Abstract
Microchips that perform single cell capture, array, and identification have become powerful tools for single cell studies, which can reveal precise underlying mechanisms among bulk cell populations. However, current single cell capture and on-chip immunostaining methods consume more time and reagent than desired. To optimize this technology, we designed a novel trap structure for single cell capture, array, and release, and meanwhile an oscillatory method was used to perform rapid on-chip cell immunostaining. The trap structure array used equal distribution of lateral flow to achieve single cell array in high velocity flows and decrease the risk of clogging. A length of glass capillary with a sealed bubble was inserted into the outlet so that it could act in a manner analogous to that of a capacitor in an RC circuit. By applying one periodic air pressure to the inlet, oscillation motion was generated, which significantly enhanced the on-chip reaction efficiency. In addition, the oscillation performance could be easily regulated by changing the length of the capillary. The trapped cells could maintain their positions during oscillation; hence, they were able to be tracked in real time. Through our trap microchip, 12 μm microbeads were successfully trapped to form a microarray with a capture efficiency of ∼92.7% and 2 μm microbeads were filtered. With an optimized oscillation condition (Ppush = 0.03 MPa, f = 1 Hz, L = 3 cm), fast on-chip immunostaining was achieved with the advantages of less time (5 min) and reagent (2 μl) consumption. The effectiveness of this method was demonstrated through quantitative microbead and qualitative Caco-2 cell experiments. The device is simple, flexible, and efficient, which we believe provides a promising approach to single cell heterogeneity studies, drug screening, and clinical diagnosis.
I. INTRODUCTION
Single cell capture and analysis have played an important role in recent years. Unlike conventional bulk analysis, single cell study can identify detailed differences among seemingly identical cell populations. This is critical to revealing precise mechanisms of complicated biological processes.1,2 With the deep analysis of single cell, breakthroughs have been made in many fields such as the studies of tumor heterogeneity,3 cancer development,4 stem cell functionality,5 drug sensitivity,6 and so on.
Early development of single cell analysis begins with flow cytometry and laser capture microdissection (LCM), which offer an extensive depth of information for single cell.7 However, flow cytometry requires a large number of isolated, labeled, and purified cells for accurate results, and target cells cannot be traced for long periods of time.7,8 LCM is limited by high cost, low visual resolution, and low throughput.7,10 Microfluidic devices have emerged as remarkable tools for single cell studies, which own great aptitude for high-throughput processing and precise control in temporal and spatial.9–11
Microchip-based single cell capture, array, and analysis is the fundamental method used in many single cell research such as circulating tumor cells (CTCs) study.10,12 By carefully designing the trap structure, cells can be immobilized in specific positions on the chip. On-chip identification and functionality tests are then performed to gain new insights into various biological processes. Weng et al. developed a microarray of butterfly shaped traps to determine the cell membrane permeability of rat hepatocytes and patient-derived CTCs across a broad temperature range, which explored the biophysics of membrane transport.13 Di Carlo et al. presented a U-shaped microfluidic device to trap single cell in large arrays for enzyme kinetics analysis of three different cell types, which could be directly applied to quantify a variety of intracellular enzymes with available fluorogenic substrates.14 However, in these reports, the trap structures are positioned on the main flow path of the chip. The trapped cells are distributed unevenly, majority of which are trapped in the frontal structures, thus easily increasing the local flow resistance. Moreover, few studies have investigated cell capture and on-chip immunostaining.
Of the analysis methods including proteome, transcriptome, genome identification, and functionality tests, immunostaining is the most common and fundamental technique used to demonstrate protein expression. For example, this method was used to identify EpCam+/CK+/CD45− cells as epithelium originated tumor cells leaking into the circulation system.12,15 However, current on-chip staining methods have some drawbacks. The widely used on-chip staining method is static staining, where antibody solutions are primed to the chip and incubated with the cells. This method shows particular advantages of handy manipulation and low reagent consumption. However, due to the very small Reynolds number (usually 10−6–10) in microscale, the fluid flows in a laminar manner, and mixing relies on molecular diffusion.16 Resulting from the difficulty of on-chip mixing, static staining generally requires 30–90 min,17–20 and long time consumption may cause potential damage to cell integrity and some substances.21 Chen et al. demonstrated that their microarray chip performed well where the intensity of SYTO13 labeling leukocytes came to saturate after ∼30 s.22 Although it successfully realized on-chip fast staining, SYTO13 is a kind of cell-permeant nucleic staining dye, which can penetrate the cell membrane and combine with the nucleus directly. The other method is flow staining, where the solutions are constantly pumped into the chip by a syringe at a low flow rate, typically at 20 μl/h lasting for 30 min.23 This method is highly effective due to the continuous supply of fresh solution and higher probability of molecular collision in the dynamic state. However, it consumes large amounts of expensive reagents. The efficiency of on-chip immunostaining has not yet been explored. It is highly desirable to design a simple microdevice that can achieve single cell capture, array, release, and on-chip fast staining.
Mixing has a critical influence on staining efficiency by affecting the probabilities of antibody and antigen molecular collision and binding. Quake et al. introduced an on-chip peristaltic pump, where solutions were actively pumped in a circulating loop to accelerate mixing process in microscale.24 Other active methods of mixing solutions include magnetic, electrical, and acoustical approaches.25–27 However, these methods require multiple microvalves or external equipment, which increase the complexity of the microfluidic system. In macroscale, mixing can be accomplished rapidly by shaking, stirring, or oscillating to cause turbulence in fluids. It is well known that the mixing efficiency can be greatly improved by introducing turbulence into the chip. Abolhasani et al. introduced an oscillatory multiphase flow mixer in which a train of droplets moved back and forth due to the application of an alternating pressure gradient to each end of the microchannel.28 This method has a simple design and does not require equipment other than pressure sources, so it can be well integrated with various microfluidic processes. However, their oscillatory multiphase flow mixer requires at least two accurately controlled pressure sources, which put forward high requirements to the equipment.
In our study, we developed an integrated microfluidic device which combined single cell separation, array, release, and fast oscillatory staining on one chip. Based on an equally distributed lateral flow design, the chip could form a single cell microarray in high velocity with a low risk of clogging. Some researchers have used sealed bubbles to act as pressure sensors due to their compressibility.29 Inspired by these works, we conducted an oscillation device using a sealed capillary at the outlet of the chip, and one periodic air pressure was applied to the inlet. The oscillation on the chip was generated alongside the compression and decompression of the bubble in the sealed capillary, which was like the charging and discharging process of an RC circuit. The oscillation performance could be adjusted simply to meet different study demands by changing the length of the capillary. Through oscillatory staining, the same staining results were achieved with less time and reagent consumption. Due to simple construction, handy operation, flexible adjustment, and high staining efficiency, we believe the microdevice provides a novel approach to single cell fast identification and has potential for application to rare cell studies like CTCs and stem cells.
II. MATERIALS AND METHODS
A. Fabrication of the microfluidic device
The microfluidic chip was designed using AutoCAD software (Autodesk, USA). First, positive photoresist (AZ 4903, MicroChemicals, Germany) with a height of 1 μm was patterned on a 3-in. silicon wafer to protect the areas that were not intended to be etched. After that, the patterns were etched 19 μm deep through NMC ICP deep silicon etching system to produce a mold with a total height of 20 μm. Then a mixture of 20 g Sylgard PDMS (Dow Corning, USA) pre-polymer and its crosslinking agent (10:1 ratio) was poured into the mold and baked at 75 °C for 1 h. The cured replica was peeled from the mold and cut into the designed shape. Then, a puncher with a 2 mm external diameter was used to drill inlet and outlet holes. Finally, the PDMS replica was permanently bonded to a glass substrate after oxygen plasma treatment.
B. IgG coated on protein A functionalized microspheres
We purchased protein A functionalized polystyrene microspheres with 12 μm diameters from Bangslabs Inc. (CP02N, Micromod, Germany). Rabbit IgG (150 μg/l, Dingguo, China) polyclonal antibodies were bound to protein A functionalized beads by a simple one-step process according to the protocol. Briefly, the beads were preliminarily washed with PBS and Tween-20 to remove various additives, including EDTA, antimicrobials, and surfactants. Then, an antibody suspension was added to the microspheres and mixed gently for 45 min at room temperature. The unattached antibodies were washed away, and the IgG coated microspheres were stored in PBS at 4 °C.
C. Fluorescence quantification
Fluorescein sodium dye (Sigma-Aldrich, USA) was used to evaluate the oscillatory mixing efficiency. Fluorescence images were captured using a microscope (IX71, Olympus, Japan) with a CCD (DP73, Olympus, Japan) at the exposure time of 500 μs. MATLAB was used to analyze the fluorescence intensity. By setting the range of intensity, the edge of the mixing area was detected, and the intensity of each pixel inside the area was measured. The ratio of the standard deviation to the average fluorescence intensity (σ/) of all the fluorescence spots was plotted as a function of time to measure the signal variance. Since the volume of the chip chamber is much smaller than that of the capillary, we defined the complete mixing time as the time to reach the σ/ value of control group, where chip was completely filled with fluorescent dye.
ImageJ was used to analyze the fluorescence intensity of the stained microbeads. After setting the same threshold, the area of interest was highlighted and then the mean intensity and standard deviation were calculated in each image. We considered staining to be complete when the average gray value of the trapped microbeads reached the maximum and the change range of the mean gray value among subsequent images was <1%.
D. Image-based oscillatory parameter analysis
We used red dye (Allura Red, Dyestuffs Research Institute, China) to investigate the influence of various parameters on oscillation. Experimental videos were recorded from the same vision using a zoom lens (Navitar 12X, USA) with a CCD (DigiRetina 16 CCD, TUCSEN, China) which takes 16 frames per second. Based on OpenCV library, each frame was extracted and analyzed. The contour of the region filled with red liquid was outlined. Then, the entire image was inverted into an image where the region of interest was presented as a bright field while other regions were visualized as a dark field. The interfacial position of the red liquid was measured in each image by its coordinate Y value.
E. Cell culture and preprocessing
Colorectal cancer cell line Caco-2 cells were cultured in a DMEM medium (Gibco, USA) containing 10% fetal bovine serum (Gibco, USA) and 1% antibiotics (penicillin-streptomycin, Gibco, USA) at a humidified 5% CO2 and 37 °C incubator. The cells were passaged by trypsin (Gibco, USA) after growing to 70%–80% confluence. The cells were fixed by 4% paraformaldehyde (PFA) for 15 min at room temperature. Before staining, the fixed cells were permeated with 0.1% TritonX-100 (Dingguo, China) and blocked with BSA (Sigma-Aldrich, USA) for 5 min.
F. Off-chip staining
The preprocessed cells (∼106/ml) were diluted to ∼105/ml. 10 μl of EpCam-PE (Proteintech, USA, 1:25) and 10 μl of CK20-FITC (Miltenyi, Germany, 1:10) were added to 10 μl of cell suspension and incubated with cells at room temperature for 30 min. Then, PBS was used to wash away the unbound antibodies. The stained cells were resuspended with 10 μl of PBS.
G. Microbead capture and release
Microbead suspension (12 μm diameter, ρ = 1.05 g/cm3, ∼2.6 × 104/ml) was diluted to ∼1000/ml with PBS solution. 1 μl solution of 2 μm microbeads (Beisile, China) was added to 300 μl of the dilute 12 μm microbead solution. After loading the mixing solution into the chip, we counted the 12 μm microbeads that had been trapped and calculated the trap efficiency. For release, a back flow was used at the outlet for 5 min, and the unreleased microbeads on the chip were counted. The release efficiency was calculated as the ratio of released microbeads (trapped microbeads minus unreleased microbeads) to trapped microbeads.
H. Tumor cell collection from blood
Peripheral blood was obtained from healthy human donors, collected in a Vacutainer tube containing the anticoagulant EDTA, and processed within 6 h. We spiked the known number of Caco-2 cells (∼200) into 2 ml blood. The blood samples were introduced into the chip with 12 or 18 μm gap directly under the pressure of 0.01 MPa. RBCs were removed by PBS. The trapped cells were detected by immunostaining. Briefly, trapped cells were fixed by 4% PFA for 15 min, permeabilized with 0.1% TritonX-100 solution for 5 min, and blocked with BSA for 5 min. Then these trapped cells were stained by a cocktail including 1 μl of CK20-FITC, 1 μl of CD45-PE (Miltenyi, Germany), and 1 μl of DAPI using the oscillatory method for 5 min (Ppush = 0.03 MPa, f = 1 Hz, L = 3 cm). Stained cells were enumerated and classified. The CK−/CD45+/DAPI+ cells were identified as WBCs and the CK+/CD45−/DAPI+ cells were identified as CTCs.12
III. RESULTS AND DISCUSSION
A. Design of the capture microchip
The microchip comprises 351 baleen-shaped trap structures [Fig. 1(a)]. The details of the geometry are shown in Figs. 1(b) and 1(c). The ratio of the side channel to main channel width is designed to be 2:1 in order to distribute the flow in all directions more uniformly. To trap cells of various sizes (generally somatic cells of human have diameters of 10–20 μm), the width of the opening is 25 μm and that of the flow microchannel decreases linearly from 10 μm to 5 μm. The height of the structure is 20 μm. Besides, all the microchannels are connected to a main opening inlet channel and share a joint outlet. A cell whose diameter matches the width of the microchannel can occupy an empty structure when the flow passes through the trap sites. Consequently, the trapped cells can be immobilized in specific positions. Because of the trap design, the solution flows in all directions on the chip, so the particles with diameters of 5 μm or larger are trapped, while those below 5 μm are filtered. Just like a wide-band filter, the widths of the trap structure entrance and exit can be adjusted to fit different applications. For the application to CTC enrichment from whole blood, the gap can be widened to trap tumor cells and filter WBCs/RBCs. In our study, we also designed chips with 12 μm or 18 μm gap [Fig. S1(a)] and used whole blood to evaluate their performance.
FIG. 1.
Design of the single cell capture microchip. (a)–(c) The detailed geometry of the trap structure. (d) The COMSOL simulation of the design. (e) The flow velocity in the red line of each trap site was calculated by COMSOL (the velocity of the inlet was set to 0.01m/s). (f) The flow velocity in each trap site [the red line in (e)] from the left to the right (1–61) was shown sequentially.
COMSOL simulation shows that the flow is distributed nearly equally in all trap sites [Figs. 1(d)–1(f)], unlike in conventional cell capture chips. The flow moves fast through the inlet and outlet, and slow as it passes through each trap site. Figure 1(f) shows that the flow velocities in each trap site calculated by COMSOL (velocity is set to 0.01 m/s at the inlet) vary little and are distributed symmetrically on the chip.
B. Isolation, capture, and release in high flow velocity
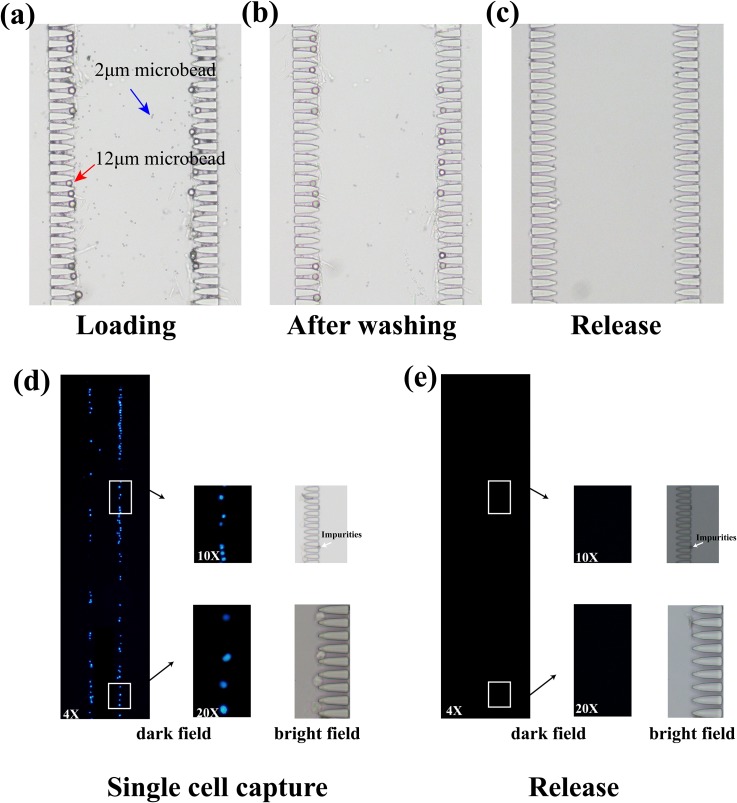
A mixing solution compound of 12 μm and 2 μm microbeads was used to validate the isolation and capture performance of the design. Due to the lateral flow distribution, 300 μl of solution was primed rapidly within 1 min by air pressure (0.03 MPa).12 μm microbeads were trapped with an efficiency of ∼92.7% and 2 μm microbeads leaked away in all directions [Figs. 2(a) and 2(b)], which could be observed in movie M1 in the supplementary material. Using a back flow at the outlet driven by the pressure of 0.03 MPa, the captured microbeads flowed back to the inlet. ∼99% of the trapped microbeads were released after 5 min of back washing, while ∼1% remained in the chip due to nonspecific absorption [Fig. 2(c)]. The released microbeads could be collected, which were available for further studies.
FIG. 2.
(a)–(c) The isolation and release of microbeads using the trap structures. The red arrow presented that the 12 μm microbead was trapped and the blue arrow presented that the 2 μm microbead was separated. (d) and (e) The capture and release of Caco-2 cells using the trap structures. Caco-2 cells stained by DAPI off the chip were used to evaluate the performance. A full view of fluorescent images under the objective magnification of 4× was shown. The images under the objective magnification of 10× and 20× were also presented to show the details inside the white boxes. The white arrows represented the unstained impurities.
For single cell capture and release, as shown in Fig. 2(d), the DAPI stained Caco-2 cells got stuck in the trap structures and were distributed evenly on the chip. Meanwhile, the trapped cells could also be released [Fig. 2(e)]. The trap efficiency of cells reached 91.6% and release efficiency was 100%. The high capture and release efficiency was quite important for downstream studies.
C. Construction of the oscillation device
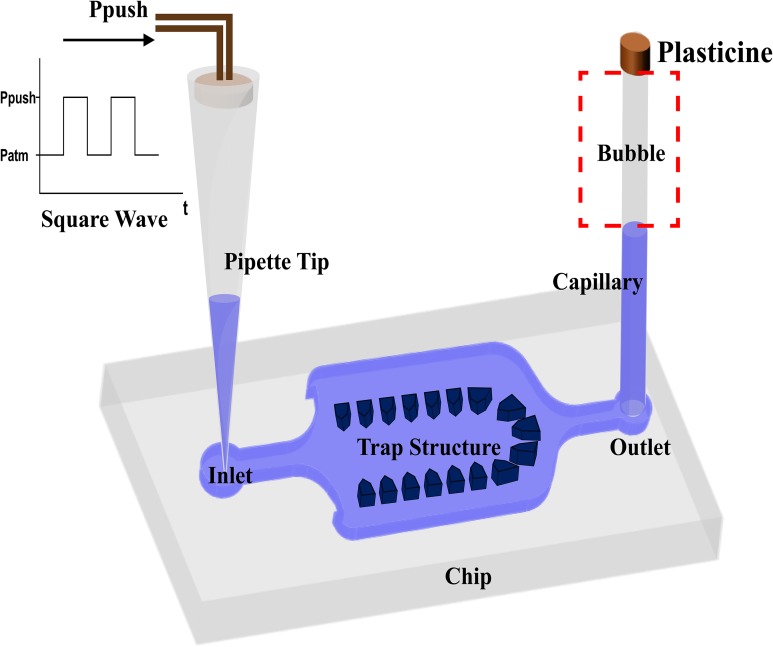
We used some simple materials to construct the oscillation system (Fig. 3). A pipette tip is placed at the inlet to avoid changing the tube when adding different liquids. This helps us avoid developing bubbles into the chip. One end of a length of glass capillary is sealed with plasticine and the other end is inserted into the outlet. The sealed capillary produces a large air bubble. At the inlet, only a pressure source is employed, which is controlled by periodically activating and deactivating a solenoid valve via square wave signals generated by an Arduino. Thus, oscillation is driven by the periodic pressure and the sealed bubble in the capillary. The capillary does not occupy the space on the chip, and it has lower permeability than other materials (like PDMS). In addition, it offers flexibility to adjust the oscillatory performance to match different conditions by simply changing the capillary length. Thus, the entire oscillation device is constructed simply and flexibly, and compatible with chips of different designs.
FIG. 3.
Schematic of the oscillation device. The sealed capillary at the outlet stores an air bubble. The inlet is connected to a square wave pressure source.
D. Working mechanism of oscillation
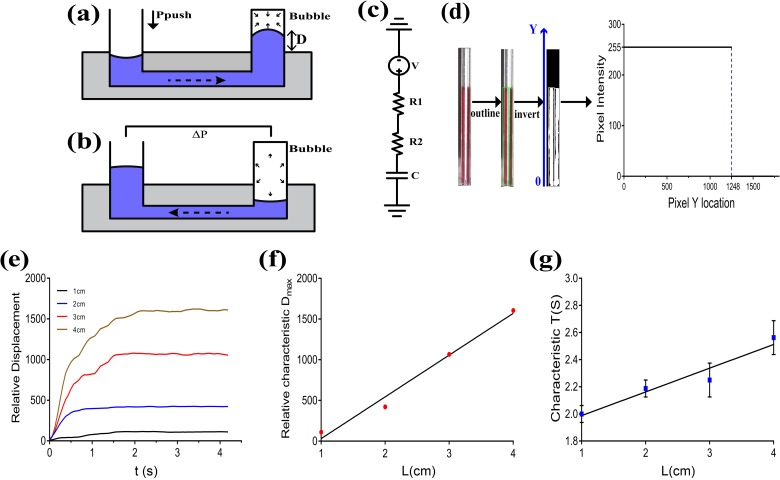
An external air pressure via square wave signals is applied to the inlet. When the pressure (Ppush) is on, the liquid in the pipette tip is primed to the chip, causing the gas bubble in the capillary to compress rapidly [Fig. 4(a)]. When the pressure (Ppush) is off, because the pressure of the bubble in the capillary is higher than atmosphere (), the bubble drives the liquid to move back [Fig. 4(b)]. The liquid in the channel moves back and forth along with the cyclical compression and decompression of the bubble. The entire fluid circuit can be simulated as an RC circuit, where the compression and decompression of the gas bubble are analogous to the charging and discharging processes of a capacitor through resistors [Fig. 4(c)].
FIG. 4.
Oscillation parameter study. (a) The compression process of the bubble when Ppush is on. The fluid is primed to the chip by Ppush. (b) The decompression process of the bubble when Ppush is off. The fluid is driven by to flow backwards. (c) Fluidic analogous RC circuit model to explain oscillation work principle. (d) Processing of experimental images. The OpenCV library was used to outline the interested region using green rectangles. Then, the image was inverted. The intensity along with the liquid position was shown, and the interfacial position was identified.30 (e) Relative displacement in capillaries with lengths of 1 cm, 2 cm, 3 cm, and 4 cm (Ppush = 0.05 MPa). (f) Relative characteristic Dmax in capillaries with lengths of 1 cm, 2 cm, 3 cm, and 4 cm (Ppush = 0.05 MPa). (g) Characteristic compression time (T) in capillaries with lengths of 1 cm, 2 cm, 3 cm, and 4 cm (Ppush = 0.05 MPa).
E. Oscillatory parameter analysis
We developed a simplified physical model to measure the displacement (D). D means the displacement of the liquid into the capillary when the external pressure is on. Because the volume of the chip chamber (∼70 nl) is much smaller than that of the capillary (1 cm: ∼720 nl), the volume of the microchip is ignored during these calculations. Assuming that the ideal gas law applies, the maximum displacement (Dmax) can be calculated using following formula:
| (1) |
where Patm is the atmosphere pressure and Ppush is the pressure applied to the inlet. L is the length of the capillary.
As shown in Figs. 4(d) and 4(e), the liquid interfacial location in the capillary rose quickly in the beginning, then slowed down, and finally reached the equilibrium in experimental parameter analysis.
Then, the characteristic maximum displacement (characteristic Dmax) and characteristic compression time (characteristic T) were measured. When the first time to reach , we identified the time as characteristic T. Characteristic Dmax was calculated averagely after characteristic T. As shown in Figs. 4(f) and 4(g), when Ppush was fixed, characteristic T and characteristic Dmax increased almost linearly along with the increasing length of the capillary (L) in the range of 1–4 cm. These results were consistent with the theoretical Eq. (1). Therefore, we can regulate the oscillation performance flexibly by changing the length of the capillary to satisfy different requirements.
F. Oscillatory mixing evaluation
To evaluate oscillation-based mixing performance, we measured the mixing efficiency of fluorescein sodium dye and PBS. The chip was filled with PBS solution (∼70 nL) and 2 μl of fluorescein sodium was held in the pipette tip. When the two solutions were mixed without oscillation, it costs a long time of ∼108 min to complete mixing, where the process mainly relied on molecular diffusion alone.
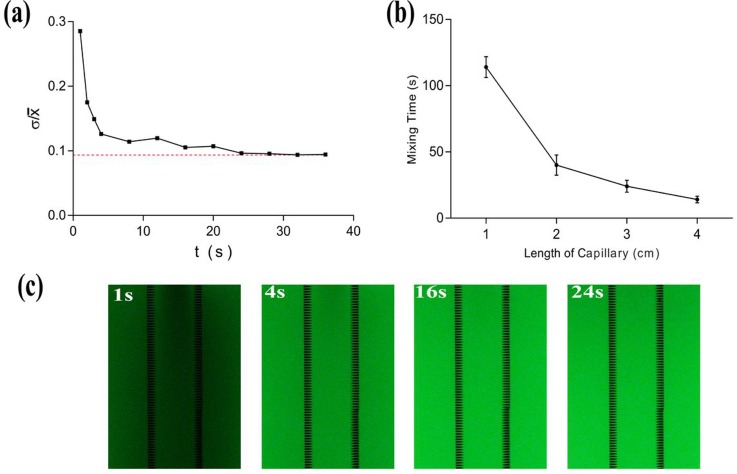
At the beginning of oscillatory mixing, the fluorescent dye was distributed unevenly, so the ratio of the standard deviation to the average fluorescence intensity (σ/) of all the fluorescence spots was high. As the oscillation proceeded, σ/ gradually deceased until mixing was complete [Figs. 5(a) and 5(c)]. The volume of the PBS in the chip could be ignored for the purpose of calculation, so we identified the chip filled with fluorescent dye as the control and defined the time to reach the σ/ value of the control group as the time needed to mix completely.
FIG. 5.
Oscillatory mixing evaluation. (a) σ/ of the solution in the chip vs. time during oscillation (Ppush = 0.03 MPa, f = 1 Hz, L = 3 cm). (b) Mixing time with capillary lengths from 1 cm to 4 cm (Ppush=0.03 MPa, f = 1 Hz). (c) Experimental fluorescence images captured at 1 s, 4 s, 16 s, and 24 s during the oscillatory mixing process (Ppush = 0.03 MPa, f = 1 Hz, L = 3 cm).
We further investigated the influence of capillary length on the mixing efficiency. When the applied frequency (1 Hz) and pressure (0.03 MPa) were fixed, complete mixing using 1 cm and 4 cm capillaries costs 112 s and 14 s, respectively. Thus, oscillatory mixing was greatly faster than mixing based on molecular diffusion. Moreover, the mixing time decreased as the length increased [Fig. 5(b)]. Within the range of 1–4 cm, longer capillary exhibited higher mixing efficiency.
However, it is difficult to insert capillaries longer than 4 cm into the chip because of their fragility. High inlet pressures lead to difficulty maintaining cell integrity in a high shear force environment. In addition, the combination of high pressures and long movement distances can cause the trapped cells to get dislodged. Low pressures cannot mix effectively in such conditions. A frequency of 1 Hz is considered mild for most cells. Thus, we employed Ppush of 0.03 MPa, a frequency of 1 Hz, and a capillary length of 3 cm in subsequent biological studies.
G. Microbead capture and immunoassay
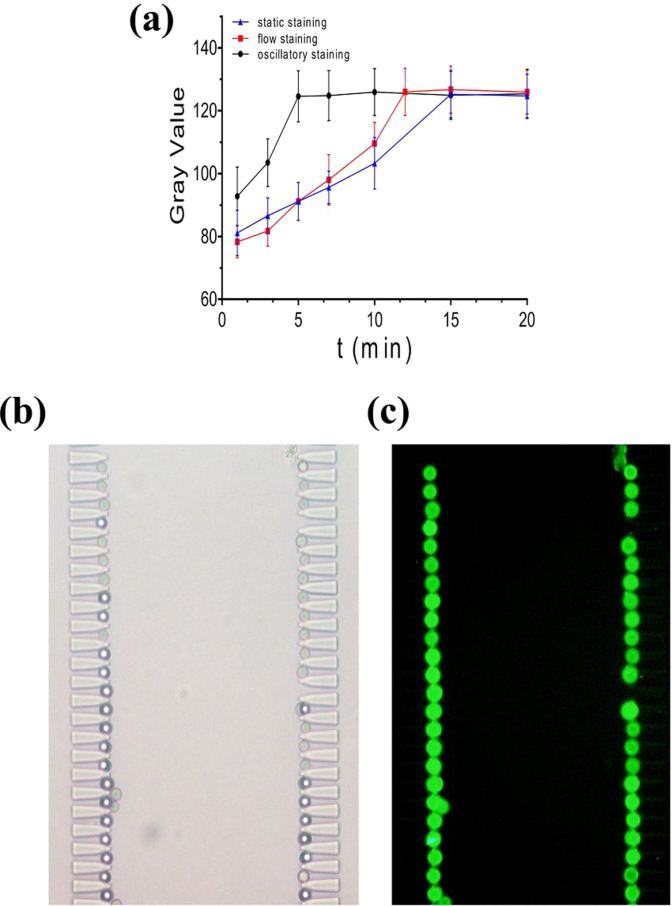
Rabbit IgG coated polystyrene microbeads (12 μm in diameter) were used to measure the oscillatory immunostaining efficiency quantitatively. First, the microbead solution was diluted to 100/μl, and 2 μl of suspension was primed. Microbeads were trapped on the chip, forming a single bead microarray [Fig. 6(b)]. Then, three separate methods were used to stain the microbeads.
FIG. 6.
Microbead immunostaining. (a) The mean gray values of microbeads using static, flow (20 μl/h), and oscillatory staining (Ppush = 0.03 MPa, f = 1 Hz, L = 3 cm). (b) Bright field image of the microbead microarray. (c) Fluorescence image of microbeads after completion of oscillatory staining (5 min).
In the static staining method, 2 μl of FITC conjugated goat anti-rabbit IgG solution was introduced into the chip and incubated with the microbeads. For flow staining, FITC-IgG solution was continuously driven into the chip at 20 μl/h using a syringe. Oscillatory staining was performed in the condition of Ppush 0.03 MPa, f 1 Hz, and L 3 cm. We detected the average microbead fluorescence intensity quantitatively at the same exposure time of 800 ms.
Static staining method is the most popular among researchers because it is easy to operate and no extra equipment is needed. However, it costs nearly 15 min using 2 μl of reagent to complete staining, which primarily resulted from the lack of molecular collisions in a static environment. Flow staining drove the beads to shake slightly in situ, increasing the collision rate between beads and reagent molecules, so complete detection required 12 min and 4 μl of IgG solution. However, the method is so reagent consuming that it is unsuitable for expensive reagents. Compared to these two methods, oscillatory staining needed less time (5 min) and less antibody consumption (2 μl) when achieved the same fluorescence intensity (Fig. 6). All experiments were performed at least three times and the results were calculated averagely. Meanwhile, the positions of the microbeads remained fixed during the oscillatory process, which made it possible to track and observe each bead on line (movie M2 in the supplementary material). Thus, oscillatory staining is a feasible staining method with the advantages of high efficiency, low reagent consumption, and simple operation.
H. Cell capture and fast detection
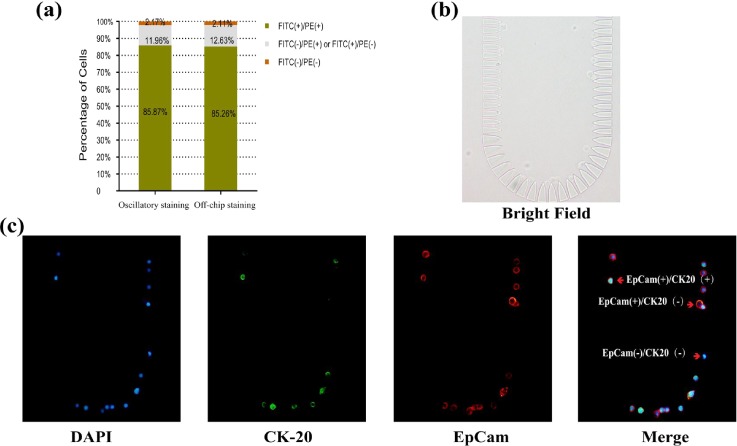
To verify whether oscillatory staining was appropriate for cell detection, we used the method to stain Caco-2 cells on chip. 1 μl suspension of Caco-2 cells (100/μl) was used, and the cells were trapped on the chip [Fig. 7(b)]. First, DAPI stained cell nuclei within about 10 s. EpCam and CK20 were detected using antibody probing via oscillatory staining. After 5 min of oscillation (Ppush = 0.03 MPa, f = 1 Hz, L = 3 cm) with 2 μl of antibody cocktail, EpCam+/CK20+ was detected on 85.87% cells (Fig. 7). Compared to off-chip staining (85.26% EpCam+/CK20+ cells detected), the difference was small enough to be ignored, which may result from the differences of the antigen expression level or antigen loss during culturing. Oscillatory staining achieved the same staining performance in less time and with less reagent cost. Besides, the positions of the cells remained fixed during oscillation (movie M3 in the supplementary material). Thus, it is applicable to the studies with constraints regarding time and consumption of expensive reagents like CTC and stem cell studies.
FIG. 7.
Fast cell detection. (a) Comparison of the staining efficiencies of oscillatory and off-chip staining. (b) Bright field image of the single cell microarray. (c) Fluorescence images of cells via oscillatory staining for 5 min (exposure times: DAPI 20 ms, CK20 307.7 ms, EpCam 79.36 ms).
Using chips with 12 or 18 μm gap, CTCs could be enriched from whole blood (movie M4 in the supplementary material). Then these trapped cells were successfully identified by the oscillatory staining method. Despite relatively high tumor cell (CK+/CD45−/DAPI+) trap efficiency (∼79%), many WBCs (CK−/CD45+/DAPI+) were trapped simultaneously via the chip with 12 μm gap (Fig. S1). This is because most of normal WBCs and CTCs have a diameter ranging from 8 μm to 20 μm, and there exists significant size overlap between CTCs and WBCs.31–33 It is reported that Lee et al. have developed a platform to capture CTCs. The chip with 10 μm gap was designed to filter RBCs out and simultaneously trap WBCs/leukemia cells, and then the researchers identified leukemia cells through fluorescence lifetime imaging microscopy.31 When the gap of our chip was widened to 18 μm, most WBCs were filtered but more tumor cells escaped. The trap efficiency was as low as ∼32.5% (Fig. S1). Thus, the performance was unsatisfactory when isolating CTCs from a large background of RBCs and WBCs only by size (movie M5 in the supplementary material). Actually, for most negative selection methods of CTC enrichment, multiple methods like using magnetic force tend to be integrated for the purpose of collecting CTCs and removing WBCs effectively.34–37 Our microdevice has shown better performance in time and reagent consumption than current CTC detection methods. Integrated with other isolation techniques, it offers a powerful and efficient tool for CTC negative enrichment and characterization.
Further we investigated whether the shortened staining process (5 min) contributed to cell viability. Unfortunately, nearly 50% of cells died after 1 min of oscillation. These unsatisfactory results may be caused by long cell preprocessing time or the high air pressure applied. So, improving cell preservation or applying air pressure via sinusoidal waveforms or other gradually increasing forms may promote cell viability, which we will study in the future. However, cell viability is not necessary for most single cell research. In these cases, our oscillatory staining method shows great superiority.
IV. CONCLUSIONS
In response to the growing demand for single cell trap and analysis in microfluidic platforms, we developed an integrated microsystem that combined hydrodynamic single cell trap, array, release, and fast oscillatory immunostaining. Using a lateral flow trap design, we achieved single cell capture and release in high flow velocity and avoided clogging. Cells were immobilized in specific positions so that we could observe and track each one easily. To enhance on-chip immunostaining, we constructed a simple oscillation device to accelerate mixing in microscale. Oscillatory motion was driven by only one periodic pressure source and a sealed capillary. Capillaries of different lengths showed different mixing performances. So, the device can be applied to various conditions by adjusting the capillary length. Oscillation increased molecular collisions between antibodies and antigens, speeding up the staining process. This method required only 2 μl of reagent to produce the same microbead staining performance in 5 min as flowing staining (12 min, 4 μl) and static staining (15 min, 2 μl). At the same time, beads or cells were able to maintain their positions during oscillation. Owing to the advantages of easy fabrication, simple operation, flexible adjustment, low reagent consumption, and high staining efficiency, we are convinced that our microdevice for single cell microarray and fast on-chip staining will be an efficient tool for single cell research.
V. SUPPLEMENTARY MATERIAL
See supplementary material for the performance of microbead isolation and capture; on-chip oscillation process of both microbeads and cells; CTC enrichment from whole blood; loss of some CTCs using the chip with 12 μm gap; and CTC capture performance via chips with 12 μm gap and 18 μm gap.
ACKNOWLEDGMENTS
This study was funded by the National High-tech R&D Program of China (863 Program 2015AA020401) and the Shanghai Committee of Science and Technology of China (Grant No. 14DZ1940305).
Contributor Information
Xiang Chen, Email: .
Zhihai Peng, Email: .
References
- 1. Navin N. E., Genome Biol. 15(8), 452 (2014). 10.1186/s13059-014-0452-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leslie M., Science 331(6013), 24–25 (2011). 10.1126/science.331.6013.24-b [DOI] [PubMed] [Google Scholar]
- 3. Puram S. V., Tirosh I., Parikh A. S., Patel A. P., Yizhak K., Gillespie S., Rodman C., Luo C. L., Mroz E. A., Emerick K. S., Deschler D. G., Varvares M. A., Mylvaganam R., Rozenblatt-Rosen O., Rocco J. W., Faquin W. C., Lin D. T., Regev A., and Bernstein B. E., Cell 171(7), 1611–1624 (2017). 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bendall S. C. and Nolan G. P., Nat. Biotechnol. 30(7), 639–647 (2012). 10.1038/nbt.2283 [DOI] [PubMed] [Google Scholar]
- 5. Wen L. and Tang F., Genome Biol. 17, 71 (2016). 10.1186/s13059-016-0941-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spaethling J. M. and Eberwine J. H., Curr. Opin. Pharmacol. 13(5), 786–790 (2013). 10.1016/j.coph.2013.04.011 [DOI] [PubMed] [Google Scholar]
- 7. Lindström S. and Andersson-Svahn H., Lab Chip 10(24), 3363–3372 (2010). 10.1039/c0lc00150c [DOI] [PubMed] [Google Scholar]
- 8. Murphy T. W., Zhang Q., Naler L. B., Ma S., and Lu C., Analyst 143(1), 60–80 (2017). 10.1039/C7AN01346A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao L., Ma C., Shen S. F., Tian C., Xu J., Tu Q., Li T. B., Wang Y. L., and Wang J. Y., Biosens. Bioelectron. 78, 423–430 (2016). 10.1016/j.bios.2015.09.055 [DOI] [PubMed] [Google Scholar]
- 10. Kim J., Cho H., Han S. I., and Han K. H., Anal. Chem. 88(9), 4857–4863 (2016). 10.1021/acs.analchem.6b00570 [DOI] [PubMed] [Google Scholar]
- 11. El-Ali J., Sorger P. K., and Jensen K. F., Nature 442(7101), 403–411 (2006). 10.1038/nature05063 [DOI] [PubMed] [Google Scholar]
- 12. Yeo T., Tan S. J., Lim C. L., Lau D. P., Chua Y. W., Krisna S. S., Iyer G., Tan G. S., Lim T. K., Tan D. S., Lim W. T., and Lim C. T., Sci. Rep. 6, 22076 (2016). 10.1038/srep22076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weng L., Ellett F., Edd J., Wong K. H. K., Uygun K., Irimia D., Stott S. L., and Toner M., Lab Chip 17(23), 4077–4088 (2017). 10.1039/C7LC00883J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Di Carlo D., Aghdam N., and Lee L. P., Anal. Chem. 78(14), 4925–4930 (2006). 10.1021/ac060541s [DOI] [PubMed] [Google Scholar]
- 15. Burinaru T. A., Avram M., Avram A., Mărculescu C., Ţîncu B., Ţucureanu V., Matei A., and Militaru M., ACS Comb. Sci. 20(3), 107–126 (2018). 10.1021/acscombsci.7b00146 [DOI] [PubMed] [Google Scholar]
- 16. Ward K. and Fan Z. H., Micromech. Microeng. 25(9), 094001 (2015) 10.1088/0960-1317/25/9/094001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin C. H., Hsiao Y. H., Chang H. C., Yeh C. F., He C. K., Salm E. M., Chen C., Chiu I. M., and Hsu C. H., Lab Chip 15(14), 2928–2938 (2015). 10.1039/C5LC00541H [DOI] [PubMed] [Google Scholar]
- 18. Chang C. L., Huang W., Jalal S. I., Chan B. D., Mahmood A., Shahda S., O'Neil B. H., Matei D. E., and Savran C. A., Lab Chip 15(7), 1677–1688 (2015). 10.1039/C5LC00100E [DOI] [PubMed] [Google Scholar]
- 19. O'Neill A. T., Monteiro-Riviere N. A., and Walker G. M., Cytotechnology 56(3), 197–207 (2008). 10.1007/s10616-008-9149-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cho W., Pradhan R., Chen H. Y., Weng Y. H., Chu H. Y., Tseng F. G., Lin C. P., and Jiang J. K., Sci. Rep. 7(1), 11385 (2017). 10.1038/s41598-017-09829-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Che J., Yu V., Garon E. B., Goldmanbc J. W., and Di Carlo D., Lab Chip 17(8), 1452–1461 (2017). 10.1039/C7LC00038C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y., Austin R. H., and Sturm J. C., Biomicrofluidics 11(5), 054107 (2017). 10.1063/1.4985771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang M., Wen C. Y., Wu L. L., Hong S. L., Hu J., Xu C. M., Pang D. W., and Zhang Z. L., Lab Chip 16(7), 1214–1223 (2016). 10.1039/C5LC01555C [DOI] [PubMed] [Google Scholar]
- 24. Chou H. P., Unger M. A., and Quake S. R., Biomed. Microdevices 3(4), 323–330 (2001). 10.1023/A:1012412916446 [DOI] [Google Scholar]
- 25. Ballard M., Owen D., Mills Z. G., Hesketh P. J., and Alexeev A., Microfluid. Nanofluid. 20(6), 88 (2016). 10.1007/s10404-016-1750-1 [DOI] [Google Scholar]
- 26. Niu X. Z., Liu L. Y., Wen W. J., and Sheng P., Appl. Phys. Lett. 88(15), 153508 (2006). 10.1063/1.2195567 [DOI] [Google Scholar]
- 27. Fan J., Li B., Xing S., and Pan T., Lab Chip 15(12), 2670–2679 (2015). 10.1039/C5LC00432B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abolhasani M., Oskooei A., Klinkova A., Kumacheva E., and Günther A., Lab Chip 14(13), 2309–2318 (2014). 10.1039/C4LC00131A [DOI] [PubMed] [Google Scholar]
- 29. Tang L. J., Liu J. Q., and Yang B., in IEEE International Electron Devices Meeting (IEDM) (2015), pp. 29.7.1–29.7.4. [Google Scholar]
- 30. Suteria N. S., Nekouei M., and Vanapalli S. A., Lab Chip 18(2), 343–355 (2018). 10.1039/C7LC00889A [DOI] [PubMed] [Google Scholar]
- 31. Lee D. H., Li X., Ma N., Digman M. A., and Lee A. P., Lab Chip 18(9), 1349–1358 (2018). 10.1039/C7LC01301A [DOI] [PubMed] [Google Scholar]
- 32. Hao S. J., Wan Y., Xia Y. Q., Zou X., and Zheng S. Y., Adv. Drug. Delivery Rev. (published online, 2018) 10.1016/j.addr.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 33. Coumans F. A., van Dalum G., Beck M., and Terstappen L. W., PloS one 8, e61770 (2013). 10.1371/journal.pone.0061770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karabacak N. M., Spuhler P. S., Fachin F., Lim E. J., Pai V., Ozkumur E., Martel J. M., Kojic N., Smith K., Chen P. I., Yang J., Hwang H., Morgan B., Trautwein J., Barber T. A., Stott S. L., Maheswaran S., Kapur R., Haber D. A., and Toner M., Nat. Protoc. 9(3), 694–710 (2014). 10.1038/nprot.2014.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ko J., Bhagwat N., Yee S. S., Black T., Redlinger C., Romeo J., O'Hara M., Raj A., Carpenter E. L., Stanger B. Z., and Issadore D., Lab Chip 17(18), 3086–3096 (2017). 10.1039/C7LC00703E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen J., Chen D., Yuan T., Xie Y., and Chen X., Biomicrofluidics 7(3), 034106 (2013). 10.1063/1.4808179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsai S. C., Hung L. Y., and Lee G. B., Biomicrofluidics 11(3), 034122 (2017). 10.1063/1.4991476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the performance of microbead isolation and capture; on-chip oscillation process of both microbeads and cells; CTC enrichment from whole blood; loss of some CTCs using the chip with 12 μm gap; and CTC capture performance via chips with 12 μm gap and 18 μm gap.