Mammal genomes retain remnants of chitinase genes, pointing to dietary shifts in the wake of the K/Pg mass extinction.
Abstract
The end-Cretaceous extinction led to a massive faunal turnover, with placental mammals radiating in the wake of nonavian dinosaurs. Fossils indicate that Cretaceous stem placentals were generally insectivorous, whereas their earliest Cenozoic descendants occupied a variety of dietary niches. It is hypothesized that this dietary radiation resulted from the opening of niche space, following the extinction of dinosaurian carnivores and herbivores. We provide the first genomic evidence for the occurrence and timing of this dietary radiation in placental mammals. By comparing the genomes of 107 placental mammals, we robustly infer that chitinase genes (CHIAs), encoding enzymes capable of digesting insect exoskeletal chitin, were present as five functional copies in the ancestor of all placental mammals, and the number of functional CHIAs in the genomes of extant species positively correlates with the percentage of invertebrates in their diets. The diverse repertoire of CHIAs in early placental mammals corroborates fossil evidence of insectivory in Cretaceous eutherians, with descendant lineages repeatedly losing CHIAs beginning at the Cretaceous/Paleogene (K/Pg) boundary as they radiated into noninsectivorous niches. Furthermore, the timing of gene loss suggests that interordinal diversification of placental mammals in the Cretaceous predates the dietary radiation in the early Cenozoic, helping to reconcile a long-standing debate between molecular timetrees and the fossil record. Our results demonstrate that placental mammal genomes, including humans, retain a molecular record of the post-K/Pg placental adaptive radiation in the form of numerous chitinase pseudogenes.
INTRODUCTION
The traditional model of mammalian evolution posits that after originating in the Triassic-Jurassic, Mesozoic mammals were typically small, nocturnal, generalized terrestrial insectivores, limited in taxonomic and ecological diversity due to predation by and/or competition with dinosaurs. After the extinction of nonavian dinosaurs at the Cretaceous/Paleogene (K/Pg) boundary, Cenozoic mammals then radiated into an enormous range of ecologically distinct niches (1, 2). Recent research suggests that this model is incomplete, with some fossil evidence for Mesozoic diversification and niche expansion in mammals (3–6). However, most Mesozoic eutherians (placental mammals + stem taxa) were small, likely terrestrial-scansorial, and had jaw and dental morphologies consistent with insectivory (1, 4, 7). Furthermore, although there is some evidence of eutherian niche diversification in the Late Cretaceous (1, 8, 9), the fossil record unequivocally points to an unprecedented adaptive radiation of placental mammals after the K/Pg boundary (1, 8–11).
Here, we test the hypothesis that this landmark evolutionary radiation left a molecular signature in placental mammal genomes, specifically in regard to diet. Jeuniaux (12) hypothesized that mammals inherited gastrointestinal (GI) chitinases from a vertebrate ancestor and subsequently lost them as they adapted to noninsectivorous diets. Molecular studies have isolated an acidic mammalian chitinase (CHIA) expressed in the mammalian GI tract (13, 14) that is stable in the presence of GI proteases (15), suggesting a role in chitin digestion. In the absence of dietary chitin, selection on chitinase genes may have been relaxed, allowing for the accumulation of formerly deleterious mutations (for example, frameshift and nonsense mutations), resulting in unitary pseudogenes and gene deletions. We tested Jeuniaux’s (12) hypothesis by analyzing patterns of acidic mammalian chitinase gene (CHIA) loss in placental mammals relative to the K/Pg boundary. We provide evidence that CHIAs retain a genomic signal of the early Cenozoic radiation of placental mammals, via the repeated, convergent decay of CHIAs in noninsectivorous mammals after the K/Pg boundary.
RESULTS AND DISCUSSION
Five CHIA paralogs in the last common ancestor of placental mammals
The original study describing CHIA reported only a single gene (13), but we found numerous placental CHIA paralogs, similar to recent analyses (16, 17). Phylogenetic analyses cluster these into five major clades (Fig. 1; figs. S1 to S3; and table S1), which we refer to as CHIA1, CHIA2, CHIA3, CHIA4, and CHIA5. All four superorders of placental mammals (Xenarthra, Afrotheria, Euarchontoglires, and Laurasiatheria) are represented by orthologs in all five CHIA clades, implying that the last common ancestor of placental mammals had five CHIAs. There is a consistent syntenic orientation of the paralogs, with multiple instances of all five being localized to the same genomic contig/scaffold (fig. S4 and table S2). Each putatively functional CHIA paralog is of nearly identical length, consists of 11 coding exons, and has canonical splice donor and acceptor sites, a catalytic domain in exon 5 (fig. S5) (18), and a chitin-binding domain in exon 11 (fig. S6) (19), suggesting that all five CHIAs encode enzymes with chitinolytic function. Furthermore, transcriptome analyses indicate that all five CHIAs can be expressed in placental mammal GI tracts (table S3) and, therefore, each potentially participates in the breakdown of ingested chitin.
Fig. 1. Placental mammal CHIA gene tree, simplified from fig. S3.
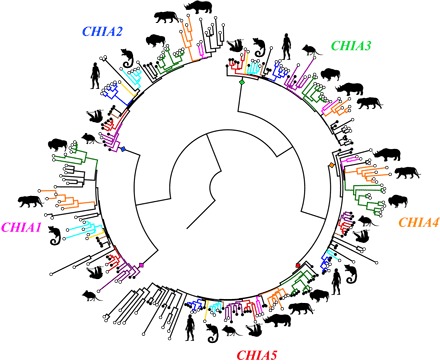
Closed circles indicate functional CHIAs, and open circles indicate pseudogenic CHIAs and/or CHIAs lacking a chitinolytic and/or chitin-binding domain. Colored branches correspond to a subset of placental mammal clades: green, Cetartiodactyla; orange, Carnivora; pink, Perissodactyla; red, Xenarthra; purple, Afrotheria; brown, Scandentia; cyan, Strepsirrhini; blue, Anthropoidea; yellow, Dermoptera. Silhouettes and licenses here and throughout are from phylopic.org.
CHIA loss and dietary shifts to carnivory and herbivory
Although we inferred gene duplications early in mammalian history (fig. S1), the only duplications in crown Placentalia resulted in paralogs likely to be nonfunctional or lacking chitinolytic function (tables S1 and S4 and figs. S2 and S3), with at least some of these duplicates appearing to have neofunctionalized into genes with immune-related functions (fig. S3) (20). By contrast, we found that CHIA loss, via pseudogenization and whole-gene deletion, has occurred very frequently, particularly in carnivorous and herbivorous lineages (table S1). Strikingly, species with five to three CHIA copies have, on average, a diet that consists almost entirely of invertebrates (five CHIAs, 88.3% diet invertebrates on average; four CHIAs, 85%; three CHIAs, 80%), species with two copies on average consume a moderate amount of invertebrates (63.3%), and species with one or no CHIAs average minimal invertebrate consumption (one CHIA, 21.9%; no CHIAs, 7%). Phylogenetic generalized least-squares (PGLS) regression analyses using CHIAs derived from both gene models (n = 73, r2 = 0.3358, P = 7.824 × 10−8) and genomic contigs (n = 70, r2 = 0.4213, P = 1.219 × 10−9) support a positive relationship between the percentage of invertebrates in the diet and the number of functional CHIAs in the genome (Fig. 2), a finding corroborated by a recent CHIA analysis focused on Primates (17). The association of high numbers of functional CHIAs with insectivory, and the inference of five CHIAs in the last common ancestor of placental mammals, suggests that this ancestor and most crown Cretaceous placental lineages were probably also insectivorous. If correct, then such a diet would likely have imposed constraints on body size and other life history characters in these lineages and should therefore help infer traits in the earliest placental mammals (21, 22).
Fig. 2. PGLS regression of the number of functional contig-derived CHIAs versus the percent of the diet consisting of invertebrates.
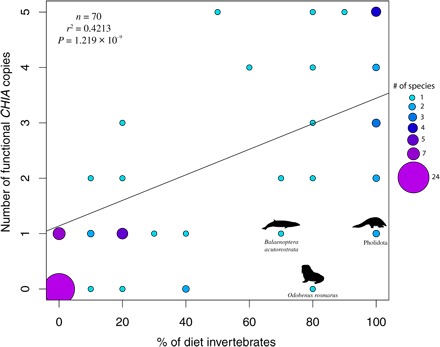
See text for discussion of highlighted species.
Whereas the earliest placental mammals had five CHIAs, inactivating mutations and gene deletions shared between multiple species point to losses of functional CHIAs during the origins of modern placental clades (Fig. 3 and table S4). Whereas some CHIAs were lost in individual species or families, other losses occurred before interordinal (Paenungulata and Ostentoria) and intraordinal (Chiroptera, Carnivora, Perissodactyla, Cetartiodactyla, and Lagomorpha) diversification. Significantly, all or nearly all CHIA genes were lost during the origin and diversification of noninsectivorous lineages, including the herbivorous sloths (Folivora), hyraxes, elephants, and sirenians (Paenungulata), Old World fruit bats (Pteropodidae), horses, rhinoceroses, and tapirs (Perissodactyla), camels, swine, and ruminants (Cetartiodactyla), colugos (Dermoptera), lemurs (Lemuriformes), monkeys and apes (Anthropoidea), rabbits and pikas (Lagomorpha), rodents (Rodentia), and the largely carnivorous false vampire bats (Megadermatidae), whales (Cetacea), and dogs, cats, and kin (Carnivora) (Fig. 1 and tables S1, S4, and S5).
Fig. 3. Examples of CHIA shared inactivating mutations.
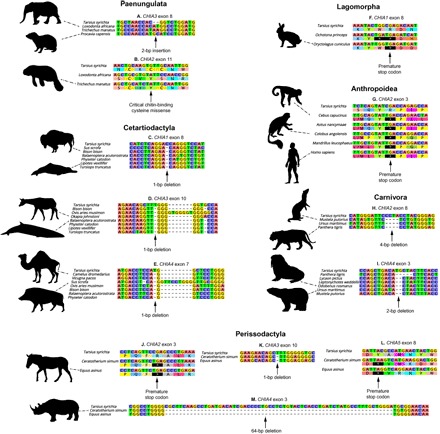
Each alignment has a Tarsius syrichta ortholog to provide a functional outgroup comparison. See Fig. 4 for gene inactivations shown in (A) to (M) mapped onto the placental mammal phylogeny.
CHIA loss and the post-K/Pg placental mammal radiation
The presence of CHIA pseudogenes in various species allowed us to more precisely date the timing of gene inactivations relative to the K/Pg boundary. We used a method (23) based on the accumulation of nonsynonymous mutations after selection is relaxed. Using divergence times between lineages estimated with a molecular clock, shared inactivating mutations provide minimum dates for pseudogenization. We then analyzed the ratio of nonsynonymous to synonymous nucleotide substitutions (dN/dS) to model the timing of the transition from purifying to relaxed selection (table S6).
Estimates of the timing of CHIA inactivations provide support for a dietary radiation of placental mammals after the K/Pg mass extinction 66 million years (Ma) ago (Fig. 4 and table S7). Assuming a densely calibrated molecular timetree with broad taxonomic representation (24), the earliest CHIA pseudogene arose just before the K/Pg boundary (67.5 Ma ago), with 6 having occurred during the Paleocene (64.2 to 57 Ma ago) and 11 in the Eocene (53.4 to 37.4 Ma ago). Although we were unable to precisely estimate some inactivation dates due to assumption violations of the codon evolution models (see below), shared inactivating mutations (Fig. 3 and table S4) provide minimum dates for gene loss in these cases. Therefore, we estimate that another two CHIA inactivations occurred at the very end of the Cretaceous (67 to 66 Ma ago), six occurred in the Paleocene (63 to 56 Ma ago), and eight in the Eocene (55.5 to 34 Ma ago; Fig. 4).
Fig. 4. Patterns of CHIA loss through time.
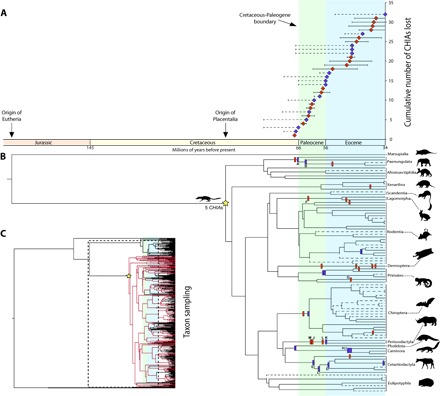
(A) The cumulative number of CHIA losses in placental mammals relative to the origin of eutherians, placental mammals, and the K/Pg boundary. Red symbols indicate mean estimates of gene inactivation dates based on four model assumptions, and purple symbols indicate minimum pseudogenization dates based on shared inactivating mutations. Solid bars indicate the range of estimates (table S7), and dashed bars indicate potential range of dates based on branch lengths. (B) The phylogenetic positioning of the CHIA inactivation estimates shown in (A). Letters correspond to gene losses shown in Fig. 3. (C) Timetree (24) indicating lineages sampled for CHIA analyses (red branches). The dashed box indicates zoomed-in portion of phylogeny shown in (B).
There remain a handful of additional gene losses that may have occurred in the Cretaceous, but dating these events is less precise. For example, multiple CHIAs in rodents are extremely fragmentary pseudogenes or appear to have been completely deleted (table S1 and S4 and fig. S4); one parsimonious solution is that there were multiple CHIA losses early in rodent history, as early as the Cretaceous. Alternatively, these CHIAs may have been lost repeatedly, paralleling the patterns we found across placental mammals (Fig. 4). Additional sequencing of rodent genomes belonging to long phylogenetic branches may provide further clarity on this issue. Another example concerns a possible shared 1–base pair (bp) deletion between cetartiodactyls and perissodactyls in exon 10 of CHIA3. If this is a synapomorphy, then such a mutation would be dated to the Cretaceous (24–26) and would help to resolve the laurasiatherian polytomy. Nonetheless, 1-bp deletions are common, the positioning of this deletion is uncertain, and this exon is missing in the earliest diverging cetartiodactyl and ostentorian lineages, indicating that this mutation plausibly occurred convergently. Hence, various uncertainties concerning the timing of specific gene deletions and the precise dating of phylogenies and gene inactivations (Fig. 4) allow for the possibility of a few Cretaceous CHIA losses. However, the available data suggest that placental mammals began reducing their number of CHIA copies and, therefore, their consumption of insects, at and following the K/Pg boundary (Fig. 4), a conclusion broadly consonant with both the fossil record (table S5) and inferences made from correlations between the rate of genomic evolution and life-history traits (27).
Our results may also help reconcile seemingly incongruent results between molecular timetrees and the fossil record (25, 26, 28). Divergence time estimates based on relaxed molecular clocks routinely provide evidence of Cretaceous interordinal diversification (25, 26), but analyses of the fossil record have yielded no unambiguous Cretaceous crown placental mammals (22, 29). Our results suggest that Cretaceous placental diversification was uncoupled from phenotypic divergence in the early Cenozoic because diversification of placental mammals substantially predates (25.8 Ma) the repeated loss of chitinase genes beginning near the K/Pg boundary (Fig. 4).
Functional CHIA copy number and diet mismatches
Although the patterns of CHIA loss and retention largely reflect patterns of dietary evolution, there exist discrepancies that suggest that more research is needed. First, some CHIA copy number and diet mismatches are likely an artifact of imprecise dietary coding. We used “percent invertebrates in the diet” as a proxy for the amount of chitin in the diet, due to the tendency of invertebrate prey of placental mammals being insects, but some mammals eat copious amounts of invertebrates that likely have minimal amounts of chitin. For instance, the walrus (Odobenus rosmarus) has no functional CHIAs, and 80% of its diet is composed of invertebrates (Fig. 2 and table S1); however, most of these prey are bivalves (table S1). Future studies may benefit from directly calculating the amount of chitin in the diet to determine whether this leads to a more precise prediction of CHIA copy number.
Shared inactivating mutations suggest that other mismatches between diet and CHIA copy number likely result from historical contingencies, with species that inherited few CHIA copies being constrained by the apparent rarity of CHIA duplications. For instance, we found that the baleen whale Balaenoptera acutorostrata has only a single CHIA despite consuming copious amounts of chitin-rich crustaceans (Fig. 2). On the basis of shared inactivating mutations, four CHIAs were pseudogenized in the ancestors that baleen whales share with herbivorous hoofed mammals and cephalopod- and fish-eating toothed whales (Fig. 3 and table S4), suggesting that historical dietary shifts predating the origin of filter feeding have limited baleen whales to a single CHIA (table S5). Similarly, the myrmecophagous (ant-/termite-eating) pangolins (Pholidota) have a single CHIA (Fig. 2), in contrast to the convergently myrmecophagous Southern tamandua (Tamandua tetradactyla, Vermilingua) and aardvark (Orycteropus afer, Tubulidentata), which have four and five functional CHIAs, respectively. We found evidence of CHIA loss in the common ancestor of pangolins and placental carnivores (table S4), which, paired with fossil evidence (table S5), suggests that pangolins may have descended from carnivorous ancestors before reverting to a strictly insectivorous diet.
Regardless of potential historical contingencies, it is unclear why secondary CHIA duplications appear to be so rare in placental mammals and limited to instances where chitinolytic ability has been lost (fig. S3), because this would seem to be a simple mechanism to increase chitinase activity when adapting to a chitin-rich diet. These chitinophagous, CHIA-limited mammals may be compensating via alternative mechanisms, such as increasing the expression of another CHIA or using chitin-digesting bacteria (30, 31), chitinolytic lysozymes (32), or an additional mammalian chitinase, chitotriosidase (CHIT1) (33). CHIT1 is generally expressed in immunity-related tissues (33, 34), but we found evidence that pangolin salivary glands express CHIT1 alongside CHIA5 (table S3), possibly suggesting co-option for digestive function.
Other mismatches seem more difficult to explain. For instance, the shrews (Soricidae) have historically been considered to be insectivores that have retained their diets from an ancestral placental mammal stock (35), yet the species represented in this study (Sorex araneus) only has two intact CHIAs (table S1). This contrasts with other members of this hypothetical ancestral stock, such as tree shrews (Scandentia; five CHIAs), elephant shrews (Macroscelidea; five CHIAs), hedgehogs (Erinaceidae; four CHIAs), tenrecs (Tenrecidae; four CHIAs), golden moles (Chrysochloridae; five CHIAs), and true moles (Talpidae; three CHIAs), which all have three to five intact CHIAs. Although classically considered insectivores, it is possible that this designation is too coarse, because soricids frequently feed on vertebrate prey or scavenge (39.8% clade average) (36), and such a dietary shift may have led to some CHIA losses. Alternatively, some gene absences may be due to assembly errors because we cannot confirm CHIA3’s deletion in S. araneus with synteny data. Analyses of additional genomes may help clarify this and other apparent deviations (table S5).
Finally, note that we are operating under the assumption that having five CHIAs increases the expression of chitinases in the GI tract, allowing for greater efficiency of chitin digestion. The relationship between functional CHIA copy number and diet suggests that this hypothesis is quite plausible, and our gene expression analyses (table S3) provide evidence that multiple CHIAs can be expressed in the same GI organ, with two CHIAs being expressed in the pancreas of an insectivorous tree shrew (Tupaia chinensis) and three in a salivary gland of an insectivorous bat (Macrotus californicus). However, it remains possible that some CHIAs are expressed in and specialized for different tissues and may have other functions outside of the GI tract. For instance, CHIA5 is expressed in both the GI tract and the lungs of mouse and human, indicating a possible defensive role against chitinous pathogens (13). Notably, only CHIA1 and CHIA2 appear to be robustly expressed in the tree shrew pancreas, suggesting that CHIA3, CHIA4, and CHIA5 may be better optimized for other tissues. Furthermore, certain CHIAs are retained more frequently than others, with nearly half of the species surveyed having CHIA5 versus approximately 12% having CHIA1, indicating a possible functional bias in CHIA loss. Future comparative studies could test for tissue-specific expression and differences in optimal pH activity and stability to understand the distinctions between these paralogs, which could lead to a better understanding of some of the mismatches in CHIA copy number and diet.
CONCLUSIONS
Together, these results suggest that chitinase genes (CHIAs) provide a genomic signal of the post-K/Pg dietary radiation of placental mammals. Consistent with the fossil record, the patterns of CHIA evolution indicate that the earliest placental mammals were highly insectivorous and that descendant lineages adapted to noninsectivorous dietary niches near and after the K/Pg boundary. Consequently, many placental mammals, including humans (16, 17), retain “genomic fossils” in the form of CHIA pseudogenes, providing a molecular record of their insectivorous past.
MATERIALS AND METHODS
Collection of annotated sequences
We quantified the total number of acidic mammalian chitinase genes (CHIAs) in the genomes of 107 placental mammal species (table S1), using both annotated gene models (88 species, 54 families, and 17 orders) and direct queries of genomic contigs (74 species, 62 families, and 19 orders; fig. S7), with 55 species overlapping between the two data sets. All genomes examined were available in National Center for Biotechnology Information (NCBI)’s nucleotide collection and whole-genome shotgun (WGS) contig database except three assemblies generated using the Discovar de novo protocol (Tamandua tetradactyla, Chaetophractus vellerosus, and Tolypeutes matacus; https://software.broadinstitute.org/software/discovar/blog/) and made available to us by the Broad Institute. The latter assemblies were imported into Geneious v. 9.1.8 (37) for analysis. For annotated gene models, we obtained sequences derived from NCBI’s Eukaryotic Genome Annotation (EGA) pipeline by BLASTing (discontiguous megablast) a human CHIA mRNA reference sequence (NM_201652) against the NCBI nucleotide collection and performing separate Basic Local Alignment Search Tool (BLAST) searches against all the placental taxonomic orders. We downloaded all relevant EGA gene models, which are denoted by an “XM_” accession number prefix and are annotated as CHIA or any annotation suggesting homology to this protein (for example, “acidic mammalian chitinase-like”). We considered the gene absent/nonfunctional under any of the following conditions: (i) we retrieved negative BLAST results; (ii) the gene model was annotated as a “low-quality protein,” a designation given if construction of the model required correction for premature stop codons and/or frameshift insertions or deletions (indels) found in the reference genomic contigs; and (iii) the sequence lacked a canonical catalytic domain (DXXDXDXE) (18).
Gene tree analyses
We discovered numerous examples of species with potential CHIA paralogs, which necessitated comparison through phylogenetic analyses to estimate their evolutionary history. We obtained additional gene models from nonplacental mammal vertebrate taxa and a CHIT1 (chitotriosidase) outgroup (NM_003465), imported all of the sequences into Geneious, aligned them using MUltiple Sequence Comparison by Log-Expectation (MUSCLE) (38), manually adjusted the alignments, and removed sequences and sequence positions determined by eye to have dubious homology (data set S1). We then estimated a gene tree using Randomized Axelerated Maximum Likelihood (RAxML) v. 8.2.10 (39) in Cyberinfrastructure for Phylogenetic Research (CIPRES) (RAxML-HPC2 on Extreme Science and Engineering Discovery Environment) (40) with 500 bootstrap replications but otherwise default settings (-m GTRCAT). Placental CHIA sequences clustered in five distinct clades, which we designated CHIA1, CHIA2, CHIA3, CHIA4, and CHIA5. On the basis of these clade assignments, sequences from all five CHIA subclades were realigned separately (that is, all CHIA1 sequences, all CHIA2 sequences, etc.), and then each subclade alignment was aligned successively using the Geneious Translation align tool implementing the MUSCLE algorithm. We then performed a RAxML analysis on this finalized alignment (data set S1) in CIPRES using the same parameters (fig. S1).
Assembly of contig-derived sequences
Given the inference of five distinct clades of CHIA within placental mammals, we directly queried the genomic contigs (WGS) of a subset of species to verify the presence of distinct CHIA genes (data set S2), determine their synteny, and validate the presence of inactivating mutations. Some genome assemblies present in WGS had not been annotated by EGA at the time of our study, so we examined additional genomes to broaden taxonomic diversity. We directly queried at least one member of each mammalian family for which there was a genome assembly available and, in some cases, examined additional species from a family if we were unable to recover a particular CHIA paralog or to check for inactivating mutations shared by confamilials. On the basis of the predicted gene models, very few mammals appeared to have all five predicted CHIA genes. We chose one of these species, the Philippine tarsier (T. syrichta), as the reference genome from which to obtain and compare all five CHIA genes (fig. S1). We BLASTed (megablast) the T. syrichta gene models against its WGS assembly and confirmed that all five T. syrichta CHIA sequences map to separate genomic regions (table S1). Each of these sequences is characterized by a functional catalytic domain in exon 5 (DXXDXDXE; fig. S5) (18), six critical cysteines in the chitin-binding domain (fig. S6) (19), intact exon-intron boundaries and the absence of inactivating mutations. Each gene model has 11 coding exons, with the exception of CHIA2, which has 10. After comparisons with gene models from other species, it became apparent that exon 2 was erroneously excluded from the T. syrichta gene model, despite being present in the assembly.
We designed a set of five in silico probes from the T. syrichta contigs, one for each CHIA gene, to capture CHIA sequences in the genome assemblies of other placental mammals. These probes encompassed the exons, introns, and flanking sequences of each of the predicted genes, allowing for greater confidence in homology and synteny between the respective paralogs. We BLASTed the probes (discontiguous megablast) against the WGS and performed local BLAST searches on the three Discovar de novo assemblies (discontiguous megablast). In instances of negative BLAST results (that is, no hits for CHIA sequences), we designed and BLASTed probes based on more closely related taxa. When we obtained BLAST hits, we downloaded the entire contiguous sequence that encompassed all the hits with the highest homology, ignoring hits that were clearly repetitive elements. As we downloaded each sequence, we recorded the accession number of each contig, the position of the BLAST hits within each contig, and the position of flanking genes to determine synteny (tables S1, S2, and S8 and fig. S4). We imported each captured sequence into Geneious, aligned the imported sequence to the T. syrichta probe reference using MUSCLE, and successively aligned additional sequences derived from the same ordinal and/or superordinal taxon, frequently using alignments of outgroup taxa to anchor the new alignments (data set S2). We examined each of the gene sequences for putative inactivating mutations, including missense mutations to the conserved catalytic domain or chitin-binding domain residues, start codon mutations, frameshift indels, premature stop codons, and splice site mutations (table S4). If a sequence had any of these mutations, it was assumed to be a pseudogene, unless its only mutation(s) is (are) (i) a start codon mutation but it had an alternative start upstream of the first exon or (ii) a GT→GC splice donor mutation because GC is a relatively common acceptable splice donor variant (41). If a sequence had a substantial amount of missing sequence (five or more exons) and its closest available relative(s) had a pseudogenic ortholog, then we assumed that it is a pseudogene.
After finalizing this data set, we removed all introns and flanking sequences, aligned each sequence to its corresponding orthologs (for example, all CHIA1s, all CHIA2s, etc.) using MUSCLE in Geneious, and then aligned each individual paralog alignment to each other successively using Geneious’ Translation align with the MUSCLE algorithm. Using this master alignment, we aligned (MUSCLE in Geneious) the following published CHIA mRNAs: Macaca fascicularis (NM_001284548, stomach), Homo sapiens (NM_201653, stomach/lung), Mus musculus (NM_023186, salivary gland), Rattus norvegicus (NM_207586, stomach), Bos taurus (NM_174699, fetal liver), and Sus scrofa (NM_001258377, lung). Next, we aligned (MUSCLE in Geneious) the following mRNAs assembled from RNA sequencing experiments (see details below): T. chinensis CHIA1 (pancreas), T. chinensis CHIA2 (pancreas), M. californicus CHIA3 (submandibular gland), and M. californicus CHIA4 (submandibular gland). Finally, we aligned (Translation align, MUSCLE in Geneious) several chitinase-like mRNA sequences expressed in M. musculus: Chil6/BYm/Chil8 (NM_178412), Chil4/Ym2/Chi3l4 (NM_145126), Chil3/Ym1/Chi3l3 (NM_009892), and Chil5/Bclp/Chi3l7 (NM_001080816), as well as a CHIT1 outgroup (NM_003465). We examined the master alignment by eye, adjusted it manually, removed sequence positions of dubious homology, and removed sequences that were likely to be phylogenetically uninformative because of missing data and/or minimal or no overlap with related species (for example, one exon only). We then executed a RAxML analysis through CIPRES on this contig-derived sequence alignment (data set S1) using the methods described above.
The resulting tree (fig. S2) largely recapitulated the results from the gene model–derived analyses (fig. S1) with the major exception of CHIA4 being nested within CHIA5. Upon closer inspection, it became apparent that the chiropteran CHIA4 and CHIA5 sequences seemed to be influencing this aberrant topology. Specifically, chiropteran CHIA4 is paraphyletic at the base of the CHIA4 clade, and chiropteran CHIA5 sequences form the sister group to the entire CHIA4 clade. Reexamining the gene model–derived phylogeny (fig. S1) shows that the chiropteran gene models do not include any CHIA5 sequences but seem to imply multiple recent gene duplications among the chiropteran CHIA4s. However, CHIA4 and CHIA5 sequences can readily be distinguished as occupying distinct positions on the same contig for multiple, distantly related species [Rousettus aegyptiacus (Pteropodidae), Hipposideros armiger (Hipposideridae), Rhinolophus sinicus (Rhinolophidae), Miniopterus natalensis (Miniopteridae), and Eptesicus fuscus (Vespertilionidae); table S2 and fig. S2].
Upon directly examining the sequences, we found evidence that gene conversion may be to blame for these discordant results (table S9). Specifically, when comparing chiropteran CHIA4 and CHIA5 sequences of the same species to a reference CHIA4 sequence (T. syrichta), we found that most exons were more similar between the CHIA4 and CHIA5 paralogs than the chiropteran CHIA4s were to the orthologous T. syrichta CHIA4 sequence. We compared the introns for two species and found a similar pattern. To give a detailed example, E. fuscus CHIA4 exons 1, 2, 4, and 11 had higher similarity to T. syrichta CHIA4 (84.6 to 96.7%; average, 91.9%) than to E. fuscus CHIA5 (66.2 to 89.5%; average, 74.8%), whereas E. fuscus CHIA4 exons 3 and 5 to 10 had higher similarity to E. fuscus CHIA5 (95.5 to 100%; average, 97.7%) than to T. syrichta CHIA4 (90.1 to 95.2%; average, 92.3%). Similarly, E. fuscus CHIA4 introns 2 to 4, 7, and 10 were more similar to T. syrichta CHIA4 (40.8 to 67%; average, 59.3%) than to E. fuscus CHIA5 (11.2 to 36.7%; average, 25%), whereas E. fuscus CHIA4 introns 5, 6, 8, and 9 had higher identity to E. fuscus CHIA5 (76.7 to 96.5%; average, 89.7%) than to T. syrichta CHIA4 (49.8 to 71.6 %; average, 60.8%). Further examination of another 10 mammals that retain functional CHIA4 and CHIA5 indicates that gene conversion for these two paralogs is likely phylogenetically widespread. This is perhaps an unsurprising conclusion given that CHIA4 and CHIA5 are in tandem (fig. S4), appear to be derived from a relatively recent (placental mammal-specific?) gene duplication (fig. S1), and that gene conversion is prone to occurring in tandemly duplicated genes that share high homology (42). We reanalyzed the contig-derived data set after removing all chiropteran CHIA4 and CHIA5 sequences (data set S1), and the resulting phylogeny (Fig. 1 and fig. S3) resolves CHIA4 and CHIA5 as separate clades.
Gene models versus contig-derived sequences
Of the 311 genes for which we directly compared both EGA gene models and sequences derived from genomic contigs (WGS), 95.5% (297) were in agreement about functionality, that is, both predicted a functional gene or both predicted a nonfunctional/absent gene, although many absent gene models could be found as pseudogenes in the contigs. The exceptions included sequences predicted to be of low quality but no inactivating mutations could be found in the contig sequence (two), instances where EGA did not predict a sequence to be present in a genome but we confirmed that the gene is present and intact (nine), and cases in which EGA predicted a functional gene but we found evidence of pseudogenization (three). Although the two data sets are highly congruent, the errors in the gene models led us to base all our results and discussion in the paper and the Supplementary Materials on the contig-derived sequences, with two exceptions: one of two PGLS regression analyses referred to in Results and fig. S1.
Regression analyses
We tested whether the number of intact CHIAs in a genome correlates with the amount of insect prey consumed by a species. We derived the latter metric from EltonTraits v. 1.0 (36), which provides the estimated percentage of invertebrate prey in the diet, a proxy for the amount of insects consumed. We performed PGLS regression analyses with the caper package in R (43, 44), testing the hypothesis separately with the gene model (EGA) and contig-derived (WGS) data sets. We assumed the timetree from Emerling et al. (24) and implemented phylogenetic corrections using maximum-likelihood estimates of Pagel’s λ. We made taxonomic substitutions when species in our data set were not represented in the Emerling et al. (24) phylogeny. Specifically, we assumed that species in the same taxon (for example, genus and family) could be interchanged if there were no other congeners or confamilials in the phylogeny. For instance, Emerling et al. (24) have only a single hipposiderid in their tree, Hipposideros commersoni, and our chitinase data set included H. armiger, which we deemed to be an acceptable substitution. Species were removed from the analyses if there were no taxonomic equivalents.
Estimation of gene inactivation dates
To estimate the temporal distribution of CHIA loss during placental history, we used the dN/dS ratio method of Meredith et al. (23) to date the inactivation of the different CHIA paralogs. This method is derived from the assumption that genes, on average, are under purifying selection throughout evolutionary history until they become inactivated. Once inactivated, these pseudogenes are expected to undergo relaxed selection on average, and the dN/dS ratio on the transitional (“mixed”) branch (that is, transitioning from purifying to relaxed selection) provides a signal of the timing of this shift in selection pressures. This can then be used to calculate the timing of gene inactivation, assuming divergence times obtained from molecular dating analyses, with shared inactivating mutations (table S4) providing a minimum branch on which pseudogenization occurred. We performed separate analyses for each CHIA paralog (data set S1) using PAML (phylogenetic analysis by maximum likelihood) v. 4.8 (45), implementing two separate codon model assumptions (F1X4 and F3X4) and assuming the topology and divergence times of Emerling et al. (24). We removed all chiropteran CHIA4 and CHIA5 sequences due to their disproportionate influence on the gene tree topology (fig. S2), and we assumed that any other CHIA gene conversion has occurred at an equal rate on average during the evolution of placental mammals.
When performing dN/dS ratio analyses on a particular CHIA paralog, we allowed for estimation of ω (dN/dS ratio) on each of the mixed category branches (purifying to relaxed selection), each individual clade of pseudogene (relaxed selection) branches (that is, each set of descendant branches that post-dates a mixed branch), and the remaining branches were grouped together as functional (purifying selection) category branches [table S6; see the study of Meredith et al. (23) for additional details]. After this initial analysis, we successively fixed each set of pseudogene branches to ω = 1 to test whether the dN/dS estimates are statistically distinguishable from an assumption of relaxed selection. Furthermore, in instances where mixed branches were estimated to have a dN/dS ratio > 1, we similarly compared them to a null model where the branch was fixed at ω = 1. Finally, we implemented a model whereby all pseudogene category branches were fixed at ω = 1 (table S6). The mixed category branches estimated during this final analysis were then entered into the calculations of Meredith et al. (23) to obtain point estimates of gene inactivation (table S7). We discarded estimates where the mixed branch ω was greater than 1 and/or the pseudogene branch dN/dS ratios significantly deviated from a model where ω was fixed at 1, due to the violation of the assumption that pseudogenization involves a transition from purifying to relaxed selection.
CHIA gene expression analyses
Because the presence of multiple CHIA paralogs in the genomes of placental mammals does not guarantee their expression in GI tissues, we queried RNA sequencing experiments to test whether all five CHIAs can be expressed in the GI tract. We examined GI tissue RNA libraries of three species that have (T. chinensis) or potentially have (M. californicus and Desmodus rotundus) three or more CHIAs in their genomes (tables S1 and S3). These libraries have been deposited into NCBI’s Sequence Read Archive, so we queried them via NCBI’s BLAST interface. We BLASTed (megablast) all five of the assembled T. chinensis CHIA paralogs against a pancreas library (46). M. californicus and D. rotundus are phyllostomid chiropterans with no available genome assemblies, so we BLASTed (discontiguous megablast) assembled CHIA3 to CHIA5 sequences from Pteronotus parnellii, given that mormoopids are the closest living relatives to phyllostomids. We BLASTed against the single available salivary gland transcriptome for M. californicus and five salivary gland transcriptomes for D. rotundus (47–49).
D. rotundus, in contrast to M. californicus, showed no evidence of robust transcription of CHIAs in its salivary glands, with BLAST hits ranging from zero to 13 for CHIA3 to CHIA5 (table S3). We tested whether an additional mammalian chitinase (CHIT1) and prolactin-induced protein (PIP), a salivary gland positive control, show evidence of gene expression in M. californicus and D. rotundus, by BLASTing (discontiguous megablast) the coding sequences of Myotis brandtii gene models for both CHIT1 (XM_005881866.2) and PIP (XM_005862784.2) against all six RNA libraries. We found evidence of robust transcription of CHIT1 in the M. californicus salivary gland (5963 hits), compared to practically no hits in D. rotundus (0 to 6 hits), but PIP returned a large amount of hits for both M. californicus (20,000 hits) and four of the five D. rotundus libraries (6317 to 12,547 hits; table S3). This suggests that D. rotundus does not express CHIAs in its salivary glands, whereas M. californicus does, an observation consistent with their respective sanguivorous and insectivorous diets.
Because of the surprisingly minimal number of CHIAs in the genomes of the highly insectivorous pangolins (table S1), possibly due to a historical contingency (table S5), we tested whether the immune chitinase gene CHIT1 is expressed in the GI tract alongside CHIA5. In NCBI, we BLASTed (megablast) the assembled Manis javanica CHIA5 and the coding sequence of an M. javanica CHIT1 gene model (XM_017667770.1) against an M. javanica salivary gland RNA library (table S3) (50).
Supplementary Material
Acknowledgments
We thank three anonymous reviewers for comments on a previous draft of the manuscript and J. Johnson and the Broad Institute for providing access to xenarthran genome assemblies. Funding: This research was supported by an NSF Postdoctoral Research Fellowship in Biology (award no. 1523943; C.A.E.), an NSF Postdoctoral Fellow Research Opportunities in Europe award (C.A.E.), the France-Berkeley Fund (F.D. and M.W.N.), a European Research Council consolidator grant (ConvergeAnt no. 683257; F.D.), and the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007-2013) under REA grant agreement no. PCOFUND-GA-2013-609102, through the PRESTIGE programme coordinated by Campus France (C.A.E.). This is contribution ISEM 2018-049 of the Institut des Sciences de l’Evolution. Author contributions: C.A.E. conceived the study, collected the data, analyzed the data, and wrote the manuscript with input from M.W.N. and F.D. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/5/eaar6478/DC1
fig. S1. Vertebrate RAxML CHIA gene tree based on CHIA gene models.
fig. S2. Placental mammal RAxML CHIA gene tree based on genomic contig–derived and mRNA sequences.
fig. S3. Placental mammal RAxML CHIA gene tree based on genomic contig–derived and mRNA sequences, after removing chiropteran CHIA4 and CHIA5 sequences.
fig. S4. Diagram demonstrating synteny of CHIA paralogs and nearby genes on their respective genomic scaffolds/contigs.
fig. S5. T. syrichta CHIA1, CHIA2, CHIA3, CHIA4, and CHIA5 gene models showing conserved chitinolytic domain (DXXDXDXE).
fig. S6. T. syrichta CHIA1, CHIA2, CHIA3, CHIA4, and CHIA5 gene models showing chitin-binding domain with six conserved cysteine residues.
fig. S7. Cladogram showing relationships of species in WGS data set and mapped gene losses.
table S1. CHIA summary.
table S2. Synteny of CHIA paralogs.
table S3. CHIA gene expression analyses.
table S4. Distribution of genetic lesions and missing data across CHIA exons.
table S5. Discussion of CHIA number, timing of loss, diets of extant taxa, and the fossil record.
table S6. dN/dS ratio models used for gene inactivation estimates.
table S7. CHIA inactivation date estimates calculated from dN/dS ratio models in table S6 and assuming divergence dates from the study of Emerling et al. (24).
table S8. Scaffold/contig coordinates of OVGP1, PIFO, and DENND2D for species in fig. S4.
table S9. Evidence of gene conversion between CHIA4 and CHIA5.
data set S1. Alignments used for analyses.
data set S2. Alignments used for constructing CHIA genes from scaffolds/contigs.
REFERENCES AND NOTES
- 1.T. S. Kemp, The Origin and Evolution of Mammals (Oxford Univ. Press, 2005). [Google Scholar]
- 2.J. D. Archibald, Extinction and Radiation: How the Fall of Dinosaurs Led to the Rise of Mammals (JHU Press, 2011). [Google Scholar]
- 3.Luo Z.-X., Transformation and diversification in early mammal evolution. Nature 450, 1011–1019 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Grossnickle D. M., Polly P. D., Mammal disparity decreases during the Cretaceous angiosperm radiation. Proc. Biol. Sci. 280, 20132110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Close R. A., Friedman M., Lloyd G. T., Benson R. B. J., Evidence for a mid-Jurassic adaptive radiation in mammals. Curr. Biol. 25, 2137–2142 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Wilson G. P., Evans A. R., Corfe I. J., Smits P. D., Fortelius M., Jernvall J., Adaptive radiation of multituberculate mammals before the extinction of dinosaurs. Nature 483, 457–460 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Luo Z.-X., Yuan C.-X., Meng Q.-J., Ji Q., A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature 476, 442–445 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Halliday T. J. D., Goswami A., Eutherian morphological disparity across the end-Cretaceous mass extinction. Biol. J. Linn. Soc. 118, 152–168 (2016). [Google Scholar]
- 9.Grossnickle D. M., Newham E., Therian mammals experience an ecomorphological radiation during the Late Cretaceous and selective extinction at the K–Pg boundary. Proc. Biol. Sci. 283, 20160256 (2016). [Google Scholar]
- 10.Smith F. A., Boyer A. G., Brown J. H., Costa D. P., Dayan T., Ernest S. K., Evans A. R., Fortelius M., Gittleman J. L., Hamilton M. J., Harding L. E., Lintulaakso K., Lyons S. K., McCain C., Okie J. G., Saarinen J. J., Sibly R. M., Stephens P. R., Theodor J., Uhen M. D., The evolution of maximum body size of terrestrial mammals. Science 330, 1216–1219 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Springer M. S., Emerling C. A., Meredith R. W., Janečka J. E., Eizirik E., Murphy W. J., Waking the undead: Implications of a soft explosive model for the timing of placental mammal diversification. Mol. Phylogenet. Evol. 106, 86–102 (2017). [DOI] [PubMed] [Google Scholar]
- 12.C. Jeuniaux, On some biochemical aspects of regressive evolution in animals, in Biochemical Evolution and the Origin of Life, E. Schoffeniels, Ed. (North-Holland Publishing Company, 1971), pp. 304–313. [Google Scholar]
- 13.Boot R. G., Blommaart E. F. C., Swart E., Ghauharali-van der Vlugt K., Bijl N., Moe C., Place A., Aerts J. M. F. G., Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J. Biol. Chem. 276, 6770–6778 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Strobel S., Roswag A., Becker N. I., Trenczek T. E., Encarnação J. A., Insectivorous bats digest chitin in the stomach using acidic mammalian chitinase. PLOS ONE 8, e72770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohno M., Kimura M., Miyazaki H., Okawa K., Onuki R., Nemoto C., Tabata E., Wakita S., Kashimura A., Sakaguchi M., Sugahara Y., Nukina N., Bauer P. O., Oyama F., Acidic mammalian chitinase is a proteases-resistant glycosidase in mouse digestive system. Sci. Rep. 6, 37756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain M., Wilson J. B., New paralogues and revised time line in the expansion of the vertebrate GH18 family. J. Mol. Evol. 76, 240–260 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Janiak M. C., Chaney M. E., Tosi A. J., Evolution of acidic mammalian chitinase genes (CHIA) is related to body mass and insectivory in primates. Mol. Biol. Evol. 35, 607–622 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Olland A. M., Strand J., Presman E., Czerwinski R., Joseph-McCarthy D., Krykbaev R., Schlingmann G., Chopra R., Lin L., Fleming M., Kriz R., Stahl M., Somers W., Fitz L., Mosyak L., Triad of polar residues implicated in pH specificity of acidic mammalian chitinase. Protein Sci. 18, 569–578 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tjoelker L. W., Gosting L., Frey S., Hunter C. L., Trong H. L., Steiner B., Brammer H., Gray P. W., Structural and functional definition of the human chitinase chitin-binding domain. J. Biol. Chem. 275, 514–520 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Nio J., Fujimoto W., Konno A., Kon Y., Owhashi M., Iwanaga T., Cellular expression of murine Ym1 and Ym2, chitinase family proteins, as revealed by in situ hybridization and immunohistochemistry. Histochem. Cell Biol. 121, 473–482 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Romiguier J., Ranwez V., Douzery E. J. P., Galtier N., Genomic evidence for large, long-lived ancestors to placental mammals. Mol. Biol. Evol. 30, 5–13 (2013). [DOI] [PubMed] [Google Scholar]
- 22.O’Leary M. A., Bloch J. I., Flynn J. J., Gaudin T. J., Giallombardo A., Giannini N. P., Goldberg S. L., Kraatz B. P., Luo Z.-X., Meng J., Ni X., Novacek M. J., Perini F. A., Randall Z. S., Rougier G. W., Sargis E. J., Silcox M. T., Simmons N. B., Spaulding M., Velazco P. M., Weksler M., Wible J. R., Cirranello A. L., The placental mammal ancestor and the post–K-Pg radiation of placentals. Science 339, 662–667 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Meredith R. W., Gatesy J., Murphy W. J., Ryder O. A., Springer M. S., Molecular decay of the tooth gene Enamelin (ENAM) mirrors the loss of enamel in the fossil record of placental mammals. PLOS Genet. 5, e1000634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emerling C. A., Huynh H. T., Nguyen M. A., Meredith R. W., Springer M. S., Spectral shifts of mammalian ultraviolet-sensitive pigments (short wavelength-sensitive opsin 1) are associated with eye length and photic niche evolution. Proc. R. Soc. B 282, 20151817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meredith R. W., Janečka J. E., Gatesy J., Ryder O. A., Fisher C. A., Teeling E. C., Goodbla A., Eizirik E., Simão T. L., Stadler T., Rabosky D. L., Honeycutt R. L., Flynn J. J., Ingram C. M., Steiner C., Williams T. L., Robinson T. J., Burk-Herrick A., Westerman M., Ayoub N. A., Springer M. S., Murphy W. J., Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 334, 521–524 (2011). [DOI] [PubMed] [Google Scholar]
- 26.dos Reis M., Inoue J., Hasegawa M., Asher R. J., Donoghue P. C., Yang Z., Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc. Biol. Sci. 279, 3491–3500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J., Yonezawa T., Kishino H., Rates of molecular evolution suggest natural history of life history traits and a post-K-Pg nocturnal bottleneck of placentals. Curr. Biol. 27, 3025–3033.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Archibald J. D., Deutschman D. H., Quantitative analysis of the timing of the origin and diversification of extant placental orders. J. Mamm. Evol. 8, 107–124 (2001). [Google Scholar]
- 29.Halliday T. J. D., Upchurch P., Goswami A., Resolving the relationships of Paleocene placental mammals. Biol. Rev. 92, 521–550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delsuc F., Metcalf J. L., Wegener Parfrey L., Song S. J., González A., Knight R., Convergence of gut microbiomes in myrmecophagous mammals. Mol. Ecol. 23, 1301–1317 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Sanders J. G., Beichman A. C., Roman J., Scott J. J., Emerson D., McCarthy J. J., Girguis P. R., Baleen whales host a unique gut microbiome with similarities to both carnivores and herbivores. Nat. Commun. 6, 8285 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., He G., Xu H., Han X., Jones G., Rossiter S. J., Zhang S., Adaptive functional diversification of lysozyme in insectivorous bats. Mol. Biol. Evol. 31, 2829–2835 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Boot R. G., Renkema G. H., Strijland A., Van Zonneveld A. J., Aerts J. M. F. G., Cloning of a cDNA encoding chitotriosidase, a human chitinase produced by macrophages. J. Biol. Chem. 270, 26252–26256 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Boot R. G., Bussink A. P., Verhoek M., de Boer P. A., Moorman A. F., Aerts J. M., Marked differences in tissue-specific expression of chitinases in mouse and man. J. Histochem. Cytochem. 53, 1283–1292 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Matthew W. D., The carnivora and the insectivora of the bridger basin, middle eocene. Mem. Am. Museum Nat. Hist. 9, 1199–203 (1909). [Google Scholar]
- 36.Wilman H., Belmaker J., Simpson J., de la Rosa C., Rivadeneira M. M., Jetz W., EltonTraits 1.0: Species-level foraging attributes of the world’s birds and mammals. Ecology 95, 2027 (2014). [Google Scholar]
- 37.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A., Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. A. Miller, W. Pfeiffer, T. Schwartz, Creating the CIPRES Science Gateway for inference of large phylogenetic trees, in 2010 Gateway Computing Environments Workshop, GCE 2010 (IEEE, 2010), pp. 1–8. [Google Scholar]
- 41.Burset M., Seledtsov I. A., Solovyev V. V., Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 28, 4364–4375 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J.-M., Cooper D. N., Chuzhanova N., Férec C., Patrinos G. P., Gene conversion: Mechanisms, evolution and human disease. Nat. Rev. Genet. 8, 762–775 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Grafen A., The phylogenetic regression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 326, 119–157 (1989). [DOI] [PubMed] [Google Scholar]
- 44.D. Orme, R. Freckleton, G. Thomas, T. Petzoldt, S. Fritz, N. Isaac, W. Pearse, “caper: Comparative analyses of phylogenetics and evolution in R,” 29 November 2013; https://cran.r-project.org/web/packages/caper/index.html.
- 45.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Fan Y., Huang Z.-Y., Cao C.-C., Chen C.-S., Chen Y.-X., Fan D.-D., He J., Hou H. L., Hu L., Hu X.-T., Jiang X.-T., Lai R., Lang Y.-S., Liang B., Liao S.-G., Mu D., Ma Y.-Y., Niu Y. Y., Sun X.-Q., Xia J.-Q., Xiao J., Xiong Z.-Q., Xu L., Yang L., Zhang Y., Zhao W., Zhao X.-D., Zheng Y.-T., Zhou J.-M., Zhu Y.-B., Zhang G.-J., Wang J., Yao Y.-G., Genome of the Chinese tree shrew. Nat. Commun. 4, 1426 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Low D. H. W., Sunagar K., Undheim E. A., Ali S. A., Alagon A. C., Ruder T., Jackson T. N., Pineda Gonzalez S., King G. F., Jones A., Antunes A., Fry B. G., Dracula’s children: Molecular evolution of vampire bat venom. J. Proteomics 89, 95–111 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Francischetti I. M. B., Assumpção T. C., Ma D., Li Y., Vicente E. C., Uieda W., Ribeiro J. M., The “Vampirome”: Transcriptome and proteome analysis of the principal and accessory submaxillary glands of the vampire bat Desmodus rotundus, a vector of human rabies. J. Proteomics 82, 288–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phillips C. D., Baker R. J., Secretory gene recruitments in vampire bat salivary adaptation and potential convergences with sanguivorous leeches. Front. Ecol. Evol. 3, 1–11 (2015). [Google Scholar]
- 50.Ma J.-E., Li L. M., Jiang H. Y., Zhang X. J., Li J., Li G. Y., Yuan L. H., Wu J., Chen J. P., Transcriptomic analysis identifies genes and pathways related to myrmecophagy in the Malayan pangolin (Manis javanica). PeerJ 5, e4140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R. M. Nowak, Walker’s Mammals of the World (Johns Hopkins Univ. Press, 1999). [Google Scholar]
- 52.Hofreiter M., Poinar H. N., Spaulding W. G., Bauer K., Martin P. S., Possnert G., Pääbo S., A molecular analysis of ground sloth diet through the last glaciation. Mol. Ecol. 9, 1975–1984 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Bargo M. S., The ground sloth Megatherium americanum: Skull shape, bite forces, and diet. Acta Palaeontol. Pol. 46, 173–192 (2001). [Google Scholar]
- 54.McAfee R. K., Feeding mechanics and dietary implications in the fossil sloth Neocnus (Mammalia: Xenarthra: Megalonychidae) from Haiti. J. Morphol. 272, 1204–1216 (2011). [DOI] [PubMed] [Google Scholar]
- 55.McKenna M. C. M., Wyss A. A. R., Flynn J. J., Paleogene pseudoglyptodont xenarthrans from central Chile and Argentine Patagonia. Am. Mus. Novit. 3536, 1–18 (2006). [Google Scholar]
- 56.Vizcaíno S. F., Cassini G. H., Fernicola J. C., Susana Bargo M., Evaluating habitats and feeding habits through ecomorphological features in glyptodonts (Mammalia, xenarthra). Ameghiniana 48, 305–319 (2011). [Google Scholar]
- 57.Gheerbrant E., Paleocene emergence of elephant relatives and the rapid radiation of African ungulates. Proc. Natl. Acad. Sci. U.S.A. 106, 10717–10721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gheerbrant E., Amaghzaz M., Bouya B., Goussard F., Letenneur C., Ocepeia (middle Paleocene of Morocco): The oldest skull of an afrotherian mammal. PLOS ONE 9, e89739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gheerbrant E., Filippo A., Schmitt A., Convergence of afrotherian and laurasiatherian ungulate-like mammals: First morphological evidence from the Paleocene of Morocco. PLOS ONE 11, e0157556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simmons N. B., Geisler J. H., Phylogenetic relationships of Icaronycteris, Archaeonycteris, Hassianycteris, and Palaeochiropteryx to extant bat lineages, with comments on the evolution of echolocation and foraging strategies in Microchiroptera. Bull. Am. Museum Nat. Hist. 235, 1–182 (1998). [Google Scholar]
- 61.Simmons N. B., Seymour K. L., Habersetzer J., Gunnell G. F., Primitive Early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature 451, 818–821 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Price E. R., Brun A., Caviedes-Vidal E., Karasov W. H., Digestive adaptations of aerial lifestyles. Phys. Chem. Chem. Phys. 30, 69–78 (2015). [DOI] [PubMed] [Google Scholar]
- 63.L. D. Martin, in Carnivore Behavior, Ecology, and Evolution, J. L. Gittleman, Ed. (Springer, 1989), pp. 536–568. [Google Scholar]
- 64.Spaulding M., Flynn J. J., Phylogeny of the Carnivoramorpha: The impact of postcranial characters. J. Syst. Palaeontol. 10, 653–677 (2012). [Google Scholar]
- 65.Storch G., Eomanis waldi, ein Schuppentier aus dem Mittel-Eozän der ‘Grube Messel’ bei Darmstadt (Mammalia: Pholidota). Senckenbergiana Lethaea 59, 503–529 (1978). [Google Scholar]
- 66.Gaudin T. J., Emry R. J., Wible J. R., The phylogeny of living and extinct pangolins (Mammalia, Pholidota) and associated taxa: A morphology based analysis. J. Mamm. Evol. 16, 235–305 (2009). [Google Scholar]
- 67.Solé F., Gheerbrant E., Amaghzaz M., Bouya B., Further evidence of the African antiquity of hyaenodontid (’Creodonta’, Mammalia) evolution. Zool. J. Linn. Soc. 156, 827–846 (2009). [Google Scholar]
- 68.Rose K. D., Holbrook L. T., Rana R. S., Kumar K., Jones K. E., Ahrens H. E., Missiaen P., Sahni A., Smith T., Early Eocene fossils suggest that the mammalian order Perissodactyla originated in India. Nat. Commun. 5, 5570 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Cooper L. N., Seiffert E. R., Clementz M., Madar S. I., Bajpai S., Hussain S. T., Thewissen J. G., Anthracobunids from the middle eocene of India and Pakistan are stem perissodactyls. PLOS ONE 9, e109232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Welker F., Collins M. J., Thomas J. A., Wadsley M., Brace S., Cappellini E., Turvey S. T., Reguero M., Gelfo J. N., Kramarz A., Burger J., Thomas-Oates J., Ashford D. A., Ashton P. D., Rowsell K., Porter D. M., Kessler B., Fischer R., Baessmann C., Kaspar S., Olsen J. V., Kiley P., Elliott J. A., Kelstrup C. D., Mullin V., Hofreiter M., Willerslev E., Hublin J.-J., Orlando L., Barnes I., MacPhee R. D. E., Ancient proteins resolve the evolutionary history of Darwin’s South American ungulates. Nature 522, 81–84 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Buckley M., Ancient collagen reveals evolutionary history of the endemic South American “ungulates.”. Proc. Biol. Sci. 282, 20142671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Westbury M., Baleka S., Barlow A., Hartmann S., Paijmans J. L. A., Kramarz A., Forasiepi A. M., Bond M., Gelfo J. N., Reguero M. A., López-Mendoza P., Taglioretti M., Scaglia F., Rinderknecht A., Jones W., Mena F., Billet G., de Muizon C., Aguilar J. L., MacPhee R. D.E., Hofreiter M., A mitogenomic timetree for Darwin’s enigmatic South American mammal Macrauchenia patachonica. Nat. Commun. 8, 15951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thewissen J. G. M., Hussain S. T., Postcranial Osteology of the most Primitive Artiodactyl: Diacodexis pakistanensis (Dichobunidae). Anat. Histol. Embryol. 19, 37–48 (1990). [DOI] [PubMed] [Google Scholar]
- 74.J. M. Theodor, in The Evolution of Artiodactyls, D. R. Prothero, S. E. Foss, Eds. (JHU Press, 2007), pp. 32–58. [Google Scholar]
- 75.Li Q., Ni X., An early Oligocene fossil demonstrates treeshrews are slowly evolving “living fossils.”. Sci. Rep. 6, 18627 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marivaux L., Bocat L., Chaimanee Y., Jaeger J.-J., Marandat B., Srisuk P., Tafforeau P., Yamee C., Welcomme J.-L., Cynocephalid dermopterans from the Palaeogene of South Asia (Thailand, Myanmar and Pakistan): Systematic, evolutionary and palaeobiogeographic implications. Zool. Scr. 35, 395–420 (2006). [Google Scholar]
- 77.Cartmill M., Rethinking primate origins. Science 184, 436–443 (1974). [DOI] [PubMed] [Google Scholar]
- 78.Williams B. A., Kay R. F., Kirk E. C., New perspectives on anthropoid origins. Proc. Natl. Acad. Sci. U.S.A. 107, 4797–4804 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ni X., Gebo D. L., Dagosto M., Meng J., Tafforeau P., Flynn J. J., Beard K. C., The oldest known primate skeleton and early haplorhine evolution. Nature 498, 60–64 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/5/eaar6478/DC1
fig. S1. Vertebrate RAxML CHIA gene tree based on CHIA gene models.
fig. S2. Placental mammal RAxML CHIA gene tree based on genomic contig–derived and mRNA sequences.
fig. S3. Placental mammal RAxML CHIA gene tree based on genomic contig–derived and mRNA sequences, after removing chiropteran CHIA4 and CHIA5 sequences.
fig. S4. Diagram demonstrating synteny of CHIA paralogs and nearby genes on their respective genomic scaffolds/contigs.
fig. S5. T. syrichta CHIA1, CHIA2, CHIA3, CHIA4, and CHIA5 gene models showing conserved chitinolytic domain (DXXDXDXE).
fig. S6. T. syrichta CHIA1, CHIA2, CHIA3, CHIA4, and CHIA5 gene models showing chitin-binding domain with six conserved cysteine residues.
fig. S7. Cladogram showing relationships of species in WGS data set and mapped gene losses.
table S1. CHIA summary.
table S2. Synteny of CHIA paralogs.
table S3. CHIA gene expression analyses.
table S4. Distribution of genetic lesions and missing data across CHIA exons.
table S5. Discussion of CHIA number, timing of loss, diets of extant taxa, and the fossil record.
table S6. dN/dS ratio models used for gene inactivation estimates.
table S7. CHIA inactivation date estimates calculated from dN/dS ratio models in table S6 and assuming divergence dates from the study of Emerling et al. (24).
table S8. Scaffold/contig coordinates of OVGP1, PIFO, and DENND2D for species in fig. S4.
table S9. Evidence of gene conversion between CHIA4 and CHIA5.
data set S1. Alignments used for analyses.
data set S2. Alignments used for constructing CHIA genes from scaffolds/contigs.


