Abstract
Phosphatidylinositol phosphate kinases (PIPKs) generate a lipid messenger phosphatidylinositol 4,5-bisphosphate (PI4,5P2) that controls essentially all aspects of cellular functions. PI4,5P2 rapidly diffuses in the membrane of the lipid bilayer and does not greatly change in membrane or cellular content, and thus PI4,5P2 generation by PIPKs is tightly linked to its usage in subcellular compartments. Based on this verity, recent study of PI4,5P2 signal transduction has been focused on investigations of individual PIPKs and their underlying molecular regulation of cellular processes. Here, we will discuss recent advances in the study of how PIPKs control specific cellular events through assembly and regulation of PI4,5P2 effectors that mediate specific cellular processes. A focus will be on the roles of PIPKs in control of the phosphoinositide 3-kinase pathway and autophagy.
Keywords: Phosphoinositide; Phosphatidylinositol phosphate kinase; Phosphatidylinositol 4,5-bisphosphate; Phosphoinositide 3-kinase; Autophagy; Signal transduction
Introduction
PI4,5P2 is a lipid messenger that has a plethora of fundamental roles in the regulation of cell physiology including survival, proliferation, motility, immune responses, gene expression and others (Choi et al., 2015; Di Paolo and De Camilli, 2006(Barlow et al. 2012)). PI4,5P2 has long been considered as a substrate for type I phosphoinositide 3-kinases (PI3K) or phospholipases to generate other signaling lipids. However, it is now well established that PI4,5P2 has its own signaling roles (Choi et al. 2015). PI4,5P2 binds to proteins targets called PI4,5P2 effectors and regulates their functions by controlling activity, subcellular localization, or protein-protein interaction (Figure 1). Due to its versatile cellular roles, dysregulation of PI4,5P2-generating enzymes are implicated in human diseases including neurological disorders and cancers (McCrea and De Camilli, 2009).
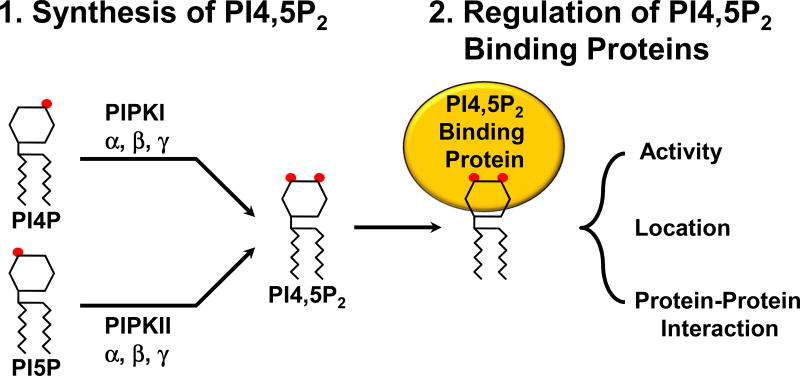
Figure 1.
A schematic representation of PI4,5P2 generation by type I vs. type II PIP kinases. Type I PIP kinase isoforms phosphorylate the 5’ hydroxyl group of the insositol ring, while type II enzymes phosphorylate the 4’ hydroxyl group. Once PI4,5P2 is generated, it binds to a set of protein targets called PI4,5P2 effectors and controls their activity, subcellular location, and PI4,5P2 effectors’ interaction with other proteins.
A majority of cellular PI4,5P2 is generated by type I and type II phosphatidylinositol phosphate kinases (PIPK) and α, β, and γ isoforms of each type of PIPK, and splice variants are expressed in higher eukaryotes (van den Bout and Divecha, 2009). Type I PIPKs (PIPKIs) phosphorylate the 5’ hydroxyl group of the inositol head group of phosphatidylinositol 4-phosphate (PI4P), whereas type II PIPKs (PIPKIIs) phosphorylate the 4’ hydroxyl group of the inositol head group of PI5P (Figure 1) to generate PI4,5P2.
The subcellular location of each PIPK isoform is diverse. For example, PIPKIα, PIPKIβ, and PIPKIγi1 localize in the plasma membrane (Choi et al., 2013; Giudici et al., 2006; Xia et al., 2011). PIPKIγi2 is found in the focal adhesion and PIPKIγi5 localizes in endosomes and lysosomes (Di Paolo et al., 2002; Ling et al., 2002; Tan et al., 2016b). Some PIPKs including PIPKIα, PIPKIγi4, PIPKIIα, and PIPKIIβ localize in the nucleus (Ciruela et al., 2000; Schill and Anderson, 2009)(Boronenkov et al., 1998). This subcellular targeting indicates that PIPK isoforms controls site-specific PI4,5P2 generation and function at each subcellular compartment (Choi et al., 2015). There is significant evidence that specific PIPK isoforms target to distinct cellular locations via interacting with specific partners. Often, these targeting proteins bind to PI4,5P2 and their functions are controlled by PI4,5P2 binding indicating that they are PI4,5P2 effectors. Thus, the physical association of PI4,5P2 effectors with PIPKs defines in part the specificity of the PI4,5P2 signal. PI4,5P2 very rapidly diffuses in membranes and phospholipid bilayers (Golebiewska et al., 2011; Golebiewska et al., 2008). As such, to define specificity of PI4,5P2 signal generation in some cases is tightly linked to its usage by an interaction between the PIPK and the PI4,5P2 effector (Choi et al., 2015). During the last 10 years, the identification and understanding of how PIPK isoforms control specific cellular processes has expanded, emphasizing the significance of the pathophysiological role of PI4,5P2 and its generating enzymes. In this review, we will summarize and discuss the most recent advances and implications.
The key role of PIPK in the PI3K pathways
PI4,5P2 regulates many proteins directly but one of its most notable roles is as a substrate for the agonist-activated PI3Ks (Chalhoub and Baker, 2009)(Kriplani et al. 2015). PIPKs generate PI4,5P2, which is used by class I PI3Ks for PI3,4,5P3 synthesis. PI3,4,5P3 is a paramount lipid messenger and, once generated, it drives cellular events that promote cell cycle progression and resistance to cell death (Chalhoub and Baker, 2009). PI3,4,5P3 binds and activates diverse protein targets including phosphoinositide-dependent kinase 1 (PDK1) and Akt. As a result, the class I PI3K nodes are frequently mutated in human cancers (Samuels and Waldman, 2010; Vanhaesebroeck et al., 2010). It has been assumed that class I PI3Ks utilize a preexisting PI4,5P2 pool for PI3,4,5P3 synthesis, as the cellular concentration of PI4,5P2 is at least two orders of magnitude higher than that of PI3,4,5P3 (Insall and Weiner, 2001; Stephens et al., 1993). However, this paradigm is challenged by findings that a substantial fraction of preexisting PI4,5P2 is sequestered by a set of PI4,5P2-binding proteins (Golebiewska et al., 2008; McLaughlin et al., 2002) and thus unavailable for use by class I PI3Ks.
This suggests that de novo PI4,5P2 generation might be coordinated with class I PI3K activation and PI3,4,5P3 generation at a specific cellular compartment. In support of this notion, PI4,5P2 colocalization with PI3,4,5P3 at the leading edge of migrating leukocytes has been reported (Sharma et al., 2008). In this and other studies, PI4,5P2-generating enzymes PIPKIα and PIPKIγ also localize at the leading edge of migrating cells (Choi et al., 2013; Thapa et al., 2011), further supporting a concerted PI4,5P2 and PI3,4,5P3 generation mechanism. Moreover, recently specific phosphatidylinositol 4-kinases and PIPKs have been shown to be required for PI3,4,5P3 generation under certain conditions (Fets et al., 2014; Nakatsu et al., 2012; Thapa et al., 2015). This indicates that specific phosphatidylinositol 4-kinases and PIPKs are required for de novo synthesis of the PI4,5P2 that is then used for PI3,4,5P3 generation, suggesting compartmentalization or synergism between these enzymes. Further, the internalization of receptors such as epidermal growth factor receptor (EGFR) results in continued signaling through the PI3K pathway indicating that the PI3,4,5P3 generation occurs in the endosomal compartments (Sorkin and Goh, 2009; Sun et al., 2007; Tomas et al., 2014). Yet, there is substantial evidence that there is little PI4P, PI4,5P2 or PI3,4,5P3 in the endosome (Di Paolo and De Camilli, 2006; Tan et al., 2015b).
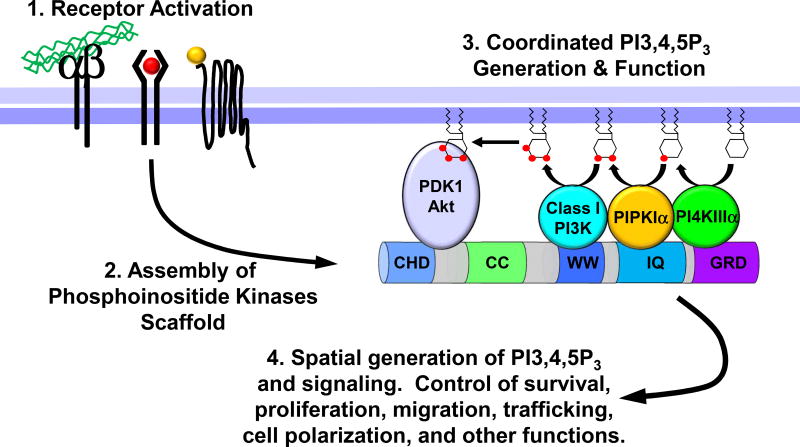
This begs the question: How does the PI3K pathway function in an endosomal compartment that lacks substrates for PI3,4,5P3 generation? This has been answered in a study that shows the molecular details of a concerted mechanism where the full PI3K pathway components are assembled downstream of agonist stimulation (Choi et al., 2016). Here, all of the PI4P-, PI4,5P2-, and PI3,4,5P3-generating enzymes bind to distinct subdomains of the IQ motif-containing GTPase Activating Protein 1 (IQGAP1). In this IQGAP1-scaffolded phosphoinositide kinase complex, PI4KIIIα, PIPKIα, and class I PI3K sequentially phosphorylate phosphatidylinositol at 4’, 5’, and 3’ positions of the inositol ring to generate PI3,4,5P3 in a processive manner (Figure 2). Furthermore, PDK1 and Akt that are activated by PI3,4,5P3 are also associated with the IQGAP1 multi-kinase complex. Association of the PI3,4,5P3 effectors in the IQGAP1 complex provides a mechanism of specificity of the PI3K-Akt pathway (Choi et al., 2016)(Rameh and Mackey, 2016). More recently, a downstream target of Akt FOXO1 is shown to bind to the IQGAP1 complex (Pan et al., 2017). This suggests that the IQGAP1 multi-enzyme complex streamlines the synthesis of the PI3,4,5P3 signal and that PI3,4,5P3 is directly transferred to Akt and FOXO1, leading to their activation. The involvement of PIPKIα in PI3,4,5P3 generation and Akt activation is further supported by a study showing that inhibition of PIPKIα using an ATP-mimetic small molecule reduces PI3,4,5P3 levels and Akt phosphorylation in prostate cancer xenografts (Semenas et al., 2014).
Figure 2.
A schematic representation of the IQGAP1-scaffolded class I PI3K-Akt signaling platform. IQGAP1 binds to PI4KIIIα, PIPKIα, and class I PI3K that generate PI4P, PI4,5P2, and PI3,4,5P3, respectively. The generated PI3,4,5P3 is directly transferred to PDK1 and Akt, which are also associated with IQGAP1. In the IQGAP1-mediated multi-kinase complex, PI3,4,5P3 generation is tightly linked to its usage.
PIPKIγ is also involved in the PI3K-Akt pathway. PIPKIγ is shown to bind to IQGAP1 (Choi et al., 2013), and depletion of PIPKIγ reduces Akt phosphorylation (Thapa et al., 2017). Akt phosphorylation is increased during epithelial to mesenchymal transition (EMT), a process by which epithelial cells lose the polarity and cell-cell contacts, and gain migratory and invasive phenotypes and thus is a fundamental step in oncogenic progression (Ye and Weinberg, 2015). Interestingly, PIPKIγ interaction with IQGAP1 and Akt phosphorylation are increased in cells undergoing EMT (Thapa et al., 2017). Also, while EMT progresses, PIPKIγ expression levels gradually increase whereas PIPKIα expression decreases. This suggests that PIPKIα might be switched to PIPKIγ in the IQGAP1 multi-kinase complex during the EMT process. In support of this isoform switch hypothesis, in some epithelial cells depletion of PIPKIα dramatically increases the expression of PIPKIγ leading to an enhanced association of PIPKIγ with IQGAP1 (Choi and Anderson, 2016). Further work is needed to fully understand how a putative switch between PIPKIα and PIPKIγ controls the PI3K-Akt pathway. A structural study reveals that the kinase domain of PIPKIα forms a side-to-side homodimer and that the dimerization is critical for catalytic activity (Hu et al., 2015). The PIPKIα homodimer is stabilized by an ionic contact and a hydrophobic interaction of the monomers and, interestingly, these sequences are conserved in PIPKIγ, raising a possibility that PIPKIγ also may form a homodimer or that PIPKIα and PIPKIγ might form a heterodimer. In support of this, PIPKII isoforms are shown to form heterodimers (Rao et al., 1998). These results in sum suggest that in the IQGAP1 multi-kinase complex PIPKIα and PIPKIγ may have a redundant role by possibly forming a heterodimer.
PIPKIIs are implicated in the PI3K-Akt pathway. PIPKIIβ germline knockout mice display enhanced skeletal muscle Akt activity and insulin sensitivity (Lamia et al., 2004). However, paradoxically, these mice are smaller compared to the wild type mice and have decreased adiposity on a high-fat diet, consistent with a report that a loss-of-mutant of a PIPKII homolog in Drosophila reduces body weight (Gupta et al., 2013). Conversely, ectopic expression of PIPKIIβ reduces cellular PI3,4,5P3 levels and Akt activity (Carricaburu et al., 2003). Recently, mice with a germline deletion of PIPKIIγ are reported to have a hyperactivation of mammalian target of rapamycin complex 1 (mTORC1) signaling, which is a downstream target of Akt. These results suggest that PIPKIIβ and PIPKIIγ does not supply PI4,5P2 for class I PI3K generation of PI3,4,5P3 and activation of Akt. Instead, PIPKIIβ and PIPKIIγ are hypothesized to control PI5P levels in cells rather than generate PI4,5P2. For example, depletion of PIPKIIβ increases PI5P levels leading to cellular senescence of cancer cells (Emerling et al., 2013) and differentiation of myoblasts (Stijf-Bultsma et al., 2015). However, the molecular details of how PI5P regulates these processes are not fully understood. Several PI5P effector proteins are identified but systematic approaches are needed to elucidate the roles of a unique lipid messenger PI5P. Contrast to PIPKIIβ and PIPKIIγ, deletion of PIPKIIα reduces PI3,4,5P3 levels and Akt and mTORC1 phosphorylation in B lymphocytes (Bulley et al., 2016). It appears that reduction of the PI3K-Akt signaling by PIPKIIα deletion is due to defective PI4,5P2 generation rather than PI5P removal. These contrasting results are intriguing as PIPKIIβ is shown to interact with a pronounced fraction of PIPKIIα (Bultsma et al., 2010). Further works are needed to elucidate the relative contribution of each PIPKII isoform in PI4,5P2 generation vs. PI5P removal.
The role of PIPKs in regulation of autophagy
Macroautophagy (referred to as autophagy in this paper) is an indispensable cellular process that degrades defective organelles, protein aggregates, and long-lived proteins via the lysosomal pathway and is thus essential for organelle/protein homeostasis and nutrient recycling, particularly during starvation and stress (Dall'Armi et al., 2013; Kaur and Debnath, 2015). Autophagy is a step-wise process that involves an orchestrated sequence of membrane remodeling and trafficking events that are controlled by a family of autophagy-related (ATG) proteins. Autophagy initiation begins with the formation of the phagophore assembly site (PAS) (Galluzzi et al., 2017; Yang and Klionsky, 2010; Yu et al., 2017). Further nucleation of PAS requires the class III PI3K complex composed of the kinase subunit vacuolar protein sorting 34 (VPS34) and its regulatory subunits VPS15, ATG14, and Beclin 1. The phagophore membrane elongates and eventually forms a spherical structure called an autophagosome with double layer membranes. Autophagosome formation is largely dependent on two unique ubiquitin-like conjugation pathways (Tanida et al., 2004). The first generates the AGT5–ATG12 conjugate, then it forms a multimeric complex with ATG16, and the second produces the conjugation of a membrane lipid phosphatidylethanolamine (PE) to the microtubule-associated protein 1 light chain 3 (LC3). PE-conjugated LC3 is required for the expansion of autophagic membranes and the fusion of autophagosomes with lysosomes via its oligomerization and membrane tethering capability (Nakatogawa et al., 2009). Finally, autophagosomes fuse with endocytic and lysosomal compartments, leading to the formation of the autolysosome.
The functions of phosphoinositides, particularly PI3P, the product of the class III PI3K are well characterized in autophagy. During autophagosome biogenesis initiation and maturation, PI3P plays critical roles. A set of autophagy regulatory proteins contains PI3P binding modules such as the PX, FYVE, and BATS domain and thus are recruited to the site of PI3P generation at autophagic membranes in a specific order (Dall'Armi et al., 2013). The origin of the phagophore membrane is controversial with evidence that the ER and Golgi provide membrane but the endosome and plasma membrane have also been implicated (Ge et al., 2013; Ktistakis and Tooze, 2016; Mari et al., 2011).
Multiple receptors are thought to control mTOR regulation of autophagy (Yang and Klionsky, 2010). Agonist activated EGFR through regulation of mTOR and directly by phosphorylation of Beclin1 inhibits autophagy (Wei et al., 2013; Yang and Klionsky, 2010). Remarkably, the inactive EGFR promotes autophagy initiation downstream of multiple signals including serum starvation (Tan et al., 2016a; Tan et al., 2015c). The agonist activated and the inactive EGFR are both controlled by the lysosomal protein transmembrane 4 beta (LAPTM4B) and a PIPKI. LAPTM4B is an oncogene that is overexpressed in many human cancers (Meng et al., 2016). LAPTM4B is located in the endosomal compartment but is concentrated in the late endosome and lysosome (Tan et al., 2015a). For the agonist activated EGFR, LAPTM4B controls the downregulation of EGFR by blocking its sorting to intraluminal vesicles of the multivesicular endosome and its subsequent degradation in the lysosome (Tan et al., 2015a). The activated EGFR does not directly associate with LAPTM4B (Tan et al., 2015a). Instead, the N-terminus of LAPTM4B binds to the unique C-terminus of PIPKIγi5 and as LAPTM4B is a PI4,5P2 binding protein the generation of PI4,5P2 controls the activity of the Nedd4 ubiquitin ligase that is associated with the C-terminus of LAPTM4B. Nedd4 then ubiquitinates Hrs which blocks Hrs’ ability to sort EGFR to intraluminal vesicles. The Nedd4 ubiquitination of Hrs inhibits its role in active EGFR intraluminal sorting and thus the lysosomal degradation of agonist activated EGFR (Tan et al., 2015a).
In serum starved cells and upon its inhibition, EGFR becomes localized to the late endosome (Tan et al., 2016a; Tan et al., 2015c). The mechanism for inactive EGFR localization is through a direct interaction with LAPTM4B that is specific for the inactive EGFR (Tan et al., 2015c). When LAPTM4B is knocked down the inactive EGFR no longer localized to the late endosome. Inactive EGFR localizes to endosomal compartments that are adjacent to the ER (Eden, 2016; Eden et al., 2012; Eden et al., 2010; Tan et al., 2015c) and also close to sites of autophagy, i.e. next to the induced LC3-II an autophagosome marker (Tan et al., 2015c). Significantly, the loss of either EGFR or LAPTM4B inhibits basal and serum starvation-induced autophagy. The combined data indicate that LAPTM4B and inactive EGFR together control autophagy. The underlying mechanism is through interactions with LAPTM4B and the Sec5 exocyst sub-complex that facilitates Rubicon association with EGFR, which promotes the release of Beclin 1 from Rubicon and initiates autophagy (Tan et al., 2015c). These data also suggest a role for the LAPTM4B associated PIPKI, the PIPKIγi5.
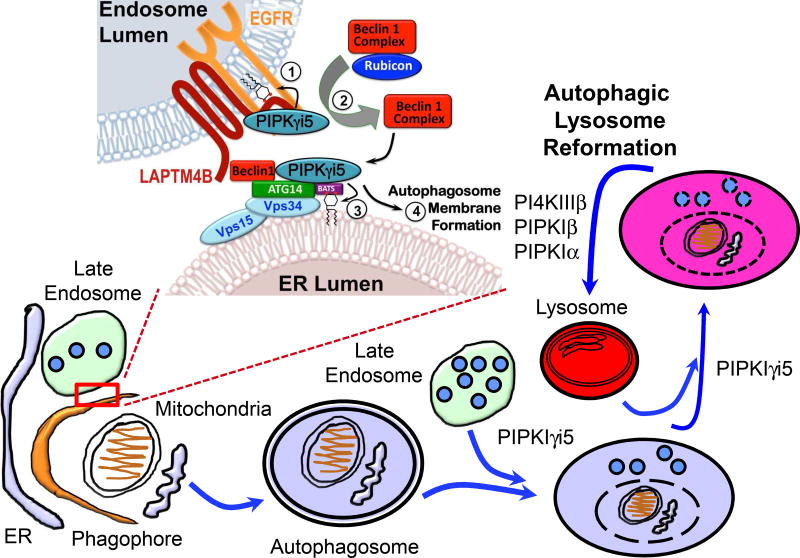
Autophagy is controlled by the synchronized actions of the autophagy-related (ATG) proteins. Barkor/ATG14-VPS34 is a class III PI3K complex and its product PI3P regulates autophagy initiation. ATG14 contains a C-terminal Barkor/ATG14 autophagosome-targeting sequence (BATS) domain that senses PI3P-containing membranes that are curved. The ATG14 BATS domain also binds PI4,5P2, but the functional is unclear. ATG14 interacts with PIPKIγi5. In addition, PIPKIγi5 and PI4,5P2 localize with autophagosomes that are associated with late endosomes and the ER (Tan et al., 2016b). PI4,5P2 generation at these sites requires PIPKIγi5. Loss of PIPKIγi5 results in a loss of ATG14, UV irradiation resistance-associated gene (UVRAG), and Beclin 1 and a block of autophagy. PI4,5P2 binding to the ATG14–BATS domain regulates ATG14 interaction with VPS34 and Beclin 1, and thus plays a key role in ATG14 complex assembly and autophagy initiation. PIPKIγi5 is required for autophagy initiation and the fusion of the autophagosome with the lysosome (Tan et al., 2016b). As depicted in Figure 3, the EGFR, LAPTM4B and PIPKIγi5 and the Beclin 1, Vps34, ATG14 and PIPKIγi5 complexes may occupy different compartments but as a large fraction of autophagosomes colocalize with late endosomes and endoplasmic reticulum and these sites have PI4,5P2 and PIPKIγi5 appears to be responsible for the autophagosome-associated PI4,5P2 generation.
Figure 3.
A schematic representation of the role of PIPK isoforms in autophagy. Each PIPK isoform has unique but sometimes redundant roles in the steps of autophagy progression. PIPKs and their product PI4,5P2 now have clear functional roles in autophagy generally by promoting membrane reformation.
It is clear that EGFR, LAPTM4B, and PIPKIγi5 are localized to the endosome that the late endosome (multivesicular endosome) is largely docked with the ER. Further, the multivesicular endosome can fuse with the autophagosome. Thus, the EGFR, LAPTM4B, and PIPKIγi5 on the endosome may regulate fusion with the autophagosome. Further, the PIPKIγi5 does control the fusion of the lysosome with the autophagosome (Tan et al., 2016b). Mechanistically, PI4,5P2 generation by PIPKIγi5 on these autophagosomes is required for ATG14 function. PIPKIγi5-mediated PI4,5P2 signaling functions upstream of PI3P by stabilizing ATG14 and Beclin 1 and promoting the ATG14-Beclin 1-VPS34 complex assembly. PI4,5P2-binding of ATG14 on its BATS domain, which also binds PI3P, regulates the association with VPS34 and Beclin 1. This is important as ATG14 controls PI3K activity and localization of VPS34 to autophagosomes (Dall'Armi et al., 2013). Thus, PI4,5P2 generation by PIPKIγi5 at phagophore initiation is critical for driving the entire autophagy progression via interactions with multiple membrane compartments. Autophagy initiates at a preautophagosomal structure (PAS) (Dall'Armi et al., 2013; Ktistakis and Tooze, 2016; Mari et al., 2011; Yang and Klionsky, 2010). The role for the PIPKIγi5 appears to be at the endosome and ER and the PI4,5P2 signaling pathway at these sites that controls ATG14 complex stability and assembly required for autophagy. These results define an unexpected role for PIPKIγi5 and PI4,5P2 in autophagic membrane trafficking and further indicates a role for the ER-late endosome contacts in autophagy regulation as a potential mammalian PAS as illustrated in Figure 3.
The small G-protein Arf6 regulates PIPKIγ isoforms and phospholipase D (PLD), further the product of PLD activity PA activates PIPKI isoforms (Funakoshi et al., 2010, 2011; Jenkins et al., 1994; Massenburg et al., 1994). Arf6 regulates phagophore formation and colocalizes with ATG12 and ATG16, which control phagophore elongation, and knockdown of Arf6 reduces ATG12- and ATG16-positive phagophore (Moreau et al., 2012). Arf6 controls phagophore formation by regulating PI4,5P2 generation in part at the plasma membrane, as depletion of the plasma membrane pool of PI4,5P2 by targeted delivery of PI4,5P2-specific 5-phosphatase or knockdown of all PIPKIγ isoforms reduced the number of ATG12-positive phagophores. Additionally, ATG12- and ATG16-positive compartments partially overlap with PI4,5P2 staining. These data are consistent with a role for Arf6 in regulating the kinase activity of PIPKIγi5 or similar kinases in promoting roles in autophagosome formation.
Roles of PI4,5P2 and PI4,5P2-metabolizing enzymes in autophagy are further validated as PI4,5P2 is found on membranes of autophagic compartments located in the late endosomes and lysosomes (De Leo et al., 2016). In this study, the PIPKIα and PIPKIβ are appear responsible for PI4,5P2 generation in these compartments (Figure 3). Importantly, knockdown of OCRL (Lowe oculocerebrorenal syndrome protein) which converts PI4,5P2 to PI4P, led to accumulation of PI4,5P2 on membranes of autophagosome-lysosome fusion sites and this dramatically increased LC3 puncta formation (Nakamura et al., 2016).
Recently, PI4,5P2 generation is shown to play an important role in a key step in autophagy, autophagic lysosome reformation (ALR). ALR that occurs during prolonged starvation refers to the de novo biogenesis of lysosomes from existing autolysosomes via tubulation of the limiting membranes (Chen and Yu, 2017). Proteomic analyses of these tubules identify clathrin, its associated molecules, and PI4,5P2-generating enzymes as key mediators of ALR (Rong et al., 2012). Further characterization of the proteomic screen with biochemical and cell biological approaches indicates that PI4,5P2 generated by PIPKIβ mediates the recruitment of clathrin via its AP2 adaptor complex to autolysosomes. Of note, PI4,5P2 plays a key role in AP2 complex formation onto membranes (Choi et al., 2015). Contrary to PIPKIβ, another PIPKI isoform PIPKIα appears to regulate the fission of ALR tubules reforming from autolysosomes. Interestingly, PI4P is uniformly distributed on membranes of autolysosomes, while PI4,5P2 (and PIPKIα) is enriched on ALR tubules emerging from autolysosomes (Figure 3).
Autophagy is triggered by diverse cellular cues (Dall'Armi et al., 2013; Kaur and Debnath, 2015; Tan et al., 2016a). In general, upon starvation or stress the concentration of cellular energy molecules including ATP and GTP diminishes. The reduction of energy molecule concentration is recognized by a few energy sensing molecules. For instance, under nutrient scarcity, a key energy sensor AMP activated protein kinase (AMPK) phosphorylates ULK1 which is a mammalian homolog of yeast ATG1, leading to autophagy initiation. In contrast, under nutrient sufficiency, high mTOR activity prevents ULK1 activation by phosphorylating different residues on ULK1 (Kim et al., 2010). Recently, PIPKII isoforms especially PIPKIIβ are identified as cellular GTP sensors (Sumita et al., 2015). PIPKIIβ preferentially utilizes GTP to produce PI4,5P2 from PI5P. Interestingly, PIPKIIβ has a Michaelis constant (KM) value for GTP that falls within the range of physiological GTP concentration and its kinase activity changes in direct proportion to the cellular GTP concentration. This suggests that PIPKIIβ is an active GTP sensor. Based on this, one can expect that in energy scarcity, reduction of cellular GTP concentration inhibits PIPKIIβ kinase activity, leading to the accumulation of cellular PI5P. Interestingly, PI5P generation is linked autophagosome biogenesis (Vicinanza et al., 2015).
Taken together, PI4,5P2 and PIPK isoforms are critical for multiple steps in autophagy. However, the molecular details of how and where PI4,5P2 effectors control the autophagic process is not well understood compared to the PI3P signaling pathway. Systematic characterization of the protein interactome of PI4,5P2 might provide global insights of the PI4,5P2 signal transduction during autophagy.
Summary and future prospects
PIPKs generate a lipid messenger PI4,5P2, and signaling specificity of PI4,5P2 is determined by PIPK association with PI4,5P2 effectors (Choi et al., 2015). A direct contribution of PI4,5P2-generating enzymes in PI3,4,5P3 generation by class I PI3Ks has long been suspected and now one mechanism is emerging. IQGAP1 scaffolds all of the phosphoinositide kinases that are need to generated PI3,4,5P3 from phosphatidylinositol. Also, PI3,45P3 effectors PDK1 and Akt are in the IQGAP1 multi-kinase complex, enabling the generated PI3,45P3 to be directly transferred to PI3,4,5P3 effectors and used for their activation (Figure 2). This seemingly complicated mechanism of PI3,4,5P3 generation and function however well fits with a key principle of signal transduction: signals are only generated when and where they are needed (Good et al., 2011). Another key feature of the IQGAP1-mediated class I PI3K-Akt pathway is that it can be spatially assembled on any membrane. Concentration of PI4P and PI4,5P2 is reported to be high in Golgi and the plasma membrane, respectively, whereas low in other membranes including endosomes (Balla, 2013). The IQGAP1 multi-kinase complex has the potential to sequentially generate PI3,4,5P3 from phosphatidylinositol. Importantly, phosphatidylinositol is ubiquitously found in all membranes (Balla, 2013; van Meer et al., 2008). We envision that assembly of the IQGAP1-scaffolded kinases will provide a route to produce PI3,4,5P3 on any membrane. Further works are needed for elucidating when and where the IQGAP1 complex is assembled.
Autophagy involves remodeling and trafficking of membranes. PI4,5P2 and its generating enzymes (PIPKs) have long been implicated in membrane dynamics. Thus it is not surprising that PI4,5P2 and PIPKs control autophagy. However, it is unanticipated that PI4,5P2 directly activates autophagy regulatory proteins (Tan et al., 2016b). Based on current understanding, PI4,5P2 appears to control essentially every step of autophagy, but only a limited number of PI4,5P2 effector autophagic proteins have been discovered. Thus, more systematic approaches are required to fully understand the functional role of PI4,5P2 and PIPKs in the control of autophagy.
PIPK isoforms, especially PIPKIα and PIPKIIα, activate the PI3K-Akt-mTORC pathway (Bulley et al., 2016; Choi et al., 2016). mTORC is reported to suppress autophagy by phosphorylating ULK1 (Kim et al., 2010). Paradoxically in another study, PIPKIα appears to promote autophagy (De Leo et al., 2016). These seemingly contradictory results can be in part explained by cell type specificity. Further works are needed to elucidate the exact role and the relative contribution of each PIPK isoform in the regulation of the PI3K-Akt-mTORC pathway vs. autophagy.
Acknowledgments
Due to space constraints some relevant studies may not have been referenced. This work was supported by NIH grants to RAA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have declared no conflict of interest.
References
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiological reviews. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulley SJ, Droubi A, Clarke JH, Anderson KE, Stephens LR, Hawkins PT, Irvine RF. In B cells, phosphatidylinositol 5-phosphate 4-kinase-alpha synthesizes PI(4,5)P2 to impact mTORC2 and Akt signaling. Proc Natl Acad Sci U S A. 2016;113:10571–10576. doi: 10.1073/pnas.1522478113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultsma Y, Keune WJ, Divecha N. PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform PIP4Kalpha. Biochem J. 2010;430:223–235. doi: 10.1042/BJ20100341. [DOI] [PubMed] [Google Scholar]
- Carricaburu V, Lamia KA, Lo E, Favereaux L, Payrastre B, Cantley LC, Rameh LE. The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc Natl Acad Sci U S A. 2003;100:9867–9872. doi: 10.1073/pnas.1734038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yu L. Recent progress in autophagic lysosome reformation. Traffic. 2017;18:358– 361. doi: 10.1111/tra.12484. [DOI] [PubMed] [Google Scholar]
- Choi S, Anderson RA. IQGAP1 is a phosphoinositide effector and kinase scaffold. Adv Biol Regul. 2016;60:29–35. doi: 10.1016/j.jbior.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Hedman AC, Sayedyahossein S, Thapa N, Sacks DB, Anderson RA. Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat Cell Biol. 2016;18:1324–1335. doi: 10.1038/ncb3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Thapa N, Hedman AC, Li Z, Sacks DB, Anderson RA. IQGAP1 is a novel phosphatidylinositol 4,5 bisphosphate effector in regulation of directional cell migration. EMBO J. 2013;32:2617–2630. doi: 10.1038/emboj.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Thapa N, Tan X, Hedman AC, Anderson RA. PIP kinases define PI4,5P(2)signaling specificity by association with effectors. Biochim Biophys Acta. 2015;1851:711–723. doi: 10.1016/j.bbalip.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruela A, Hinchliffe KA, Divecha N, Irvine RF. Nuclear targeting of the beta isoform of type II phosphatidylinositol phosphate kinase (phosphatidylinositol 5-phosphate 4-kinase) by its alpha-helix 7. Biochem J. 2000;346(Pt 3):587–591. [PMC free article] [PubMed] [Google Scholar]
- Dall'Armi C, Devereaux KA, Di Paolo G. The role of lipids in the control of autophagy. Curr Biol. 2013;23:R33–45. doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leo MG, Staiano L, Vicinanza M, Luciani A, Carissimo A, Mutarelli M, Di Campli A, Polishchuk E, Di Tullio G, Morra V, et al. Autophagosome-lysosome fusion triggers a lysosomal response mediated by TLR9 and controlled by OCRL. Nat Cell Biol. 2016;18:839–850. doi: 10.1038/ncb3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, Chang S, Guo J, Wenk MR, De Camilli P. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature. 2002;420:85–89. doi: 10.1038/nature01147. [DOI] [PubMed] [Google Scholar]
- Emerling BM, Hurov JB, Poulogiannis G, Tsukazawa KS, Choo-Wing R, Wulf GM, Bell EL, Shim HS, Lamia KA, Rameh LE, et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell. 2013;155:844–857. doi: 10.1016/j.cell.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudici ML, Lee K, Lim R, Irvine RF. The intracellular localisation and mobility of Type Igamma phosphatidylinositol 4P 5-kinase splice variants. FEBS Lett. 2006;580:6933–6937. doi: 10.1016/j.febslet.2006.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiewska U, Kay JG, Masters T, Grinstein S, Im W, Pastor RW, Scarlata S, McLaughlin S. Evidence for a fence that impedes the diffusion of phosphatidylinositol 4,5-bisphosphate out of the forming phagosomes of macrophages. Mol Biol Cell. 2011;22:3498–3507. doi: 10.1091/mbc.E11-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiewska U, Nyako M, Woturski W, Zaitseva I, McLaughlin S. Diffusion coefficient of fluorescent phosphatidylinositol 4,5-bisphosphate in the plasma membrane of cells. Mol Biol Cell. 2008;19:1663–1669. doi: 10.1091/mbc.E07-12-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Toscano S, Trivedi D, Jones DR, Mathre S, Clarke JH, Divecha N, Raghu P. Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates TOR signaling and cell growth during Drosophila development. Proc Natl Acad Sci U S A. 2013;110:5963–5968. doi: 10.1073/pnas.1219333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Yuan Q, Kang X, Qin Y, Li L, Ha Y, Wu D. Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled. Nat Commun. 2015;6:8205. doi: 10.1038/ncomms9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall RH, Weiner OD. PIP3, PIP2, and cell movement--similar messages, different meanings? Dev Cell. 2001;1:743–747. doi: 10.1016/s1534-5807(01)00086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2010;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Peroni OD, Kim YB, Rameh LE, Kahn BB, Cantley LC. Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta−/− mice. Mol Cell Biol. 2004;24:5080–5087. doi: 10.1128/MCB.24.11.5080-5087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- McCrea HJ, De Camilli P. Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology (Bethesda) 2009;24:8–16. doi: 10.1152/physiol.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Wang J, Gambhir A, Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Puri C, Rubinsztein DC. Arf6 promotes autophagosome formation via effects on phosphatidylinositol 4,5-bisphosphate and phospholipase D. J Cell Biol. 2012;196:483–496. doi: 10.1083/jcb.201110114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Pan CW, Jin X, Zhao Y, Pan Y, Yang J, Karnes RJ, Zhang J, Wang L, Huang H. AKT-phosphorylated FOXO1 suppresses ERK activation and chemoresistance by disrupting IQGAP1-MAPK interaction. EMBO J. 2017;36:995–1010. doi: 10.15252/embj.201695534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh LE, Mackey AM. IQGAP1 makes PI(3)K signalling as easy as PIP, PIP2, PIP3. Nat Cell Biol. 2016;18:1263–1265. doi: 10.1038/ncb3440. [DOI] [PubMed] [Google Scholar]
- Rao VD, Misra S, Boronenkov IV, Anderson RA, Hurley JH. Structure of type IIbeta phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation. Cell. 1998;94:829–839. doi: 10.1016/s0092-8674(00)81741-9. [DOI] [PubMed] [Google Scholar]
- Rong Y, Liu M, Ma L, Du W, Zhang H, Tian Y, Cao Z, Li Y, Ren H, Zhang C, et al. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat Cell Biol. 2012;14:924–934. doi: 10.1038/ncb2557. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol. 2010;347:21–41. doi: 10.1007/82_2010_68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schill NJ, Anderson RA. Two novel phosphatidylinositol-4-phosphate 5-kinase type Igamma splice variants expressed in human cells display distinctive cellular targeting. Biochem J. 2009;422:473–482. doi: 10.1042/BJ20090638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenas J, Hedblom A, Miftakhova RR, Sarwar M, Larsson R, Shcherbina L, Johansson ME, Harkonen P, Sterner O, Persson JL. The role of PI3K/AKT-related PIP5K1alpha and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc Natl Acad Sci U S A. 2014;111:E3689–3698. doi: 10.1073/pnas.1405801111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VP, DesMarais V, Sumners C, Shaw G, Narang A. Immunostaining evidence for PI(4,5)P2 localization at the leading edge of chemoattractant-stimulated HL-60 cells. J Leukoc Biol. 2008;84:440–447. doi: 10.1189/jlb.0907636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens LR, Jackson TR, Hawkins PT. Agonist-stimulated synthesis of phosphatidylinositol(3,4,5)-trisphosphate: a new intracellular signalling system? Biochim Biophys Acta. 1993;1179:27–75. doi: 10.1016/0167-4889(93)90072-w. [DOI] [PubMed] [Google Scholar]
- Stijf-Bultsma Y, Sommer L, Tauber M, Baalbaki M, Giardoglou P, Jones DR, Gelato KA, van Pelt J, Shah Z, Rahnamoun H, et al. The basal transcription complex component TAF3 transduces changes in nuclear phosphoinositides into transcriptional output. Mol Cell. 2015;58:453–467. doi: 10.1016/j.molcel.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumita K, Lo YH, Takeuchi K, Senda M, Kofuji S, Ikeda Y, Terakawa J, Sasaki M, Yoshino H, Majd N, et al. The Lipid Kinase PI5P4Kbeta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis. Mol Cell. 2015;61:187–198. doi: 10.1016/j.molcel.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Lambert PF, Rapraeger AC, Anderson RA. Stress-Induced EGFR Trafficking: Mechanisms, Functions, and Therapeutic Implications. Trends Cell Biol. 2016a;26:352–366. doi: 10.1016/j.tcb.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Thapa N, Liao Y, Choi S, Anderson RA. PtdIns(4,5)P2 signaling regulates ATG14 and autophagy. Proc Natl Acad Sci U S A. 2016b;113:10896–10901. doi: 10.1073/pnas.1523145113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa N, Sun Y, Schramp M, Choi S, Ling K, Anderson RA. Phosphoinositide signaling regulates the exocyst complex and polarized integrin trafficking in directionally migrating cells. Dev Cell. 2011;22:116–130. doi: 10.1016/j.devcel.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa N, Tan X, Choi S, Wise T, Anderson RA. PIPKIgamma and talin couple phosphoinositide and adhesion signaling to control the epithelial to mesenchymal transition. Oncogene. 2017;36:899–911. doi: 10.1038/onc.2016.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bout I, Divecha N. PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J Cell Sci. 2009;122:3837–3850. doi: 10.1242/jcs.056127. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nature reviews Molecular cell biology. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- Vicinanza M, Korolchuk VI, Ashkenazi A, Puri C, Menzies FM, Clarke JH, Rubinsztein DC. PI(5)P regulates autophagosome biogenesis. Mol Cell. 2015;57:219–234. doi: 10.1016/j.molcel.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Irvine RF, Giudici ML. Phosphatidylinositol 4-phosphate 5-kinase Igamma_v6, a new splice variant found in rodents and humans. Biochem Biophys Res Commun. 2011;411:416–420. doi: 10.1016/j.bbrc.2011.06.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Weinberg RA. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronenkov IV, Loijens JC, Umeda M, Anderson RA. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol Biol Cell. 1998;9:3547–3560. doi: 10.1091/mbc.9.12.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annual review of pathology. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Hedman AC, Sayedyahossein S, Thapa N, Sacks DB, Anderson RA. Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat Cell Biol. 2016;18:1324–1335. doi: 10.1038/ncb3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Thapa N, Tan X, Hedman AC, Anderson RA. PIP kinases define PI4,5P(2)signaling specificity by association with effectors. Biochim Biophys Acta. 2015;1851:711–723. doi: 10.1016/j.bbalip.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Armi C, Devereaux KA, Di Paolo G. The role of lipids in the control of autophagy. Curr Biol. 2013;23:R33–45. doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Eden ER. The formation and function of ER-endosome membrane contact sites. Biochim Biophys Acta. 2016;1861:874–879. doi: 10.1016/j.bbalip.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden ER, Burgoyne T, Edgar JR, Sorkin A, Futter CE. The relationship between ER-multivesicular body membrane contacts and the ESCRT machinery. Biochemical Society transactions. 2012;40:464–468. doi: 10.1042/BST20110774. [DOI] [PubMed] [Google Scholar]
- Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nature cell biology. 2010;12:267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- Fets L, Nichols JM, Kay RR. A PIP5 kinase essential for efficient chemotactic signaling. Curr Biol. 2014;24:415–421. doi: 10.1016/j.cub.2013.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi Y, Hasegawa H, Kanaho Y. Activation mechanisms of PIP5K isozymes by the small GTPase ARF6. Adv Enzyme Regul. 2010;50:72–80. doi: 10.1016/j.advenzreg.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Funakoshi Y, Hasegawa H, Kanaho Y. Regulation of PIP5K activity by Arf6 and its physiological significance. Journal of cellular physiology. 2011;226:888–895. doi: 10.1002/jcp.22482. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, et al. Molecular definitions of autophagy and related processes. The EMBO journal. 2017;36:1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GH, Fisette PL, Anderson RA. Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J Biol Chem. 1994;269:11547–11554. [PubMed] [Google Scholar]
- Ktistakis NT, Tooze SA. Digesting the Expanding Mechanisms of Autophagy. Trends in cell biology. 2016;26:624–635. doi: 10.1016/j.tcb.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Mari M, Tooze SA, Reggiori F. The puzzling origin of the autophagosomal membrane. F1000 biology reports. 2011;3:25. doi: 10.3410/B3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenburg D, Han JS, Liyanage M, Patton WA, Rhee SG, Moss J, Vaughan M. Activation of rat brain phospholipase D by ADP-ribosylation factors 1,5, and 6: separation of ADP-ribosylation factor-dependent and oleatedependent enzymes. Proc Natl Acad Sci U S A. 1994;91:11718–11722. doi: 10.1073/pnas.91.24.11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Wang L, Chen D, Chang Y, Zhang M, Xu JJ, Zhou R, Zhang QY. LAPTM4B: an oncogene in various solid tumors and its functions. Oncogene. 2016;35:6359–6365. doi: 10.1038/onc.2016.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Hasegawa J, Yoshimori T. Regulation of lysosomal phosphoinositide balance by INPP5E is essential for autophagosome-lysosome fusion. Autophagy. 2016;12:2500–2501. doi: 10.1080/15548627.2016.1234568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, Pirruccello M, Hao M, Ingolia NT, Wenk MR, et al. PtdIns4P synthesis by PI4KIIIalpha at the plasma membrane and its impact on plasma membrane identity. J Cell Biol. 2012;199:1003–1016. doi: 10.1083/jcb.201206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Experimental cell research. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- Sun Y, Ling K, Wagoner MP, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase is required for EGF-stimulated directional cell migration. J Cell Biol. 2007;178:297–308. doi: 10.1083/jcb.200701078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Lambert PF, Rapraeger AC, Anderson RA. Stress-Induced EGFR Trafficking: Mechanisms, Functions, and Therapeutic Implications. Trends Cell Biol. 2016a;26:352–366. doi: 10.1016/j.tcb.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Sun Y, Thapa N, Liao Y, Hedman AC, Anderson RA. LAPTM4B is a PtdIns(4,5)P2 effector that regulates EGFR signaling, lysosomal sorting, and degradation. The EMBO journal. 2015a;34:475–490. doi: 10.15252/embj.201489425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Thapa N, Choi S, Anderson RA. Emerging roles of PtdIns(4,5)P2 – beyond the plasma membrane. Journal of Cell Science. 2015b;128:4047–4056. doi: 10.1242/jcs.175208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Thapa N, Liao Y, Choi S, Anderson RA. PtdIns(4,5)P2 signaling regulates ATG14 and autophagy. Proc Natl Acad Sci U S A. 2016b;113:10896–10901. doi: 10.1073/pnas.1523145113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Thapa N, Sun Y, Anderson RA. A kinase-independent role for EGF receptor in autophagy initiation. Cell. 2015c;160:145–160. doi: 10.1016/j.cell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa N, Choi S, Tan X, Wise T, Anderson RA. Phosphatidylinositol Phosphate 5-Kinase Igamma and Phosphoinositide 3-Kinase/Akt Signaling Couple to Promote Oncogenic Growth. J Biol Chem. 2015;290:18843–18854. doi: 10.1074/jbc.M114.596742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends in cell biology. 2014;24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nature cell biology. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2017 doi: 10.1080/15548627.2017.1378838. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 2012;20(1):25–35. doi: 10.1016/j.tcb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Thapa N, Tan X, Hedman AC, Anderson RA. PIP kinases define PI4,5P(2)signaling specificity by association with effectors. Biochim Biophys Acta. 2015;1851(6):711–723. doi: 10.1016/j.bbalip.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriplani N, Hermida MA, Brown ER, Leslie NR. Class I PI 3-kinases: Function and evolution. Adv Biol Regul. 2015;59:53–64. doi: 10.1016/j.jbior.2015.05.002. [DOI] [PubMed] [Google Scholar]