Abstract
Early clinical trials using murine double minute 2 (MDM2) inhibitors demonstrated proof-of-concept of p53-induced apoptosis by MDM2 inhibition in cancer cells, however, not all wild-type TP53 tumors are sensitive to MDM2 inhibition. Therefore, more potent inhibitors and biomarkers predictive of tumor sensitivity are needed. The novel MDM2 inhibitor DS-3032b is 10-fold more potent than the first-generation inhibitor nutlin-3a. TP53 mutations were predictive of resistance to DS-3032b, and allele frequencies of TP53 mutations were negatively correlated with sensitivity to DS-3032b. However, sensitivity to DS-3032b of TP53 wild-type tumors varied greatly. We thus used two methods to create predictive gene signatures: First, by comparing sensitivity to MDM2 inhibition with basal mRNA expression profiles in 240 cancer cell lines, a 175-gene signature was defined and validated in patient-derived tumor xenograft models and ex vivo human acute myeloid leukemia (AML) cells. Second, an AML-specific 1532-gene signature was defined by performing random forest analysis with cross validation using gene expression profiles of 41 primary AML samples. The combination of TP53 mutation status with the two gene signatures provided the best positive predictive values (81 and 82%, compared to 62% for TP53 mutation status alone). In addition, the top-ranked 50 genes selected from the AML-specific 1532-gene signature conserved high predictive performance, suggesting that a more feasible size of gene signature can be generated through this method for clinical implementation. Our model is being tested in ongoing clinical trials of MDM2 inhibitors.
Keywords: MDM2, TP53, gene signature, DS-3032b, biomarker
Introduction
The p53 protein plays a pivotal role in tumor suppression and maintenance of genome integrity, acting as a transcription factor that can induce apoptosis, cell cycle arrest, DNA repair and/or senescence in response to cellular stresses. Although approximately half of all cancers harbor TP53 mutations, wild-type p53 is also frequently inactivated by other mechanisms. The E3 ligase murine double minute 2 (MDM2, also known as HDM2) is frequently overexpressed in various cancer (1,2), directly inactivating p53 function by binding to its transactivation domain and also inducing its degradation through p53 ubiquitination. Pharmacological inhibition of the p53-MDM2 interaction has been developed into a therapeutic approach for exerting p53-mediated anti-tumor effects.
Several preclinical (3–8) and early clinical (9–11) studies of MDM2 inhibitors demonstrated the proof-of-concept of their anti-tumor effects via p53 activation in both solid and hematologic tumors. There are multiple ongoing clinical studies (12,13) of MDM2 inhibitors in several cancers, including the novel MDM2 inhibitor DS-3032b, still in an early phase (solid tumors and lymphoma, NCT01877382; myeloma, NCT02579824_DS3032b; leukemia NCT02319369). Clinical responses in these trials have been limited overall, but some patients clearly achieve clinical benefit by monotherapy with MDM2 inhibitors (9–13). Therefore, there is a need for improved MDM2 inhibitors, or efficacious combination therapies, as well as predictive biomarkers for monotherapy with current MDM2 inhibitors.
We report here that DS-3032b exerts highly potent p53-dependent anti-tumor effects both in vitro and in vivo. In addition, to test the hypothesis that gene signatures can predict sensitivity to MDM2 inhibition, we measured sensitivities to MDM2 inhibition in a broad range of cancer cell lines, patient-derived xenograft models and primary leukemia specimens and used these to derive predictive signatures from gene expression profiles determined before treatment. We report two different bioinformatic approaches to define predictive gene signatures: one used a panel of 240 molecularly-annotated cancer cell lines to find a common signature among various tumor types, and the other focused primarily on acute myeloid leukemia (AML) as an example to define tumor-type-specific signatures. Considering that TP53 mutation status is a predictive but not a sufficient biomarker, we also combined the gene signatures with TP53 mutation status, aiming at improved prediction of response.
Materials and Methods
Reagents
DS-5272 (8), DS-3032b and DS-3032a (a salt-free form of DS-3032b) were developed and synthesized by Daiichi Sankyo Co., Ltd. (Figure 1A). (−)-Nutlin-3a was purchased from Cayman Chemical Company.
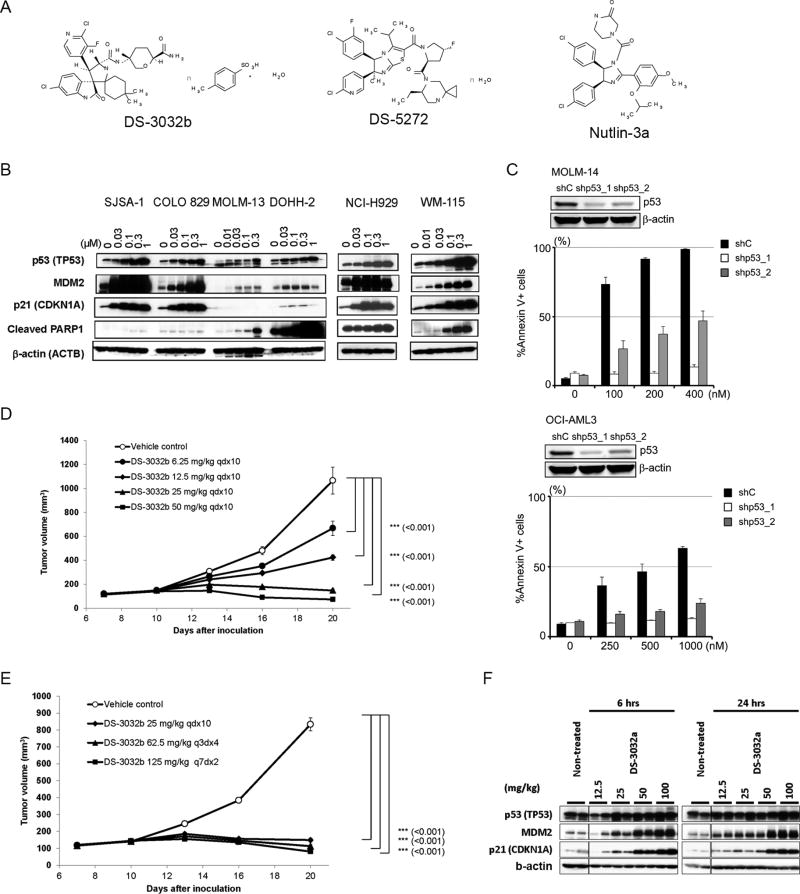
Figure 1. DS-3032b induces p53-dependent apoptosis in tumor cells by activating p53 via inhibition of MDM2-p53 interaction in culture and in mice.
(A) Chemical structures of DS-3032b, DS-5272 and nutlin-3a. (B) Immunoblot of p53, MDM2, p21, cleaved PARP1 and b-actin in tumor cells treated with DS-3032b for 24 hours. (C) Apoptosis induced by DS-3032b treatment for 48 hours in TP53 knock-down or intact MOLM-14 and OCI-AML3 cells. (n=3 experiments for each cell line). p53 knockdown was confirmed. (D) Time-course measurement of tumor xenograft volumes in mice inoculated with SJSA-1 cells treated with DS-3032b. Each data point and bar represent the mean and standard error of the estimated tumor volume in each group, respectively (n = 6). (E) Time-course measurement of tumor xenograft volumes in mice inoculated with SJSA-1 cells treated with DS-3032b. Each data point and bar represent the mean and standard error of the tumor volume in each group, respectively (n = 10). (F) Immunoblot of p53, MDM2 and p21 in xenograft tumors at 6 and 24 hours after a single administration of DS-3032a to mice bearing MOLM-13-derived tumors. β-actin was used as an internal control.
Cell lines and cell culture
SJSA-1, Saos-2, WM-115 and DLD-1 cells were purchased from the American Type Culture Collection (Manassas, VA). MOLM-14, OCI-AML3, DOHH-2, and Z-138 cells were purchased from the Leibniz-Institut Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). The authenticity of the cell lines was confirmed by DNA fingerprinting with the short tandem repeat method, using a PowerPlex 16 HS System (Promega, Madison, WI) within six months before the experiments. Mycoplasma was tested by PCR method and confirmed the 30 negativity for all the cell lines used (14). Cryopreserved cells, or fresh patient samples, were cultured as described previously (14) and used at passage four to six for the in vitro and in vivo experiments.
Apoptosis assay and flow cytometric analysis
For 48-hour drug treatment experiments, cell lines were harvested at log-phase growth, plated at a density of 1.5 × 105/ml for MOLM-14, OCI-AML3 and Z-138, or 1.0 × 106/ml for primary AML and MCL cells, and treated with DS-3032b (0, 25, 50, 100, 250, 500, 1000 nM). Annexin V and PI (purchased from Sigma-Aldrich) binding assays were performed to assess apoptosis as described previously (14). Annexin V- and PI-negative cells were counted as live cells, and % live cell number was calculated by normalizing to untreated controls. Based on the live cell counts measured at each concentration, IC50 was calculated by using GraphPad Software (ver. 6). Calculations of specific apoptosis and area under the curve (AUC) based on the %live cell number are in supplemental information.
Analysis of patient derived tumor xenograft mice
In vivo study in patient derived tumor xenograft mice were performed by Champions Oncology Inc., MD, USA. 13 tumor models including melanoma, NSCLC, colorectal and pancreatic, were selected from Champions Tumorgraft™ library. Gene expression data and TP53 genotype information were provided by Champions Oncology. Female nu/nu mice (Harlan) between 6 – 9 weeks of age were implanted bilaterally in the flank region with tumor fragments harvested from host animals, each implanted from a specific passage lot. When tumors reached approximately 125–250 mm3, animals were matched by tumor volume into treatment and control groups and dosing initiated (Day 0). Animals bearing tumors were grouped into treatment and vehicle groups (n=10, respectively) and DS-3032b was administered at 10mg/kg/day for 10 days. Tumor dimensions were measured twice weekly by digital caliper. Tumor volume (TV) was calculated using the formula (1): TV= (width)2 × (length) × 0.52. Percent tumor growth inhibition (%TGI) values were calculated for each treatment group (T) versus control (C) using tumor measurements of initial (i) and at the end of dosing or the closest tumor measurement (f) by the formula (2): %TGI= 1−Tf−Ti / Cf−C. All experimental procedures were performed in accordance with the in-house guideline of the Institutional Animal Care and Use Committee of Champions Oncology Inc.
Animal studies and experiments with human samples
For all the animal studies in the present study, the study protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of Champions Oncology Inc. (patient-derived xenograft mice) and Covance Laboratories Inc. (SJSA-1 and MOLM-13 xenograft mice). Studies with primary human leukemia samples were approved by the institutional review board at MD Anderson Cancer Center. Forty-one peripheral blood or bone marrow samples from 38 patients with AML, newly diagnosed, or relapsed / refractory after chemotherapy were collected.
Generation of the 175-gene signature and sensitivity score
A differential expression analysis of the mRNA datasets derived from sensitive and resistant cell lines was performed. Genes were ranked by p-value according to Student’s two class t-test in each cell line in Oncopanel. Genes differentially upregulated in sensitive cell lines, defined by GI50s of DS-5272 (< 1.5 µM), contributed to the sensitivity signature. The signature contained the top one percent of ranked genes (n=175). The robustness of the gene signatures was inspected in several ways. First, heat maps of the entire 1% gene lists were generated. Visual inspection of the heat map was performed to characterize the extent and consistency of the differential expression across cell lines and down the gene list. Second, the Q-value was calculated as (p-value/p-value rank)*number of genes measured. As a guideline, all signatures were required to have ≥ 10 genes with Q-value ≤ 0.1 to support further analysis. Lastly, the sensitivity signature was compared against other drug signatures by comparing the total number of genes at the Q-value threshold 0.05 and the Q-value of the least significant gene in the top 1% signature.
The expression values of the genes within the signature were Z-score normalized. Once normalized, the average Z-score of the sensitivity signature genes was calculated on an individual cell line, murine or human sample basis to generate a sensitivity score, respectively. The same calculation was used in GEP data derived from Oncomine and primary AML samples. Data from the microarray studies has been submitted to the Gene Expression Omnibus. The accession number is GSE110087.
Statistical analyses
For random forest methods with cross validation in primary AML samples, gene expression profiling of the 41 primary AML samples was used. Eleven each among 33 TP53 wild-type samples, or 14 each among 41 (including 8 TP53 mutant samples) were selected as sensitive or resistant to DS-3032 based on AUC based on %live cell number. Classification was performed using the Random Forest method to identify a predictive algorithm with a particular gene set. Forests were created with 10000 decision trees. Performance was assessed by AUC of a receiver-operating characteristic (ROC) curve, from internal out-of-bag (OOB) testing results. Each tree uses a different random bootstrap sample of two-thirds of cases from the original data. A test set classification is obtained for each case in about one-third of the trees. By default, random forests method performs a binary classification (in the present study, sensitive vs resistant to DS-3032b). However it also reports a probability for being sensitive to DS-3032b. The threshold for predicting sensitive or resistant was set as 0.5 probability. Overall accuracy was defined as a ratio of the cases correctly predicted as sensitive or resistant to the total cases. Positive predictive value was defined as a ratio of the cases correctly predicted as sensitive to the total cases predicted as sensitive. Negative predictive value was defined as a ratio of the cases correctly predicted as resistant to the cases predicted as resistant. Sensitivity was defined as a ratio of the cases correctly predicted as sensitive to the total sensitive cases.
Results
DS-3032b inhibits MDM2-p53 interaction and activates p53 at nanomolar concentrations
The chemical structure of DS-3032b is shown in Figure 1A. To examine the in vitro inhibitory effects of DS-3032b on MDM2-p53 interaction, we utilized homogeneous time resolved fluorescence (HTRF). DS-3032b inhibited the MDM2-p53 interaction with IC50 values of 5.57 nM (Figure S1A). Consistently, nanomolar concentrations (30–1000 nM) of DS-3032b increased p53 levels in a dose-dependent manner by 24 hours, along with induction of the p53 targets MDM2 and p21, in various cancer cell lines (SJSA-1, Colo829, MOLM-13, DOHH-2, NCI-H929 and WM-115) harboring wild-type TP53 (Figure 1B). Treatment with DS-3032b also induced PARP cleavage, indicating apoptosis induction (Figure 1B). Dose-dependent increase in expressions of the p53 target genes CDKN1A (p21) and BBC3 (PUMA) were also confirmed in SJSA-1 cells (Figure S1B). These results proved that DS-3032b is a potent inhibitor of MDM2 and indeed activates p53 function.
DS-3032b causes p53-dependent anti-tumor effects both in vitro and in vivo
We next treated 6 human cancer cell lines derived from various tumor types (MOLM-13, acute myeloid leukemia; DOHH-2, non-Hodgkin’s B-cell lymphoma; SJSA-1, osteosarcoma; WM-115, melanoma; DLD-1, colon adenocarcinoma; Saos-2, osteosarcoma) with DS-3032b or nutlin-3a, and determined their GI50 values (Table S1). DLD-1 and Saos-2 cells with TP53 mutations were resistant to either of the drugs, while the other four cell lines with wild-type TP53 were more sensitive with nanomolar values of GI50s. Compared to GI50 values of nutlin-3a, DS-3032b was approximately 10-fold more potent. We then used a TP53 wild-type AML and mantle cell lymphoma (MCL) cell lines (MOLM-14, OCI-AML3, and Z-138), and knocked down TP53 expression using short hairpin RNA to test for p53-dependency of DS-3032b. As expected, TP53 knocked-down cells are significantly resistant to DS-3032b- and nutlin-3a-induced apoptosis (Figure 1C, Supplemental Figure S1C), confirming the p53-dependency of apoptosis induction of these two inhibitors. Of note, we also treated circulating MCL cells obtained from five patients. All samples had wild-type TP53 as determined by Sanger sequencing. DS-3032b induced apoptosis in a dose-dependent manner in all of the samples (Figure S1D).
We further tested the in vivo activity of DS-3032b using tumor xenograft models with TP53 wild-type MOLM-13 and SJSA-1 cells. In both models, DS-3032b inhibited tumor growth in a dose-dependent manner, especially with oral administration at 25 and 50 mg/kg/day (Figures 1D, 1E and S1E, and Table S2), indicating the potential of clinical use of this agent. Daily and intermittent dosing (every 3 or 7 days) (Figure 1E) both showed in vivo efficacy, suggesting relatively long lasting efficacy of DS-3032b in vivo. We also confirmed induction of p53, MDM2 and p21 protein in tumor cells 6 and 24 hours after in vivo treatment with DS-3032b (Figure 1F).
We also treated a panel of 240 cell lines from various cancers (OncoPanel, Eurofins Panlabs) with DS-3032a (a free form of DS-3032b), DS-5272 (another prototypic MDM2 inhibitor developed previously) (8), and nutlin-3a. As expected, GI50 profile of DS-3032b is highly correlated with that of DS-5272 (r = 0.98, p = 2.2e–16) (Figure S2A) (Supplemental File S1).
TP53 mutation status alone is not a sufficient biomarker to determine anti-tumor effects of DS-3032b
We have shown in clinical studies of the MDM2 inhibitor RG7112 that therapeutic response to RG7112 as a single drug varies from patient to patient in cancer patients harboring wild-type TP53 tumors (10). We established first that sensitivity to MDM2 inhibition is in part dependent on baseline MDM2 levels (4) and confirmed this finding in the AML clinical trial of RG7112, where both baseline and induced MDM2 levels were correlated with response (10). The heterogeneous response in p53 wild type cell lines had also been observed in cell lines from a single disease entity (5,15).
Indeed, the data from OncoPanel cell lines treated with the three MDM2 inhibitors showed that TP53 wild-type cell lines have a large range of GI50s, (e.g., varying from < 0.1 µM to > 10 µM for both DS-5272 and DS-3032b) (Figure S2B, Table S3 and Supplemental File S1). Cell lines that were TP53 wild-type but were resistant to all the three inhibitors included HeLa, C4I and C32 cells, which may be related to human papilloma virus infection (HeLa and C4I) and ARF gene deletion (C32) that inactivate p53. However, the 24% of TP53 wild-type cell lines that were deemed resistant did not all have known causes of resistance (Figure 2A). Also, our in vitro data using cell lines or primary MCL cells with wild-type TP53 (Table S1 and Figure S1D) showed that sensitivity varied over a certain range (GI50 < 100 nM to > 250 nM).
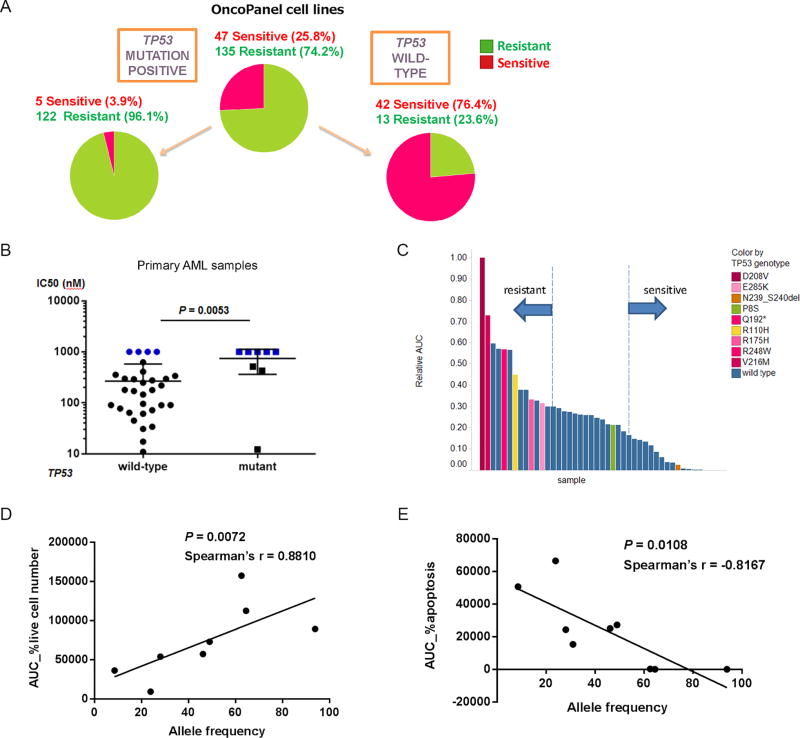
Figure 2. p53 mutation status and cell susceptibility to MDM2 inhibition.
(A) Pie-charts showing the proportion of cancer cell lines sensitive or resistant to DS-5272 and TP53 mutation status. The following cell lines were excluded due to unclear annotation on TP53; A172, DOHH-2, HT-1197, SW-48, MALME3M, A673, CAOV3, CAPAN-2, EB3, HS-766T, K-562, L428, SKNAS and DLD1. (B) GI50s of DS-3032b in 41 primary AML cells and TP53 mutation status (P = 0.0053, Mann Whitney test). (C) Sensitivity of 41 primary AML cells to DS-3032b and TP53 mutation status. Y axis represents the area under the curve (AUC) calculated according to %live cell number at each concentration of DS-3032b in each sample. (D and E) Correlation between allele frequencies of TP53 mutations and sensitivities to DS-3032b in 8 primary AML samples. Y axes represent AUC calculated according to %live cell number (D) or %apoptosis (E) at each concentration of DS-3032b in each sample.
To confirm the variance of sensitivities to DS-3032b of wild-type TP53 tumor cells through ex vivo experiments, we collected 41 primary acute myeloid leukemia (AML) specimens obtained from 38 patients, which were all clinically annotated (Table S4). Eight (19.5%) had TP53 point mutations, detected by next generation sequencing and Sanger sequencing. Samples with TP53 wild-type were significantly more sensitive to DS-3032b than those with TP53 mutations (Figure 2B). Since exact GI50 values could not be calculated in highly sensitive (< 25 nM) or highly resistant (> 1000 nM) samples, we also used the area under the curve (AUC) based on percentages of live cell number (normalized to untreated controls) at each concentration (Figures 2C and S2C, and Supplemental File S2). We then focused on the outer one third of sensitive or resistant samples, and assessed how efficiently TP53 mutation status alone could predict the sensitivity to DS-3032b (Figure 2C and Table S5). Reflecting that some of the wild-type TP53 samples were as resistant or even more resistant than TP53 mutated samples, the positive predictive value (PPV) (62%) and overall accuracy (68%) were relatively low, compared to high sensitivity of 93% (Table 1). This indicates that predictive biomarkers with higher PPV than that of TP53 mutation status alone are desired for more effectively predicting tumor sensitivity to MDM2 inhibition.
Table 1.
Predictive performance of 14 sensitive/resistant samples among 41 primary AML samples using different algorithms.
| Overall accuracy (%) |
Positive predictive value (%) |
Negative predictive value (%) |
Sensitivity | ||
|---|---|---|---|---|---|
| A: | TP53 mutation status alone | 68 | 62 | 86 | 93 |
| B: | 4-gene signature | 68 | 63 | 78 | 86 |
| C: | 175-gene signature alone | 79 | 72 | 90 | 93 |
| D: | Combination of A & C | 82 | 81 | 85 | 86 |
| E: | Random forest (1532 genes) | 68 | 67 | 69 | 72 |
| F: | Combination of A & E | 75 | 82 | 71 | 68 |
TP53 mutation status can also be a continuous variable, based on the allele frequency of mutant TP53. Of note, AUC values based on live cell numbers (Figure 2D) and apoptosis (Figure 2E) are both correlated with allele frequencies of TP53 mutations in a positive and negative manner, respectively. This indicates that in cases with mutated TP53, the allele frequency of TP53 mutations could potentially serve to predict sensitivity to MDM2 inhibition.
A cell line-based sensitivity gene signature better predicts the sensitivity of tumor cells to MDM2 inhibition than TP53 mutation status alone
Among the 240 OncoPanel cell lines, we defined 62 sensitive and 164 resistant cell lines, based on GI50 values of DS-5272 (cut-off values of < 1.5µM, and > 10 µM, respectively) (Figure S2B and Table S6). This categorization was almost identical to that for DS-3032b. Specifically, only three (786-o, HS294T, and RPMI8226) among 240 cell lines on the panel were categorized discordantly. As to 786-o and HS294T, the dose-response curves are essentially similar (Figure S2D), and we therefore concluded that the sensitivities of these lines to DS-3032b and DS-5272 are basically similar. The reason for discordant data for RPMI8226 cells remains unclear.
Using publicly-available gene expression profiling data for all the cell lines, we then selected 175 genes ranked at the top one percent of the genes of higher expression level with lowest p-value in the 62 sensitive cell lines than resistant cell lines (Supplemental File S3). We thus defined the average of Z-scores of the 175 genes as a “sensitivity score”. The sensitivity score similarly predicted responses to 3 different MDM2 inhibitors (DS-5272, nutlin3a, and DS-3032a (free form of DS-3032b)) (Figure S2E), indicating its robustness as predictive gene signature for different MDM2 inhibitors. Of note, we also defined a “resistance score” by selecting the top one percent of genes of higher expression level with lowest p-value in resistant lines (Supplemental File S3), however, it did not predict the sensitivity/resistance well (worse than predictive performance by TP53 mutation status alone), thus we decided to further develop the sensitivity score.
A combination of TP53 genotype and gene signature showed further enrichment of sensitive cell lines compared to using either of them alone (Figure S3A). Specifically, by applying the sensitivity signature, 41 of 55 p53 wild-type cell lines had high sensitivity scores and the rest of 14 lines had low scores. 39 of 41 (95%) cell lines with high scores were sensitive while 11 of 14 (79%) cell lines with low scores are resistant to DS-5272, suggesting potentially high predictive performance of this signature.
Cell line-based 175-gene signature is validated in different datasets in tumors ex vivo and in vivo
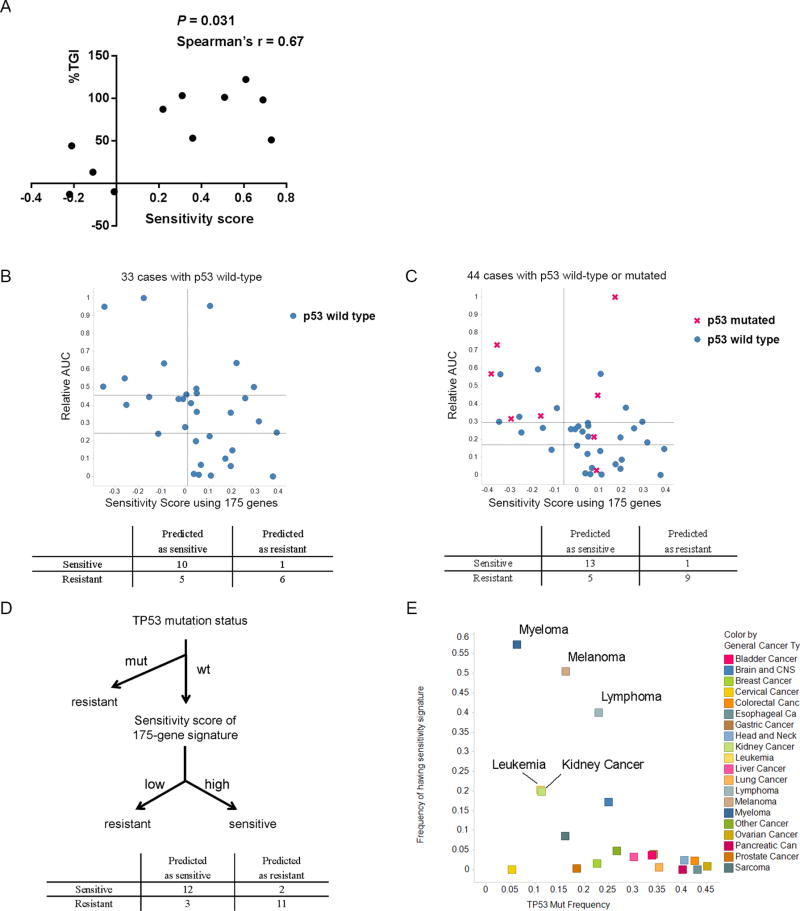
To validate the 175-gene signature, we used patient-derived tumor xenografts. In vivo anti-tumor activities of DS-3032b were evaluated in 13 total tumor models, derived from melanoma, NSCLC, colorectal and pancreatic cancers (Table S7A). A positive correlation was observed between the sensitivity scores and TGI values (Figure 3A, Spearman’ r = 0.67, P = 0.031). The prediction accuracy, sensitivity and PPV were 85, 88 and 88% respectively (Table S7B).
Figure 3. Validation of the 175-gene signature for predicting tumor sensitivity to MDM2 inhibition.
(A) Correlation between sensitivity scores based on the 175-gene signature and tumor growth inhibition (%TGI, see also Methods) by DS-3032b in patient-derived xenograft mice. Spearman’s r = 0.67, P = 0.031. see also Table S7. (B and C) Validation of the sensitivity score based on the 175-gene signature for predicting sensitivity to DS-3032b of primary AML samples with wild-type TP53 (B, left), or irrespective of TP53 mutation status (C, left). Confusion matrices (B and C, right) correspond to the point with best sensitivity/specificity tradeoff on each ROC curve. (D) Schema of predicting sensitivity of primary AML samples to DS-3032b by combining TP53 mutation status and sensitivity scores of 175-gene signature. (E) Correlation between the frequency of specimens with high sensitivity scores and that of specimens with TP53 mutations in various tumor types. Sensitivity scores > 0 were counted as high scores.
Performing gene expression profiling in addition to ex vivo cell viability assays of AML samples described above (Table S4), we tested whether the sensitivity score of each sample predicts the sensitivity to DS-3032b. First, we focused only on the 33 cases with wild-type TP53. As the outer third of sensitive or resistant samples, eleven p53 wild-type samples each were selected as sensitive or resistant to DS-3032 based on AUC of %live cell number, and the 175-gene signature was tested. The prediction accuracy, sensitivity and PPV were 73, 91 and 67%, respectively (Figure 3B and Figure S3B). Next, we also tested the 175-gene signature in all 41 samples including additional 8 cases with TP53 mutations. Focusing on the outer third (fourteen each) sensitive or resistant samples in the 41 samples, the prediction accuracy, sensitivity and positive predictive values (PPV) were 79, 93 and 72% (Figure 3C and Figure S3C) respectively. Compared to results for TP53 mutation status alone described above (68, 93 and 62%, respectively) (Table S5), the 175-gene signature improved PPV (Table 1). Using the same dataset of our primary AML samples, we also tested a recently reported 4-gene predictive signature (16), which showed relatively low predictive performance (prediction accuracy, sensitivity and PPV were 68, 86 and 63%, respectively) (Table S8 and Table 1), compared to the 175-gene signature (Figure S3D).
We further tested the predictive performance when the 175-gene signature was applied after TP53 mutation status was incorporated as a first predictive factor for resistance. Specifically, we predicted samples with TP53 mutations as resistant, and sensitivities to DS-3032b of the rest of samples with wild-type TP53 was predicted using the 175-gene signature (Figure 3D, upper). The accuracy, sensitivity and PPV for predicting the top 14 sensitive or resistant samples were 82, 86, 81%, respectively (Figure 3D, lower, and Table 1), showing potentially more balanced performance with higher PPV compared to using TP53 mutation status or the gene signature alone.
Of note, the top-ranked genes in the signature were well-known p53-inducible genes (Table S9), suggesting that the gene signature may reflect high basal activity of p53 which may lower the threshold for inducing p53-mediated apoptosis by MDM2 inhibition. With this notion, supporting the superiority of our gene signature to TP53 mutation status alone, the 175-gene signature applied to genomic data of more than 15,000 clinical samples obtained from the OncoMine database revealed that only a subset of tumors relatively enriched with wild-type TP53 (melanoma, lymphoma, myeloma, renal cell cancer, and leukemia) contains high sensitivity scores (Figure 3E, Table S10). On the other hand, cervical cancer has low p53 mutation frequency and a low sensitivity score, probably owing to p53 inactivation via HPV infection in cervical cancer.
Generating a tumor-type-specific gene signature for predicting drug sensitivity of MDM2 inhibitors
Whereas a common gene signature regardless of tumor types is useful in terms of its broad application, tumor-type-specific gene signatures perhaps need to be explored to achieve a higher performance for predicting drug sensitivity in a particular tumor type. Thus, we examined another bioinformatics approach by utilizing only the ex vivo dataset of primary AML samples described above.
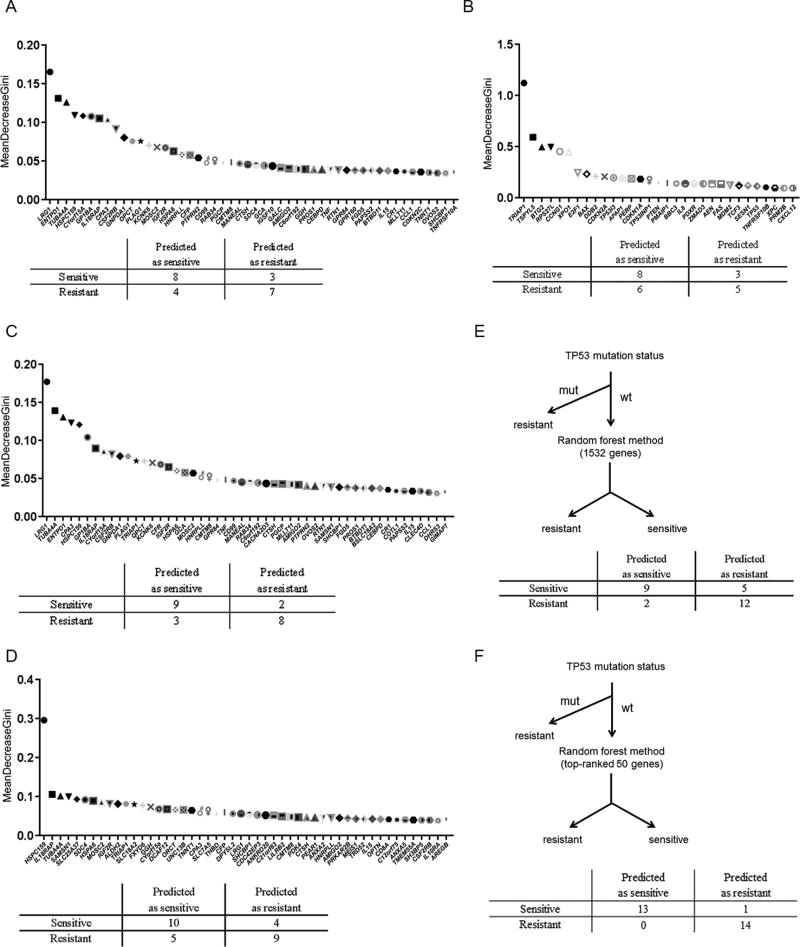
We first hypothesized that genes with high expression variance have highest predictive value for determining drug sensitivity. Using a fairly routine bioinformatics method for classifying gene signatures (17), we first selected the 1500 top-ranked genes based on expression variance among 33 primary AML samples with wild-type TP53 (Supplemental File S2). We used these genes’ expression values to predict the results of ex vivo treatment with DS-3032b with the random forest method (18), ranking every gene based on the significance for the prediction (referred to as “variable importance”) (Figure 4A, upper). Cross validation was performed by holding out one third of the total samples for testing and the learners (decision tree) ensembles were trained on only the remaining two thirds of the data. The overall accuracies, sensitivity and PPV of the identified algorithm with the 1500 genes for predicting the top 11 sensitive or resistant samples among 33 were 68, 73 and 67%, respectively (the confusion matrix is shown in Figure 4A, lower), showing similar accuracy and PPV to the 175-gene signature described above. Of note, only 8 out of 175 genes in our cell line-based signature (Supplemental File S1) and none of 13 genes (19) or 4 genes (16), previously reported gene signatures, were ranked in these 1500 genes. These results indicate that genes expressed with high variance in a particular tumor type, regardless of the relationship to p53, also participate in determining the drug sensitivity to a certain extent, perhaps in different way(s) from the other gene sets enriched with p53 transcriptional targets. In contrast to the unbiased approach based on gene expression variance above, we next selected 32 genes (Table S11) based on previous reports suggesting the potential predictive values of the genes. Those included 13 genes previously published in predictive gene signatures for the sensitivity to MDM2 inhibitors (16,19), 22 genes that were upregulated or downregulated by an MDM2 inhibitor (RG7112) determined in a phase 1 clinical trial (10), TRIAP1 and E2A (TCF3) which are reported to be transcriptional co-factors of p53 (20), and Drosha which is reported to be downstream of MDM2 (21). Various cell-intrinsic mechanisms of p53 inactivation are known (22), and most of them (e.g., MDM2 overexpression, ARF deletion, NPM1 mutation, FLT3-ITD, PML-RARalpha, activation of PI3K or MAPK pathways) result in hyper-activation of MDM2 which inhibits p53-mediated signaling. XPO1, a nuclear exporter of p53, contributes another mechanism of p53 inactivation. Therefore, we included these two key genes in the list. We then applied these 32 genes in the random forest method, which showed the accuracy of 59.1%, sensitivity of 73%, and PPV of 57% (Figure 4B). Next, we combined the 1500 genes selected by the unbiased approach above and the referenced 32 genes. The random forest method created the algorithm with accuracy of 77%, sensitivity of 82%, and PPV of 75% (Figure 4C, Table 1), indicating that combining the two gene selection methods improved the predictive performance.
Figure 4. Generating a predictive gene signature using random forest methods with cross validation.
(A) Unbiased approach: 50 genes ranked with highest significance in the random forest method are listed as examples from 1500 genes which showed high expression variances in 33 primary AML samples with wild-type TP53 (upper). (B) Referenced approach: The selected 32 genes are shown in order according to the significance for prediction (upper). (C) Combined approach: 1500 and 32 genes used in (A) and (B) were used and 50 genes assessed with highest significance in the random forest method are listed (upper). Confusion matrices were generated at the point of the ROC curve which provided highest predictive performance of random forest methods using each gene set to predict 11 of most sensitive or resistant specimens among 33 wild-type TP53 AML samples to DS-3032b (A to C, lower). (D) 1500 which showed high expression variances in all the 41 primary AML samples were selected and the 32 genes used in (B) were used and 50 genes assessed with highest significance in the random forest method are listed (upper). (E) Schema of predicting sensitivity of primary AML samples to DS-3032b by combining TP53 mutation status and random forest method with the same gene set as in (C). (F) Schema of predicting sensitivity of primary AML samples to DS-3032b by combining TP53 mutation status and random forest method with the 50 top-ranked genes in (C). Confusion matrices were generated at the point of the ROC curve which provided highest predictive performance of random forest methods using each gene set to predict 14 of most sensitive or resistant specimens among 41 wild-type TP53 AML samples to DS-3032b (D, E and F, lower).
We then tested the algorithm of combined genes on all 41 samples including ones with TP53 mutations. Specifically, another set of 1500 genes was selected according to expression variance among all the 41 AML samples, combined with the same 32 referenced genes, then the random forest method was performed. The accuracy, sensitivity and PPV for predicting the top 14 sensitive or resistant samples were 68, 72, 67%, respectively (Figure 4D and Table 1), showing reduced predictive performance than when focusing on only TP53 wild-type samples.
Finally, we combined this algorithm with TP53 mutation status. Specifically, we predicted samples with TP53 mutations as resistant, and then applied the random forest method using 1532 genes to the remaining wild-type TP53 samples (Figure 4E, upper). The accuracy, sensitivity and PPV for predicting the top 14 sensitive or resistant samples were 75, 68, 82%, respectively (Figure 4E and Table 1), showing better accuracy and PPV than the random forest method alone. This indicated that combining the random forest method with the prediction by TP53 mutation status provides a better algorithm compared to each alone. Of note, when we followed the same flow chart and ran the random forest method with the 50 top-ranked genes in the 1532 genes (Figure 4F), the high accuracy, sensitivity and PPV were conserved (96, 93, 100%) (Figure 4F). Although re-selecting the 50 genes defined by the first learning iteration has a possibility of overfitting, it suggests that narrowing down gene numbers based on the rank defined by the random forest method could be an option for defining a more feasible scale of gene set for further clinical implementation.
Discussion
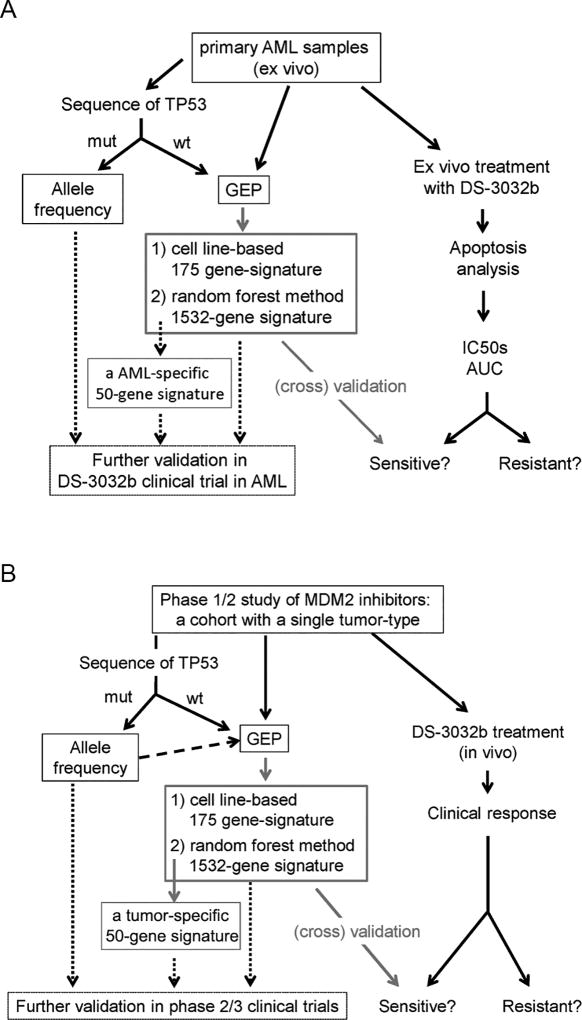
Our study demonstrated that DS-3032b is a highly potent MDM2 inhibitor and induces p53-mediated apoptogenic effects in various cancer cells. In addition, to overcome insufficient prediction of tumor cell sensitivities to MDM2 inhibition by TP53 mutation status alone, two different approaches for defining genetic biomarkers were developed: 1) a 175-gene signature defined by a cell line-based approach; and 2) An AML-specific 1532-gene signature defined by the random forest method. We propose that a combination of the classical factor TP53 mutation status and the gene signatures provide the best algorithms for predicting tumor sensitivities to MDM2 inhibitors (Figure 5A).
Figure 5. Proposal of further investigation of predictive gene signatures.
(A) A schematic figure of generating predictive gene signatures in the present study and further validation planned in future. (B) A proposal of utilizing similar algorithms analyzed in the present study for future clinical trials.
Our 175-gene signature showed a robust predictive performance of the sensitivities to DS-3032b in validation datasets of human ex vivo specimens and patient-derived xenograft murine models. Importantly, although the 175 genes in the present study included all of the 4 genes which were previously reported as predictive gene signature for other MDM2 inhibitors (16), our signature of 175 genes had higher predictive power. Of note, although another 13-gene signature has also been reported (19), it was found later not to be predictive due to the revised annotation of TP53 mutations in the cell lines (23). These findings collectively suggest that incorporating a broader spectrum of p53 transcriptional targets into the gene signature is superior to focusing on fewer genes. A concern for further developing this signature is that it is derived from cell lines of various cancers, not a single disease entity. Considering that a variety of p53 target genes are differently regulated depending on tissue types (24,25), the cell line panel-based identification of key predictive genes may not be the best approach.
Therefore, an AML-specific gene signature was developed for correcting the potential weakness of the 175-gene signature. However, this approach is also challenging due to the limited availability of independent datasets for validation. As a partial solution, we utilized the random forest method (18), an ensemble classifier that leverages the predictive power of multiple learners through an ensemble approach, for two reasons: 1) it is capable of handling a large number of features (i.e., > 1500 genes analyzed in an algorithm with 10000 trees in the present study), 2) it provides intrinsic cross-validation during the testing and assessment procedure. Thus, the process of three-fold cross validation, intrinsic in the random forest procedure, maximized the generalizability of the defined signature. However, we recognize the limitation of the present study because we lack another independent AML dataset for complete validation of the developed gene signature. A matched clinical cohort from recent MDM2 inhibitor trials would be optimal, but only one such database exists (from the completed phase 1 study of RG7112) (10), which is not publicly available. However, the ongoing clinical trial of DS-3032b in AML (NCT02319369) will definitely provide such a dataset in near future. One could also argue the feasibility of 1532-gene expression profiling for clinical implementation. In this regard, we could narrow down the number of selected genes since every gene is ranked according to its predictive significance. For example, the 50 top-ranked genes used in Figure 4F will be also tested in the trial dataset (Figure 5A).
The cell line-based 175-gene signature and the AML-specific 1532-gene signature share only a limited number of genes (26 genes), for at least two reasons: 1) the signatures were developed through entirely different algorithms (one selected highly expressed genes in sensitive cell lines, while the other used random forest machine learning), 2) cell types used for gene expression profiling were different (AML-specific or not). Considering that only about 60 genes were found to be common p53 target genes in different tissue types in the meta-analysis of 16 genome-wide datasets (24,25), the 26-gene overlap in our signatures is not surprising, and may rather well reflect AML-specific characteristics. Therefore, even though validation is more advanced for the 175-gene signature, we believe that it will be important to develop these two signatures in parallel. Specifically, we plan to validate the two signatures in the ongoing clinical study of DS-3032b in AML patients (NCT02319369) (Figure 5A). Furthermore, because the tumor-type specific approach can be applied to relatively small datasets (e.g. 33 samples as in the present study), ongoing clinical studies of DS-3032b and other MDM2 inhibitors for any cancers could additionally define improved tumor-type-specific gene signatures (Figure 5B). Moreover, a similar tumor-type-specific approach may be potentially applied to other molecularly targeted agents, if a referenced gene set related to the agent’s mechanism of action (corresponding to the 32 referenced genes for MDM2 inhibition) can be identified.
Supplementary Material
Statement of Significance.
This study demonstrates that gene expression profiling combined with TP53 mutational status predicts anti-tumor effects of MDM2 inhibitors in vitro and in vivo.
Acknowledgments
MA received partial funding for this project from Daiichi Sankyo.
Financial Support: This work was supported in part by grants from the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad Award (to JI); the Ministry of Education, Culture, Sports, Science and Technology in Japan (26461425), the Princess Takamatsu Cancer Research Fund (14-24610), the Osaka Cancer Research Foundation (to KK); and the National Institutes of Health USA (CA49639, 100632, CA136411, and CA16672), and the Paul and Mary Haas Chair in Genetics (to MA).
We thank Jairo A Matthews for clinical data collection. We thank Dr. N. Hail and Mr. D.R. Norwood for editorial assistance. We also thank Kenji Watanabe for technical support. This work was supported in part by grants from the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad Award (to JI); the Ministry of Education, Culture, Sports, Science and Technology in Japan (26461425), the Princess Takamatsu Cancer Research Fund (14-24610), the Osaka Cancer Research Foundation (to KK); and the National Institutes of Health USA (CA49639, 100632, CA136411, and CA16672), and the Paul and Mary Haas Chair in Genetics (to MA).
Footnotes
Conflict of interest: KN, TS, KT, and AT are/were employees of and own stock in Daiichi Sankyo. JI has ownership interests in Daiichi Sankyo.
Author contributions: JI, KN, AZ, KK, RED, RA and MA conceived and designed the study. JI, TS, KT, DC, RZ and LH acquired the data. JI, KN, KK, TS, KT, WM, MCJM, CD, RED and RA analyzed and interpreted the data. JI, KN, TS, KT, KK, RED and MA wrote, reviewed and/or revised the manuscript.
References
- 1.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26(15):3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 4.Kojima K, Konopleva M, Samudio IJ, Shikami M, Cabreira-Hansen M, McQueen T, et al. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood. 2005;106(9):3150–3159. doi: 10.1182/blood-2005-02-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long J, Parkin B, Ouillette P, Bixby D, Shedden K, Erba H, et al. Multiple distinct molecular mechanisms influence sensitivity and resistance to MDM2 inhibitors in adult acute myelogenous leukemia. Blood. 2010;116(1):71–80. doi: 10.1182/blood-2010-01-261628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Q, Zhang Z, Liu JJ, Jiang N, Zhang J, Ross TM, et al. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem. 2013;56(14):5979–5983. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 7.Tovar C, Graves B, Packman K, Filipovic Z, Higgins B, Xia M, et al. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013;73(8):2587–2597. doi: 10.1158/0008-5472.CAN-12-2807. [DOI] [PubMed] [Google Scholar]
- 8.Miyazaki M, Uoto K, Sugimoto Y, Naito H, Yoshida K, Okayama T, et al. Discovery of DS-5272 as a promising candidate: A potent and orally active p53-MDM2 interaction inhibitor. Bioorg Med Chem. 2015;23(10):2360–2367. doi: 10.1016/j.bmc.2015.03.069. [DOI] [PubMed] [Google Scholar]
- 9.Ray-Coquard I, Blay JY, Italiano A, Le Cesne A, Penel N, Zhi J, et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol. 2012;13(11):1133–1140. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 10.Andreeff M, Kelly KR, Yee K, Assouline S, Strair R, Popplewell L, et al. Results of the Phase I Trial of RG7112, a Small-Molecule MDM2 Antagonist in Leukemia. Clin Cancer Res. 2016;22(4):868–876. doi: 10.1158/1078-0432.CCR-15-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reis B, Jukofsky L, Chen G, Martinelli G, Zhong H, So V, et al. Acute myeloid leukemia patient clinical response to idasanutlin (RG7388) is associated with pre-treatment MDM2 protein expression in leukemic blasts. Haematologica. 2016 doi: 10.3324/haematol.2015.139717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yee K, Martinelli G, Vey N. Phase 1/1b study of RG7388, a potent MDM2 antagonist in acute myelogenous leukemia patients. Blood. 2014;124 abstract #116. [Google Scholar]
- 13.Bauer T, Hong D, Somaiah N, Cai C, Song C, Kumar P, et al. A phase I dose escalation study of the MDM2 inhibitor DS-3032b in patients with advanced solid tumors and lymphomas. Mol Cancer Ther. 2015;14 Abstract B27. [Google Scholar]
- 14.Ishizawa J, Kojima K, Chachad D, Ruvolo P, Ruvolo V, Jacamo RO, et al. ATF4 induction through an atypical integrated stress response to ONC201 triggers p53-independent apoptosis in hematological malignancies. Sci Signal. 2016;9(415):ra17. doi: 10.1126/scisignal.aac4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishizawa J, Kojima K, McQueen T, Ruvolo V, Chachad D, Nogueras-Gonzalez GM, et al. Mitochondrial Profiling of Acute Myeloid Leukemia in the Assessment of Response to Apoptosis Modulating Drugs. PLoS One. 2015;10(9):e0138377. doi: 10.1371/journal.pone.0138377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong H, Chen G, Jukofsky L, Geho D, Han SW, Birzele F, et al. MDM2 antagonist clinical response association with a gene expression signature in acute myeloid leukaemia. Br J Haematol. 2015;171(3):432–435. doi: 10.1111/bjh.13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 19.Jeay S, Gaulis S, Ferretti S, Bitter H, Ito M, Valat T, et al. A distinct p53 target gene set predicts for response to the selective p53-HDM2 inhibitor NVP-CGM097. Elife. 2015;4 doi: 10.7554/eLife.06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrysik Z, Kim J, Tan AC, Espinosa JM. A genetic screen identifies TCF3/E2A and TRIAP1 as pathway-specific regulators of the cellular response to p53 activation. Cell Rep. 2013;3(5):1346–1354. doi: 10.1016/j.celrep.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye P, Liu Y, Chen C, Tang F, Wu Q, Wang X, et al. An mTORC1-Mdm2-Drosha axis for miRNA biogenesis in response to glucose- and amino acid-deprivation. Mol Cell. 2015;57(4):708–720. doi: 10.1016/j.molcel.2014.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prokocimer M, Molchadsky A, Rotter V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: projections on diagnostic workup and therapy. Blood. 2017;130(6):699–712. doi: 10.1182/blood-2017-02-763086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonkin D. Expression signature based on TP53 target genes doesn't predict response to TP53-MDM2 inhibitor in wild type TP53 tumors. Elife. 2015;4 doi: 10.7554/eLife.10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170(6):1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36(28):3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.