Abstract
Recent evidence indicates a link between gut pathology and microbiome with hypertension (HTN) in animal models. However, whether this association exists in humans is unknown. Thus, our objectives in the present study were to test the hypotheses that high blood pressure (BP) patients have distinct gut microbiomes and that gut–epithelial barrier function markers and microbiome composition could predict systolic BP (SBP). Fecal samples, analyzed by shotgun metagenomics, displayed taxonomic and functional changes, including altered butyrate production between patients with high BP and reference subjects. Significant increases in plasma of intestinal fatty acid binding protein (I-FABP), lipopolysaccharide (LPS), and augmented gut-targetting proinflammatory T helper 17 (Th17) cells in high BP patients demonstrated increased intestinal inflammation and permeability. Zonulin, a gut epithelial tight junction protein regulator, was markedly elevated, further supporting gut barrier dysfunction in high BP. Zonulin strongly correlated with SBP (R2 = 0.5301, P<0.0001). Two models predicting SBP were built using stepwise linear regression analysis of microbiome data and circulating markers of gut health, and validated in a separate cohort by prediction of SBP from zonulin in plasma (R2 = 0.4608, P<0.0001). The mouse model of HTN, chronic angiotensin II (Ang II) infusion, was used to confirm the effects of butyrate and gut barrier function on the cardiovascular system and BP. These results support our conclusion that intestinal barrier dysfunction and microbiome function are linked to HTN in humans. They suggest that manipulation of gut microbiome and its barrier functions could be the new therapeutic and diagnostic avenues for HTN.
Introduction
Hypertension (HTN) is the most prevalent modifiable risk factor for cardiovascular disease. Despite multipronged community-based efforts to influence lifestyle and recent pharmacological and surgical advances in the management of high blood pressure (BP), cases of number of HTN have doubled globally between 1990 and 2015 [1]. Moreover, recent SPRINT (Systolic Blood Pressure Intervention Trial) and American Heart Association/American College of Cardiology guidelines lowering the pressures defining high BP cohort (HBP) indicate efforts to reduce it will continue as serious health foci [2]. While more aggressive intervention of HTN is to be expected, response rates to monotherapy with any antihypertensive drug remain only ~50% [3]. In addition, ~15% of this rapidly growing population is resistant to all interventions, underscoring the need for diverse and innovative strategies for HTN [1].
The mosaic theory of HTN by Dr Irvin Page suggested that HTN is associated with multiple factors and organs including environmental, genetic, neural, endocrine, humoral, and immune [4,5]. While roles of the brain, kidney, bone marrow, and immune cells in BP regulation have been thoroughly investigated, involvement of the gut, one of the largest endocrine, immune, and neural organs in the body [6,7], in HTN has been underestimated. There are multiple reasons to investigate gut and its microbiome in HTN; (i) gut is the single most important organ for adsorption of nutrients and ions, which impacts BP greatly [8–10], (ii) dysregulation of microbiota is related to various metabolic diseases and HTN risk factors such as obesity, heart failure, and diabetes mellitus [11], indicating a major role in BP regulation, (iii) gut pathophysiology in animal HTN models [12], and (iv) fecal transplantation from HTN subjects can elevate BP of normal animals [13,14]. These observations suggest the gut as a promising new therapeutic target for human HTN. However, the exact roles of bacteria and gut health in human HTN have not been elucidated. Therefore, the present study’s goals were to clarify functional changes in the microbiome and pathophysiology in patients with HBP and to validate these findings using the chronic angiotensin II (Ang II) infusion mouse model of HTN.
Methods
Human study cohort
Institutional Review Board (IRB) at the University of Florida reviewed and approved all protocols and informed consent form for the study. Subjects, age >18 years meeting the inclusion/exclusion criteria and who were competent and willing to provide written consent were recruited. Exclusion criteria were known pregnancy at the time of recruitment or within 6 months before enrolment, antibiotic treatment within 2 months of enrolment, taking certain medications (e.g. probiotic pills, anti-inflammatory agents, glucocorticoids, other immune-modulating medications) and history of intestinal surgery, inflammatory bowel disease, celiac disease, lactose intolerance, chronic pancreatitis, or other malabsorption disorders. Study participants did not change their diet for the study. During clinic visits, nurses or physicians measured BP of the study participants by averaging two to three sphygmomanometer readings in the seated participant, after 5 min rest with both feet on the floor. Using pre-2017 HTN guidelines, we divided subjects into three groups based on their clinic systolic BP (SBP) readings. The high BP cohort (HBP) was defined as SBP ≥140 mmHg, the prehypertensive cohort as SBP from 120 to 139 mmHg and the reference cohort (REF) as SBP ≤130 mmHg irrespective of subject’s antihypertensive drug regimen. After BP measurements, stool and blood samples were collected using the OMNIgene GUT kit (DNA Genotek) and BD Vacutainer® EDTA coated tubes (BD). Samples were kept at −80°C until further analysis. Characteristics of study cohorts are summarized in Supplementary Table S1. There was no significant difference in serum glucose/HbA1c levels and body mass index between the two groups. We analyzed only REF and HBP cohorts in support of our stated hypothesis; prehypertensive cohort was omitted.
Whole genome sequencing, quality assessment, and filtering of human stool
DNA in preservation fluid from OMNIgene GUT kit was extracted and sequenced in one batch following the instructions provided with PowerFecal DNA extraction kit (MoBio). DNA sequencing and analyses were performed by WrightLabs, LLC. One nanogram DNA per sample (n=40) underwent Nextera XT (Illumina) tagmentation and library preparation. All samples underwent dual index barcoding. Quality of final libraries was assessed using a high sensitivity bioanalyzer chip (Agilent). Equimolar amounts of the library were pooled and purified using QIAquick gel purification kit (Qiagen). Purified libraries underwent sequencing on the Illumina HiSeq4000 following a 2 × 150 index run. Raw read quality was assessed using the program FastQC to obtain average Q-scores across the read length of all R1 and R2 fastq files. The program Trimmomatic was employed to quality filter and pair the raw sequence data. A sliding window filtration was utilized to cut reads at a 4-base average Q-score of 28 or lower and reads trimmed below 80 bp discarded. Trimmomatic filtered reads were then paired and all unpaired reads discarded. Paired R1 and R2 files were concatenated and run through the Kneaddata pipeline to remove human DNA reads from filtered sequence data.
Taxonomic/functional gene profiling and comparative analysis
The metagenome analysis tool MetaPhlAn was implemented to quantitate the bacterial taxonomic profile within each sample. The taxonomic outputs from all metagenomes were merged into a single tsv table, which was converted into a biom file for downstream analyses. To obtain functional gene profiles, filtered data were annotated using the Uniref90 database within Humann2. Uniref90 annotations were regrouped as Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO) terms, which consequently underwent counts per million (CPM) normalization within Humann2 for stratified barplot analysis. Both taxonomic and functional count data were uploaded to the online analysis tool METAGENassist for partial least squares discriminant analysis (PLS-DA). Interquartile range filtering was utilized on the profiled datasets. Variables with over 85% zeros were removed and abundance were then normalized using an autoscaling algorithm within METAGENassist. KOs were further summarized by function using the KEGG database. Relative abundances of taxonomic profiles and functional pathways were multiplied by 1 million and formatted as previously described [15]. Comparisons were made with ‘HBP’ as the main categorical variable (‘REF’). α levels of 0.05 were used for both the Kruskal–Wallis and pairwise Wilcoxon tests. Linear Discriminant Analysis (LDA) scores greater than 2.5 were displayed for taxonomy, and 0.5 for functional pathways. Abundance of prevalent taxa were organized into a heat map using the R package pheatmap. CPM normalized KO data were converted into a QIIME formatted biom file to generate principal co-ordinate analysis (PCoA) plots using the Bray–Curtis distance metric. The ANOSIM and PERMANOVA tests were employed to determine the significance of clustering between sample cohorts (REF compared with HBP). CSS-normalized counts tables were separated into REF and HBP groups. These were aligned to KEGG pathways of interest using the R package, Pathview [16]. A volcano plot was generated by plotting −log10 of the P-value against the log2 fold change of a KO term in REF relative to HBP examples with the R package, ggplot2 [17]. A quasi-likelihood F-test transformed P-value threshold of 13 (P<0.05) was utilized to highlight significantly differentiating data points. Classification of bacterial taxonomy based on SCFA production was performed as previously described [18–20]. All networks were generated with CoNet and visualized in Cytoscape. Input filtering constrained the minimum occurrence of operational taxonomic units (OTUs) and considered only those present in at least one sample. Networks were constructed using Spearman’s correlation methods with threshold setting at 0.9, Bray–Curtis dissimilarity at the automatic threshold setting, and Kullback–Leibler dissimilarity at the automatic threshold setting; the edge selection parameter was set to 50 for the strongest positive and negative correlations. Randomization steps included permutations and bootstraps with filtering of unstable edges and Benjamini–Hochberg procedure with a P-value of 0.05. Node clusters with less than or equal to three edges were not shown in the final network. Edge coloration indicated co-presence in blue or mutual exclusion in red.
16S rRNA gene sequencing and analysis of mice feces
Fecal pellets were collected without environmental contamination, immediately frozen in liquid nitrogen, and stored at −80°C until analysis, within 3 months of collection. Nucleic acid extractions were performed on ~40 mg of each sample using Powerfecal DNA Isolation kit following manufacturer’s instructions (MoBio). Total DNA was quantitated with Qubit 2.0 (Invitrogen) fluorometer with the dsDNA high sensitivity option, and 10 ng of each sample was amplified by PCR using the Illumina iTag kit. Twenty-five microliter PCRs were carried out with a MJ Research PTC-200 thermocycler (Bio–Rad, Hercules, CA) and included 1× PCR buffer, 0.8 mM dNTPs, 0.625 U Taq polymerase, 0.2 μM amplification primers targetting V3–V4 region of the 16S ribosomal subunit (DNA Genotek). The cycling conditions were 98°C for 3 min; 35 cycles of 98°C for 1 min, 55°C for 40 s, and 72°C for 10 min, then kept at 4°C. PCR products were visualized on a 1% SYBRsafe E-gel (Life Technologies), then pooled and purified from the gel with the Qiagen Gel Purification Kit (Qiagen). Purified samples were quantitated using the Qubit 2.0 fluorometer and combined in equimolar amounts. Quality of the libraries was checked with the 2100 Bioanalyzer DNA 100 chip (Agilent Technologies). Library pools were size verified using the Fragment Analyzer CE (Advanced Analytical Technologies Inc.) and quantitated using the Qubit High Sensitivity dsDNA kit (Life Technologies). After dilution to a final concentration of 1 nM and a 10% spike of PhiX V3 library (Illumina), pools were denatured for 5 min in an equal volume of 0.1 N NaOH then further diluted to 12 pM in Illumina’s HT1 buffer. The denatured and PhiX-spiked 12 pM pool was loaded on an Illumina MiSeq V2 300 cycle kit cassette with 16S rRNA library sequencing primers and set for 150 bases, paired-end reads. Sequences were trimmed at a length of 150 bp and quality filtered at an expected error of less than 0.5% using USEARCH v71. The FLASh algorithm was used for read merging and automated rejection of low quality sequences [21]; quality screening for length and ambiguous bases was performed in mothur [22]. After quality filtering, reads were analyzed using the QIIME 1.9.0 software package [23,24]. Chimeric sequences were identified using USEARCH61.4. Open reference OTUs were picked using the USEARCH61 algorithm, and taxonomy assignment was performed using the Greengenes 16S rRNA gene database (13–7 release, 97%) [25]. Assigned taxonomy were organized into a BIOM formatted OTU table, which was summarized within Qiime-1.9. High quality sequence data were obtained for all 40 samples. All samples were incorporated into a CSS normalized OTU table, as sequencing depth exceeded 1000 sequences for each sample [26]. Rarefaction curves were generated within the QIIME-1.9.0 sequence analysis package using an unrarefied OTU table. Multiple rarefactions were conducted on sequences across all samples from a minimum depth of zero sequences, to a maximum depth of 110000 sequences, with a step size of 11000 sequences/sample for 20 iterations per step. In taxonomic comparisons, OTUs that were not classified at the kingdom level were discarded. All β diversity analyses were generated from a CSS normalized OTU table within QIIME-1.9.0. A weighted UniFrac distance matrix was first generated and visualized in 3D space using Emperor. The ANOSIM test for significance was calculated within QIIME-1.9.0, with the weighted UniFrac distance matrix serving as the input. Taxonomy was summarized at the genus level within QIIME-1.9.0 and uploaded to the Galaxy platform to generate LDA effects size (LEfSe)/cladogram enrichment plots considering significant enrichment at a P<0.05, LDA score > 2.8.
Animals
All protocols involving animals were approved by the University of Florida Institutional Animal Care and Use Committee (IACUC) and complied with the standards stated in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Rats were housed in a temperature-controlled room (22–23°C) with a 12:12-h light-dark cycle, in specific pathogen-free cages, and had access to standard irradiated rodent chow (7912, Envigo) and water ad libitum. Fecal samples were collected between 8 and 9 AM.
Animal procedures
Adult C57BL6 mice and Sprague–Dawley (SD) rats were purchased from Charles River Laboratories and used for all the experiments. Eight-week-old male C57BL6 mice were used for experiments involving Ang II infusion. HTN was induced by chronic infusion of Ang II (1000 ng/kg/min, Bachem) using mini-osmotic pumps (1004, ALZET) implanted subcutaneously. Control C57BL6 animals received 0.9% saline in osmotic pumps. For butyrate administration, sodium butyrate (Sigma) was diluted in PBS injected intraperitoneally (1 g/kg body weight in 150 μl PBS) daily at 9 AM after BP measurement for 28 days. The dose of butyrate was determined by a pilot experiment with different butyrate concentrations (0.2–1 g/kg). For in vivo imaging of whole animals and gut permeability measurements, mice were anesthetized and shaved from neck to lower torso to visualize fluorescence without interference from fur. On the next day, mice were administered with FITC-dextran (Sigma, 44 mg/100 g body weight) by oral gavage with a needle attached to a 1-ml syringe. Fluorescence of abdomen area was measured using IVIS Imaging System 200 under isoflurane anesthesia at 2 and 4 h after injection and analyzed for fluorescence based on the manufacturer’s recommendation (Xenogen). At 4 h after imaging, blood was collected from the facial vein, and FITC-dextran level in the serum was measured in a 96-well plate in duplicate. For intestinal tissue hypoxia measurement, the hypoxia probe pimonidazole hydrochloride (Hypoxyprobe-1 kit; HPI) was used according to the manufacturer’s (HPI) protocol.
Telemetry
Seven to ten days before Ang II and/or butyrate infusion, radiotransmitters were implanted in mice as recommended by the manufacturer (HD-X11, Data Sciences International) [27] to measure mean arterial pressure (MAP) by telemetry. Briefly, isoflurane-anesthetized mice were laid on their backs on the surgical platform, and ~1.5 cm midline incision was made on the neck while retracting the salivary glands. The left common carotid artery was exposed at the bifurcation toward the heart, without injuring the vagal nerve. The BP monitoring catheter was positioned into the aortic arch via the carotid artery. The catheter was fixed with 6/0 non-absorbable braided surgical silk sutures (Ethicon), and the transmitter was placed in a subcutaneous pocket in the left flank. The skin was closed with a continuous 6/0 non-absorbable monofilament polypropylene suture (Ethicon). Analyses of ECG, HR, and systolic, diastolic, and mean BP were performed using Dataquest A.R.T. 4.3 software (Data Sciences International). Spectral analysis of the SBP and pulse interval waveforms was performed as previously described using the Hey Presto software [28,29].
Real-time RT-PCR
RNA was isolated from ileum (small intestine) and proximal colon (large intestine) using TRIzol reagent (Ambion) per manufacturer’s protocols. Genomic DNA was removed using DNase I treatment (2 U/sample, Ambion). The purity of RNA was evaluated spectrophotometrically by 260/280 ratio. Reverse transcription was accomplished using High Capacity Reverse Transcription kit (Applied Biosystems) and 1 μg RNA. Real-time RT-PCR was performed using Taqman Universal PCR Master Mix and Taqman Gene Expression Assay primers (Applied Biosystems): Mm99999915 g1 (GAPDH), Mm04225243 g1 (prostaglandin-endoperoxide synthase (Ptgs) 1 (Ptgs1), Mm03294838 g1 (Ptgs2), Mm00661045 m1 (monocarboxylate transpoter (MCT) 1 (MCT1)), Mm00446102 m1 (MCT4), Mm00500912 m1 (Occludin), Mm00493699 m1 (zonula occludens-1 (ZO-1)), Mm00515514 s1 (Claudin 4). RT-PCR was run using StepOnePlus (Applied Biosystems) sequence detection system. All cDNA samples were assayed in duplicate. Data were normalized to GAPDH, which showed no significant changes in any treatments.
Isolated rat heart preparation
SD rats weighing 250–300 g were administered 250 IU heparin intraperitoneally. Twenty minutes later, animals were killed by decapitation. Hearts were immediately excised and mounted on to a Langendorff apparatus (130101EZ, Radnoti). Hearts were perfused with Krebs–Ringer bicarbonate solution (KRS; 118.4 mM NaCl, 4.7 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 2.5 mM CaCl2, 26.5 mM NaHCO3, and 11.7 mM glucose) at a constant pressure of 95 cm of H2O. The perfusate was maintained at 37°C and bubbled with 95% O2/5% CO2. A balloon catheter connected to a pressure transducer (AD Instruments) filled with degassed water interfaced with a PowerLab data acquisition unit (AD Instruments) was inserted into the left ventricle (LV) to measure intraventricular pressure. Extreme care was taken to remove any air by repeatedly flushing the balloon and tubing with degassed water. After 30 min, the functional parameters LV developed pressure (DP), coronary flow rate, heart rate, LV + dp/dt and LV − dp/dt were recorded for each butyrate dose (prepared in KRS). Each heart was exposed to two doses of butyrate interspersed by washing with KRS. Data were analyzed using Chart software (AD Instruments). A minimum of 150 cycles was used for analysis, and results were averaged over the perfusion period. The coronary flow rate was measured by collecting the coronary effluent into a measuring cylinder for 1 min [30–32].
Assessment of mouse cardiac function
Echocardiograms were performed using a GE vivid7 ultrasound machine with a 12-Hz transducer (GE Healthcare). After 4 weeks of treatment, C57BL6 mice were anesthetized with 2% isoflurane. M-mode echocardiography was performed in the parasternal short-axis view at the level of the papillary muscles. LV systolic function was determined as fractional shortening (FS) (%) = (LVEDD − LVESD/LVEDD) × 100 in the short axis. LV diastolic function was assessed by analyzing the mitral valve E/A ratio using Doppler in apical four-chamber view. The E/A ratio was the peak velocity flow in early diastole (E-wave) to peak velocity flow in late diastole caused by atrial contraction (A-wave). All measurements were based on the average of at least three consecutive cardiac cycles.
Assessment of resistance artery function using pressure myography
Two sets of experiments were performed to measure artery function in mice. The C57BL6 mice were humanely killed with ketamine/xylazine/acepromazine after 4 weeks of Ang II/butyrate treatment as described above and aortae were harvested from the thorax. Aortae were cleaned of extraneous tissue and mounted on a myograph in Krebs–Henseleit buffer containing (in mmol/l) NaCl 119, KCl 4.7, CaCl2 2.5, MgSO4 0.289, NaHCO3 25, KH2PO4 1.18, glucose 5.5, and gassed with 95% O2/5% CO2. The aortic rings were suspended by two L-shaped stainless steel wires inserted into the lumen followed by immersion in 10-ml organ chambers filled with Kreb’s buffer. For dose-dependent relaxation experiments (Supplementary Figure S4D), the aortae were mounted on a myograph, preconstricted with phenylephrine (2.0 μM), and treated with graded doses of acetylcholine (10−9–10−3 mol/l) in a cumulative manner, starting at the lowest dose and recording each response for 3 min before application of the next highest dose. Changes in isometric tension were recorded using a force-displacement transducer (Grass FT 0.3, Quincy, MA) connected to a Power Lab system 400 (ML 118, PowerLab, AD Instruments, Medford, MA) and saved in a computer. Percentage relaxation was calculated at each dose and a dose–response curve constructed. For the maximal relaxation measurements (Supplementary Figure S4E), aortae preconstricted with 2.0 μM phenylephrine were treated with a single acute dose of acetylcholine (50 μM). Vessels were allowed to relax for 15 min and percentage of maximum relaxation calculated.
Measurement of circulating zonulin, intestinal fatty acid binding protein, lipopolysaccharides, and butyrate
Plasma samples were used in duplicate and manufacturer recommended methods were followed for all assays. Plasma intestinal fatty acid binding protein (I-FABP) (R&D Systems) concentrations were quantitated using a specific ELISA. Zonulin (catalog number K5601, Immundiagnostik AG) in plasma was estimated by competitive ELISA. Plasma lipopolysaccharide (LPS) (antibodies online) was estimated by sandwich ELISA. Interassay coefficients of variance were 6.5, 6.3, 2.1% for IFABP, LPS, and zonulin assays, respectively. For butyrate measurement, snap-frozen plasma samples were sent to the Michigan Regional Comprehensive Metabolomics Resource Core. Samples were dispersed in acidified water spiked with stable-isotope labeled short chain fatty acid standards, and then extracted with diethyl ether. The ether layer was immediately analyzed by GC-MS using a Phenomenex ZB-WAX column on an Agilent 6890 GC with a 5973 MS detector. Quantitation was performed by calibration to internal butyrate standards.
Flow cytometry
Mouse blood was collected from the submandibular vein into EDTA-coated tubes (Sarstedt). Human blood was obtained from subjects in a recumbent position with a 22-gauge needle for a clean venipuncture of an antecubital vein. Samples were collected into EDTA coated vacutainers (Becton Dickinson). Peripheral blood mononuclear cells (PBMCs) were prepared in a Ficoll density gradient. After washing, PBMCs were treated with Fixation/Permeabilization buffer for 40 min (eBioscience) and then FC block (BioLegend) for 10 min on ice for blocking. Cells were incubated with following antibodies for 30 min in ice to analyze T helper 17 (Th17) cells from mouse and human PBMCs; mouse CD4 (clone RM4-5, FITC), mouse Il-17 (clone eBio17B7, eFlour660), mouse CCR2 (chemokine receptor 2) (clone 475301, PE), human CD4 (clone OKT4, FITC), human IL-17 (BL168, Alexa Flour 647), CCR6 (G034E3, PE/Cy7), integrin β7 (clone FIB27, PE). Antibodies were purchased from BioLegend, eBioscience, and R&D Systems. Each population was sorted by BD FACS LSR II (BD bioscience).
Image and statistical analyses
Positive areas were measured and analysed by two researchers, blind to treatment group identity, using Fiji (Figure 7A and Supplementary Figure S4A) [33]. Statistical methods for analyses of microbiome data are described as above. Analyses of non-microbiome data were performed with Prism software version 5.0 (GraphPad) or SPSS version 24 (IBM). Two-tailed Student’s t test, Mann–Whitney U test, or ANOVA with Tukey’s multiple comparison tests were done, as appropriate, for comparisons between human cohorts or amongst mouse cohorts. Data are expressed as a mean ± S.E.M. P-values <0.05 were considered statistically significant. Stepwise linear regression analysis was used to determine the most parsimonious set of predictors effectively predicting the dependent variable (SBP) using SPSS. We used eight markers as independent variables, which were strictly related to gut microbiome and gut barrier functions (details in Supplementary Table S3). The Durbin–Watson statistic was used to ensure the independence of observations. The distance that accounts for correlation between variables was described using Mahalanobis distance. Normality of residuals was examined with a normal probability–probability (P–P) plot.
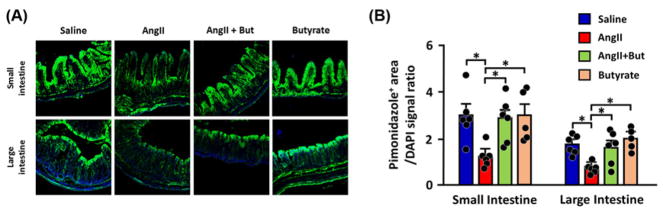
Figure 7. Butyrate treatment restored hypoxia in the gut epithelia of Ang II-infused mice.
(A) Representative images of pimonidazole staining showing hypoxia in the gut barrier; greener the tissue the more it is hypoxic.
(B) Quantitation of pimonidazole-positive areas in small and large intestines. *P<0.05.
Results
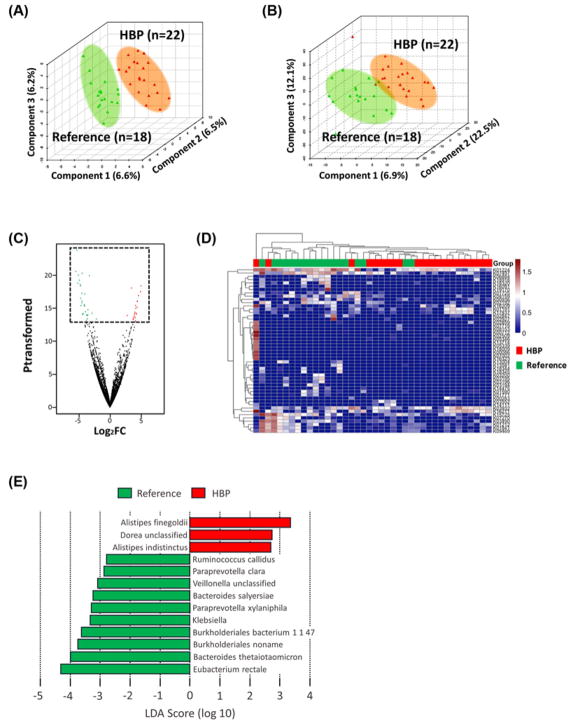
We studied fecal samples from 22 patients with HBP (average ± S.E.M., SBP 155.8 ± 3.4 mmHg) and 18 REF (121.1 ± 1.5 mmHg) for their microbial composition and plasma for biomarkers of gut epithelial permeability. Supplementary Table S1 shows human subjects’ characteristics. PLS-DA plots of overall differences in taxonomy (Figure 1A) and functional genes from KO profiles (Figure 1B) showed defined clustering of samples, indicating differential taxonomic and functional gene profiles between HBP and REF cohorts. Altered functional ortholog changes in the metagenomic analysis and heat map based on the most altered KO abundances of the volcano plot showed clustering of the HBP patients within each cohort confirming functional gene differences (Figure 1C,D). The most enriched bacterial taxa within each cohort from MetaPhlAn taxonomic hits (LDA score >2.0, P<0.05) were shown using LEfSe (Figure 1E). These data indicate that the HBP cohort has a distinct microbiome composition containing different functional genes compared with the REF cohort.
Figure 1. Gut microbiome in patients with HBP is taxonomically and functionally different.
(A) PLS-DA plot of the two cohorts based on bacterial taxonomy (REF, n=18; HBP, n=22). (B) PLS-DA plot based on KO for functional differences. (C) Volcano plot displaying significantly enriched functional genes (KO terms) in REF cohort (green) or HBP (red). The x-axis is log2 fold change and the y-axis is transformed P-value. Genes not different between cohorts are shown in black, A quasi-likelihood F-test transformed P-value threshold of 13 (P<0.05) was utilized to highlight significantly differentiating data points. (D) Significantly enriched KO terms from the volcano plot displayed within a heat map. The top dendrogram, and top row colored red (HBP) and green (REF), shows that the subjects tend to cluster by BP; abundances of the KO terms also cluster, dendrogram on the left. Rows are KO terms, columns individual subjects, and abundance is shown in individual cells on a sliding color scale of blue (low) to high (red). (E) LEfSe identified significantly different bacterial taxa enriched in each cohort at LDA score > 2.5, P<0.05 (red bars HBP enriched, green bars reference enriched). Bars represent bacterial species, except Klebsiella (genus).
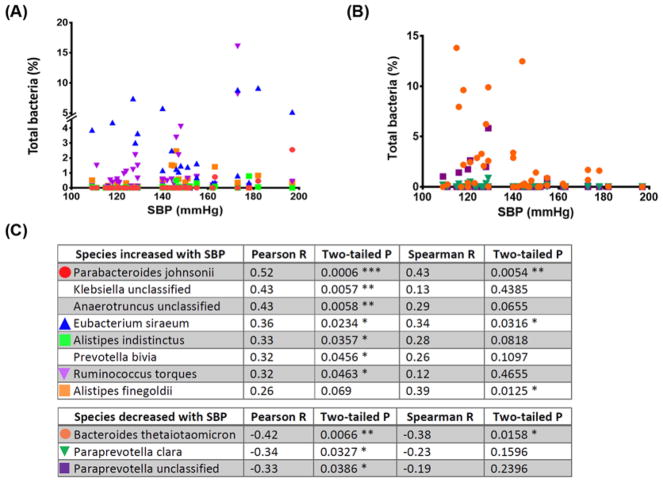
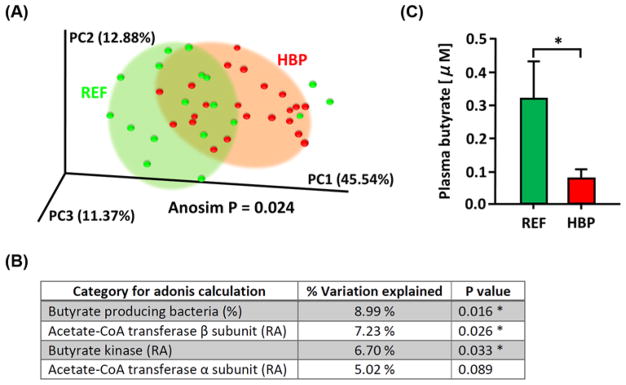
To investigate species that are more closely related to SBP, correlation analyses were done with SBP and bacterial abundance using Pearson correlation coefficients with P<0.05 (except Alistipes finegoldii, Figure 2A,B). This resulted in identification of eight positively and three negatively correlated bacterial species with SBP (Figure 2C). LEfSe of KEGG orthologs combined with bacterial taxa also showed most of the significant functional changes were within these taxa (Supplementary Figure S1 and Table S2). Many of the HBP enriched bacterial taxa were associated with intestinal inflammation and gut barrier dysfunction, while REF abundant (or relatively depleted in HBP) taxa had a huge repertoire of enzymes that degrade complex carbohydrates such as polysaccharides (see ‘Discussion’ section) [34,35]. As microbial fermentation of glycans and polysaccharides stimulates short chain fatty acid production [36] and animal studies have shown a decrease in butyrate-producing bacterial population in HTN [18,37], we examined the existence of such a relationship in HBP patients. First, we determined the level of butyrate generating enzymes (acetate-CoA transferase and butyrate kinase) and abundance of butyrate-producing bacteria (Butyricimonas, Butyricicoccus, Eubacterium, Anaerostipes, Coprococcus, Roseburia, Faecalibacterium, Subdoligranulum, Fusobacterium) in each sample. PCoA plot of these data found that individual samples were grouped significantly into REF and HBP cohorts (Figure 3A,B). These data indicate that higher capacity for bacterial butyrate production is associated with lower SBP. Indeed, Eubacterium rectale, the most significant butyrate-producing bacterium of the human gut [38], was depleted in the HBP cohort (Figure 1E) and levels of other major butyrate-producing genera such as Rosebria and Eubacterium were 49–67% of the REF cohort (data not shown). This was further validated by measurement of plasma butyrate using LC-MS, which was significantly lower in the HBP cohort (Figure 3C).
Figure 2. Bacterial taxa positively and negatively correlated with SBP.
(A) Bacterial species that were significantly positively correlated with SBP were analyzed using the Pearson correlation coefficient (except A. finegoldii). Please note the break in the y-axis. (B) Bacterial species significantly negatively correlated with SBP (Pearson correlation coefficient). (C) Symbols from panels (A,B), and correlation coefficients are explained in the tables. *P<0.05; **P<0.01; ***P<0.001.
Figure 3. Gut microbiome of the HBP cohort has reduced capacity for butyrate production.
(A) PCoA plot was generated based on the abundance of the butyrate-producing bacteria and butyrate generating enzymes (butyrate kinase and acetate-CoA transferase) in each fecal sample. (B) Adonis summary from the PCoA plot was shown. (C) Plasma butyrate measured by GC (n=7 for each cohort, P=0.0175 by Mann–Whitney test). *P<0.05. Abbreviation: RA; relative abundance in fecal bacterial genes.
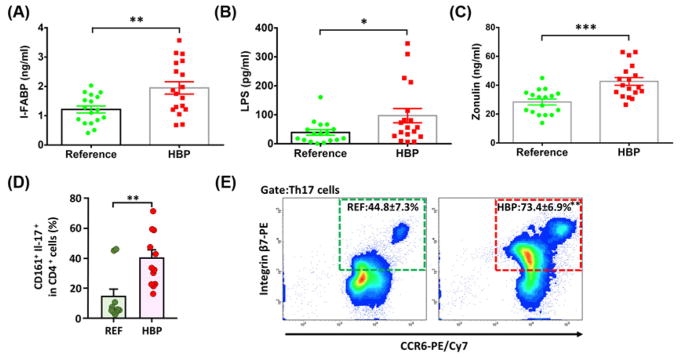
Based on these observations, the association of intestinal inflammation and gut epithelial barrier dysfunction with the HBP cohort was examined, as many studies showed increased incidence of these with butyrate depletion [39]. We measured I-FABP, LPS, and soluble form of zonulin in plasma, biomarkers for gut epithelial health [40]. We observed marked increases in I-FABP (1.22 ± 0.12 ng/ml REF compared with 1.95 ± 0.21 ng/ml HBP, P=0.0052), LPS (39.04 ± 9.52 pg/ml REF compared with 97.11 ± 24.72 pg/ml HBP, P=0.0395), and zonulin (28.40 ± 2.06 ng/ml REF compared with 42.64 ± 2.67 ng/ml HBP, P=0.0002) in the HBP cohort (Figure 4A–C). The increase in zonulin, an important regulator of gut epithelial tight junction proteins, was the most significant (Figure 4C). In addition, we observed a significant increase in Th17 cells expressing CD161 and CCR6/integrin β7 in the HBP cohort, which are markers for gut homing and inflammation (Figure 4D,E) [41,42]. Together, these observations indicated decreased gut barrier function and increased inflammation in patients with HBP.
Figure 4. Increased gut epithelial barrier markers and gut-homing proinflammatory Th17 cells in HBP patients.
(A) Plasma I-FABP. (B) Plasma LPS. (C) Plasma zonulin. n=17 and 18 for REF and HBP cohorts, respectively for panels (A–C). (D) Increased peripheral blood CD161+ Th17 cells, imprinted for gut-homing and inflammation [41], in the HBP cohort. (E) Increased expression of integrin β7 and CCR6, homing receptors for migration of lymphocytes to the intestinal mucosa [41], n=10 and 12 for REF and HBP cohorts, respectively for panels (D,E). *P<0.05; **P<0.01; ***P<0.001 with Student’s t tests.
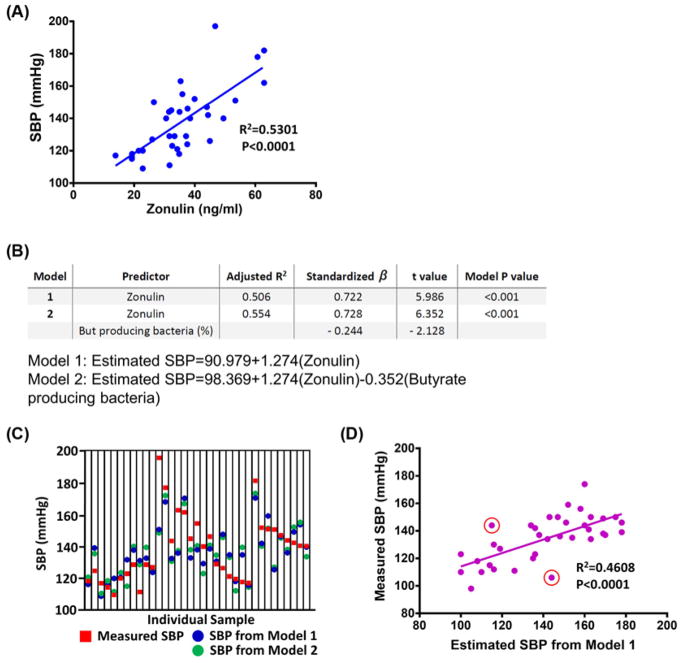
Next, we performed correlation analyses of various gut-derived markers to SBP and found plasma zonulin was most strongly and positively correlated (Figure 5A and Supplementary Table S3). Two stepwise linear regression models were used to determine whether circulating markers for gut health and genes from bacterial shotgun metagenomic analysis could predict SBP (Figure 5B). The first model used plasma zonulin as a single predictor (adjusted R2 = 0.506), and the second model had two predictors, zonulin and abundance of butyrate-producing bacteria (adjusted R2 = 0.554). These results indicate approximately half of SBP variation could be associated with the plasma zonulin level. The inclusion of butyrate-producing bacterial abundance further improved the correlation between observed and predicted SBP values by ~5% (Figure 5B). Comparison of SBP calculated by the two prediction models and measured SBP is summarized in Figure 5C. Model one (zonulin alone) was further validated by prediction of SBP using zonulin in a separate validation cohort (n=36, R2 = 0.4608, Figure 5D). Samples for this validation cohort were previously unexamined, samples from our recruited subjects and samples from a separate clinical trial (ClinicalTrials.gov identifier: NCT02133872), none were included in the metagenomic stool analysis. The validation cohort came from subjects with the complete range of BPs to investigate the clinical applicability of the modeling. Of 36 samples examined, 23 were correctly categorized, 11 were slightly miscategorized (such as REF categorized as Pre-HBP, or Pre-HBP as HBP) and 2 samples were wrongly categorized (one REF predicted as HBP, and one HBP predicted as REF, these are highlighted by red circles in Figure 5D).
Figure 5. Stepwise linear regression analysis to predict subject’s SBP based on gut factors.
(A) The relationship between SBP and plasma zonulin (REF, n=17; HBP, n=18). (B) Stepwise linear regression analysis was performed using SBP as a dependent value and various gut/microbiome factors as independent values to build a model for SBP prediction (details in Supplementary Table S3). (C) A graph showing SBP predicted for each individual using models 1 (blue symbols) and 2 (green symbols) from the stepwise linear regression analyses and actual SBP measured in the clinic (red symbols) for validation. (D) Comparison of clinically measured SBP and estimated SBP from plasma zonulin in a separate validation cohort for model 1 (n=36). SBP from the validation cohort ranged from 98 to 174 mmHg (average: 134 mmHg). Only two samples were incorrectly categorized by the model (REF predicted as HBP and HBP as REF, marked in red circles). See ‘Results’ section for details.
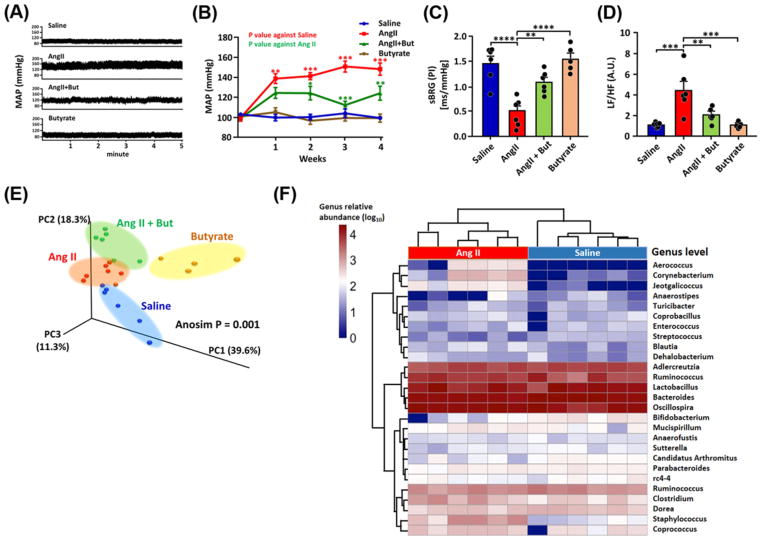
Since butyrate-producing bacteria and plasma butyrate were relatively depleted in HBP patients, we tested the hypothesis that butyrate supplementation would ameliorate HBP in mice chronically infused with Ang II. Butyrate co-administration to C57Bl6 mice infused with Ang II significantly reduced MAP (147.3 ± 6.2 mmHg, Ang II compared with 122.9 ± 7.0 mmHg, Ang II + butyrate at week 4, Figure 6A,B). Power spectral analysis of the BP telemetry data showed decreased spontaneous cardiac baroreceptor reflex gain (sBRG) and increased cardiac sympathetic tone in Ang II-treated mice, that was ameliorated by butyrate co-treatment (Figure 6C,D) confirming the MAP lowering effects of butyrate. Fecal 16S rRNA gene sequencing analysis with unweighted UniFrac PCoA revealed distinct microbiome compositions in each cohort (Figure 6E). Genus level heat maps also demonstrated clear clustering of saline compared with Ang II mice and Ang II compared with Ang II + Butyrate treated mice (Figure 6F and Supplementary Figure S2A). The saline control group was enriched for Bifidobacteriaceae, a well-known beneficial bacterial family (Figure 6F and Supplementary Figure S2) [43]. In contrast, the Ang II group had different enriched taxa, such as Staphylococcaceae, which can impair gut barrier functions [44,45]. These data demonstrate that Ang II-HTN mice have taxonomically distinct microbiota. Furthermore, Akkermansia muciniphila, that reverse diet-induced colonic mucosal barrier dysfunction [46], were enriched in the Ang II + Butyrate group when compared with the Ang II group (Supplementary Figure S2D). Thus, the taxonomic analysis indicates that butyrate-induced microbial changes may result in faster mucus turnover, thus improving gut barrier function and suppressing gut inflammation [46,47].
Figure 6. Butyrate treatment attenuated Ang II-induced HTN and altered gut microbiome in C57BL6 mice.
(A) Representative radiotelemetry tracings of MAP in saline, Ang II, butyrate, and Ang II + Butyrate treated mice; (B) weekly MAP in each cohort. (C,D) Spectral analysis of SBP and pulse interval waveforms of the telemetry data. Cardiac sBRG was attenuated in the Ang II-infused animals but reversed by butyrate co-treatment (C). Increased cardiac sympathetic tone in Ang II mice accompanied this as shown by the low frequency (LF):high frequency (HF) ratio; this was also attenuated by co-treatment with butyrate (D). (E) PCoA plot with weighted UniFrac metric showing distinct microbiome composition in each group. (F) Heat map analysis of the most abundant bacterial genera detected in Saline and Ang II-treated groups showing the distinct separation of the Ang II and saline-treated mice. The heat map colors represent the relative percentage of microbial genus assigned within each sample. Comparisons of other groups were shown in Supplementary Figure S4. Oval shades surrounding the majority of symbols of each group in panel (E) are visual aids. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001, ANOVA with Tukey’s multiple comparison tests. n=6 per cohort except for the butyrate, n=5, throughout all panels. **P<0.01; ***P<0.001; ****P<0.0001. Abbreviation: A.U., arbitrary unit.
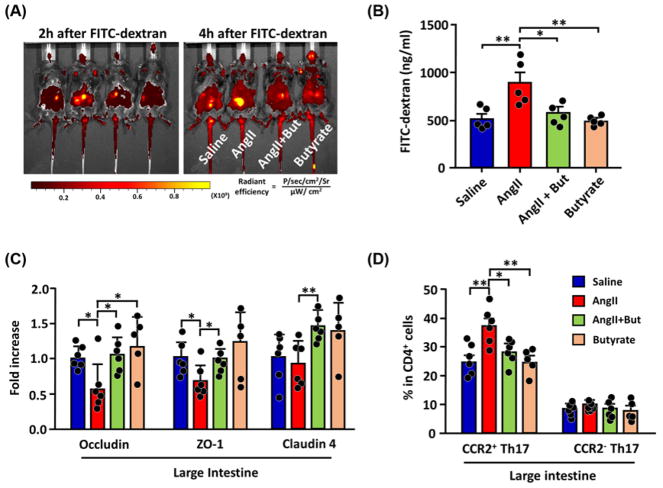
The mammalian digestive tract epithelial cells create a tight barrier in the gut, contributing to the hypoxic environment of the lumen. Damage to this barrier makes the environment less hypoxic, conducive to aerobic bacterial growth [39,48]. Microbial-derived butyrate stabilizes gut epithelial tight junction proteins, depletes oxygen, and activates/stabilizes HIF-1 in epithelial cells [49,50]. Oxygenation state and effects of butyrate in intestines of Ang II-HTN mice were studied using pimonidazole, a marker for hypoxia. Intestines of Ang II-HTN mice were significantly less hypoxic, while butyrate co-treatment restored hypoxia (Figure 7A,B). Also, there were increased aerobic bacteria in Ang II mice feces, again reversed by butyrate co-treatment (Supplementary Figure S2). Damage to gut epithelial barrier function in HTN was visualized in vivo by feeding FITC-dextran (Figure 8A) and finding significantly increased circulating FITC-dextran in plasma (Figure 8B); both were attenuated by butyrate. We observed lowered levels of mRNA for occludin, ZO-1, and claudin 4 by qPCR in Ang II mice intestine. Butyrate co-treatment prevented these alterations (Figure 8C). Increased mRNAs for gut inflammation (Ptgs2) and decreased membrane transporter specific for butyrate (MCT4) were observed in gut epithelial cells of Ang II-HTN mice (Supplementary Figure S3). We characterized laminar propria lymphocytes to confirm gut inflammation in these mice. FACS analysis indicated a marked increase in Th17 cells in HTN that was reversed by butyrate (Figure 8D). Th17 cells have critical roles in chronic inflammation in both animals and human [51–53]. Interestingly, most of the increased Th17 cells in mice intestine were CCR2+ cells known to be more proinflammatory, producing high levels of GM-CSF/IFN-γ and IL-17 [54,55]. These data demonstrate increased gut inflammation in both human and mouse HBP (Figures 4D,E and 8D).
Figure 8. Butyrate treatment normalized gut barrier dysfunction in Ang II-induced HTN mice.
(A) Representative in vivo fluorescent images of the abdomen at 2 and 4 h after oral feeding of FITC-dextran (0.44 mg/g body weight). (B) Serum concentration of FITC-dextran measured the permeability of the intestine. n=5 per mouse cohort. (C) Evaluation of intestinal wall integrity by qPCR of tight junction proteins in gut epithelial cells. (D) FACS analysis of proinflammatory CCR2+ CD4+ Il-17+ Th17 cells in the blood after 4 weeks of treatment. *P<0.05; **P<0.01.
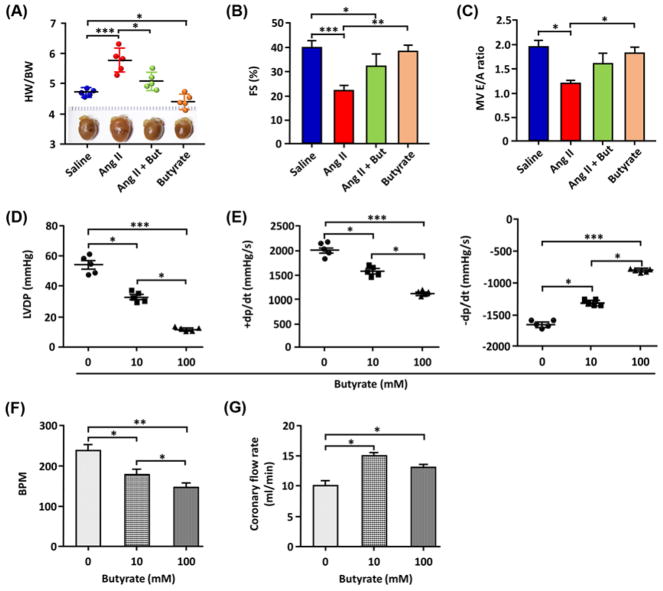
We studied effects of butyrate on HTN-induced cardiac pathophysiology to understand its effects on the heart. HTN-induced cardiac hypertrophy and decreased FS were both reversed (Figure 9A,B). Decreased early (E) to late (A) ventricular filling velocity in Ang II mice was reversed by butyrate (Figure 9C). These beneficial myocardial effects appeared to be direct since butyrate caused dose-dependent decreases in LV diastolic pressure and +dp/dt, and an increase in −dp/dt in isolated heart preparations (Figure 9D–F). Butyrate decreased heart rate and increased coronary flow (Figure 9F,G). Beneficial effects of butyrate were associated with decreased oxidative stress and inflammatory status in blood vessels (Supplementary Figure S4A–C). In vitro data showed that butyrate augmented acetylcholine-induced vasorelaxation of phenylephrine-preconstricted aortic rings (Supplementary Figure S4D,E). Therefore, the animal study supports findings in human subjects that gut microbiota combined with bacterial metabolites such as butyrate link gut leakiness and HBP.
Figure 9. Cardiac functions impaired at 4 weeks of Ang II treatment were restored by butyrate treatment.
Measurements shown in panels (A–C) were made after 4 weeks of Ang II treatment (five mice/group). (A) Cardiac hypertrophy, as the ratio of heart weight (HW) to body weight (BW). (B) Echocardiographic measurements of LV systolic function assessed by FS. (C) LV diastolic function measured using mitral valve E wave /A wave (MV E/A) ratio. (D–G) Measurements shown in panels (D–G) were made in hearts isolated from SD rats using the Langendorff technique (n=5/group). LVDP and rate of LV pressure development (+dp/dt) and decline (–dp/dt) of the SD rats’ hearts were averaged over the duration of perfusion in isovolumetric Langendorff preparation with 0, 10, and 100 mM of sodium butyrate (D,E). Heart rate, beats per min (BPM), (F) and coronary flow (G) following perfusion with 0, 10, or 100 mM sodium butyrate. n=5 for each group, *P<0.05; **P<0.01; ***P<0.001.
Discussion
Evidence has been accumulating in recent years emphasizing the involvement of gut microbiota in HTN [56,57]. Changes in microbiota have been associated with alterations in gut pathology in animal models of HTN [12]. However, little is known about gut microbiota and its relationship with gut barrier function in patients with HBP. In the present study, we present evidence of such relationships in humans. The most significant findings of the present study are: (i) taxonomic and functional changes in gut microbiome with altered butyrate production in patients with HBP, (ii) plasma biomarkers of gut epithelial barrier dysfunction and altered immune status in HBP patients, (iii) depletion of bacterial communities important in carbohydrate metabolism impacting butyrate production in HBP patients, (iv) possible prediction of SBP variations in human by plasma zonulin and gut microbiome composition. These findings demonstrate a novel relationship between gut barrier, gut bacteria functions, and SBP in human.
Using shotgun metagenomic analysis, we investigated both taxa and gene level changes in the human gut microbiome, revealing that HBP is associated with shaping of microbial communities within the microbiome and with changes in functional potential of the microbial consortia (Figure 1). We hypothesized that analyzing the correlation of SBP and taxa abundance would be a better, more precise way to distinguish bacteria that had significant impacts on HBP. We observed that Ruminococcus torques, involved in mucus degradation and gut barrier dysfunction [58], positively correlated with the increase in SBP. Furthermore, both Eubacterium siraeum and A. finegoldii, known to trigger intestinal inflammation [59,60], positively correlated with SBP. Although further validation is needed, this suggests that certain gut bacteria may drive intestinal gut barrier dysfunction and inflammation, which was confirmed by measurement of plasma markers and gut-targetted proinflammatory cells (Figure 4). In contrast, bacterial taxa such as Bacteroides thetaiotaomicron, critical for foraging host mucosal glycan as well as reinforcing mucosal barrier and immune system against pathogens, were inversely correlated with SBP (Figure 2) [34,61–64]. Another bacterium, E. rectale was the most significantly enriched taxa in the REF cohort by LEfSe analysis (Figure 1E). A study by Mahowald et al. [65] showed what might happen in the normotensive gut by using a simplified microbiota model only comprising B. thetaiotaomicron and E. rectale. They demonstrated that B. thetaiotaomicron adapts to the presence of E. rectale by increasing expression of various genes involved in the harvest of host-derived mucin glycans that E. rectale cannot utilize. Exchange of bacterial signals between the two strains caused B. thetaiotaomicron to generate more mucosal glycans in the intestine and E. rectale to decrease production of glycan-degrading enzymes and increase butyrate production, which strengthened the intestinal epithelium barrier [65]. This model and our data showing B. thetaiotaomicron and E. rectale as the most enriched taxa in the REF cohort suggest that reciprocal beneficial effects observed in a healthy bacterial ecosystem are hampered in the HBP cohort [66].
Previous studies indicated decreased butyrate-producing bacteria in HTN animal models [18,67]. This study, for the first time, demonstrates that butyrate-producing bacteria and butyrate-producing enzymes significant clustered and separated human REF and HBP cohorts (ANOSIM, P=0.024). This indicates that the capacity for butyrate production is one of the metabolic characteristics differentiating the two microbiomes. This observation was further supported by the significant decrease in plasma butyrate in humans and amelioration of HBP in the butyrate-supplemented mouse cohort. Butyrate supplementation produced beneficial effects on Ang II-HTN by shifting microbiome populations, influencing cardiac and vascular functions, suggesting potential mechanisms linking butyrate-producing bacteria to SBP in human HTN.
One important finding of our study is the understanding of bacterial ecology and bacterial gene regulation in normal and HTN conditions. Bacteria constantly monitor environmental cues and fine-tune gene regulation in response to change. The most REF enriched (and HBP depleted) KO alterations occurred in B. thetaiotaomicron. The impact of KO changes was broad, with carbohydrate metabolism, transport system, and transcription/translation the most impacted functional pathways, suggesting differences in function as well as abundance (Supplementary Figure S1 and Table S2). B. thetaiotaomicron has an elaborate apparatus for acquiring and hydrolyzing otherwise indigestible dietary polysaccharides [34,61]. The result is fortification of the mucosal barrier, mediated by butyrate-producing bacteria. It is notable that the abundance of glycan-foraging B. thetaiotaomicron was very significantly and negatively associated with SBP (P=0.0066 for B. thetaiotaomicron, Figure 2C). This indicates that abundance of glycan-foraging bacteria such as B. thetaiotaomicron and Paraprevotella or reciprocal beneficial interactions within the microbiome are also important for gut-mediated BP regulation.
HBP-enriched KOs were related to more diverse bacterial strains. The four major taxa were A. finegoldii, Fae-calibacterium prausnitzii, E. siraeum, and R. torques (Supplementary Figure S1 and Table S2). The numbers of all, except butyrate producing F. prausnitzii, were positively associated with SBP (Figure 2A,C). It is of interest that many functions of F. prausnitzii were altered, but the percentage of F. prausnitzii bacteria, in the total microbiome was not different in HBP compared with REF (3.40 ± 0.77% in REF and 3.49 ± 0.60% in HBP, P=0.9249). REF enriched B. thetaiotaomicron and F. prausnitzii are metabolically complementary bacteria. F. prausnitzii attenuates effects of B. thetaiotaomicron on goblet cell differentiation, mucin glycosylation, and oligosaccharide formation, thus diminishing fortification of the mucosal barrier by B. thetaiotaomicron [68]. A significantly reduced number of B. thetaiotaomicron combined with gut pathophysiological changes in the HBP cohort may have triggered these functional alterations in F. prausnitzii, suggesting the importance of interactions amongst gut bacteria and their environment. Overall, these data show that the HBP cohort had depleted B. thetaiotaomicron, increased number and functional genes of A. finegoldii, E. siraeum, and R. torques, as well as functional gene (KO) changes in F. prausnitzii. These findings illustrate the close and complex interactions amongst gut bacteria, intestinal environment, bacterial metabolism, and BP regulation.
Although our human and mouse studies reveal important roles of gut microbiome and pathophysiology in HTN, there are several limitations. First, although plasma zonulin is strongly correlated with SBP, it cannot be used solely to predict HBP as other diseases such as celiac disease or mental disorders are associated with increased zonulin [69,70]. Rather, it may be used as a marker to understand the importance of gut pathology/inflammation to etiology of HTN in an individual. It also suggests which patients might respond to the judicial use of pre- and probiotics. Second, the sample size is limited due to the high cost of whole genome sequencing and sample availability. Study subjects were recruited from a single clinic within a small geographic area and were divided based on SBP alone, without consideration of possible confounding factors such as diet, culture, demographics, or race. Therefore, our data need to be interpreted with caution. Finally, the study does not address whether changes in zonulin and butyrate-producing bacteria could be detected before the onset of HTN.
The present study demonstrates previously unknown correlations between decreased gut epithelial barrier functions, increased inflammation and altered microbiome ecology, and HBP. Gut and its microbiome constantly communicate with the environment that is known to profoundly influence BP. Thus, dysfunctional host–microbiome cross-talk potentially plays a key role in the initiation and establishment of HTN. Our results demonstrate a strong correlation of SBP with gut bacteria and barrier dysfunction, which may provide important clues to the role of microbiome in HTN. We have previously shown that increased gut sympathetic nerve activity and reduced gut tight junction proteins precede development of HTN in spontaneously hypertensive rats [12]. Together, it is tempting to speculate that prohypertensive signals (diet, salt, environment, toxins etc.) increase gut sympathetic activity and the influx of proinflammatory Th17 cells, initiating pathology of the gut leading to increased BP. They provide the rationale for a large-scale clinical trial to validate this concept and to explore therapeutic and biomarker potentials.
Supplementary Material
Clinical perspectives.
Gut is one of the first organs that encounters environmental factors (diet, stress, toxin, exercise, pollutants, smoking etc.) which are all important risk factors for HTN. However, links amongst gut pathology, microbial ecology, and BP remained elusive.
The present study provides evidence for these relationships and demonstrates that gut microbiota and bacterial metabolites, such as butyrate, link gut leakiness, and high BP.
Imbalanced host–microbiome cross-talk may be an important cause of HTN, which is relevant for the development of new therapeutic targets using probiotics or antibiotics.
Acknowledgments
Funding
This work was supported by the National Institute of Health [grant numbers HL33610, HL132448]
Abbreviations
- Ang II
angiotensin II
- BP
blood pressure
- CCR2
chemokine receptor 2
- CPM
counts per million
- HBP
high BP cohort
- HTN
hypertension
- I-FABP
intestinal fatty acid binding protein
- IL-17
Interleukin 17
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KO
KEGG orthology
- KRS
Krebs–Ringer bicarbonate solution
- LDA
linear discriminant analysis
- LEfSe
LDA effects size
- LPS
lipopolysaccharide
- LV
left ventricle
- MAP
mean arterial pressure
- MCT
monocarboxylate transporter
- OTU
operational taxonomic unit
- PBMC
peripheral blood mononuclear cell
- PCoA
principal co-ordinate analysis
- PLS-DA
partial least squares discriminant analysis
- Ptgs
prostaglandin-endoperoxide synthase
- REF
reference cohort
- SBP
systolic BP
- SCFA
short chain fatty acid
- SD
Sprague–Dawley
- Th17
T helper 17
- ZO-1
zonula occludens-1.
Footnotes
Author contribution
S.K., R.G., A.K., Y.Q., G.L., and K.H. performed the experiments. S.K., M.M., E.M.H., and C.J.P. recruited and evaluated human study subjects. S.K., E.M.R., C.J.P., and M.K.R. wrote the manuscript and analyzed the data. C.J.P. and M.K.R. conceived and supervised the work.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, et al. Heart Disease and Stroke Statistics-2017 update: a report from the American Heart Association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. https://doi.org/10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okin PM, Kjeldsen SE, Devereux RB. The relationship of all-cause mortality to average on-treatment systolic blood pressure is significantly related to baseline systolic blood pressure: implications for interpretation of the Systolic Blood Pressure Intervention Trial study. J Hypertens. 2017 doi: 10.1097/HJH.0000000000001620. https://doi.org/10.1097/HJH.0000000000001620. [DOI] [PubMed]
- 3.Materson BJ, Reda DJ, Cushman WC, Massie BM, Freis ED, Kochar MS, et al. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo The Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. N Engl J Med. 1993;328:914–921. doi: 10.1056/NEJM199304013281303. https://doi.org/10.1056/NEJM199304013281303. [DOI] [PubMed] [Google Scholar]
- 4.Page IH. The mosaic theory of arterial hypertension–its interpretation. Perspect Biol Med. 1967;10:325–333. doi: 10.1353/pbm.1967.0031. https://doi.org/10.1353/pbm.1967.0031. [DOI] [PubMed] [Google Scholar]
- 5.Raizada MK, Joe B, Bryan NS, Chang EB, Dewhirst FE, Borisy GG, et al. Report of the National Heart, Lung, and Blood Institute Working Group on the role of microbiota in blood pressure regulation: current status and future directions. Hypertension. 2017 doi: 10.1161/HYPERTENSIONAHA.117.09699. https://doi.org/10.1161/HYPERTENSIONAHA.117.09699. [DOI] [PMC free article] [PubMed]
- 6.Furness JB. Integrated neural and endocrine control of gastrointestinal function. Adv Exp Med Biol. 2016;891:159–173. doi: 10.1007/978-3-319-27592-5_16. https://doi.org/10.1007/978-3-319-27592-516. [DOI] [PubMed] [Google Scholar]
- 7.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. https://doi.org/10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato A, Romero MF. Regulation of electroneutral NaCl absorption by the small intestine. Annu Rev Physiol. 2011;73:261–281. doi: 10.1146/annurev-physiol-012110-142244. https://doi.org/10.1146/annurev-physiol-012110-142244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linz D, Wirth K, Linz W, Heuer HO, Frick W, Hofmeister A, et al. Antihypertensive and laxative effects by pharmacological inhibition of sodium-proton-exchanger subtype 3-mediated sodium absorption in the gut. Hypertension. 2012;60:1560–1567. doi: 10.1161/HYPERTENSIONAHA.112.201590. https://doi.org/10.1161/HYPERTENSIONAHA.112.201590. [DOI] [PubMed] [Google Scholar]
- 10.Madero M, Perez-Pozo SE, Jalal D, Johnson RJ, Sanchez-Lozada LG. Dietary fructose and hypertension. Curr Hypertens Rep. 2011;13:29–35. doi: 10.1007/s11906-010-0163-x. https://doi.org/10.1007/s11906-010-0163-x. [DOI] [PubMed] [Google Scholar]
- 11.Boulange CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8:42. doi: 10.1186/s13073-016-0303-2. https://doi.org/10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santisteban MM, Qi Y, Zubcevic J, Kim S, Yang T, Shenoy V, et al. Hypertension-linked pathophysiological alterations in the gut. Circ Res. 2017;120:312–323. doi: 10.1161/CIRCRESAHA.116.309006. https://doi.org/10.1161/CIRCRESAHA.116.309006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karbach SH, Schonfelder T, Brandao I, Wilms E, Hormann N, Jackel S, et al. Gut microbiota promote angiotensin II-induced arterial hypertension and vascular dysfunction. J Am Heart Assoc. 2016:5. doi: 10.1161/JAHA.116.003698. https://doi.org/10.1161/JAHA.116.003698. [DOI] [PMC free article] [PubMed]
- 14.Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14. doi: 10.1186/s40168-016-0222-x. https://doi.org/10.1186/s40168-016-0222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. https://doi.org/10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo W, Pant G, Bhavnasi YK, Blanchard SG, Jr, Brouwer C. Pathview Web: user friendly pathway visualization and data integration. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx372. https://doi.org/10.1093/nar/gkx372. [DOI] [PMC free article] [PubMed]
- 17.Ito K, Murphy D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst Pharmacol. 2013;2:e79. doi: 10.1038/psp.2013.56. https://doi.org/10.1038/psp.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, et al. Gut dysbiosis is linked to hypertension. Hypertension. 2015;65:1331–1340. doi: 10.1161/HYPERTENSIONAHA.115.05315. https://doi.org/10.1161/HYPERTENSIONAHA.115.05315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol. 2013;51:2884–2892. doi: 10.1128/JCM.00845-13. https://doi.org/10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5:e00889. doi: 10.1128/mBio.00889-14. https://doi.org/10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. https://doi.org/10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. https://doi.org/10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. https://doi.org/10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. https://doi.org/10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. https://doi.org/10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costea PI, Zeller G, Sunagawa S, Bork P. A fair comparison. Nat Methods. 2014;11:359. doi: 10.1038/nmeth.2897. https://doi.org/10.1038/nmeth.2897. [DOI] [PubMed] [Google Scholar]
- 27.Szalai G, Romero R, Chaiworapongsa T, Xu Y, Wang B, Ahn H, et al. Full-length human placental sFlt-1-e15a isoform induces distinct maternal phenotypes of preeclampsia in mice. PLoS ONE. 2015;10:e0119547. doi: 10.1371/journal.pone.0119547. https://doi.org/10.1371/journal.pone.0119547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waki H, Katahira K, Polson JW, Kasparov S, Murphy D, Paton JF. Automation of analysis of cardiovascular autonomic function from chronic measurements of arterial pressure in conscious rats. Exp Physiol. 2006;91:201–213. doi: 10.1113/expphysiol.2005.031716. https://doi.org/10.1113/expphysiol.2005.031716. [DOI] [PubMed] [Google Scholar]
- 29.Zubcevic J, Waki H, Diez-Freire C, Gampel A, Raizada MK, Paton JF. Chronic blockade of phosphatidylinositol 3-kinase in the nucleus tractus solitarii is prohypertensive in the spontaneously hypertensive rat. Hypertension. 2009;53:97–103. doi: 10.1161/HYPERTENSIONAHA.108.122341. https://doi.org/10.1161/HYPERTENSIONAHA.108.122341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci USA. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. https://doi.org/10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Du J, Zhao YT, Zhang L, Lv G, Zhuang S, et al. Histone deacetylase (HDAC) inhibition improves myocardial function and prevents cardiac remodeling in diabetic mice. Cardiovasc Diabetol. 2015;14:99. doi: 10.1186/s12933-015-0262-8. https://doi.org/10.1186/s12933-015-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006;113:51–59. doi: 10.1161/CIRCULATIONAHA.105.559724. https://doi.org/10.1161/CIRCULATIONAHA.105.559724. [DOI] [PubMed] [Google Scholar]
- 33.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. https://doi.org/10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. https://doi.org/10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 35.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. https://doi.org/10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. https://doi.org/10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durgan DJ, Ganesh BP, Cope JL, Ajami NJ, Phillips SC, Petrosino JF, et al. Role of the gut microbiome in obstructive sleep apnea-induced hypertension. Hypertension. 2016;67:469–474. doi: 10.1161/HYPERTENSIONAHA.115.06672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. https://doi.org/10.1128/AEM.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konig J, Wells J, Cani PD, Garcia-Rodenas CL, MacDonald T, Mercenier A, et al. Human intestinal barrier function in health and disease. Clin Transl Gastroenterol. 2016;7:e196. doi: 10.1038/ctg.2016.54. https://doi.org/10.1038/ctg.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fasano A. Zonulin, regulation of tight junctions, and autoimmune diseases. Ann NY Acad Sci. 2012;1258:25–33. doi: 10.1111/j.1749-6632.2012.06538.x. https://doi.org/10.1111/j.1749-6632.2012.06538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleinschek MA, Boniface K, Sadekova S, Grein J, Murphy EE, Turner SP, et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J Exp Med. 2009;206:525–534. doi: 10.1084/jem.20081712. https://doi.org/10.1084/jem.20081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cosmi L, De Palma R, Santarlasci V, Maggi L, Capone M, Frosali F, et al. Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor. J Exp Med. 2008;205:1903–1916. doi: 10.1084/jem.20080397. https://doi.org/10.1084/jem.20080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kechagia M, Basoulis D, Konstantopoulou S, Dimitriadi D, Gyftopoulou K, Skarmoutsou N, et al. Health benefits of probiotics: a review. ISRN Nutr. 2013;2013:48. doi: 10.5402/2013/481651. https://doi.org/10.5402/2013/481651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwak YK, Vikstrom E, Magnusson KE, Vecsey-Semjen B, Colque-Navarro P, Mollby R. The Staphylococcus aureus alpha-toxin perturbs the barrier function in Caco-2 epithelial cell monolayers by altering junctional integrity. Infect Immun. 2012;80:1670–1680. doi: 10.1128/IAI.00001-12. https://doi.org/10.1128/IAI.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hocke AC, Temmesfeld-Wollbrueck B, Schmeck B, Berger K, Frisch EM, Witzenrath M, et al. Perturbation of endothelial junction proteins by Staphylococcus aureus alpha-toxin: inhibition of endothelial gap formation by adrenomedullin. Histochem Cell Biol. 2006;126:305–316. doi: 10.1007/s00418-006-0174-5. https://doi.org/10.1007/s00418-006-0174-5. [DOI] [PubMed] [Google Scholar]
- 46.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. https://doi.org/10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneeberger M, Everard A, Gomez-Valades AG, Matamoros S, Ramirez S, Delzenne NM, et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. 2015;5:16643. doi: 10.1038/srep16643. https://doi.org/10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Earley ZM, Akhtar S, Green SJ, Naqib A, Khan O, Cannon AR, et al. Burn injury alters the intestinal microbiome and increases gut permeability and bacterial translocation. PLoS ONE. 2015;10:e0129996. doi: 10.1371/journal.pone.0129996. https://doi.org/10.1371/journal.pone.0129996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. https://doi.org/10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Correa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol. 2016;5:e73. doi: 10.1038/cti.2016.17. https://doi.org/10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leppkes M, Becker C, Ivanov II, Hirth S, Wirtz S, Neufert C, et al. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–267. doi: 10.1053/j.gastro.2008.10.018. https://doi.org/10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 52.Itani HA, McMaster WG, Jr, Saleh MA, Nazarewicz RR, Mikolajczyk TP, Kaszuba AM, et al. Activation of human T cells in hypertension: studies of humanized mice and hypertensive humans. Hypertension. 2016;68:123–132. doi: 10.1161/HYPERTENSIONAHA.116.07237. https://doi.org/10.1161/HYPERTENSIONAHA.116.07237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ji Q, Cheng G, Ma N, Huang Y, Lin Y, Zhou Q, et al. Circulating Th1, Th2, and Th17 levels in hypertensive patients. Dis Markers. 2017;2017:7146290. doi: 10.1155/2017/7146290. https://doi.org/10.1155/2017/7146290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kara EE, McKenzie DR, Bastow CR, Gregor CE, Fenix KA, Ogunniyi AD, et al. CCR2 defines in vivo development and homing of IL-23-driven GM-CSF-producing Th17 cells. Nat Commun. 2015;6:8644. doi: 10.1038/ncomms9644. https://doi.org/10.1038/ncomms9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato W, Aranami T, Yamamura T. Cutting edge: human Th17 cells are identified as bearing CCR2+CCR5− phenotype. J Immunol. 2007;178:7525–7529. doi: 10.4049/jimmunol.178.12.7525. https://doi.org/10.4049/jimmunol.178.12.7525. [DOI] [PubMed] [Google Scholar]
- 56.Richards EM, Pepine CJ, Raizada MK, Kim S. The gut, its microbiome, and hypertension. Curr Hypertens Rep. 2017;19:36. doi: 10.1007/s11906-017-0734-1. https://doi.org/10.1007/s11906-017-0734-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qi Y, Kim S, Richards EM, Raizada MK, Pepine CJ. Gut microbiota: potential for a unifying hypothesis for prevention and treatment of hypertension. Circ Res. 2017;120:1724–1726. doi: 10.1161/CIRCRESAHA.117.310734. https://doi.org/10.1161/CIRCRESAHA.117.310734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. https://doi.org/10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 59.Moschen AR, Gerner RR, Wang J, Klepsch V, Adolph TE, Reider SJ, et al. Lipocalin 2 protects from inflammation and tumorigenesis associated with gut microbiota alterations. Cell Host Microbe. 2016;19:455–469. doi: 10.1016/j.chom.2016.03.007. https://doi.org/10.1016/j.chom.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 60.Yassour M, Lim MY, Yun HS, Tickle TL, Sung J, Song YM, et al. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 2016;8:17. doi: 10.1186/s13073-016-0271-6. https://doi.org/10.1186/s13073-016-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. https://doi.org/10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 62.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. https://doi.org/10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 63.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. https://doi.org/10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. https://doi.org/10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 65.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. https://doi.org/10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maier E, Anderson RC, Roy NC. Understanding how commensal obligate anaerobic bacteria regulate immune functions in the large intestine. Nutrients. 2015;7:45–73. doi: 10.3390/nu7010045. https://doi.org/10.3390/nu7010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L, Zhu Q, Lu A, Liu X, Zhang L, Xu C, et al. Sodium butyrate suppresses angiotensin II-induced hypertension by inhibition of renal (pro)renin receptor and intrarenal renin-angiotensin system. J Hypertens. 2017;35:1899–1908. doi: 10.1097/HJH.0000000000001378. https://doi.org/10.1097/HJH.0000000000001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013;11:61. doi: 10.1186/1741-7007-11-61. https://doi.org/10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fasano A, Not T, Wang W, Uzzau S, Berti I, Tommasini A, et al. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet. 2000;355:1518–1519. doi: 10.1016/S0140-6736(00)02169-3. https://doi.org/10.1016/S0140-6736(00)02169-3. [DOI] [PubMed] [Google Scholar]
- 70.Stevens BR, Goel R, Seungbum K, Richards EM, Holbert RC, Pepine CJ, et al. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety or depression. Gut. 2017 doi: 10.1136/gutjnl-2017-314759. https://doi.org/10.1136/gutjnl-2017-314759. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.