Abstract
The significance of lipid droplets in lipid metabolism, cell signaling, and regulating longevity is increasingly recognized, yet the lipid droplet’s unique properties and architecture make it difficult to size and study using conventional methods. To begin to address this issue, we demonstrate the capabilities of nanoparticle tracking analysis (NTA) for sizing of lipid droplets. NTA was found to be adequate to assess lipid droplet stability over time, indicating that lipid droplet preparations are stable for up to 24 hours. NTA had the ability to compare the size distributions of lipid droplets from adult and geriatric mouse liver tissue, suggesting an age-related decrease in lipid droplet size. This is the first report on the use of NTA to size intracellular organelles.
Keywords: lipid droplets, nanoparticle tracking analysis, aging, liver, organelles
Graphical abstract
Light scattering reveal the temporal positions of individual lipid droplets, which are recorded with a camera. The two-dimensional diffusion constant of each lipid droplet is extracted from the data set, which is then used to calculate a hydrodynamic radius using the Stokes-Einstein equation
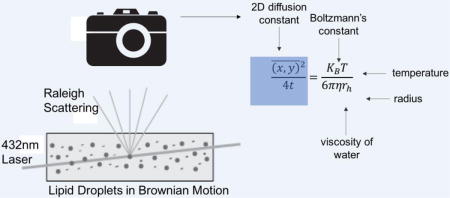
1. Introduction
Intracellular lipid droplets, composed of a neutral lipid core surrounded by a phospholipid monolayer, are found in most cell types. These dynamic organelles play roles in many cellular processes including lipid metabolism, cell signaling, immune function, membrane trafficking, and regulation of longevity[1, 2]. Larger triacylglycerol rich lipid droplets (10-200 μm diameter) are present in adipocytes [3, 4] while smaller lipid droplets (1 μm diameter or less) enriched in cholesteryl esters exist in brown adipose tissue, liver, muscle, heart, and connective tissues [5]. The accumulation of excess lipids in lipid droplets is associated with a multitude of lipid storage associated disorders, including lipodystrophies, obesity, cachexia, atherosclerosis, and non-alcoholic fatty liver disease (NAFLD)[6]. In these disease states, the accumulation of lipid droplets is both a hallmark and etiological factor in disease development and progression[7, 8]. Although numerous studies have defined the molecular and physiological changes that occur in the progression of these diseases, techniques to study individual lipid droplets are limited. To begin to address this, it is imperative to investigate new methods that enable the measurement of individual lipid droplets.
The most commonly employed method of measuring lipid droplet size is confocal fluorescent microscopy [9–11]. Lipid droplets in cells and tissue sections are visualized by a number of commercially available lipophilic dyes [12–15] and the area and number of lipid droplets in the field of view are quantified using image analysis software [16]. Lipophilic dyes with improved selectivity, improved photostability, and reduced cytotoxicity have been developed in recent years [17, 18]. Measuring the size of individual lipid droplets from microscopy images is not high-throughput and microscopy experiments with lipophilic dyes must be interpreted with caution, as fluorescence signal is influenced by variations in the cellular distribution of the dye and its incorporation into other lipid rich structures. Colocalization experiments with bona-fide lipid droplet markers are required for correct identification of lipid droplets, but often these lipid droplet proteins do not coat all lipid droplets. Further, in order to evaluate lipid droplets in tissues by microscopy, the sample of interest must be embedded, sectioned, and stained prior to visualization [14], requiring a major time investment. Recently developed anti-Stokes Raman Scattering (CARS) [19] and Stimulated Raman Scattering (SERS) [20] imaging methods have been used to size lipid droplets in cells. While these label-free methods represent an improvement over fluorescent microscopy, a sophisticated instrumental setup is required and these methods are further limited by the relatively low signal intensity and long acquisition time required for Raman-based imaging techniques. Recently, three dimensional quantitative phase imaging of lipid droplets in hepatocytes was reported [21], but this method also requires a sophisticated instrumental setup and so far has only been applied to individual cells. Despite existing imaging methods for measuring lipid droplet size in cells and tissues, a label free method based on individual lipid droplet size measurements would reduce the bias introduced by limited throughput, alleviate the necessity of complex instrumental setup, and the use of lipophilic dyes, and allow the lipid droplets to be recovered for downstream analysis.
Flow cytometry and capillary cytometry are well-defined methods for determining heterogeneity among individual organelles[22–24]. To our knowledge, there has been no report of flow cytometry of individual lipid droplets, possibly due to their unique physiochemical properties including low specific gravity and tendency to adsorb to surfaces. Size distribution of individual lipid droplets isolated from cells and tissues has been measured by dynamic light scattering (DLS) [9], but this method is an ensemble technique based on the assumption that dispersed particles are uniform in composition, making it inappropriate for heterogeneous biological samples. Furthermore, DLS measurements are highly influenced by the presence of large particles or aggregates in the particle suspension.
Nanoparticle tracking analysis (NTA) is a single-particle measurement technique, as opposed to the ensemble approach of DLS, enabling it to measure individual nanoparticle size and concentration in a given sample[25]. In NTA, a particle dispersion is placed within an optical cell and irradiated with an intense laser source. The light scattered by individual particles within the cell is collected using a microscope objective and recorded with a camera. While the particles are too small to image optically, their scattered light reveals the position and Brownian motion of each particle in the field of view. The recorded motion is analyzed to derive the two-dimensional diffusion constant of each particle, which is then used to calculate the particle’s sphere equivalent hydrodynamic radius. Unlike DLS, NTA provides a direct number-weighted particle size distribution and increased size resolution for non-uniform samples. NTA has been used previously to determine the size and concentration of biological particles, including exosomes [26–29], virus particles [30] and gold nanoparticle protein conjugates [31], but has never been used to measure the size of lipid droplets.
This article introduces a new method for monitoring lipid droplet size in tissues using NTA. Analysis of isolated lipid droplet dispersions via NTA takes less than ten minutes, and can be applied to lipid droplets isolated from any tissue or cell of interest. Further, NTA is non-destructive, and the lipid droplets can be recovered after measurement for downstream analysis. We assessed the ability of a Nanosight LM-10 instrument to measure size and determine its suitability to measure lipid droplets isolated from mouse liver. We also applied the method to monitor lipid droplet preparation stability over time and measure the lipid droplet size distributions in adult and geriatric mice. This work represents the first size measurements of isolated lipid droplets and intracellular organelles. In the future, NTA of lipid droplets could be used to examine lipid droplets obtained from tissue and cell lines of any model system, model organism, or clinical sample of interest.
2. Experimental Section
2.1 Materials, Reagents, Buffers, and Solutions
Sucrose and ethylenediaminetetraacetic acid (EDTA) were obtained from Sigma Aldrich (St. Louis, MO). Tris base was obtained from Thermo Fisher (Waltham, MA). Water was purified with a Millipore Synergy UV system (18.2mΩ/cm, Bedford MA). Liver homogenization buffer (Buffer H) consisted of 250mM sucrose, 10mM Tris, pH 7.0. Buffer A, 2X concentration, consisted of 500mM sucrose, 2mM EDTA, and 40mM Tris base, pH 7.8. Sucrose gradient step solutions (25% and 35% sucrose v/v) were mixed with Millipore water to a final concentration of 1X Buffer A. Pierce Protease Inhibitor Mini Tablets (4-benzenesulfonyl fluoride hydrocholoride, aprotinin, bestatin, E-64, leupeptin and pepstatin A, Thermo Fisher, Waltham MA) were added to all buffers in accordance with manufacturer’s instructions.
2.2 Human and Animal Rights Statement
All mice were housed in a designated clean facility and treated in accordance with protocols approved by the University of Minnesota Institutional Animal Care and Use Committee.
2.3 Lipid droplet isolation from mouse liver
Lipid droplets were prepared from the livers of C57BL6 female mice, as described previously [32, 33]. Mice were euthanized by intraperitoneal injection of pentobarbital (200 mg/Kg) and the liver was excised. All isolation steps were done at 4°C. The liver was weighed and minced in a petri dish using a flat razor blade into small (1mm) pieces and resuspended in 3mL of Buffer H. The liver homogenate was transferred into a nitrogen cavitation chamber and charged to 200 psi. After 15 minutes on ice, the cell lysate was released from the nitrogen cavitation chamber into a 50 mL conical tube and centrifuged at 1,000g for 10 minutes. The post nuclear supernatant was loaded onto a sucrose step gradient (4mL 35% sucrose/Buffer A, 4mL 25% sucrose/Buffer A) and centrifuged at 36,000 rpm in a Beckman SW 41 swinging bucket rotor for four hours. The lipid droplet fraction appeared as white film at the top of the tube, which was removed with a Pasteur pipette. The lipid droplets suspension was mixed by pipetting and diluted 1:90 in Buffer H prior to loading into the Nanosight LM-10 analysis chamber.
For stability measurements, lipid droplets were isolated from a 35 week old C57BL/6 mouse. The lipid droplet sample was measured initially after isolation using NTA (Day 0). The lipid droplet suspension was stored in an microcentrifuge tube on a rotator in a 4°C cold room. The lipid droplet suspension was sampled at 24 hours, 48 hours, and one week later (samples designated Day 1, Day 2, and Day 7). To investigate the differences in lipid droplet size distributions between geriatric mice and adult mice, lipid droplets were isolated from n=3 livers from geriatric mice and n=3 livers from adult mice prior to NTA. Mice in the adult group were ~35 weeks and mice in the geriatric group were ~115 weeks.
2.4 Nanoparticle Tracking Analysis
The size distributions of lipid droplets, along with those of particle size standards, were measured using a Nanosight LM-10 Nanoparticle Tracking Analyzer (Malvern, Salisbury, UK). The instrument was equipped with a 405nm laser and a high sensitivity CMOS camera. Quadruplicate videos of each sample were captured for 60 seconds in static mode. The frame rate was 25 frames per second, and up to 1,500 frames were captured per video. The chamber temperature was controlled at 25°C, camera gain was set to “automatic” and the viscosity was set to “water”. Videos were analyzed with NTA 3.2 Dev Build 3.2.16 software to yield a number-weighted size distribution in the size range of interest (between about 30 and 1000nm), along with several statistical measures of that distribution, including its median, mean, and standard deviation. To verify particle size accuracy, polystyrene latex nanosphere standards were also measured at three sizes: 203nm ± 6 nm standard deviation (#3K-200), 102nm ± 8 nm standard deviation (#3K-100), and 31nm (#3030A). All particle standards and their specifications (when available) were obtained from Thermo Fisher Scientific (Waltham, MA). Dilutions were made in sterile filtered distilled water.
2.5 Data Analysis
Each particle’s motion is tracked individually from frame to frame during the 60 second measurement period. The NTA software examines each track to determine if it meets the quality criterion, i.e., staying within the field of view for a sufficient number of frames to accurately reconstruct the particle motion. Using the subset of the raw track data that meets this criterion, the NTA software determines the particle’s diffusion constant (D, m2/s) from Brownian motion. The diffusion coefficient is subsequently used to calculate a sphere equivalent hydrodynamic radius (rh), through the Stokes-Einstein relationship (Equation 1)
| Equation 1 |
where KB is Boltzmann’s constant (1.38064852 × 10−23 m2 kg s−2 K−1), T is temperature (25°C), and η is viscosity of water (0.89 N s/m2 at 25°C). While Brownian motion occurs in three dimensions, the Nanosight observes the individual particles in two dimensions and the NTA software derives D using the equation below (Equation 2)
| Equation 2 |
where t is time and x and y are distance on two orthogonal axes. As diffusion is isotropic in space and occurs independently in all directions, 2D measurement of movement is an accurate representation of 3D Brownian motion [34, 35]. Size distribution (number weighting), size percentiles, summary statistics and videos were exported from NTA 3.2.
Raw lipid droplet size measurements were corrected using particle standard data via the procedure described below. The % error (e) in NTA measurement was calculated using Equation 3:
| Equation 3 |
Where μ is the mean particle size reported by TEM, and is is the mean size of the particle standard reported by NTA. The % error e at each lipid droplet size was calculated from the particle standard measurements using the approach described in the Electronic Supplementary Material (ESM) in Section “Supplementary Data Analysis”.
As each error in measurement was in the positive direction, a correction factor for each size was calculated using Equation 4:
| Equation 4 |
The corrected LD particle size (LD1) was calculated using Equation 5.
| Equation 5 |
where LD0 is the observed LD particle size and c is the correction factor.
Histograms and quantile-quantile (QQ plots) were used to compare distributions of lipid droplet sizes. QQ plots are created by plotting the fifth through 95th percentiles from two sample distributions. If the two distributions are similar in shape, their QQ plot approaches an X=Y line. A single preparation of lipid droplets was measured at days 0, 1, 2, and 7. For visual evaluation of the stability of the preparation, the QQ plot was produced by plotting 19 quantiles (representing the 5th–95th percentiles) of the size distributions from days 1, 2, and 7 versus the respective 5th through 95th percentiles of the size distributions from day 0. For visual analysis of the difference in LD size distribution between old and young mice, the QQ plot was produced by plotting each of the 5th–95th percentiles (avg ± SEM) of three biological replicate old mice vs the respective percentile (avg ± standard error of measurement, referred hereafter as “SEM”) of three biological replicate young mice. ANOVA and Tukey post-hoc testing was done in Prism 7 (GraphPad, San Diego, CA). Wilcoxan Rank Sum test, linear regression, size data correction, and data plotting was done in MATLAB 2016b (MathWorks, Natick, MA).
3. Results and Discussion
3.1 Figures of Merit
In order to evaluate the suitability of the Nanosight LM-10 in lipid droplet analysis, we assessed the ability to determine size accurately over size ranges expected to be found in lipid droplets. According to the manufacturer’s specifications, the Nanosight LM-10 is suitable to measure concentrations of ~106 to ~109 particles/ml in the size range from 10nm to 2000nm. These values are dependent on the nature of the particles being measured. We selected polystyrene latex nanospheres (refractive index (1.615) [36]) over metallic nanoparticles (refractive index > 2.0) because the former’s refractive index is similar to that of individual organelles, such as mitochondria (1.41) [37], and triglycerides (1.43-1.48) [38], the main component of lipid droplets.
We used polystyrene latex nanospheres of different sizes to determine the size accuracy of measuring biological particles on the Nanosight LM-10 (Table 1). The expected mean size of the smallest polystyrene latex nanoparticles used in this study, determined by the manufacturer using DLS, was 31 nm. At a particle concentration of (1.66 ± 0.08) × 109 particles/mL, the mean particle size as measured by NTA was 38 ± 1 nm (± SEM) The size distributions of the particle standards obtained via NTA can be visualized in a histogram, Figure 1A. We sized the nominal 31nm particles by TEM (ESM Fig. S1) to provide individual particle size measurements to compare with NTA measurement. TEM analysis indicated that the mean diameter of the 31 nm standards was 25 nm ± 5 nm (± SEM). Therefore, at a 25 nm polystyrene nanosphere size, NTA reports a 52% higher mean particle size than TEM. We subsequently measured the 102 nm ± 8 nm (± SEM) and 203 nm ± 5 nm (± SEM) particle standards by NTA (Figures 1B and 1C). The Nanosight reported a 5% higher mean particle size than TEM at 102 nm, and was is in agreement with TEM measurement at 203 nm. We used the measurement of these three standards to correct the sizes of lipid droplets reported by NTA. The correction factor was different for each size and was calculated using Equations 3 and Equation 4.
Table 1. Analysis of Polystyrene Latex Nanosphere Standards.
Values are representedas mean ± SEM. NTA measurements had n=5 technical replicates.
| Size Measurement | |||
|---|---|---|---|
| Particles/ml | NTA | TEM | DLS |
| (1.66 ± 0.08) × 109 | 38 ± 1 nm | 25 ± 5 nm* | 31nm |
| (2.13 ± 0.07) × 109 | 108 ± 1 nm | 102 ± 8 nm | N/A |
| (1.59 ± 0.01) × 109 | 203 ± 1 nm | 203 ± 5 nm | N/A |
indicates TEM done in this study, n=60 nanospheres. TEM measurements for additional standards are as reported by the manufacturer (Thermo Fisher, Waltham, MA).
Figure 1. Size distributions of nanosphere standards determined by NTA.
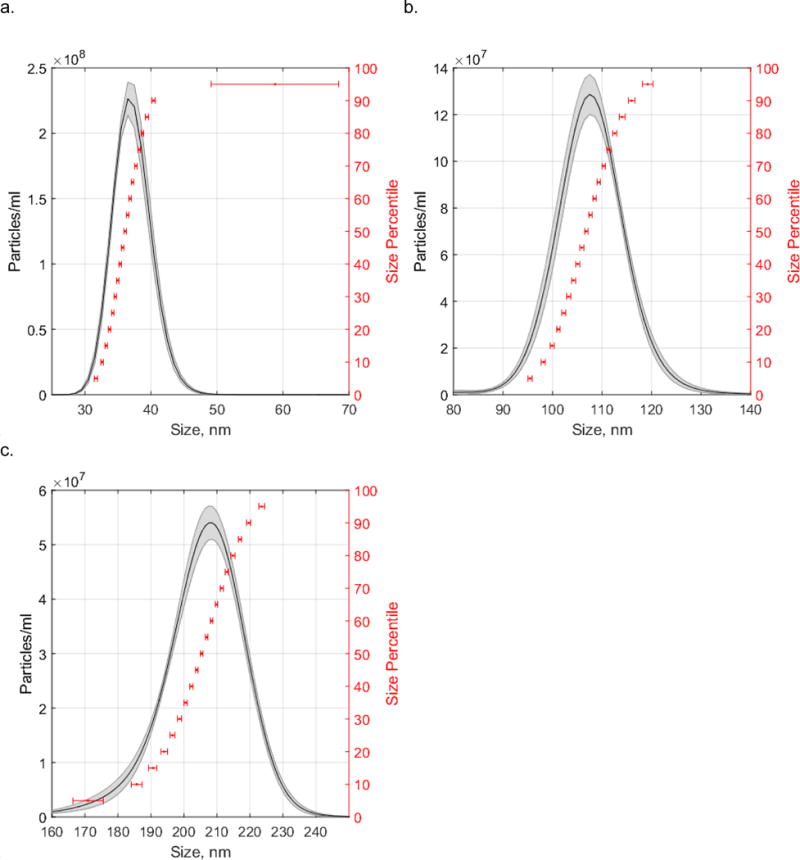
(a) 31nm, (b) 102nm, (c) 203nm. Shaded area of histogram represents particle density (average ± SEM, n=5 technical replicates). Horizontal error bars represent size error (± SEM, n=5 technical replicates) at percentiles ranging from 5th to 95th percentile at 5% increments.
We also measured a dilution series of the 203nm polystyrene latex nanospheres to establish a low concentration limit for biological particles on the Nanosight LM-10. The results from measurements of the dilution series is summarized in Table 2. At the highest particle concentration measured, 1.59 × 109 particles/mL, the mean particle size was 203 ± 1 nm (± SEM). At the lowest particle concentration measured, (1.43 ± 0.66) × 107 particles/mL, the mean particle size was 174 ± 24 nm. The tabulated data suggest that NTA decreases in measurement accuracy at concentrations below (2.58 ± 0.18) × 108 particles/mL. While the manufacturer indicates the Nanosight LM-10 can accurately size measure ~106 to ~109 particles/mL, our data suggests that NTA is only appropriate for biological particle concentrations near the high end of the manufacturer’s recommended range.
Table 2. Dilution series of 203nm standard polystyrene latex nanospheres.
Values are represented as mean ± SEM, n=5 technical replicates. The second column gives the mean number of particles tracked by NTA software per frame of recorded video.
| Mean Particles/ml | Mean Particle Size, nm | Mean Particles/frame |
|---|---|---|
| (1.59 ± 0.01) × 109 | 203 ± 1 | 81 ± 1 |
| (2.58 ± 0.18) × 108 | 218 ± 2 | 13 ± 1 |
| (1.60 ± 0.07) × 108 | 218 ± 1 | 8 ± 0.4 |
| (1.43 ± 0.66) × 107 | 174 ± 24 | 1 ± 0.3 |
3.2 Nanoparticle Tracking Analysis of Individual Lipid Droplets
The next focus of this study was to determine the suitability of NTA for measuring the size distributions of lipid droplets. Prior work demonstrated the use of NTA to measure size distributions of biological particles, most notably exosomes (20-100 nm in size) [26–29]. It is unclear whether these prior studies used size standards to assess and correct for systematic sizing errors. Furthermore, NTA has never been applied to lipid droplets.
Lipid droplets were isolated from mouse liver tissue and the purity of the cellular fractions were validated by Western blot (ESM Fig. S2). In the Nanosight chamber, lipid droplets act as individual point scatterers when illuminated with 405 nm light. Therefore, the lipid droplets appeared as small points of light undergoing random, Brownian motion (see ESM, Movie S1) from which their individual motion paths were recorded wth a CMOS video camera operating at 25 frames per second. The diameter of the individual particles was calculated from their diffusion constant by the Nanosight NTA software using Equation 1 and Equation 2 as described above, and corrected using the standard particle measurements with Equation 3 and Equation 4.
A histogram was used to visualize the corrected size distribution of lipid droplets isolated from mouse liver tissue (Figure 2). The mean corrected particle size was 168 ± 2 nm (mean ± SEM, n=5 technical replicates). In non-adipocytes, lipid droplets do not exceed 1μm in diameter under basal conditions [39]. In lipid loaded conditons or disease models, non-adipocyte lipid droplets grow in size (8-20μm) and usually form heterogenous populations varying in size and lipid composition[40, 41]. In the current study, the liver lipid droplet sizes ranged from 105 to 295nm (5th–95th percentiles) in diameter, which was representative of the lipid droplet size heterogeneity present in this particular tissue under basal conditions.
Figure 2. Mouse liver lipid droplet size distribution determined by NTA.
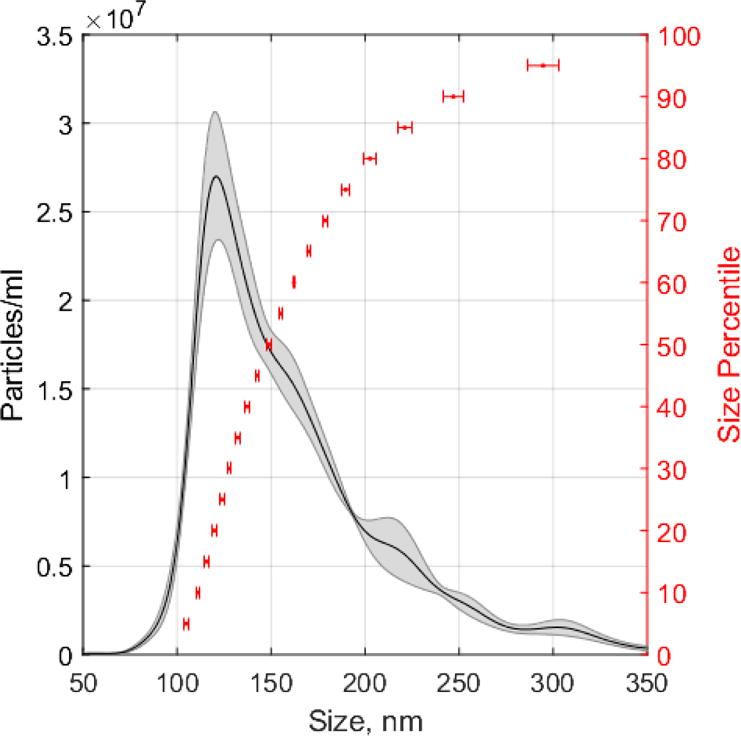
The particle concentration was (2.13 ± 0.14) × 109 particles/mL (mean ± SEM, n=5 technical replicates). Shaded area of histogram represents particle density (average ± SEM, n=5 technical replicates). Horizontal error bars represent size error (± SEM, n=5 technical replicates) at percentiles ranging from 5th to 95th percentile at 5% increments.
3.3 Lipid Droplet Stability Analysis
To investigate the stability of lipid droplets during storage, we compared the individual lipid droplet size distributions resulting from uncorrected measurements of a single preparation of lipid droplets over the course of a week. The mean lipid droplet size and mean lipid droplet concentration for each day are summarized in Table 3. The shift in distribution shape apparent on each day can be further appreciated by histograms (ESM Fig. S3). One way ANOVA analysis indicated statistically significant differences in mean LD size (ESM Table S1A) and mean LD concentration (ESM Table S2A) sampled on all four days. This suggests that the lipid droplet preparation was not stable over the one week period. Tukey post-hoc testing indicated that mean LD size was not significantly different from Day 0 to Day 1 or Day 2 (ESM Table S1A). Similary, Tukey post-hoc testing indicated only mean LD concentration Day 0 and Day 1 (ESM Table S1B) are not significantly different. Thus, LD preparations were stable in terms of size distribution and concentration for up to one day.
Table 3. Lipid Droplet Stability Study.
Each row represents the average LD concentration (particles/ml) and mean raw LD size (nm) as given by NTA, n=5 technical replicates, mean ± SEM.
| Day | Mean LD Concentration particles/ml | Mean LD size, nm |
|---|---|---|
| 0 | (2.13 ± 0.14) × 109 | 170 ± 2 |
| 1 | (2.11 ± 0.09) × 109 | 167 ± 4 |
| 2 | (1.38 ± 0.07) × 109 | 180 ± 3 |
| 7 | (2.35 ± 0.14) × 109 | 221 ± 3 |
We chose to use QQ plots to visualize changes among LD size distributions (Figure 3). The percentiles of Day 1 and Day 2 (y axis) were visually similar to Day 0, while the percentiles of Day 7 shifted in the positive y-direction. If two distributions are similar in shape, a QQ plot comparing them approaches an X=Y line. Thus, the slope of the linear fit of their QQ plot will be ~1. To investigate if this was the case, we applied linear regression analysis and made pairwise comparisons of the slopes. The distributions between slopes were not statistically significant for Day 1 vs Day 0. The statistical significant difference between the Day 2 vs Day 0 slope is indicative of changes in the lipid droplet size distribution. Not surprisingly, the comparison of Day 7 vs Day 0 displayed the largest changes and cannot be fit to a linear model. Thus, based on the ANOVA, post-hoc Tukey, QQ plots and linear regression analysis, lipid droplets maintain their stability for 24 hours. These results suggest size analysis of isolated lipid droplets must be completed within 24 hours to ensure accurate representation of the lipid droplet distribution isolated from the sample of interest.
Figure 3. Stability of uncorrected lipid droplet size distribution.
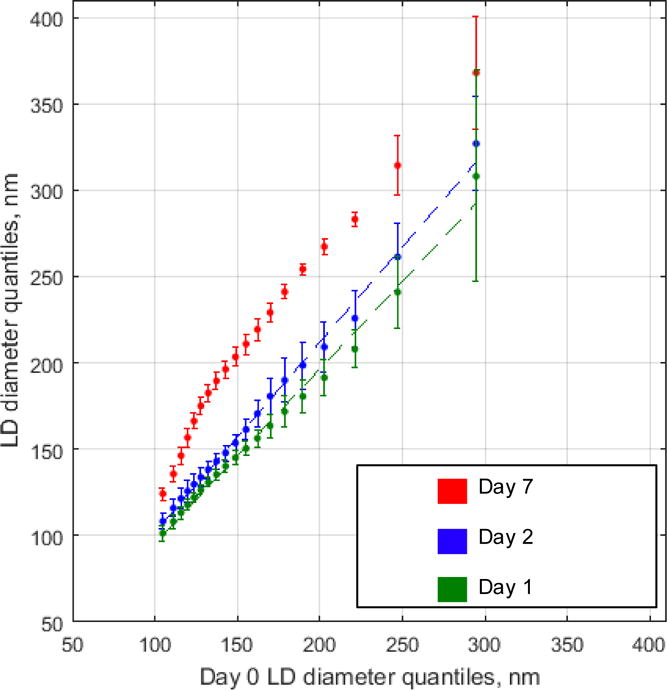
LD diameter (nm) for Days 1, 2, and 7 (y axis) vs Day 0 (x-axis). Markers represent percentiles ranging from 5th to 95th percentile at 5% increments. Dotted lines represent linear regression of each Day vs Day 0. Day 1 vs Day 0 slope confidence interval is [0.96 1.1] and Day 2 vs Day 0 slope confidence interval is [1.1 1.2]. R2 is 1.0 for both fits. Day 7 vs Day 0 fit was non-linear and not represented in the plot. Linear regression analysis was further investigated by graphing of the residuals (ESM Fig. S3).
3.4 Lipid Droplet Size Analysis in Adult and Geriatric Mice
The population of lipid droplets in any given cell is size heterogeneous, and evidence suggests that LDs of various sizes differ in in lipid composition and the recruitment of cytosolic proteins[42]. Lipid droplet size changes resulting from different membrane composition and protein recruitment have been reported previously [31], but have never been described in a comparative aging study. Aging is associated with lipid accumulation in multiple tissues, including the liver [45, 46], but the effect of age-related lipid accumulation on lipid droplet size or number has not yet been investigated. This work represents the first measurements of lipid droplet size distribution in geriatric mice.
The mean corrected LD size is summarized in Table 4. It indicates a larger variation in mean lipid droplet size among the geriatric mice, as expected. The distributions in corrected lipid droplet sizes were further compared using a QQ plot (Figure 4A). The general trend of the geriatric quantiles was a shift towards the x-axis from the adult quantiles, indicative of a smaller size. Furthermore, the clear deviation from the 5th–65th percentiles suggested an increased abundance of smaller lipid droplet sizes. The uniqueness in the distribution of each aging mouse is captured by plotting the lipid droplet size distribution from each geriatric mouse versus the average lipid droplet size distribution from the adult mice (ESM Fig. S5) as well as the respective relative variation of size distributions (ESM Fig. S6).
Table 4. Lipid Droplets from Adult and Geriatric Mice.
Each row represents the mean corrected LD size (nm), n=5 technical replicates, mean ± SEM.
| Mouse | Mean LD size, nm |
|---|---|
| Geriatric 1 | 149 ± 20 |
| Geriatric 2 | 184 ± 6 |
| Geriatric 3 | 84 ± 6 |
| Adult 1 | 168 ± 2 |
| Adult 2 | 240 ± 7 |
| Adult 3 | 227 ± 4 |
Figure 4. Comparison of the lipid droplet size distributions of adult and geriatric mice.
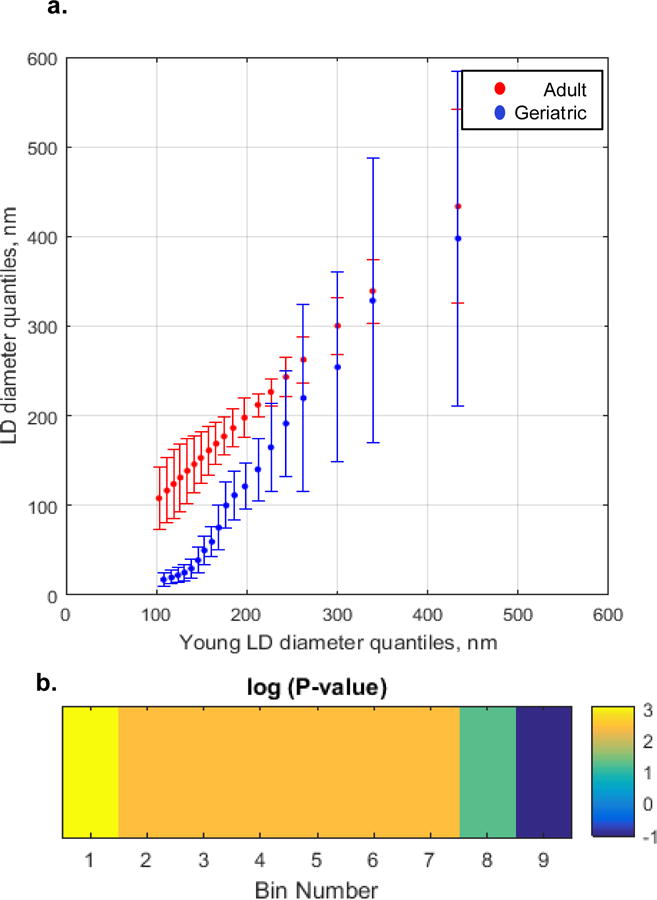
(a) QQ plot. LD diameter (nm) geriatric (y axis, blue) and adult (y axis, red) vs adult (x-axis). Geriatric and adult mice were ~ 115 and ~ 35 weeks old, respectively. Markers represent percentiles ranging from 5th to 95th percentile at 5% increments. Each percentile is represented as average ± SEM, n = 3 mice. (b) Significance plot from hypothesis testing of average adult and average geriatric LD size distributions. Color coded areas of significance are shown in the QQ lot, Part (a). A Wilcoxan Rank Sum test compared the percentiles of each distribution (for each 10% of the data set, Bins 1-9) from 1% to 99%. Each p-value was transformed by calculating −1.3-log10(P-value). This was done for ease of visualization, such that all P-values <0.05 are transformed as above 0 and all P-values that are >0.05 are transformed as below zero[47].
A statistical significance profile was necessary to ascertain the average size distribution disparities that exist in the geriatric mice as compared to the adult mice (Figure 4B). Statistically significant (p<0.05) differences existed in all regions except for the last 10% of the distribution (representing 90th–99th percentiles, 302nm-528nm for geriatric mice and 322-619nm for adult mice). The most significant differences were at first 10% (representing 1st–9th percentiles, 14-19nm for geriatric mice and 86-109nm for adult mice), representing the smallest lipid droplets. Thus, geriatric mice have a statistically significant different distribution of lipid droplet sizes relative to the adult mice, characterized by lipid droplets of smaller size.
4.0 Conclusion
While NTA has been successfully applied to exosomes [26–29], virus particles [30] and gold nanoparticle protein conjugates[31], here we introduce NTA for the size measurements of lipid droplets isolated from tissues. To our knowledge, lipid droplets have not been sized in isolation. This method is capable of measuring the size distributions of lipid droplets from any tissue or cell type amenable to lipid droplet isolation, including human tissue biopsies. The complete procedure from tissue excision, organelle isolation and NTA analysis can be completed in approximately 6 hours, and the lipid droplet sample can be recovered and used in downstream analysis. This represents a vast improvement over time consuming and low throughput imaging based lipid droplet size measurements.
Supplementary Material
Acknowledgments
Funding
This work was supported by NIH AG020866. K.A.M. acknowledges support through a University of Minnesota Doctoral Dissertation Fellowship and National Institutes of Health (NIH) AG029796. C.P.N. acknowledges support from NIH DK007203. N.M.L. acknowledges support from NIH GM008700. D.G.M acknowledges support from NIH DK114401 and NIH AG055452. A portion of this work was carried out in the Minnesota Nano Center, which receives partial support from the National Science Foundation (NSF) through the NNCI program. A portion of this work was carried out in the Characterization Facility, University of Minnesota, which receives partial support from NSF through the MRSEC program.
Biographies

Katherine A. Muratore is a postdoctoral researcher in the Department of Microbiology, Immunology, and Molecular Genetics at University of California Los Angeles. She investigates the subcellular organization of protozoan parasites relevant to human health. She obtained her Ph.D. from the Biochemistry, Molecular Biology and Biophysics Program at the University of Minnesota, while working in Dr. Edgar Arriaga’s laboratory, where she worked on lipid droplet analysis.

Charles P. Najt is a post-doctoral fellow in Dr. Douglas Mashek’s laboratory. He is characterizing the role of various lipid droplet proteins in lipid droplet signaling and how it relates to age related diseases such as non-alcoholic fatty liver disease, diabetes, cardiovascular disease, and dyslipidemia.

Nick M. Livezey is a Ph.D. student in the Chemistry Program at the University of Minnesota. He is part of Dr. Valerie Pierre’s laboratory. He is developing siderophore-based tools for studying bacteria, ranging from nano-materials to small molecules. He utilizes multiple techniques for nano-materials characterization.

James Marti is senior scientist and Outreach Director for the Minnesota Nano Center, where he manages two research laboratories focusing on the synthesis, functionalization, and characterization of nanoparticles and on the biological applications of nanotechnology. A physicist by training, Dr. Marti’s primary scientific interest has been the physics and chemistry of micron-and nanometer-scale particles, particle systems, and related materials.

Doug G. Mashek is an Associate Professor in the Department of Biochemistry, Molecular Biology and Biophysics. His research laboratory focuses on lipid droplet biology with an emphasis on their regulation and role in cell signaling and disease etiology.

Edgar A. Arriaga is a Professor in the Department of Chemistry and faculty of the Department of Biomedical Engineering and the Department of Biochemistry, Molecular Biology and Biophysics. His research program focuses on the development and application of technologies for subcellular analysis.
Footnotes
Compliance with Ethical Standards
All mice were housed in a designated clean facility and treated in accordance with protocols approved by the University of Minnesota Institutional Animal Care and Use Committee.
Conflict of Interest
The authors have no potential conflicts of interest to disclose.
References
- 1.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/S0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AA, Bourque SD, Kyryakov P, Boukh-Viner T, Gregg C, Beach A, Burstein MT, Machkalyan G, Richard V, Rampersad S, Titorenko VI. A novel function of lipid droplets in regulating longevity. Biochem Soc Trans. 2009;37:1050–1055. doi: 10.1042/BST0371050. [DOI] [PubMed] [Google Scholar]
- 3.Mcintosh AL, Storey SM, Atshaves BP. Intracellular Lipid Droplets Contain Dynamic Pools of Sphingomyelin: ADRP Binds Phospholipids with High Affinity. Lipids. 2010;45:465–477. doi: 10.1007/s11745-010-3424-1.Intracellular. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Londos C, Brasaemle DL, Schultz CJ, Segrest JP, Kimmel AR. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin Cell Dev Biol. 1999;10:51–58. doi: 10.1006/scdb.1998.0275. [DOI] [PubMed] [Google Scholar]
- 5.Chanderbhan R, Noland BJ, Scallen TJ, Vahouny GV. Sterol carrier protein2. Delivery of cholesterol from adrenal lipid droplets to mitochondria for pregnanolone synthesis. J Biol Chem. 1982;257:8928–8934. [PubMed] [Google Scholar]
- 6.Krahmer N, Farese RV, Walther TC. Balancing the fat: Lipid droplets and human disease. EMBO Mol Med. 2013;5:905–915. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol. 2013;10:330–344. doi: 10.1038/nrgastro.2013.41. [DOI] [PubMed] [Google Scholar]
- 8.Day CP, James OFW. Steatohepatitis: A tale of two “Hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/S0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 9.Ding Y, Zhang S, Yang L, Na H, Zhang P, Zhang H, Wang Y, Chen Y, Yu J, Huo C, Xu S, Garaiova M, Cong Y, Liu P. Isolating lipid droplets from multiple species. Nat Protoc. 2012;8:43–51. doi: 10.1038/nprot.2012.142. [DOI] [PubMed] [Google Scholar]
- 10.Nishimoto Y, Nakajima S, Tateya S, Saito M, Ogawa W, Tamori Y. Cell death-inducing DNA fragmentation factor A-like effector A and fat-specific protein 27β coordinately control lipid droplet size in brown adipocytes. J Biol Chem. 2017;292:10824–10834. doi: 10.1074/jbc.M116.768820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi X, Li J, Zou X, Greggain J, Rødkær SV, Færgeman NJ, Liang B, Watts JL. Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J Lipid Res. 2013;54:2504–2514. doi: 10.1194/jlr.M039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenspan P, Mayer E, Fowler S. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang HJ, Hsu CL, Yang JY, Yang WY. Monodansylpentane as a blue-fluorescent lipid-droplet marker for multi-color live-cell imaging. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehlem A, Hagberg CE, Muhl L, Eriksson U, Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat Protoc. 2013;8:1149–1155. doi: 10.1038/nprot.2013.055. [DOI] [PubMed] [Google Scholar]
- 15.Klapper M, Ehmke M, Palgunow D, Böhme M, Matthäus C, Bergner G, Dietzek B, Popp J, Döring F. Fluorescence-based fixative and vital staining of lipid droplets in Caenorhabditis elegans reveal fat stores using microscopy and flow cytometry approaches. J Lipid Res. 2011;52:1281–1293. doi: 10.1194/jlr.D011940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutsch MJ, Schriever SC, Roscher AA, Ensenauer R. Digital image analysis approach for lipid droplet size quantitation of Oil Red O-stained cultured cells. Anal Biochem. 2014;445:87–89. doi: 10.1016/j.ab.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Wu A, Kolanowski JL, Boumelhem BB, Yang K, Lee R, Kaur A, Fraser ST, New EJ, Rendina LM. A Carborane-Containing Fluorophore as a Stain of Cellular Lipid Droplets. Chem - An Asian J. 2017;12:1704–1708. doi: 10.1002/asia.201700423. [DOI] [PubMed] [Google Scholar]
- 18.Gao M, Su H, Li S, Lin Y, Ling X, Qin A, Tang BZ. An easily accessible aggregation-induced emission probe for lipid droplet-specific imaging and movement tracking. Chem Commun. 2017;53:921–924. doi: 10.1039/C6CC09471F. [DOI] [PubMed] [Google Scholar]
- 19.Smus JP, Moura CC, McMorrow E, Tare RS, Oreffo ROC, Mahajan S. Tracking adipogenic differentiation of skeletal stem cells by label-free chemically selective imaging. Chem Sci. 2015;6:7089–7096. doi: 10.1039/C5SC02168E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao C, Zhou D, Chen T, Streets AM, Huang Y. Label-Free Digital Quantification of Lipid Droplets in Single Cells by Stimulated Raman Microscopy on a Microfluidic Platform. Anal Chem. 2016;88:4931–4939. doi: 10.1021/acs.analchem.6b00862. [DOI] [PubMed] [Google Scholar]
- 21.Kim K, Lee S, Yoon J, Heo J, Choi C, Park Y. Three-dimensional label-free imaging and quantification of lipid droplets in live hepatocytes. Sci Rep. 2016;6:1–8. doi: 10.1038/srep36815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniele JR, Heydari K, Arriaga EA, Dillin A. Identification and Characterization of Mitochondrial Subtypes in Caenorhabditis elegans via Analysis of Individual Mitochondria by Flow Cytometry. Anal Chem. 2016;88:6309–6316. doi: 10.1021/acs.analchem.6b00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor TH, Frost NW, Bowser MT, Arriaga EA. Analysis of individual mitochondria via fluorescent immunolabeling with Anti-TOM22 antibodies. Anal Bioanal Chem. 2014;406:1683–91. doi: 10.1007/s00216-013-7593-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Degtyarev M, Reichelt M, Lin K. Novel quantitative autophagy analysis by organelle flow cytometry after cell sonication. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malloy A, Carr B. Nanoparticle tracking analysis - The Halo System. Part Part Syst Charact. 2006;23:197–204. doi: 10.1002/ppsc.200601031. [DOI] [Google Scholar]
- 26.Soo CY, Song Y, Zheng Y, Campbell EC, Riches AC, Gunn-Moore F, Powis SJ. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136:192–197. doi: 10.1111/j.1365-2567.2012.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oosthuyzen W, Sime NEL, Ivy JR, Turtle EJ, Street JM, Pound J, Bath LE, Webb DJ, Gregory CD, Bailey MA, Dear JW. Quantification of human urinary exosomes by nanoparticle tracking analysis. J Physiol. 2013;23:5833–5842. doi: 10.1113/jphysiol.2013.264069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatischeff I, Larquet E, Falcon-Perez JM, Turpin PY, Kruglik SG. Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. J Extracell Vesicles. 2012;1 doi: 10.3402/jev.v1i0.19179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardiner C, Ferreira YJ, Dragovic RA, Redman CWG, Sargent IL. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kramberger P, Ciringer M, Štrancar A, Peterka M. Evaluation of nanoparticle tracking analysis for total virus particle determination. Virol J. 2012;9:265. doi: 10.1186/1743-422X-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James AE, Driskell JD. Monitoring gold nanoparticle conjugation and analysis of biomolecular binding with nanoparticle tracking analysis (NTA) and dynamic light scattering (DLS) Analyst. 2013;138:1212. doi: 10.1039/c2an36467k. [DOI] [PubMed] [Google Scholar]
- 32.Atshaves BP, Storey SM, McIntosh AL, Petrescu AD, Lyuksyutova OI, Greenberg AS, Schroeder F. Sterol carrier protein-2 expression modulates protein and lipid composition of lipid droplets. J Biol Chem. 2001;276:25324–35. doi: 10.1074/jbc.M100560200. [DOI] [PubMed] [Google Scholar]
- 33.Storey SM, McIntosh AL, Senthivinayagam S, Moon KC, Atshaves BP. The phospholipid monolayer associated with perilipin-enriched lipid droplets is a highly organized rigid membrane structure. AJP Endocrinol Metab. 2011;301:E991–E1003. doi: 10.1152/ajpendo.00109.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandrasekhar S. Brownian Motion, Dynamical Friction and Stellar Dynamics. Rev Mod Phys. 1949;21:383–388. doi: 10.1111/j.1746-8361.1949.tb00859.x. [DOI] [Google Scholar]
- 35.Einstein A. On the Motion of Small Particles Suspended in a Stationary Liquid, as Required by the Molecular Kinetic Theory of Heat. Ann Phys. 1905;322:549–560. doi: 10.1002/andp.19053220806. [DOI] [Google Scholar]
- 36.Marx E, Mulholland GW. Size and Refractive Index Determination of Single Polystyrene Spheres. J Res Natl Bur Stand (1934) 1983;88:321. doi: 10.6028/jres.088.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haseda K, Kanematsu K, Noguchi K, Saito H, Umeda N, Ohta Y. Significant correlation between refractive index and activity of mitochondria: single mitochondrion study. Biomed Opt Express. 2015;6:859. doi: 10.1364/BOE.6.000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gouw TH, Vlugter JC. Physical Properties of Triglycerides. !. Density and Refractive Index. Eur J Lipid Sci Technol. 1966;68:544–569. [Google Scholar]
- 39.Suzuki M, Shinohara Y, Ohsaki Y, Fujimoto T. Lipid droplets: Size matters. J Electron Microsc (Tokyo) 2011;60 doi: 10.1093/jmicro/dfr016. [DOI] [PubMed] [Google Scholar]
- 40.Najt CP, Senthivinayagam S, Aljazi MB, Fader KA, Olenic SD, Brock JRL, Lydic TA, Jones AD, Atshaves BP. Liver-specific loss of Perilipin 2 alleviates diet-induced hepatic steatosis, inflammation, and fibrosis. Am J Physiol - Gastrointest Liver Physiol. 2016;310:G726–G738. doi: 10.1152/ajpgi.00436.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McManaman JL, Bales ES, Orlicky DJ, Jackman M, MacLean PS, Cain S, Crunk AE, Mansur A, Graham CE, Bowman TA, Greenberg AS. Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J Lipid Res. 2013;54:1346–59. doi: 10.1194/jlr.M035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang S, Wang Y, Cui L, Deng Y, Xu S, Yu J, Cichello S, Serrero G, Ying Y, Liu P. Morphologically and Functionally Distinct Lipid Droplet Subpopulations. Sci Rep. 2016;6:29539. doi: 10.1038/srep29539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen B-C, Shamay A, Argov-Argaman N. Regulation of lipid droplet size in mammary epithelial cells by remodeling of membrane lipid composition-a potential mechanism. PLoS One. 2015;10:e0121645. doi: 10.1371/journal.pone.0121645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonda-Ostaszewska E, Włostowski T, Krasowska A, Kozłowski P. Seasonal and photoperiodic effects on lipid droplet size and lipid peroxidation in the brown adipose tissue of bank voles (Myodes glareolus) Acta Theriol (Warsz) 2012;57:289–294. doi: 10.1007/s13364-012-0083-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slawik M, Vidal-Puig AJ. Lipotoxicity, overnutrition and energy metabolism in aging. Ageing Res Rev. 2006;5:144–164. doi: 10.1016/j.arr.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI. Mitochondrial Dysfunction in the Elderly: Possible Role in Insulin Resistance. Science (80−) 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daniele JR, Esping DJ, Garcia G, Parsons LS, Arriaga EA, Dillin A. High-Throughput Characterization of Region-Specific Mitochondrial Function and Morphology. Sci Rep. 2017;7:6749. doi: 10.1038/s41598-017-05152-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


