Abstract
Reversible immobilization of DNA and RNA is of great interest to researchers who seek to manipulate DNA or RNA in applications such as microarrays, DNA hydrogels and gene therapeutics. However, there is no existing system that can rapidly capture and release intact nucleic acids. To meet this unmet need, we develop a functional hydrogel for rapid DNA/RNA capture and release based on the reversible photo-cycloaddition of psoralen and pyrimidines. The functional hydrogel can be easily fabricated through copolymerization of acrylamide with the synthesized allylated psoralen. The psoralen-functionalized hydrogel exhibits effective capture and release of nucleic acids spanning a wide length range in a rapid fashion: > 90 % of the capture process is completed within 1 min; ~100 % of the release process is completed within 2 min. We observe no deleterious effects on the hybridization to the captured targets.
Keywords: functional hydrogel, oligonucleotide, photo-cycloaddition, psoralen, reversible immobilization
Reversible capture scaffold for DNA and RNA
Polyacrylamide gel is modified with psoralen through copolymerization of acrylamide with allylated psoralen to form an efficient DNA/RNA capture scaffold. The reversible photo-cycloaddition of psoralen and pyramidine allows DNA/RNA capture in 1 min and DNA/RNA release in 2 min under UV irradiation of distinct wavelengths. The capture and release applies to a wide length range of DNA/RNA and is compatible with hybridization.
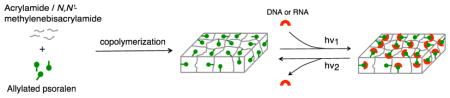
Beyond encoding genetic information, nucleic acids are now widely exploited as functional materials. Oligonucleotide probes capture complementary strands, or detect proteins and other biomolecules in the form of aptamers or ribozymes.[1] In chemical engineering, sequence-specific nucleic acids are being explored as building blocks for fabrication of functional hydrogels.[2] In synthetic biology, RNA ribozymes and DNA toe-holds constitute the basis of genetic circuits.[3] Most of these applications require nucleic acids to be robustly immobilized, and for applications where oligonucleotide probes or nucleic acid targets are to be reused, reversible immobilization is often desired.
Covalent bonds between an amine or sulfhydryl groups and their reactive groups (and some strong non-covalent bonds such as biotin-streptavidin binding) are often harnessed for immobilization of nucleic acids.[1e–1g, 4] But the linking chemistry requires chemical modifications to the nucleic acids prior to immobilization, which is inconvenient or even inapplicable to some experiments where the target nucleic acids are in small quantity,[5] or are prone to degradation or contamination.[6] Moreover, most of these immobilization assays are irreversible under normal conditions,[7] thus not suited for applications where the nucleic acids are to be recovered for subsequent analysis or efficiently delivered as gene therapeutics.[8] Recent studies on the azobenzene-based photoisomerization provide an alternative strategy for nucleic acid capture.[9] Because the capture of the nucleic acid is realized through electrostatic interaction with the photo-responsive complex, the immobilization is reversible.[10] But the capture/release process usually requires 10 to 20 min to complete, which is irrelevant for high-speed operations that take place on the order of seconds to minutes, such as microfluidic reactions and fast biological processes.[3a] Moreover, the release of high-molecular-mass targets is not effective in such systems, presumably due to the increased multivalency and the increased kinetic stability for those targets.[10b] The large size and the non-neutral charge of the supramolecular complex also limit the compatibility of this system with the gel electrophoresis which is widely used in nucleic-acid research. As such, a novel system is needed to realize rapid capture and release of nucleic acids.
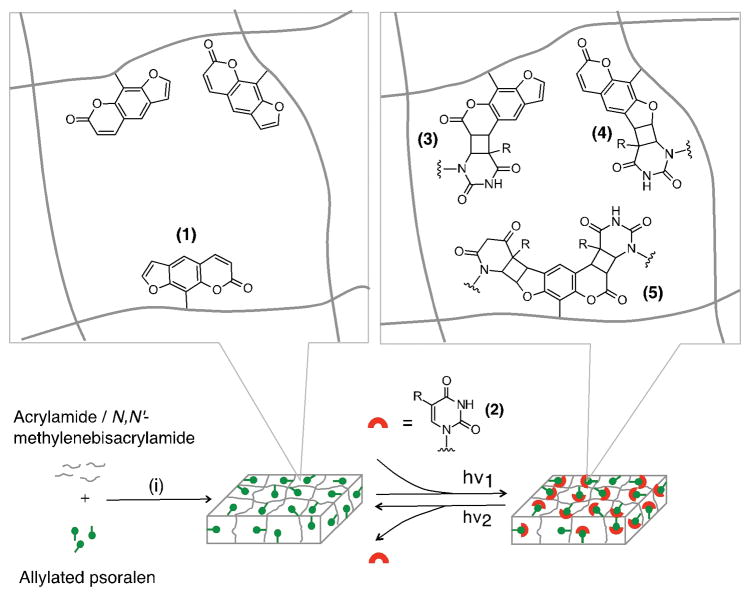
Herein we report a functional hydrogel to rapidly capture and release DNA or RNA targets based on the reversible photo-cycloaddition reaction of psoralen and pyrimidines (Figure 1). The psoralen-functionalized polyacrylamide gel was prepared through copolymerization of allylated psoralen, synthesized via an NHS ester-amine reaction, with acrylamide. The psoralen-functionalized polyacrylamide gel exhibits effective photo-responsive capture and subsequent release of DNA or RNA spanning a wide length range (80 to 500 bp) in a rapid fashion: > 90 % of the capture reactions can be completed within 1 min; ~100 % of the release reactions can be completed within 2 min. We observe no deleterious effects on the hybridization to the captured targets.
Figure 1.
Schematic illustration of the capture and release of DNA or RNA via a psoralen-modified hydrogel. Conditions: (i) photopolymerization: 2,2-azobis[2-methyl-N-(2-hydroxyethyl) propionamide] (0.2 %(w/v)), UV irradiation (> 390 nm). (1) psoralen. (2) DNA or RNA: R = H for RNA; R = CH3 for DNA. (3) pyrone-side monoadduct. (4) furan-side monoadduct. (5) diadduct. hν1: > 350 nm; hν2: < 325 nm.
This study assesses psoralen as a means to reversibly capture oligonucleotides. Psoralens are a class of planar, three-ring heterocyclic compounds that can form covalent bonds via cycloaddition reactions with pyrimidine bases of the nucleic acids (primarily thymine or uracil) upon irradiation by long-wavelength UV (320 – 410 nm).[11] The photo-cycloaddition of a psoralen molecule to the pyrimidine base can occur at the furan side (the 4′, 5′ double bond) or the pyrone side (the 3, 4 double bond) of the psoralen, resulting in three possible products (furan-side monoadduct, pyrone-side monoadduct, and diadduct).[12] Under irradiation by short-wavelength UV (240 – 313 nm), the photo-cycloaddition proceeds in the reverse direction, leading to the dissociation of the adducts.[13] This reversible, photo-activated reaction offers an ideal platform for nucleic acid capture. We selected polyacrylamide gels as scaffolds due to the wide range of applications for the material ranging from gel electrophoresis to gene therapy.[14] However, the chemical inertness of the polyacrylamide gel creates a barrier to chemically incorporating psoralen functional groups to the gel matrix.[15]
To chemically incorporate psoralen functional groups into the gel matrix, we utilize allylated psoralen. The allylated psoralen is copolymerized with the polyacrylamide gel.[14c,16] Allylated psoralen was synthesized using an NHS ester-amine reaction between the NHS ester-conjugated psoralen derivative, succinimidyl-[4-(psoralen-8-yloxy)]butyrate (SPB), and allylamine (Supporting Information, Scheme S1). The reaction proceeded in DMSO for 12 h at room temperature. The resulting allylated psoralen, N-allyl-4-((7-oxo-7H-furo[3,2-g]chromen-9-yl)oxy)butanamide, was purified and characterized by 1D 1H NMR, 2D 1H-13C HSQC (heteronuclear single quantum correlation) NMR and mass spectrometry (Supporting Information, Figure S1, S2). The synthesized allylated psoralen was subsequently added as the comonomer to the polyacrylamide gel precursor containing acrylamide and N,N′-methylenebisacrylamide to produce the psoralen-modified polyacrylamide gel (Experimental details in the Supporting Information).
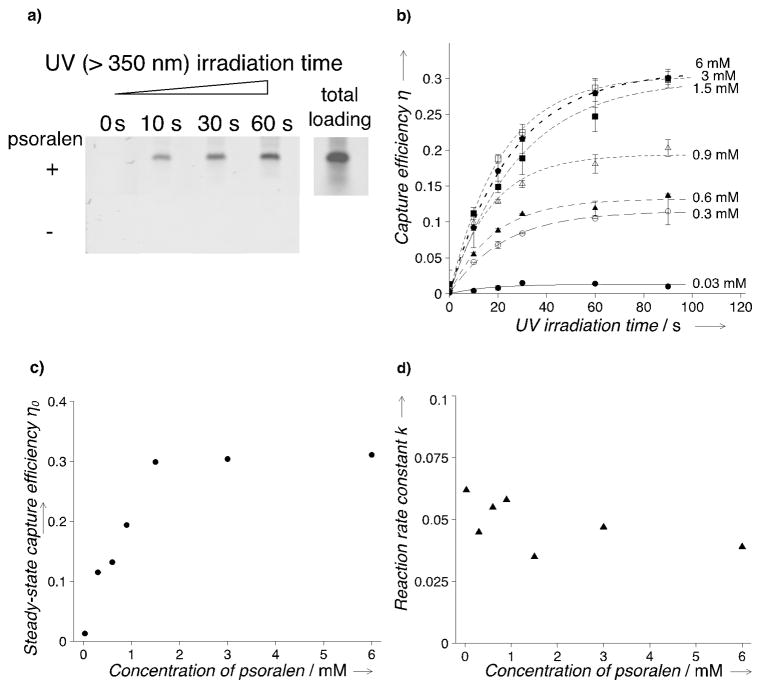
To examine the oligonucleotide-capture performance of the system, a target DNA molecule, 100-bp double-stranded DNA (dsDNA) (sequence information in the Supporting Information, Table S1), was introduced into the gel through electrophoresis and irradiated with long-wavelength (> 350 nm) UV light (Supporting Information, Figure S3). The uncaptured targets were separated from the captured targets through a post-capture electrophoretic wash. A non-specific fluorescent stain, SYBRTM Gold, was utilized to visualize the target dsDNA (Figure 2a). The capture efficiency (η) is characterized by the ratio of the fluorescence intensity before and after the electrophoretic wash. We observe that η increases with the irradiation time in a logistic pattern and reaches distinct plateaus for different psoralen concentrations in the gel (Figure 2b). Given the fact that no intermediate products were reported for the reaction of psoralen and nucleobase,[11, 12] the steady-state approximation can be applied in the fitting to the kinetics data. Thus, the relationship of η and the UV irradiation time (t) can be described as: η = η0(1 − e−kt), where η0 is the maximum capture efficiency with the increase of the irradiation duration, and k is related to the reaction rate (Fitting results in the Supporting Information, Figure S4). For ease of discussion, we refer to η0 as the steady-state capture efficiency. The steady-state capture efficiency exhibits a strong positive correlation with the concentration of the psoralen (Spearman’s ρ = 1.0000, p < 0.001): η0 increases monotonically with the increase of the psoralen concentration before leveling off at the saturation concentration of about 1.5 mM (Figure 2c). We attribute the observed psoralen-concentration dependence of the steady-state capture efficiency to the combinatorial result of changes in the number of nucleobase-reacting sites and pore structure of the gel as the molar ratio of acrylamide and allylated psoralen changes. We hypothesize that when psoralen concentration is below 1.5 mM, acrylamide prevails in the psoralen-modified polyacrylamide gel as the dominant monomer. Thus, increasing psoralen concentrations in gel precursor leads to an increase of the nucleobase-reacting sites in the polyacrylamide gel, corresponding to the enhanced capture capacity. When psoralen concentration is above 1.5 mM, the addition of allylated psoralen starts to disrupt the structure of the gel, resulting in a reduction in pore number. The structure change induced by comonomers in polymerization was also reported for the copolymer of acrylamide with sodium allylsulfonate and sodium acrylate.[17] The decrease of the pore number counteracts the increase of the psoralen density, and hence maintains the saturated level of the nucleobase-reacting sites in the polyacrylamide gel, corresponding to the saturation of the capture capacity at higher psoralen concentrations. The observed reaction rate constant k does not reveal a similarly strong correlation (Spearman’s ρ = −0.6071, p = 0.1667) to the psoralen concentration (Figure 2d), suggesting that the psoralen concentration does not critically affect the capture kinetics of the system.
Figure 2.
Photo-capture performance of the psoralen-modified polyacrylamide gel for 100-bp double-stranded DNA (dsDNA). a) Representative fluorescence images indicating the capture capacity of the psoralen-modified gel. DNA targets were visualized using SYBRTM Gold stain after photo-capture. The fluorescence images at indicated UV irradiation durations were taken after electrophoretic wash. Psoralen concentration in the ‘+’ experiments is 3 mM. Target loading: 40 ng/well. Stain: 2x SYBRTM Gold. Ex: 488 nm. Em: 520 nm BP 40 nm. Experimental conditions were the same for all the following experiments unless otherwise specified. b) Capture efficiency (η) measured at various UV irradiation durations for different psoralen concentrations. n = 3 for each data point; error bars: standard deviation. c) The steady-state capture efficiency (η0) versus the psoralen concentration. d) The reaction rate constant (k) versus the psoralen concentration.
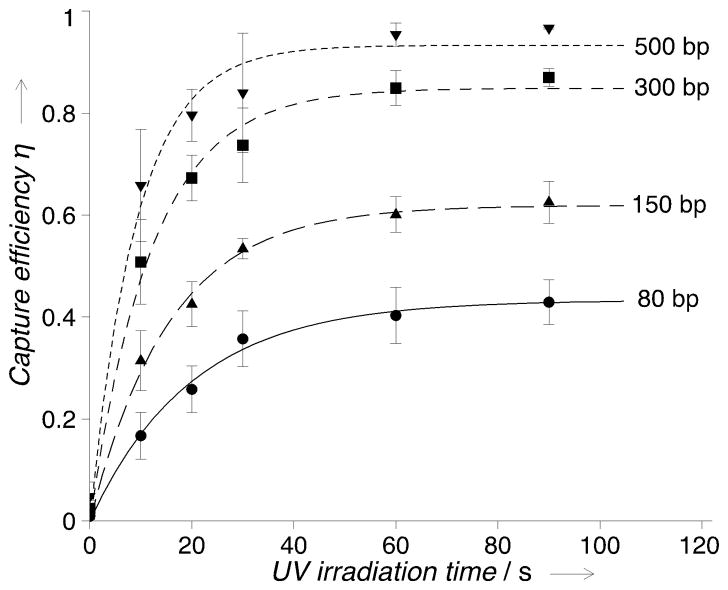
To investigate the applicable range of the capture system, we expanded the target pool by incorporating double-stranded RNA (dsRNA) of varied lengths (80 to 500 bp) to the capture experiments. All targets were captured (Figure 3), and the capture kinetics follows the same exponential pattern η = η0(1 − e−kt) regardless of the length and the sequence (Supporting Information, Figure S5). We observe that the time required to achieve the maximum capture efficiency remains ~90 s with all the DNA and RNA targets when psoralen concentration is 3 mM. This time scale is probably related to the reaction kinetics of photo-cycloaddition of psoralen and pyrimidines. We further observe that the capture performance depends on the target. Despite similar uridine percentages across the targets, longer dsRNA exhibit higher maximum capture efficiency: η0 is 0.433, 0.619, 0.849 and 0.933 for dsRNA of 80, 150, 300 and 500 bp. We attribute this length-dependence of η0 to steric effect, where longer targets have a higher chance of proximally accessing psoralen with pyrimidines. We also performed capture experiments with single-stranded DNA and single-stranded RNA. The results exhibit similar capture kinetics and lower capture efficiencies for single-stranded targets (Supporting Information, Figure S6, Table S2). We attribute the lower capture efficiency for single-stranded targets to a lack of intercalation. Taken together, the psoralen-modified polyacrylamide gels exhibit successful capture for DNA and RNA of a wide length range in both duplex and single-stranded form. Moreover, we observe no deleterious effects of the UV-induced photo-capture (< 90 s, > 350 nm of irradiation) on hybridization to the captured oligonucleotides (Supporting Information, Figure S7, S8), in agreement with studies on other psoralen derivatives where furan-side monoadducts were included in synthesized oligonucleotides to facilitate hybridization or to study kinetics of hybridization reactions.[12b, 18]
Figure 3.
Photo-capture performance of the psoralen-modified polyacrylamide gel for double-stranded RNA (dsRNA) with varied lengths. Psoralen concentration: 3 mM. The connecting curves are the exponential fitting to the experimental data (fitting results in Supporting Information). n = 3 for each data point; error bars: standard deviation.
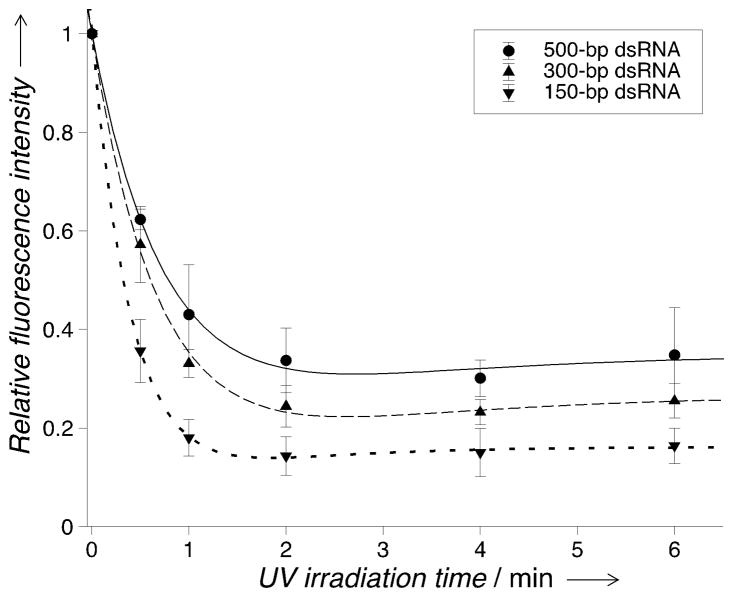
After assessing the oligonucleotide-capture characteristics of psoralen-modified polyacrylamide gels, we next sought to assess the photo-release behavior of the captured targets. We evaluated release of 150-, 300- and 500-bp dsRNA. To release the captured dsRNA, the gels were irradiated by short-wavelength UV light (< 325 nm) for various time intervals, and visualized using SYBRTM Gold stain (Supporting Information, Figure S9, S10). Electrophoretic wash was implemented following UV irradiation to remove the released dsRNA. To characterize the photo-release performance, we calculated the relative fluorescence intensity of the captured dsRNA measured before and after the < 325 nm UV irradiation. The relative fluorescence intensity decreases upon the < 325 nm irradiation and reaches the minimum after ~2 min for all the dsRNA targets (Figure 4; Supporting Information, Table S3). The maximum target-release level, characterized by the relative released fluorescence intensity, is about 0.663 (n = 4, SD = 0.066) for 500-bp dsRNA, 0.756 (n = 4, SD = 0.042) for 300-bp dsRNA, and 0.857 (n = 4, SD = 0.039) for 150-bp dsRNA. We notice that the unreleased dsRNA level increases with the dsRNA length, in accordance with the relationship of the capture efficiency and target length for dsRNA (Figure 3). We hypothesize that in addition to the dissociation of psoralen-pyrimidine adducts at short-wavelength UV (< 325 nm), there may exist concurrent reactions responsible for the capture of released targets. In fact, we observed a low level of dsRNA capture (η = ~20 %) in non-modified polyacrylamide gels under irradiation of the full-spectrum UV (containing irradiation wavelengths spanning from 280 to 410 nm, Supporting Information, Table S4), but detected no capture under irradiation of > 350 nm UV (Figure S11), suggesting possible crosslinking of polyacrylamide gel and nucleic acids under short-wavelength UV irradiation (see further discussions in Supporting Information).
Figure 4.
Photo-release of the captured dsRNA (150 to 500 bp) measured at varied UV (< 325 nm) irradiation time. The connecting curves are the fitting to the experimental data (fitting model and results in Supporting Information). Psoralen concentration: 3 mM. Photo-capture condition: 1 min irradiation by long-wavelength UV (> 350 nm). n = 4 for each data point. Error bars: standard deviation.
In summary, we designed a psoralen-functionalized polyacrylamide hydrogel that can capture nucleic acids in a rapid and reversible fashion. The psoralen-modified polyacrylamide gel can be easily fabricated through copolymerization of acrylamide with the synthesized allylated psoralen. The UV-activated cycloaddition of psoralen and nucleobases ensures the robust and controllable target capture. Using the functionalized hydrogel, we successfully captured and released DNA and RNA of a wide length range, with no observed deleterious effects on the hybridization to the captured oligonucleotides. Although polyacrylamide gel was used in this work as the capture scaffold, we envision that other hydrogels or materials, upon simple modifications, can be similarly employed for psoralen-based capture of nucleic acid targets to meet different application needs.
Supplementary Material
Acknowledgments
This work was supported by the grant from the United States National Institutes of Health (NIH Grant R21EB019880 to A.E.H.). P.P.Y.C. was supported by Australian Research Council Discovery Project Grant (DP160101591). The 900 MHz instrument was funded by NIH grant GM68933. We thank Dr. Jeff Pelton at the 900 MHz NMR facility at California Institute for Quantitative Biosciences (QB3) - Berkeley for the assistance in NMR analysis, Dr. Rita Nichiporuk and Dr. Tony Iavarone at QB3/Chemistry Mass Spectrometry Facility at UC Berkeley for the assistance in mass spectrometry analysis, Mr. Hector D. Neira in Herr lab for critical reading of the manuscript, and all the other Herr lab members for helpful discussions and suggestions for the experiments and data analysis.
Footnotes
Supporting information for this article is available at: http://dx.doi.org/10.1002/anie.201711441.
Contributor Information
Dr. Yizhe Zhang, Department of Bioengineering, University of California, Berkeley, Berkeley, CA, 94720, USA
Dr. Peggy P. Y. Chan, Department of Bioengineering, University of California, Berkeley, Berkeley, CA, 94720, USA. Faculty of Science Engineering & Technology, Swinburne University of Technology, Melbourne, VIC 3122, Australia
Prof. Amy E. Herr, Department of Bioengineering, University of California, Berkeley, Berkeley, CA, 94720, USA
References
- 1.a) Breaker RR. Nature. 2004;432:838–845. doi: 10.1038/nature03195. [DOI] [PubMed] [Google Scholar]; b) Willner I, Zayats M. Angew Chem Int Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2007;119:6528–6538. [Google Scholar]; c) Castañeda AD, Brenes NJ, Kondajji A, Crooks RM. J Am Chem Soc. 2017;139:7657–7664. doi: 10.1021/jacs.7b03648. [DOI] [PubMed] [Google Scholar]; d) Twite AA, Hsiao SC, Onoe H, Mathies RA, Francis MB. Adv Mater. 2012;24:2380–2385. doi: 10.1002/adma.201104336. [DOI] [PubMed] [Google Scholar]; e) Hsiao SC, Shum BJ, Onoe H, Douglas ES, Gartner ZJ, Mathies RA, Bertozzi CR, Francis MB. Langmuir. 2009;25:6985–6991. doi: 10.1021/la900150n. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Rodriguez MC, Kawde AN, Wang J. Chem Commun (Cambridge, U K) 2005:4267–4269. doi: 10.1039/b506571b. [DOI] [PubMed] [Google Scholar]; g) Seetharaman S, Zivarts M, Sudarsan N, Breaker RR. Nat Biotechnol. 2001;19:336–341. doi: 10.1038/86723. [DOI] [PubMed] [Google Scholar]
- 2.a) Hu Y, Guo W, Kahn JS, Aleman-Garcia MA, Willner I. Angew Chem Int Ed. 2016;55:4210–4214. doi: 10.1002/anie.201511201. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2016;128:4282–4286. [Google Scholar]; b) Liao WC, Lilienthal S, Kahn JS, Riutin M, Sohn YS, Nechushtai R, Willner I. Chem Sci. 2017;8:3362–3373. doi: 10.1039/c6sc04770j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Penchovsky R, Breaker RR. Nat Biotechnol. 2005;23:1424–1433. doi: 10.1038/nbt1155. [DOI] [PubMed] [Google Scholar]; b) Frezza BM, Cockroft SL, Ghadiri MR. J Am Chem Soc. 2007;129(48):14875–14879. doi: 10.1021/ja0710149. [DOI] [PubMed] [Google Scholar]; c) Qian L, Winfree E. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 4.a) Pirrung MC. Angew Chem Int Ed. 2002;41:1276–1289. doi: 10.1002/1521-3773(20020415)41:8<1276::aid-anie1276>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2002;114:1326–1341. [Google Scholar]; b) Du H, Strohsahl CM, Camera J, Miller BL, Krauss TD. J Am Chem Soc. 2005;127(21):7932–7940. doi: 10.1021/ja042482a. [DOI] [PubMed] [Google Scholar]
- 5.Marcus JS, Anderson WF, Quake SR. Anal Chem. 2006;78(9):3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 6.Hagan KA, Reedy CR, Uchimoto ML, Basu D, Engel DA, Landers JP. Lab Chip. 2011;11:957–961. doi: 10.1039/c0lc00136h. [DOI] [PubMed] [Google Scholar]
- 7.a) Savage MD, Mattson G, Desai S, Nielander GW, Morgensen S, Conklin EJ. Avidin-Biotin Chemistry : A Handbook. Pierce Chemical Co; 1992. [Google Scholar]; b) Tong X, Smith L. Anal Chem. 1992;64:2672–2677. doi: 10.1021/ac00046a004. [DOI] [PubMed] [Google Scholar]
- 8.a) Hong JW, Studer V, Hang G, Anderson WF, Quake SR. Nat Biotechnol. 2004;22:435–439. doi: 10.1038/nbt951. [DOI] [PubMed] [Google Scholar]; b) Li Y, Gao J, Zhang C, Cao Z, Cheng D, Liu J, Shuai X. Top Curr Chem. 2017;375:27. doi: 10.1007/s41061-017-0119-6. [DOI] [PubMed] [Google Scholar]
- 9.a) Schmidt BVKJ, Barner-Kowollik C. Angew Chem Int Ed. 2017;56:8350–8369. doi: 10.1002/anie.201612150. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2017;129:8468–8488. [Google Scholar]; b) Lubbe AS, Szymanski W, Feringa BL. Chem Soc Rev. 2017;46:1052–1079. doi: 10.1039/c6cs00461j. [DOI] [PubMed] [Google Scholar]; c) Wang Y, Ma N, Wang Z, Zhang X. Angew Chem Int Ed. 2007;46:2823–2826. doi: 10.1002/anie.200604982. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2007;119:2881–2884. [Google Scholar]; d) Tamesue S, Takashima Y, Yamaguchi H, Shinkai S, Harada A. Angew Chem Int Ed. 2010;49:7461–7464. doi: 10.1002/anie.201003567. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2010;122:7623–7626. [Google Scholar]; e) Nalluri SKM, Ravoo BJ. Angew Chem Int Ed. 2010;49:5371–5374. doi: 10.1002/anie.201001442. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2010;122:5499–5502. [Google Scholar]; f) Liu Y, Yu C, Jin H, Jiang B, Zhu X, Zhou Y, Lu Z, Yan D. J Am Chem Soc. 2013;135:4765–4770. doi: 10.1021/ja3122608. [DOI] [PubMed] [Google Scholar]
- 10.a) Liu Y-C, Le Ny A-LM, Schmidt J, Talmon Y, Chmelka BF, Lee CT. Langmuir. 2009;25:5713–5724. doi: 10.1021/la803588d. [DOI] [PubMed] [Google Scholar]; b) Nalluri SKM, Voskuhl J, Bultema JB, Boekema EJ, Ravoo BJ. Angew Chem Int Ed. 2011;50:9747–9751. doi: 10.1002/anie.201103707. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2011;123:9921–9925. [Google Scholar]
- 11.a) Hearst JE, Isaacs ST, Kanne D, Rapoport H, Straub KQ. Rev Biophys. 1984;17:1–44. doi: 10.1017/s0033583500005242. [DOI] [PubMed] [Google Scholar]; b) Deans AJ, West SC. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Takasugi M, Guendouz A, Chassignol M, Decout JL, Lhomme J, Thuong NT, Hélène C. Proc Natl Acad Sci U S A. 1991;88:5602–5606. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Spielmann HP, Dwyer TJ, Hearst JE, Wemmer DE. Biochemistry. 1995;34:12937–12953. [PubMed] [Google Scholar]; e) Kurz M, Gu K, Lohse Pa. Nucleic Acids Res. 2000;28:E83. doi: 10.1093/nar/28.18.e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Tessman JW, Isaacs ST, Hearst JE. Biochemistry. 1985;24:1669–1676. doi: 10.1021/bi00328a015. [DOI] [PubMed] [Google Scholar]; b) Shi YB, Hearst JE. Biochemistry. 1987;26:3792–3798. doi: 10.1021/bi00387a009. [DOI] [PubMed] [Google Scholar]
- 13.a) Cimino GD, Shi YB, Hearst JE. Biochemistry. 1986;25:3013–20. doi: 10.1021/bi00358a042. [DOI] [PubMed] [Google Scholar]; b) Shi YB, Hearst JE. Biochemistry. 1987;26:3786–3792. doi: 10.1021/bi00387a008. [DOI] [PubMed] [Google Scholar]
- 14.a) Nguyen DN, Green JJ, Chan JM, Langer R, Anderson DG. Adv Mater. 2009;21:847–867. doi: 10.1002/adma.200801478. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kopeček J, Yang J. Angew Chem - Int Ed. 2012;51:7396–7417. doi: 10.1002/anie.201201040. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2012;124:7512–7535. [Google Scholar]; c) Cobo I, Li M, Sumerlin BS, Perrier S. Nat Mater. 2015;14:143–159. doi: 10.1038/nmat4106. [DOI] [PubMed] [Google Scholar]; d) Liao WC, Lilienthal S, Kahn J, Riutin M, Sohn YS, Nechushtai R, Willner I. Chem Sci. 2017;8:3362–3373. doi: 10.1039/c6sc04770j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Burnham MR, Turner JN, Szarowski D, Martin DL. Biomaterials. 2006;27:5883–5891. doi: 10.1016/j.biomaterials.2006.08.001. [DOI] [PubMed] [Google Scholar]; b) Tse JR, Engler AJ. Curr Protoc Cell Biol. 2010:1–16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- 16.Rehman FN, Audeh M, Abrams ES, Hammond PW, Kenney M, Boles TC. Nucleic Acids Res. 1999;27:649–655. doi: 10.1093/nar/27.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao KJ, Zhou WJ. J Appl Polym Sci. 1994;53:1533–1538. [Google Scholar]
- 18.a) Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Annu Rev Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]; b) Hearst JE. Chem Res Toxicol. 1989;2:69–75. doi: 10.1021/tx00008a001. [DOI] [PubMed] [Google Scholar]; c) Kobertz WR, Essigmann JM. J Am Chem Soc. 1997;119:5960–5961. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.