DFOB-NBD) to avoid fluorescence quenching by iron. As shown in Fig. 4 (A and B), Ga-DFOB-NBD was taken up and accumulated inside iron-stressed P. tricornutum cells in vesicles juxtaposed to the chloroplast. This process was not observed in iron-stressed ISIP1 knockdown lines, where no intracellular fluorescence from Ga-DFOB-NBD could be detected, an observation consistent with our findings presented in Fig. 2. It should be noted that both wild-type and ISIP1 knockdown cells were alive and photosynthetically active, as shown by their intact chloroplasts and fusiform shape. Stressed and dying P. tricornutum cultures have a high proportion of ovate cells (32), which were not observed in this experiment.
Fig. 4. Siderophores are taken up into the cell by endocytosis.
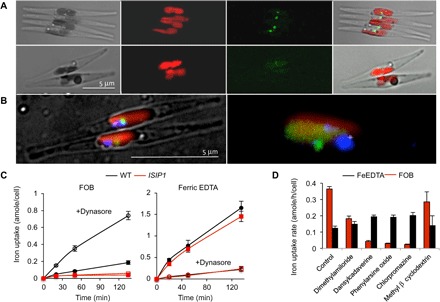
(A) Cells of the wild-type (top) and ISIP1 knockdown line (bottom) were grown for 5 days in iron-deficient medium, concentrated to about 5 × 107 cells/ml, and then incubated for 15 hours with 1 μM of the fluorescent conjugate of desferrioxamine B complexed with gallium (Ga-DFOB-NBD). Intracellular accumulation of the fluorescent siderophore analog was only observed in wild-type cells. Scale bar, 5 μm. (B) DAPI (4′,6-diamidino-2-phenylindole) staining of the cells (scale bar, 5 μm) to reveal that nuclear localization confirmed siderophore localization in a vesicle close to the chloroplast (red, chloroplast; blue, nucleus; green, fluorescent siderophore). (C) Iron uptake from 1 μM FOB (left) and from 1 μM ferric EDTA (right) by wild-type (black circles) and ISIP1 knockdown cells (red squares) in the presence (closed symbols) or absence (open symbols) of 10 μM of the endocytosis inhibitor dynasore. Dynasore inhibited iron uptake from FOB and enhanced iron uptake from ferric EDTA. (D) Effect of other endocytosis inhibitors on the rate of iron uptake from FOB and ferric EDTA (1 μM). Cells were preincubated for 10 min with the following inhibitors before adding 55Fe: dynasore (10 μM), dansylcadaverine (100 μM), phenylarsine oxide (10 μM), dimethyl amiloride (50 μM), chlorpromazine (50 μM), or methyl-β-cyclodextrin (1 mM) [added from 1000× stock solutions in dimethyl sulfoxide (DMSO); pure DMSO was added as a control].
The vesicular localization of Ga-DFOB-NBD (Fig. 4, A and B) suggested an endocytosis process. Therefore, we checked the effect of endocytosis inhibitors on the uptake of iron added in the form of siderophores or as ferric EDTA. Addition of the endocytosis inhibitor dynasore decreased siderophore uptake by more than 75% while markedly increasing iron uptake from ferric EDTA (Fig. 4C). We confirmed this finding by using a range of other endocytosis inhibitors with the strongest effects recorded using dansylcadaverine, phenylarsine oxide, and chlorpromazine (Fig. 4D). All endocytosis inhibitors had a stimulatory effect on iron uptake from ferric EDTA, which was proportional to their inhibitory effect on FOB uptake (Fig. 4D). It is possible that this increased uptake is due to the inhibition of intracellular cycling of iron transporters. For example, stabilization of the iron transporter IRT1 due to endocytosis inhibition has been shown experimentally in Arabidopsis thaliana, with an apparent increase in iron uptake at the cell surface (33). We suggest that a similar scenario is at play in P. tricornutum.
Inhibition of siderophore uptake by three endocytosis inhibitors was confirmed microscopically using Ga-DFOB-NBD. No vesicles containing the fluorophore-tagged siderophores could be detected in the presence of the endocytosis inhibitors (fig. S4A). Finally, as a further control for our findings and validation of our methods, we measured the rate of iron uptake in the presence of strong chelators of ferrous iron (ferrozine and bathophenanthroline disulfonate). As expected, addition of these chelators decreased the rate of 55Fe uptake from ferric EDTA but not from siderophores, further consolidating the view that siderophores were not reduced before uptake (fig. S4B).
Our data indicate that in P. tricornutum, hydroxamate siderophores are taken up by endocytosis in a nondissociative mechanism involving ISIP1. According to this hypothesis, the intracellular vesicle in which the siderophores accumulate should originate from endocytosis vesicles forming at the cell surface. We tested whether ISIP1 knockdown lines showed signs of aborted endocytosis by staining iron-starved cells with the amphiphilic dye FM4-64, which has been used to investigate endocytosis and vesicle trafficking in other eukaryotes (34). As expected, in wild-type cells, we observed the formation of organelles originating from the plasma membrane by endocytosis, such as vacuoles and uncharacterized vesicles, which were observed throughout the cell interior (an indication of vesicular flux). This is to be contrasted with ISIP1 knockdown cell lines, where we only observed “sickle-shaped” vesicles attached to the membrane (fig. S5). These structures are suggestive of aborted endocytic vesicles that remain attached to the plasma membrane. Accordingly, we observed less intracellular fluorescent signal of NBD in knockdown cells than in wild-type cells incubated in the presence of Ga-DFOB-NBD (fig. S5).
Siderophores are delivered to the chloroplast where Fe is released
Because most of the imaged vesicles containing the siderophores were observed near the diatom chloroplast, we hypothesized that this may be the ultimate destination for siderophore-complexed iron. This is reasonable because iron deprivation is particularly detrimental to photosynthesis, where so many proteins rely on Fe as a cofactor. When we used the nonfluorescent, iron-loaded FOB-NBD in a time course experiment, visually summarized in movie S1, we observed that iron was released rapidly from the siderophore in these vesicles once it was in the vicinity of the chloroplast. The video shows live imaged cells that have been pulsed with FOB-NBD. After just under 5 min, there is a clear bright intracellular signal of fluorescence near/at the chloroplast. We speculate that the signal is due to the unloading of Fe from the siderophore, releasing the fluorescent desferri-siderophore (DFOB-NBD; movie S1). Our observation of iron released intracellularly is further evidence that iron uptake from siderophores does not involve a reduction step at the cell surface, strengthening the findings presented in Fig. 2, and is contrary to the model proposed by Shaked and Lis (15), in which it is proposed that diatoms are able to take up organic-bound iron indiscriminately by reduction on the cell surface.
We then used a “Trojan horse” approach to test whether siderophores were taken into the chloroplast of P. tricornutum. We coupled FOB chemically to inhibitors of chlorophyll biosynthesis, which would be fatal to the cell if they reached the chloroplast interior. Specifically, we tested two such conjugates, FOB coupled to a diphenyl ether moiety (FOB-DPE) and FOB coupled to acifluorfen (FOB-AF), both inhibitors of protoporphyrinogen oxidase, which performs the ninth step in the tetrapyrrole biosynthesis pathway, and is thus essential for chlorophyll and heme biosynthesis (35). We incubated iron-starved cells with these constructs and measured their effects on cell growth and cell chlorophyll content in both wild-type cells and the ISIP1 knockdown line. Figure 5 summarizes our findings and relevant controls. Addition of both FOB-DPE and FOB-AF (at two different concentrations, 1 and 5 μM) to the growth medium caused a marked decrease in the chlorophyll content of wild-type cells, leading to cell death. Remarkably, ISIP1 knockdown cell lines were insensitive to these toxic agents, displaying no loss of chlorophyll and continued growth in their presence (red lines in Fig. 5, A to C), indicating that mutants deficient in ISIP1 were unable to deliver FOB to the chloroplast. Furthermore, the effects of the toxic conjugates could be reversed in wild-type cells by adding an excess of unconjugated FOB, acting as a competitive inhibitor to FOB-DPE and FOB-AF (Fig. 5E). This confirms that the uptake of the toxic conjugated agents was due to uptake of FOB, and not due to uptake or permeability of the cell to AF and DPE. As a further validation, we tested whether the unconjugated forms of AF and DPE were toxic to the cells, and found that they were significantly less toxic than the FOB conjugates for wild-type cells (Fig. 5D). Inhibition of growth and chlorophyll biosynthesis by the FOB conjugates of AF and DPE in P. tricornutum supports the hypothesis that internalized siderophores reach the chloroplast interior.
Fig. 5. Siderophores are taken into the chloroplast.
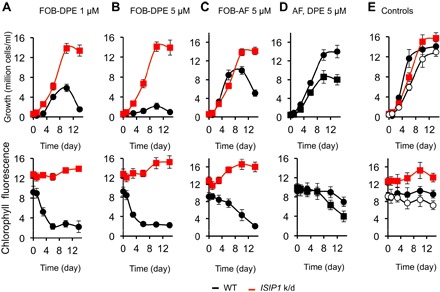
Effect of FOB coupled to protoporphyrinogen oxidase inhibitors on the growth and chlorophyll content of wild-type and ISIP1 knockdown cells shows that siderophores taken up by the cells can reach the chloroplast. In these experiments, chemical derivatives of the siderophore FOB were used in a Trojan horse approach to reach (or not) the target of the inhibitors coupled to FOB (inhibitors of protoporphyrinogen oxidase, one of the last enzymes of heme and chlorophyll biosynthesis). Wild-type and ISIP1 knockdown cells were inoculated at about 5 × 105 cells/ml in the Mf medium containing 1 to 5 μM of FOB-DPE (with a 10% excess iron, that is, an Fe/ligand ratio of 1.1 Fe for 1 DFOB) coupled to diphenyl-ether (FOB-DPE) (A and B) or to acifluorfen (FOB-AF) (C). Cell growth (top; black circles, wild-type; red squares, ISIP1 knockdown) and cell chlorophyll content (bottom; black circles, wild type; red squares, ISIP1 knockdown) were monitored by flow cytometry (see Materials and Methods; chlorophyll fluorescence is expressed in 105× FL3 units). (D) Wild-type cells were grown in the presence of 5 μM of the unconjugated forms of AF (circles) or DPE (squares). In control experiments (E), wild-type (black circles) and ISIP1 knockdown cells (red squares) were grown in the presence of 1 μM FOB (uncoupled, with a 10% excess iron; closed symbols), and wild-type cells were grown with 1 μM FOB-DPE plus an excess of FOB (10 μM, uncoupled; open circles). Data are means ± SD from four experiments (biological replicates).
ISIP1 localizes to the cell surface and chloroplast, is largely diatom-specific and globally abundant
We have shown that ISIP1 is required for the delivery of siderophore-complexed iron to the chloroplast (Fig. 5) and that its knockdown results in an abortive endocytosis process at the cell surface (fig. S5) and inability to grow on siderophore iron as a sole Fe source (Fig. 2). Iron uptake experiments confirmed that ISIP1 knockdown affected the capacity for iron uptake from FOB but not its binding (fig. S3). Although identifying the precise function of ISIP1 is the subject of ongoing investigation, we are able to comment on the intracellular localization of the protein, its phylogeny, and distribution in the environment.
We engineered a P. tricornutum line expressing a yellow fluorescent protein (YFP) gene fusion to the 3′ terminus of ISIP1 driven by the native ISIP1 promoter (ISIP1-YFP). Imaging of the cells under conditions of iron deprivation revealed that the ISIP1 protein localized to both the cell surface and the chloroplast (fig. S6). More specifically, we observed that prolonged Fe deprivation led to the accumulation of the protein on the chloroplast surface, perhaps the periplastidal compartment as seen in the study of Bullmann et al. (36), whereas in the early stages of Fe limitation, or during incubation with siderophore analogs (Ga-DFCH), ISIP1-YFP was also detected on the cell membrane. As a follow-up to these observations, we used bioinformatics to determine whether the protein had a predicted signal peptide or a chloroplast targeting sequence. The combined algorithms of SignalP v.3.0 (using neural networks) and ASAFind suggest that ISIP1 has a predicted N-terminal signal peptide but that it is not targeted to the chloroplast because it lacks the FWYL residue at +1 of the post-SP cleavage site typically associated with plastid-targeted proteins (see fig. S7 for protein sequence and annotations) (37).
In P. tricornutum, ISIP1 encodes a ~62-kDa protein composed of 569 amino acids (Phatr3 annotations) (38). Computational comparison of the peptide sequence against protein sequence databases predicts the following motifs (fig. S7): an N-terminal cleaved signal peptide, expected to direct ISIP1 to the secretory pathway, a C-terminal transmembrane helix anchoring it to the cell membrane, and a highly conserved endocytosis motif (D/ExxxL) 22 amino acids C-terminal to the transmembrane helix, also predicted by Lommer et al. (10). This endocytosis motif is recognized by the adaptor protein-2 (AP-2) complex, an abundant adaptor in plasma membrane–derived clathrin-coated vesicles with predicted orthologs in P. tricornutum (39). The extracellular bulk of the protein contains no known functional domains but is predicted to fold (SWISS-MODEL, ExPASy) into a seven-bladed β-propeller, likely composed of two lobes on each side of a cysteine-rich region, reminiscent of but not related to motifs that result in Fe-S clusters. A range of functionally unrelated proteins are known to contain β-propellers at their core; the structurally rigid surface created is often implicated in protein-protein recognition (40).
We investigated the distribution and evolution of ISIP1 across the tree of life. First, we searched for homologs of the P. tricornutum ISIP1 sequence using a reciprocal BLAST best-hit strategy against a composite library consisting of UniRef, other genomic data sets hosted separately on Joint Genome Institute (JGI), and eukaryotic transcriptomes sequenced through the MMETSP initiative (see Materials and Methods for details; Fig. 6A). We did not find any ISIP1 sequences in prokaryotes, animals/fungi, viridiplantae, and most nondiatom eukaryotic algae. Conversely, we found that ISIP1 is largely a diatom-specific gene, with 28 of 99 diatom species identified as expressing this gene, including members of the pennate and centric species, including the basally divergent genus Corethron (Fig. 6A). It is important to note that our estimate is likely to be conservative, because most of the databases queried provide information on transcribed genes, and we know that for ISIP1 transcription, a prerequisite is iron limitation.
Fig. 6. ISIP1 presence in eukaryotes and distribution/expression in the global ocean.

(A) Heatmap showing the presence of homologs of P. tricornutum ISIP1, identified by reciprocal BLAST best-hit search with a threshold e value of 1 × 10−5, against different eukaryotic species contained in a curated library consisting of decontaminated sequences from MMETSP, supplemented with a further 79 eukaryotic genome and transcriptome libraries (table S1) (41). Cells in color correspond to the presence of a homologous sequence in a particular strain; gray cells indicate species queried where no homolog was detected. Lineages are shown per the consensus topologies obtained from recent phylogenomic studies of nuclear genomes and are shaded by origin. Two transfer events, labeled with curved red arrows, demonstrate (A) a proposed plastid-associated gene transfer event that occurred from a common ancestor of pelagophytes and dictyochophytes into a common ancestor of the haptophyte lineage (41), and (B) the inferred origin point of the haptophyte-derived plastids found in fucoxanthin-containing dinoflagellates. (B) Analysis of abundance of ISIP1 sequences in the Tara Oceans metagenome and metatranscriptome data set, determined for seven ocean provinces [North Pacific Ocean (NPO), Southern Ocean (SO), South Pacific Ocean (SPO), North Atlantic Ocean (NAO), South Atlantic Ocean (SAO), Mediterranean Sea (MS), and Indian Ocean (IO)], and given as RPKM (indicated by size of circles; normalized against a set of housekeeping genes and transcripts; see Materials and Methods). The position of the circles on the diatom 18S phylogeny tree reflects the degree of taxonomic assignation from the sequence data.
We noted some possible homologs of ISIP1 in eight species of pelagophytes, haptophytes, and fucoxanthin-containing dinoflagellates within the genus Karenia (Fig. 6A). It is noteworthy that the lineages of pelagophyte, haptophyte, and fucoxanthin-containing dinoflagellate species, which appear to have a homologous copy of ISIP1, have been inferred to be connected to one another through historical plastid endosymbiosis events (41). This endosymbiosis-related connection among these other ISIP1-containing species would support the hypothesis that ISIP1 is a plastid-related protein with a plastid-associated distribution across the tree of life. Finally, we constructed a phylogeny of all ISIP1 sequences (fig. S8; see table S1 for alignments). The diatom sequences formed a robustly supported monophyletic clade (MrBayes posterior probability = 1; RAxML bootstrap support > 95%, both under three substitution matrices), to the exclusion of the pelagophyte, haptophyte, and dinoflagellate sequences. However, we noted that the branching order of the diatom ISIP1 sequences did not reflect the underlying evolutionary relationships between different diatom species. We identified multiple cases in which, for example, ISIP1 sequences from pennate diatom lineages were more closely related to sequences from centric diatoms than to other pennate species (fig. S8). We additionally found several diatom species (for example, Fragilariopsis kerguelensis and Thalassiosira antarctica) that appear to have multiple, distantly related copies of the ISIP1 gene. This distribution might reflect horizontal gene transfer events within the diatoms, or repeated paralogy events and differential loss of individual ISIP1 copies in different diatom species (fig. S8).
Using the reference sequences obtained, we assessed the environmental presence and expression of diatom ISIP1 across seven ocean provinces (North and South Atlantic, Mediterranean Sea, Indian Ocean, North and South Pacific, and Southern Ocean) by querying the Tara Oceans diatom metagenome and metatranscriptome atlas, which is composed of data collected from 68 geographical locations across all the major oceanic provinces except the Arctic (29). Our findings, summarized in Fig. 6B, show that ISIP1 is most highly expressed in the Southern Ocean and the Southern Pacific Ocean, with the majority of transcripts originating from the orders of Thalassionemales and Chaetocerotales (80 and 14%, respectively; table S1). The transcript or gene abundances, which are directly proportional to the size of the circles on the phylogenetic tree, are given as RPKM (reads per kilobase million) and normalized against a set of housekeeping genes and transcripts (see Materials and Methods). The position of the circles on the tree reflects the degree of taxonomic assignation from the retrieved sequence data. Proximity to the tips of the branches indicates a more refined taxonomic assignation achieved by the model. Where the circles are at the base of the tree, the model confidently allocated the ISIP1 sequences into the diatom phylum “Bacillariophyceae,” but further phylogenetic assignation was not supported statistically. It is of note that both Southern Ocean and the Southern Pacific Ocean, from which the majority of ISIP1 genes and transcripts were retrieved, are known to be chronically iron-limited, so our finding that this is where ISIP1 transcripts are most abundant is congruent with our physiological laboratory-based experiments.
DISCUSSION
In the present manuscript, we corroborate previous observations that some diatoms are able to use iron complexed to siderophores, which are some of the most tenacious organic chelators of iron. We compared the preferences of three sequenced species—P. tricornutum, T. oceanica, and T. pseudonana—for siderophore type and found significant differences. Particularly noteworthy is the inability of T. pseudonana to uptake rapidly either catecholate or hydroxamate siderophores compared with ferric EDTA, despite belonging to the same genus as T. oceanica, which can take up catecholate-bound Fe efficiently. Phylogenetic proximity is therefore unlikely to be an indicator of functional capacity for different modes of Fe uptake. Instead, we link the ability for siderophore uptake with the presence/absence of the ISIP1 gene, which is the most highly up-regulated gene under conditions of iron stress, defined by low availability of labile ferrous iron [Fe(II)] in laboratory experiments (9, 10). It is also noteworthy that, unlike P. tricornutum and T. oceanica, T. pseudonana is not a low–iron quota diatom because it cannot grow in the chronically low iron concentrations that are commonly found in the most severely Fe-limited regions of the world’s ocean (7, 42).
We used the MMETSP and UniProt databases to conduct a phylogenetic survey of ISIP1 distribution and found it to be largely confined to diatoms, with some transcripts detected in pelagophytes, haptophytes, and fucoxanthin-containing dinoflagellate species, linked together by historical plastid endosymbiosis events (41). Although it remains to be shown whether ISIP1 can be used as a biomarker for siderophore uptake in diatoms through more extensive analyses, no published reports would contradict this observation. For example, an influential study by Hutchins et al. (43) compared the preferences of two centric diatom species, T. weissflogii (CCMP 1049) and Skeletonema costatum (CCMP 1332), with two cyanobacterial species (Synechococcus sp. CCMP 1334 and PCC 7002) for organic chelates of Fe, including a range of siderophores and porphyrins, to conclude that the diatoms were poor competitors for siderophores. However, the data are biased by the observed inability of S. costatum to use siderophores, in sharp contrast to T. weissflogii, which readily took up alterobactins, ferrioxamine, and FCH. Our analysis of ISIP1 presence/absence underscores this finding because T. weissflogii has ISIP1 in its genetic portfolio, whereas S. costatum does not. Conversely, a recent study by Cohen et al. (8) found that ISIP1 was strongly up-regulated in natural phytoplankton communities enriched in Pseudo-nitzschia and Thalassiosira diatoms in microcosm experiments supplemented with desferrioxamine B (200 nM), a concentration sufficient to chelate any available iron in the seawater into siderophore-bound FOB.
Although the presence of ISIP1 may confer an ability to take up and use a range of siderophores, it is likely to be only a part of the molecular machinery required for the process. We found that although its synthesis is strongly up-regulated under conditions of iron starvation and that its localization to the cell membrane is induced by siderophore analogs, ISIP1 knockdown did not affect siderophore binding at the cell surface of P. tricornutum, so it is unlikely to be a binding protein for siderophores. On the other hand, a good candidate is the ferrichrome-binding protein FBP1 annotated in the P. tricornutum genome. In T. oceanica, which contains ISIP1 and lives in low-Fe oceanic regions, the catecholate siderophore ENT is taken up preferentially to hydroxamate siderophores, so it is tempting to speculate that the same ISIP1-dependent process is involved in siderophore uptake in this species, which likely has different siderophore receptors at the cell surface than P. tricornutum. T. oceanica also contains FBP1, which suggests that a third, as yet unknown, gene is responsible for the differences in physiology.
A previous study by Maldonado and Price (25) focused on the uptake of FOB by T. oceanica and concluded that it occurred through reduction of the FOB complex at the cell surface. In our hands, uptake of FOB by T. oceanica was very poor and the species had a clear preference for the catecholate siderophore ENT. We do not believe the two studies to be directly comparable (due to a range of methodological differences), although the disparity in conclusions of each study highlights the need to identify a mechanistic connection between siderophore groups and diatom species capable of their uptake.
FOB uptake kinetics were remarkable in P. tricornutum. To our knowledge, the siderophore uptake system we have uncovered shows the highest affinity described for any iron transport system in any eukaryotic cell. Our observations also question the previous hypothesis that removal of the chelated Fe occurs on the surface of the cell, by reduction, in a process that does not discriminate between siderophore product (15). We present multiple lines of evidence in support of a nonreductive mechanism for siderophore uptake. The most marked is our microscopy-based demonstration of siderophore analogs inside P. tricornutum and the unloading of Fe from a fluorescently tagged FOB in the vicinity of the chloroplast. Further evidence is the observation that, in P. tricornutum, uptake of iron from siderophores was not inhibited by ferrous chelators, which, by acting on the cell surface, would immediately remove any Fe′ released from the siderophores.
Our evidence for the process of siderophore uptake in P. tricornutum is reminiscent of what has been reported in yeast, rather than bacteria. Here, genes encoding siderophore transporters are induced by iron deprivation, whereas induction of siderophore uptake requires the sensing by the cells of siderophores (or siderophore analogs) in the medium, meaning that siderophore uptake is regulated through a delicate balance between the sensing of iron as a metal and siderophores as specific iron complexes required for inducing the uptake process (44). A similar process could be at play in P. tricornutum, which we are currently trying to determine. The role of endocytosis, a process confined to eukaryotes, further supports this connection. We found that inhibitors of endocytosis had the effect of inhibiting Fe uptake from siderophores. Endocytosis of siderophores was also aborted in P. tricornutum lines with decreased ISIP1 protein. It is possible that it is the link between ISIP1 and endocytosis, a function that bacteria are not capable of, that gives diatoms a competitive edge in natural communities for the scavenging of iron. Siderophore uptake by an endocytosis pathway targeted to the chloroplast, where iron limitation would be most acute in photosynthetic cells, is a novel and efficient pathway for surviving iron stress in an alga.
Our study highlights the importance of working with model species, which allow a detailed dissection of cell physiology and can bring to light the molecular underpinnings of environmentally important processes. However, it also cautions against unqualified extrapolations of results obtained in one species as trends that go beyond the functional group described, or into environments where the findings are irrelevant. In terms of quantifying functional relevance, the Tara Oceans data set represents an invaluable resource. Although P. tricornutum is not an abundant diatom in the ocean, the ISIP1 gene identified in this species is ubiquitous globally, and both this study and others have found its expression to be a biomarker for iron stress (8–11). Although iron speciation and bioavailability are widely recognized to be important drivers of ocean ecosystems (45), measuring the pool of iron using chemical methods alone remains a major challenge (17). We propose that routine measurements of in situ ISIP1 expression during ocean studies could provide a significant improvement on current practices. A composite biomarker for iron stress has been proposed by Marchetti et al. (46) based on the relative expression of ferritin (FTN; a measure of iron storage, when iron is replete) compared to phytotransferrin ISIP2a (a measure of iron starvation). Adding to this a measure of ISIP1 expression would serve two purposes: It would provide a more diatom-specific measure of iron status and provide a possible indicator of plankton community interactions because it signals the use of siderophores in the water column by diatoms. The latter interpretation requires caution, because it remains an open question whether ISIP1 has the unique function of mediating endocytosis-driven uptake of siderophore iron in diatoms, or whether it is involved in additional cellular processes.
Furthermore, the importance of the siderophore pool in driving diatom dynamics in natural communities requires experimental assessment in situ. This is becoming increasingly more likely as chemical techniques for identification of siderophores in seawater are improving. Although marine siderophores produced in laboratory cultures have been found to contain all major chelating groups (hydroxamate, catecholate, and carboxylate) in a staggering variety of forms (47), until recently, only hydroxamate siderophores (which are hydrophilic) have been detected in seawater. For example, in the Atlantic Ocean, ferrioxamine-type siderophores were detected ubiquitously and were found to be present at concentrations of up to 20 pM, which accounts for between 0.5 and 5% of the total dissolved Fe pool (48). Using state-of-the-art liquid chromatography–inductively coupled plasma mass spectrometry and electrospray ionization mass spectrometry with metal isotope pattern detection algorithms, Boiteau et al. (49) successfully detected amphibactins in the high-nutrient, low-chlorophyll region of a transect along a nutrient gradient in the eastern Pacific Ocean. They further detected ferrioxamine-type siderophores not only in coastal regions, which were characterized by Fe limitation, but also in oligotrophic waters, which were characterized by limitation in all nutrients. The authors proposed that changes in siderophore abundances could be characterized by a changing bacterial community, which, in turn, is driven by a trade-off between the requirements for nitrogen and Fe.
In conclusion, our finding that some diatoms demonstrate a specific preference for siderophores has consequences for the study of microbial ecology of natural communities exchanging organic products. It is clear that reliance on model species, while remaining imperative, should also be nuanced with a better understanding of the natural environment and metabolic capacities of individual species. We have used P. tricornutum as a model for low–iron quota diatoms and have found a candidate gene, ISIP1, which relates the ability of this species to take up and use hydroxamate siderophores through a mechanism that requires endocytosis and appears to not require reduction of these siderophore complexes at the cell surface. This adds to the repertoire of uptake pathways known in diatom species and highlights their adaptive/competitive abilities for obtaining Fe from the heterogeneous and dynamic pool of natural Fe complexes in the ocean. However, further physiological studies involving studies in the field will be required to determine the extent to which diatoms have the relevant molecular machinery to take up environmentally relevant siderophores. If ocean models are to accurately predict global changes to phytoplankton productivity, then they will have to incorporate an understanding of both the chemistry and speciation of Fe in seawater as well as information on the interaction between these Fe complexes, phytoplankton functional groups, and Fe cycling within the microbial communities. Given the sensitivity of these processes to oxygen, pH, and temperature, their incorporation into biogeochemical models will likely improve model performance in predicting the consequences on marine biota of a future ocean affected by climate change (50). Finally, because ISIP1-dependent siderophore uptake is nonreductive, as previously shown for ferric iron uptake mediated by ISIP2a (12), these combined results infer that our current view of iron uptake in diatoms as occurring principally through reductive mechanisms requires revision.
MATERIALS AND METHODS
Transgenic diatoms
P. tricornutum ISIP2a knockdown lines were constructed as previously described (12). P. tricornutum ISIP1 knockdown lines were constructed under the control of the FCPB promoter, as described by Siaut et al. (51), using the primers ISIP1-antisense-Fw (5′-GAATTCGATCGACCCCAAGTCTAGCA-3′) and ISIP1-antisense-Rv (5′-TCTAGATGCGCTCGTAGTTTTGAACA-3′).
Expression of the transgenic constructs was maintained under zeocin selection (50 μg/ml). To generate ISIP1-YFP lines, the ISIP1 gene was cloned into the vector plasmid pDEST-C-YFP using the Gibson Assembly Kit (New England Biolabs). A DNA fragment of ISIP1 with its promoter region [827 base pairs (bp) upstream the starting codon] was polymerase chain reaction (PCR)–amplified using the primers ISIP1-Gb-Fw and ISIP1-FP-Gb-Rv and genomic DNA from P. tricornutum as template. This DNA fragment was assembled into pDEST-C-YFP previously PCR-amplified using the primers pDEST-FP-Gb-Fw and pDEST-Gb-Rv and the plasmid pDEST-C-YFP as a template. The ISIP1 PCR-DNA fragment was designed with 20-bp overlaps (underlined region of the primers) to the flanking regions of the linearized pDEST-C-YFP. The pDEST-C-YFP PCR-DNA fragment was designed without the original constitutive FcpB promoter to study the expression of ISIP1:YFP under the native promoter of ISIP1. The resulting vector was introduced into P. tricornutum by microparticle bombardment, as described elsewhere (52). Positive transformants were selected using zeocin plates (50 μg/ml), and colonies were transferred to liquid media before screening with a microscope.
Molecular methods
For RNA and complementary DNA analysis, the nucleic acids were extracted as described by Siaut et al. (51). Quantitative PCR (qPCR) of ISIP1 was performed using the primers ISIP1-qPCR-Fw (5′-TACGAGGACGACAACGATATTGACGCA-3′) and ISIP1-qPCR-Rv (5′-GAAGATGTAGGCTGGCCATGAGACTGC-3′) following the previously optimized protocols described by Siaut et al. (51).
Expression of ISIP1 was normalized to three housekeeping genes: the 30S ribosomal protein subunit (RPS), the TATA box–binding protein (TBP), and glyceraldehyde-3-phosphate dehydrogenase C2 (gapC2), which were found to be the most constant of 12 previously analyzed housekeeping genes in P. tricornutum (51). The normalization was calculated using the ΔΔCt method described by Livak and Schmittgen (53) incorporating the refinement described by Pfaffl et al. (54), which also takes into account primer efficiencies.
For Western blots, proteins were extracted by adding 50 μl of lysis buffer [50 mM tris (pH 6.8), 2% SDS] to frozen cell pellets, and cell lysates were incubated for 30 min at room temperature. Protein extracts (10 μg) from wild-type and ISIP1 knockdown lines were resolved on 10% SDS–polyacrylamide gel electrophoresis gels and transferred to nitrocellulose transfer membranes (Whatman) using the Biometra semi-dry method. The ISIP1 protein was detected by incubating the membranes with a 1:5000 dilution of rabbit polyclonal anti-ISIP1 antibody (gift of A. E. Allen, J. Craig Venter Institute/Scripps Institution of Oceanography/University of California, San Diego) after 2 hours at 4°C, followed by a 1-hour incubation with a 1:10,000 dilution of horseradish peroxidase–conjugated goat anti-rabbit secondary antibody (Promega) in 1× phosphate-buffered saline, 1% milk, and 0.1% Tween 20. The anti-H2B antibody was used as a loading control (1:5000 dilution). The signals were visualized using the enhanced chemiluminescence kit of Amersham Biosciences, and protein levels were quantified by densitometry using the Quantity One 4.1.1 software (Bio-Rad).
Culturing conditions and diatom growth measurements
For growth comparisons (Fig. 2), steady-state cultures of wild-type axenic P. tricornutum and ISIP1 knockdown mutant were maintained under constant light (100 μmol photons m−2 s−1) at 18°C and a state of iron deprivation. Axenicity was confirmed by examining the cultures under the microscrope and by standard plating analyses. The average cell concentration reached at steady state was 5 × 104 cells/ml (with an average doubling time of 1.13 days−1 and σ2 < 0.3). These were used as precultures to inoculate the experiment described in Fig. 2 at a density of 2 × 103 cells/ml at t = 0. For precultures, the growth medium contained 15 pM of dissolved iron, Fe′ (where Fe′ is estimated as the sum of all Fe species not complexed to EDTA), determined to be sufficient to starve P. tricornutum (55). The growth medium was a modified recipe of Aquil [first described by Sunda et al. (56)] containing 100 μM nitrate, 10 μM phosphate and 100 μM silicate, standard f/2 vitamin concentrations, and 300 μM EDTA with free Zn, Mn, Co, Cu, MoO4, and SeO3 concentrations expressed as negative algorithms of 10.93, 8.03, 10.77, 12.63, 7.00, and 8.00. The total Fe concentration added was 7.5 nM Fe in the form of FeCl3, and Fe′ was calculated assuming a 2 nM background of Fe in the Aquil preparation and the equations of Sunda et al. (56), considering temperature and pH (buffered at 8.0). All culturing was carried out in polycarbonate tubes, with a culture volume of 20 ml. Reagents, media, and cultures were handled only using plasticware in a trace metal clean environment. Cell counts were taken daily in triplicate using an Accuri C6 flow cytometer (BD Biosciences) calibrated against cell density measured using a Malassez chamber and gated for P. tricornutum based on cell autofluorescence (“FL3” channel, excitation 488 nm, emission ≥ 670 nm) as well as forward and side scatter. Fluorescence measurements were also taken daily (10AU fluorometer by Turner Designs) to corroborate the cell count values. For the FOB treatment, Aquil medium equilibrated at 15 pM Fe′ was spiked with DFOB (desferrioxamine B) to bring the medium to a final concentration of 10 nM FOB. The medium was equilibrated overnight at 18°C in the dark before inoculation with P. tricornutum. The original Aquil solution was prepared with 10 nM Fe-EDTA (which was calculated to dissociate into 15 pM Fe′ under the experimental conditions).
Iron uptake measurements
Iron uptake measurements were carried out according to the methodology described previously by Morrissey et al. (12), specifically adapted to P. tricornutum. Briefly, cells were harvested at mid-exponential growth from iron-deplete cultures grown in EDTA-free artificial seawater (Mf medium) that did not contain added Fe and buffered at pH 7.8 [Hepes (1 g/liter), tris (0.3 g/liter)]. The cell concentration reached at steady state was approximately 1 × 105 cells/ml. Harvested cells were concentrated by centrifugation (discarding the supernatant) to a density of approximately 1 × 106 cells/ml, washed with iron-free Mf medium followed by 0.1 mM EDTA to chelate any iron, followed by another round of washing in iron-free media to remove the EDTA. Following these wash steps, cells were inoculated into the wells of a 96-well microtiter plate (200 μl per well) containing fresh iron-free Mf medium and kept in the dark for the duration of the experiment. At the start of each time course, wells were spiked with a source of 55Fe, and measurements were collected at regular intervals (12). For each measurement, cells were soaked for 5 min in an oxalate/EDTA mixture and then washed three times in fresh oxalate/EDTA solution, and the remaining iron was associated to the cells counted by scintillation after bleaching of the cell fluorescence with hypochlorite, which does not interfere with Fe content (57). The contaminating iron in “-Fe” cultures (with no added iron) was estimated to be less than 1 nM (58). Our plate assay for 55Fe uptake is a high-throughput alternative to the traditional 59Fe manual uptake assays. By processing and washing on a filter 24 samples at once, it enables a comparison of different conditions rapidly and with several replicates. It decreases significantly the statistical deviation observed between replicates. However, for key significant findings, we repeated our work with the more laborious traditional method, working with lower cell densities and larger volumes. For this, diatoms were grown in iron-deficient Mf medium in 1-liter plastic flasks and diluted in exponential phase of growth to 106 cells/ml. The cell suspension was distributed in 50-ml Falcon tubes, and 55Fe was then added. The cells were incubated and harvested and washed by centrifugation (2000g, 5 min) periodically, as required by the experiment, before counting. Because none of our findings were contradicted by the large-volume, lower–cell density experiments, we have presented our data from the plate assays, which provide a finer-resolution analysis.
Stock solutions of 55Fe-labeled siderophores were prepared as follows: A 10% excess of desferri-siderophore solution was added to 55FeCl3 in 0.1 M HCl to give a 5 mM 55Fe solution with a Fe/ligand ratio of 1:1.1; after 10 min at room temperature, the pH was adjusted to 7 with 1 M Hepes, and the final 1 mM stock solution was kept at −20°C until use. For ferric EDTA, the ligand-to-iron ratio was 20, and we checked by ultracentrifugation that the complex remained fully soluble during the time of experiments.
Siderophore constructs and reagents
Gallium analogs of FOB and FCH and siderophores coupled to the fluorescent moiety NBD were synthesized as previously described by van Gisbergen et al. (34). FOB coupled to inhibitors of protoporphyrinogen oxidase were synthesized as previously described by Matringe et al. (35). All other chemicals and reagents were purchased from Sigma-Aldrich, unless otherwise stated.
Microscopy
Epifluorescence microscopy was performed on a Zeiss Axioskop with an HBO 50W UV lamp using 490-nm excitation and 515- to 585-nm emission (to detect FOB-NBD), 500-nm excitation and 545-nm emission (to detect ISIP1-YFP), and 365-nm excitation and 417- to 477-nm emission (to detect DAPI to label the nucleus), and the chlorophyll autofluorescence overlay was generated using an AF filter (615- to 700-nm emission).
Confocal images were performed with a Zeiss LSM 780 microscope. We used a 63× oil immersion objective (numerical aperture, 1.4). Cells labeled with FM4-64 and with FOB-NBD were excited with the 488-nm laser line, and the emission signals were collected by detection windows ranging from 606 to 650 nm and 499 to 544 nm, respectively. Chlorophyll autofluorescence was evidenced by excitation with the 633-nm laser line, and the emission signal was collected by a detection window ranging from 637 to 758 nm. For these three fluorescent signals, we used a 488/561/633-nm multiple beam splitter. ISIP1-YFP lines grown in low-Fe Aquil media shown in Fig. 6A were visualized additionally on a Leica SP5 microscope using the same laser settings and detection filters as above.
Phylogenetic analysis
The ISIP1 protein sequence (Phatr3_J55031) was used as the seed sequence for BLASTp analysis against a curated library comprising all protein sequences in UniRef, the MMETSP (28), and all additional sequences hosted on the JGI portal. This composite database, described by Dorrell et al. (41), is more extensive than any individual resource. Sequences retrieved with a threshold e value of 1 × 10−05 were verified to be ISIP1 by a BLASTp best hit against version 3 of the P. tricornutum genome (38). The resulting set of sequences was aligned using the first 20× iterations of MAFFT v8 and then the inbuilt alignment builder in Geneious v.4.76 under the default criteria. Where multiple sequences were retrieved for individual species, a species consensus was generated. Only 36 species were found to contain ISIP1, of which 28 were from diatom transcriptomes and 8 were from dinoflagellates, pelagophytes, and haptophytes (table S1). These were realigned using Geneious and trimmed at the N terminus to the first methionine present and at the C terminus to the last residue of the protein (with 70% identity). Trees were inferred using the MrBayes and RAxML programs inbuilt into the CIPRES webserver (table S1). A Bayesian tree was inferred using three substitution models (GTR, Jones, and WAG), a minimum of 600,000 generations, and an initial burn-in discard value of 0.5. Trees were only used if the final convergence statistic between the two chains run was <0.1, and tree calculation was automatically stopped if the final convergence statistic between the two chains run was below 0.01. RAxML was inferred using three substitution models (GTR, JTT, and WAG) with automatic bootstrapping, as previously described by Dorrell et al. (41).
Querying of Tara Oceans diatom eukaryotic gene atlas
We curated the Tara Oceans data set to consider only data collected from surface samples, which were obtained consistently from the top 10 m of the ocean (excluding samples collected from the deep chlorophyll maximum) across 68 stations in the global ocean transect, for which both metagenome and metatranscriptome data are available. We pooled the data from four different size fractions (0.8 to 5 μm, 5 to 20 μm, 20 to 180 μm, and 180 to 2000 μm) to get near-exhaustive recovery of total eukaryotic biodiversity in each sample. Note that a detailed description of all Tara Oceans field sampling strategy and protocols is available in the study of Carradec et al. (29). Next, using the homologs retrieved by the reciprocal BLAST best-hit search (described in the previous section), we built a profile hidden Markov model (pHMM) with the most conserved homologs. That is, we kept only sequences fully covered by the multiple sequence alignment produced by the phylogenetic analysis, because ISIP1 does not belong to a particular protein family in any pHMM database, such as PFAM. We used the pHMM along with the software package HMMER (59) to retrieve all sequences from the Tara Oceans metagenome and metatranscriptome that matched our model of ISIP1. Only sequences taxonomically assigned as diatoms and not previously associated with any function (all Tara Oceans sequences were functionally annotated by using PFAM and HMMER) (29) were considered as true positive. For the true positives retrieved, taxonomic assignment was available and is accessible at www.genoscope.cns.fr/tara/. We have provided the full catalog of hits and their phylogenetic assignation, which was used to build Fig. 6B, in table S1. The expression values and genomic occurrences were computed in RPKM, normalized against the 20 most stable diatom ribosomal proteins in the Tara Oceans samples (a robust data set of 30,000 sequences), computed in C++. For the graphical representation, statistical analyses were conducted in R (v 3.1.2) and were plotted using the R package ggplot2 (v 2.1.0) onto a reference National Center for Biotechnology Information diatom tree (60).
Supplementary Material
Acknowledgments
We thank A. E. Allen and J. B. McQuaid for sharing the ISIP1 antibody. E.K. thanks T. Coale and J. Turnsek (both from the Allen laboratory) for assistance during experiments and useful discussions. Funding: Funding is acknowledged from the European Research Council “Diatomite” (to C.B.) and Agence Nationale de la Recherche “PhytoIron” and “Phytomet” (ANR-11-BSV7-018-02 and ANR-16-CE01-0008 to C.B. and E.L.) projects, as well as the EU MicroB3 project (to C.B.). C.B. additionally thanks the French Government “Investissements d’Avenir” programs MEMO LIFE (ANR-10-LABX-54), PSL* Research University (ANR-11-IDEX-0001-02), and Oceanomics (ANR-11-BTBR-0008), as well as the Gordon and Betty Moore Foundation and the LouisD Foundation of the Institut de France. E.K. was supported by a Marie-Curie personal fellowship (EU project 748581—IRONCOMM). R.S. acknowledges the Czech Science Foundation (18-07822S and 13-25349S), LQ1604 NPU II provided by Ministry of Education, Youth and Sports of the Czech Republic (MEYS), and CZ.1.05/1.1.00/02.0109 BIOCEV and CZ.02.1.01/0.0/0.0/16_019/0000759 CePaViP provided by The European Regional Development Fund and MEYS. J.P.-Y. was supported with a Marie-Curie postdoctoral fellowship from the 7th research program of the European Union FP7/2007-2013 under grant agreement no. PIOF-GA-2011-301466—GenMarPhyto. R.G.D. was supported with the European Molecular Biology Organization Early Career Fellowship (ALTF 1124/2014). We acknowledge the ImagoSeine core facility of the Institut Jacques Monod, also associated with IBiSA and France BioImaging infrastructures, and the IMCF at BIOCEV, Faculty of Sciences, Charles University, supported by the MEYS CR (LM2015062 Czech-BioImaging). This article is contribution number 74 of the Tara Oceans project. Author contributions: R.S., J.-M.C., and C.B. conceived the study. E.L. and C.B. supervised the work. R.S., J.P.-Y., E.K., J. Morrissey, and E.L. performed most of the experimental work. S.L., F.L., J.P.-Y., E.K., and J. Mach performed microscopy. R.G.D. and E.K. performed the phylogenetic analysis. E.P. and F.R.J.V. performed the bioinformatics analysis of Tara Oceans metagenomes and metatranscriptomes. E.L., E.K., R.S., and C.B. interpreted the data and wrote the manuscript. Competing interests: All authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/5/eaar4536/DC1
fig. S1. Siderophore uptake in P. tricornutum fits simple Michaelis-Menten kinetics.
fig. S2. Construction of an ISIP1 knockdown in P. tricornutum and expression of ISIP1 transcript and protein.
fig. S3. Siderophore uptake involves a binding step.
fig. S4. Siderophore uptake involves endocytosis.
fig. S5. ISIP1 knockdown lines are defective in endocytosis.
fig. S6. ISIP1-YFP localization and abundance under different Fe supplementation regimes.
fig. S7. ISIP1 predicted protein sequence and domain features.
fig. S8. ISIP1 phylogenetic tree.
movie S1. Live cell imaging of the uptake of NBD conjugate of FOB (FOB-NBD) by P. tricornutum.
table S1. Data used to construct Fig. 6 and fig. S8.
Reference (61)
REFERENCES AND NOTES
- 1.Morrissey J., Bowler C., Iron utilization in marine cyanobacteria and eukaryotic algae. Front. Microbiol. 3, 43 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raven J. A., Evans M. C. W., Korb R. E., The role of trace metals in electron transport in O2-evolving organisms. Photosynth. Res. 60, 111–150 (1999). [Google Scholar]
- 3.Boyd P. W., Jickells T., Law C. S., Blain S., Boyle E. A., Buesseler K. O., Coale K. H., Cullen J. J., de Baar H. J. W., Follows M., Harvey M., Lancelot C., Levasseur M., Owens N. P. J., Pollard R., Rivkin R. B., Sarmiento J., Schoemann V., Smetacek V., Takeda S., Tsuda A., Turner S., Watson A. J., Mesoscale iron enrichment experiments 1993–2005: Synthesis and future directions. Science 315, 612–617 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Coale K. H., Johnson K. S., Chavez F. P., Buesseler K. O., Barber R. T., Brzezinski M. A., Cochlan W. P., Millero F. J., Falkowski P. G., Bauer J. E., Wanninkhof R. H., Kudela R. M., Altabet M. A., Hales B. E., Takahashi T., Landry M. R., Bidigare R. R., Wang X., Chase Z., Strutton P. G., Friederich G. E., Gorbunov M. Y., Lance V. P., Hilting A. K., Hiscock M. R., Demarest M., Hiscock W. T., Sullivan K. F., Tanner S. J., Gordon R. M., Hunter C. N., Elrod V. A., Fitzwater S. E., Jones J. L., Tozzi S., Koblizek M., Roberts A. E., Herndon J., Brewster J., Ladizinsky N., Smith G., Cooper D., Timothy D., Brown S. L., Selph K. E., Sheridan C. C., Twining B. S., Johnson Z. I., Southern Ocean iron enrichment experiment: Carbon cycling in high- and low-Si waters. Science 304, 408–414 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Granum E., Raven J. A., Leegood R. C., How do marine diatoms fix 10 billion tonnes of inorganic carbon per year? Can. J. Bot. 83, 898–908 (2005). [Google Scholar]
- 6.Bowler C., Allen A. E., Badger J. H., Grimwood J., Jabbari K., Kuo A., Maheswari U., Martens C., Maumus F., Otillar R. P., Rayko E., Salamov A., Vandepoele K., Beszteri B., Gruber A., Heijde M., Katinka M., Mock T., Valentin K., Verret F., Berges J. A., Brownlee C., Cadoret J.-P., Chiovitti A., Choi C. J., Coesel S., De Martino A., Detter J. C., Durkin C., Falciatore A., Fournet J., Haruta M., Huysman M. J. J., Jenkins B. D., Jiroutova K., Jorgensen R. E., Joubert Y., Kaplan A., Kröger N., Kroth P. G., La Roche J., Lindquist E., Lommer M., Martin-Jézéquel V., Lopez P. J., Lucas S., Mangogna M., McGinnis K., Medlin L. K., Montsant A., Oudot-Le Secq M.-P., Napoli C., Obornik M., Parker M. S., Petit J.-L., Porcel B. M., Poulsen N., Robison M., Rychlewski L., Rynearson T. A., Schmutz J., Shapiro H., Siaut M., Stanley M., Sussman M. R., Taylor A. R., Vardi A., von Dassow P., Vyverman W., Willis A., Wyrwicz L. S., Rokhsar D. S., Weissenbach J., Armbrust E. V., Green B. R., Van de Peer Y., Grigoriev I. V., The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Kustka A. B., Allen A. E., Morel F. M. M., Sequence analysis and transcriptional regulation of iron acquisition genes in two marine diatoms. J. Phycol. 43, 715–729 (2007). [Google Scholar]
- 8.Cohen N. R., Ellis K. A., Lampe R. H., McNair H., Twining B. S., Maldonado M. T., Brzezinski M. A., Kuzminov F. I., Thamatrakoln K., Till C. P., Bruland K. W., Sunda W. G., Bargu S., Marchetti A., Diatom transcriptional and physiological responses to changes in iron bioavailability across ocean provinces. Front. Mar. Sci. 4, 360 (2017). [Google Scholar]
- 9.Allen A. E., LaRoche J., Maheswari U., Lommer M., Schauer N., Lopez P. J., Finazzi G., Fernie A. R., Bowler C., Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. U.S.A. 105, 10438–10443 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lommer M., Specht M., Roy A.-S., Kraemer L., Andreson R., Gutowska M. A., Wolf J., Bergner S. V., Schilhabel M. B., Klostermeier U. C., Beiko R. G., Rosenstiel P., Hippler M., LaRoche J., Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol. 13, R66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchetti A., Schruth D. M., Durkin C. A., Parker M. S., Kodner R. B., Berthiaume C. T., Morales R., Allen A. E., Armbrust E. V., Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl. Acad. Sci. U.S.A. 109, E317–E325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrissey J., Sutak R., Paz-Yepes L., Tanaka A., Moustafa A., Velucham A., Thomas Y., Botebol H., Bouget F.-Y., McQuaid J. B., Tirichine L., Allen A. E., Lesuisse E., Bowler C., A novel protein, ubiquitous in marine phytoplankton, concentrates iron at the cell surface and facilitates uptake. Curr. Biol. 25, 364–371 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Anderson M. A., Morel F. M. M., The influence of aqueous iron chemistry on the uptake of iron by the coastal diatom Thalassiosira weissflogii. Limnol. Oceanogr. 27, 789–813 (1982). [Google Scholar]
- 14.McQuaid J. B., Kustka A. B., Oborník M., Horák A., McCrow J. P., Karas B. J., Zheng H., Kindeberg T., Andersson A. J., Barbeau K. A., Allen A. E., Carbonate sensitive phytotransferrin controls high affinity iron uptake in diatoms. Nature 555, 534–537 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Shaked Y., Lis H., Disassembling iron availability to phytoplankton. Front. Microbiol. 3, 123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morel F. M. M., Kustka A. B., Shaked Y., The role of unchelated Fe in the iron nutrition of phytoplankton. Limnol. Oceanogr. 53, 400–404 (2008). [Google Scholar]
- 17.Gledhill M., Buck K. N., The organic complexation of iron in the marine environment: A review. Front. Microbiol. 3, 69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buck K. N., Selph K. E., Barbeau K. A., Iron-binding ligand production and copper speciation in an incubation experiment of Antarctic Peninsula shelf waters from the Bransfield Strait, Southern Ocean. Mar. Chem. 122, 148–159 (2010). [Google Scholar]
- 19.King A. L., Buck K. N., Barbeau K. A., Quasi-Lagrangian drifter studies of iron speciation and cycling off Point Conception, California. Mar. Chem. 128–129, 1–12 (2012). [Google Scholar]
- 20.Sutak R., Lesuisse E., Tachezy J., Richardson D. R., Crusade for iron: Iron uptake in unicellular eukaryotes and its significance for virulence. Trends Microbiol. 16, 261–268 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Lesuisse E., Simon-Casteras M., Labbe P., Siderophore-mediated iron uptake in Saccharomyces cerevisiae: The SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 144, 3455–3462 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Chu B. C., Garcia-Herrero A., Johanson T. H., Krewulak K. D., Lau C. K., Peacock R. S., Vogel H. J., Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. Biometals 23, 601–611 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Kustka A. B., Jones B. M., Hatta M., Field M. P., Milligan A. J., The influence of iron and siderophores on eukaryotic phytoplankton growth rates and community composition in the Ross Sea. Mar. Chem. 173, 195–207 (2015). [Google Scholar]
- 24.Soria-Dengg S., Horstmann U.. Ferrioxamines B and E as iron sources for the marine diatom Phaeodactylum tricornutum. Mar. Ecol. Prog. Ser. 127, 269–277 (1995). [Google Scholar]
- 25.Maldonado M. T., Price N. M., Reduction and transport of organically bound iron by Thalassiosira oceanica (Bacillariophyceae). J. Phycol. 37, 298–310 (2001). [Google Scholar]
- 26.Shaked Y., Kustka A. B., Morel F. M. M., A general kinetic model for iron acquisition by eukaryotic phytoplankton. Limnol. Oceanogr. 50, 872–882 (2005). [Google Scholar]
- 27.Griffin A. S., West S. A., Buckling A., Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Keeling P. J., Burki F., Wilcox H. M., Allam B., Allen E. E., Amaral-Zettler L. A., Armbrust E. V., Archibald J. M., Bharti A. K., Bell C. J., Beszteri B., Bidle K. D., Cameron C. T., Campbell L., Caron D. A., Cattolico R. A., Collier J. L., Coyne K., Davy S. K., Deschamps P., Dyhrman S. T., Edvardsen B., Gates R. D., Gobler C. J., Greenwood S. J., Guida S. M., Jacobi J. L., Jakobsen K. S., James E. R., Jenkins B., John U., Johnson M. D., Juhl A. R., Kamp A., Katz L. A., Kiene R., Kudryavtsev A., Leander B. S., Lin S., Lovejoy C., Lynn D., Marchetti A., McManus G., Nedelcu A. M., Menden-Deuer S., Miceli C., Mock T., Montresor M., Moran M. A., Murray S., Nadathur G., Nagai S., Ngam P. B., Palenik B., Pawlowski J., Petroni G., Piganeau G., Posewitz M. C., Rengefors K., Romano G., Rumpho M. E., Rynearson T., Schilling K. B., Schroeder D. C., Simpson A. G. B., Slamovits C. H., Smith D. R., Smith G. J., Smith S. R., Sosik H. M., Stief P., Theriot E., Twary S. N., Umale P. E., Vaulot D., Wawrik B., Wheeler G. L., Wilson W. H., Xu Y., Zingone A., Worden A. Z., The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLOS Biol. 12, e1001889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carradec Q., Pelletier E., Da Silva C., Alberti A., Seeleuthner Y., Blanc-Mathieu R., Lima-Mendez G., Rocha F., Tirichine L., Labadie K., Kirilovsky A., Bertrand A., Engelen S., Madoui M.-A., Méheust R., Poulain J., Romac S., Richter D. J., Yoshikawa G., Dimier C., Kandels-Lewis S., Picheral M., Searson S.; Tara Oceans Coordinators, Jaillon O., Aury J.-M., Karsenti E., Sullivan M. B., Sunagawa S., Bork P., Not F., Hingamp P., Raes J., Guidi L., Ogata H., de Vargas C., Iudicone D., Bowler C., Wincker P., A global ocean atlas of eukaryotic genes. Nat. Commun. 9, 373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosman D. J., Molecular mechanisms of iron uptake in fungi. Mol. Microbiol. 47, 1185–1197 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Ouchetto H., Dias M., Mornet R., Lesuisse E., Camadro J.-M., A new route to trihydroxamate-containing artificial siderophores and synthesis of a new fluorescent probe. Bioorg. Med. Chem. 13, 1799–1803 (2005). [DOI] [PubMed] [Google Scholar]
- 32.De Martino A., Bartual A., Willis A., Meichenin A., Villazán B., Maheswari U., Bowler C., Physiological and molecular evidence that environmental changes elicit morphological interconversion in the model diatom Phaeodactylum tricornutum. Protist 162, 462–481 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G., Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. U.S.A. 108, E450–E458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Gisbergen P. A. C., Esseling-Ozdoba A., Vos J. W., Microinjecting FM4-64 validates it as a marker of the endocytic pathway in plants. J. Microsc. 231, 284–290 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Matringe M., Camadro J. M., Labbe P., Scalla R., Protoporphyrinogen oxidase as a molecular target for diphenyl ether herbicides. Biochem. J. 260, 231–235 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullmann L., Haarmann R., Mirus O., Bredemeier R., Hempel F., Maier U. G., Schleiff E., Filling the gap, evolutionarily conserved Omp85 in plastids of chromalveolates. J. Biol. Chem. 285, 6848–6856 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gruber A., Rocap G., Kroth P. G., Armbrust E. V., Mock T., Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 81, 519–528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rastogi A., Maheswari U., Dorrell R. G., Vieira F. R. J., Maumus F., Kustka A., McCarthy J., Allen A. E., Kersey P., Bowler C., Tirichine L., Integrative analysis of large scale transcriptome data draws a comprehensive landscape of Phaeodactylum tricornutum genome and evolutionary origin of diatoms. Sci. Rep. 8, 4834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamb C. A., Dooley H. C., Tooze S. A., Endocytosis and autophagy: Shared machinery for degradation. Bioessays 35, 34–45 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Springer T. A., An extracellular β-propeller module predicted in lipoprotein scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J. Mol. Biol. 283, 837–862 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Dorrell R. G., Gile G., McCallum G., Méheust R., Bapteste E. P., Klinger C. M., Brillet-Guéguen L., Freeman K. D., Richter D. J., Bowler C., Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. eLife 6, e23717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchetti A., Maldonado M. T., Lane E. S., Harrison P. J., Iron requirements of the pennate diatom Pseudo-nitzschia: Comparison of oceanic (high-nitrate, low-chlorophyll waters) and coastal species. Limnol. Oceanogr. 51, 2092–2101 (2006). [Google Scholar]
- 43.Hutchins D. A., Witter A. E., Butler A., Luther G. W. III, Competition among marine phytoplankton for different chelated iron species. Nature 400, 858–861 (1999). [Google Scholar]
- 44.Kim Y., Yun C.-W., Philpott C. C., Ferrichrome induces endosome to plasma membrane cycling of the ferrichrome transporter, Arn1p, in Saccharomyces cerevisiae. EMBO J. 21, 3632–3642 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tagliabue A., Bowie A. R., Boyd P. W., Buck K. N., Johnson K. S., Saito M. A., The integral role of iron in ocean biogeochemistry. Nature 543, 51–59 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Marchetti A., Moreno C. M., Cohen N. R., Oleinikov I., deLong K., Twining B. S., Armbrust E. V., Lampe R. H., Development of a molecular-based index for assessing iron status in bloom-forming pennate diatoms. J. Phycol. 53, 820–832 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Vraspir J. M., Butler A., Chemistry of marine ligands and siderophores. Ann. Rev. Mar. Sci. 1, 43–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mawji E., Gledhill M., Milton J. A., Tarran G. A., Ussher S., Thompson A., Wolff G. A., Worsfold P. J., Achterberg E. P., Hydroxamate siderophores: Occurrence and importance in the Atlantic Ocean. Environ. Sci. Technol. 42, 8675–8680 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Boiteau R. M., Mende D. R., Hawco N. J., McIlvin M. R., Fitzsimmons J. N., Saito M. A., Sedwick P. N., DeLong E. F., Repeta D. J., Siderophore-based microbial adaptations to iron scarcity across the eastern Pacific Ocean. Proc. Natl. Acad. Sci. U.S.A. 113, 14237–14242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tréguer P., Bowler C., Moriceau B., Dutkiewicz S., Gehlen M., Aumont O., Bittner L., Dugdale R., Finkel Z., Iudicone D., Jahn O., Guidi L., Lasbleiz M., Leblanc K., Levy M., Pondaven P., Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 11, 27–37 (2017). [Google Scholar]
- 51.Siaut M., Heijde M., Mangogna M., Montsant A., Coesel S., Allen A., Manfredonia A., Falciatore A., Bowler C., Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406, 23–35 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Falciatore A., Casotti R., Leblanc C., Abrescia C., Bowler C., Transformation of nonselectable reporter genes in marine diatoms. Mar. Biotechnol. 1, 239–251 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P., Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Smith S. R., Gillard J. T. F., Kustka A. B., McCrow J. P., Badger J. H., Zheng H., New A. M., Dupont C. L., Obata T., Fernie A. R., Allen A. E., Transcriptional orchestration of the global cellular response of a model pennate diatom to diel light cycling under iron limitation. PLOS Genet. 13, e1006688 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sunda W. G., Price N. M., Morel F. M. M., Trace metal ion buffers and their use in culture studies. Algal Cult. Tech. 4, 35–63 (2005). [Google Scholar]
- 57.Tang D., Morel F. M. M., Distinguishing between cellular and Fe-oxide-associated trace elements in phytoplankton. Mar. Chem. 98, 18–30 (2006). [Google Scholar]
- 58.Sutak R., Botebol H., Blaiseau P.-L., Léger T., Bouget F.-Y., Camadro J.-M., Lesuisse E., A comparative study of iron uptake mechanisms in marine microalgae: Iron binding at the cell surface is a critical step. Plant Physiol. 160, 2271–2284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mistry J., Finn R. D., Eddy S. R., Bateman A., Punta M., Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41, e121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sorhannus U., A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Mar. Micropaleontol. 65, 1–12 (2007). [Google Scholar]
- 61.Kozik P., Francis R. W., Seaman M. N. J., Robinson M. S., A screen for endocytic motifs. Traffic 11, 843–855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/5/eaar4536/DC1
fig. S1. Siderophore uptake in P. tricornutum fits simple Michaelis-Menten kinetics.
fig. S2. Construction of an ISIP1 knockdown in P. tricornutum and expression of ISIP1 transcript and protein.
fig. S3. Siderophore uptake involves a binding step.
fig. S4. Siderophore uptake involves endocytosis.
fig. S5. ISIP1 knockdown lines are defective in endocytosis.
fig. S6. ISIP1-YFP localization and abundance under different Fe supplementation regimes.
fig. S7. ISIP1 predicted protein sequence and domain features.
fig. S8. ISIP1 phylogenetic tree.
movie S1. Live cell imaging of the uptake of NBD conjugate of FOB (FOB-NBD) by P. tricornutum.
table S1. Data used to construct Fig. 6 and fig. S8.
Reference (61)


