Abstract
Given the frequent and largely incurable occurrence of multiple myeloma (MM), identification of germline genetic mutations that predispose cells to MM may provide insight into disease etiology and the developmental mechanisms of its cell of origin, the plasma cell. Here we identified familial and early-onset MM kindreds with truncating mutations in lysine-specific demethylase 1 (LSD1/KDM1A), an epigenetic transcriptional repressor that primarily demethylates histone H3 on lysine 4 and regulates hematopoietic stem cell self-renewal. Additionally, we found higher rates of germline truncating and predicted deleterious missense KDM1A mutations in MM patients unselected for family history compared to controls. Both monoclonal gammopathy of unknown significance (MGUS) and MM cells have significantly lower KDM1A transcript levels compared with normal plasma cells. Transcriptome analysis of MM cells from KDM1A mutation carriers shows enrichment of pathways and MYC target genes previously associated with myeloma pathogenesis. In mice, antigen challenge followed by pharmacological inhibition of KDM1A promoted plasma cell expansion, enhanced secondary immune response, elicited appearance of serum paraprotein, and mediated upregulation of MYC transcriptional targets. These changes are consistent with the development of MGUS. Collectively, our findings show KDM1A is the first autosomal dominant MM germline predisposition gene, providing new insights into its mechanistic roles as a tumor suppressor during post-germinal center B cell differentiation.
Keywords: Exome sequencing, Multiple Myeloma, KDM1A, predisposition, tumor suppressor
INTRODUCTION
In multiple myeloma (MM), monoclonal plasma cells derived from post–germinal-center B cells abnormally proliferate and produce high amounts of immunoglobulin or paraprotein, which leads to lytic bone lesions, anemia and renal failure(1). MM is preceded by a pre-malignant condition, monoclonal gammopathy of undetermined significance (MGUS). Consistent with genetic predisposition, having a first-degree relative with MM elevates MM risk 2–5.5-fold(2).
Identifying MM predisposition genes can provide mechanistic insights into MGUS, myeloma pathogenesis and plasma cell differentiation. Genome wide association studies have identified 16 common variants at loci significantly associated with MM risk(3–8). However, the role of rare MM high-penetrance predisposing genes is poorly understood. To identify MM predisposition genes, we performed germline whole exome sequencing (WES) for familial MM probands and identified kindreds that carry germline truncating mutations in Lysine (K)-specific demethylase 1 (KDM1A, also called LSD1)(9). KDM1A is an epigenetic transcriptional repressor that primarily demethylates mono-methylated and di-methylated histone H3 on lysine 4 (H3K4me1/me2) to repress target gene promoters and enhancers(10–12).
We used CRISPR to introduce a “second hit” mutation in lymphoblastoid B cells from a germline KDM1A mutation carrier, which increased H3K4me1 levels. MGUS and MM cells have significantly lower KDM1A transcript levels compared with normal plasma cells, and may be particularly sensitive to KDM1A mutations causing loss of function or haploinsufficiency. We also performed mutation burden test analysis of MM patients unselected for family history and controls, which showed higher rates of germline KDM1A mutations in MM patients. Mice treated with a KDM1A small molecule inhibitor, GSK-LSD1, have enhanced secondary immune response with expansion of plasma cells, increased immunoglobulin production and appearance of serum paraprotein. RNAseq analysis of these abnormal mouse plasma cells shows enrichment of MYC oncogene transcriptional targets. Transcriptomic analysis of MM cells from KDM1A mutation carriers shows upregulation of the MYC target oncogene Cyclin D2 and enrichment of pathways associated with both intrinsic MM pathogenesis and extrinsic MM-bone marrow microenvironment interactions. Our findings show that KDM1A is a novel germline predisposition gene for multiple myeloma and provide new insights into its mechanistic roles as a tumor suppressor in B cells.
METHODS
Patient Inclusion Criteria
All patient studies were conducted in accordance with the U.S. Common Rule, after approval by an IRB at the respective recruiting institution. Informed written consent was obtained from all subjects. Familial MM probands (n=50) (Supplementary Table S1) analyzed by exome sequencing met inclusion criteria: (a) confirmed diagnosis meeting revised criteria of the International Myeloma Working Group, (b) IgG heavy/light chain analyzed, and (c) ≥1 first-degree or ≥2 second-degree relatives diagnosed with MM. KDM1A-Sanger sequencing EA validation cohort (n=400) inclusion criteria were: (a-c) (N=200) or (a), (b) and (d) MM onset younger than age 60 (n=200).
Whole-Exome Sequencing
Germline DNA extracted from peripheral blood was used for whole exome capture using Agilent SureSelect 38Mb paired-end sequencing and ran on Illumina HiSeq 2000s/2500s. FASTQ files were aligned to human reference genome (GRCh37) to generate BAM files using BWA v0.7.12. Picard tools was used for quality metric calculation and marking duplicate reads. GATK version 3.5-0-g36282e4 was used for variant calling using the haplotype caller algorithm. Variant quality score recalibrated (VQSR) data was used for filtering variants. Variant level and interval level annotations used SNPEff, ANNOVAR, and CAVA programs. Downstream analysis consisted of filtering out low quality variant calls and common variants. Average coverage depth was 80X-100X. Variants with read depth (DP) of 10 or greater and a genotype quality (GQ) score of 20 or greater were included in analyses. Variant, exon, and gene level data were obtained using information from the 1000 Genomes Project, NHBLI GO Exome Sequencing Project Exome Variant Server (EVS), Exome Aggregation Consortium (ExAC), and the combined annotation dependent depletion (CADD) server (13). Deleterious variants were defined as loss-of-function (frameshift insertion or deletion, stop-gain, splice-site change) or missense variants with CADD score >15. We performed segregation analysis using either exomes from family members or targeted Sanger sequencing. Co-segregating qualifying variants in Family 1 (Figure 1A) shared by exomes are listed in Supplementary Table S2. Exome sequence data are accessioned as NCBI SRR5641111.
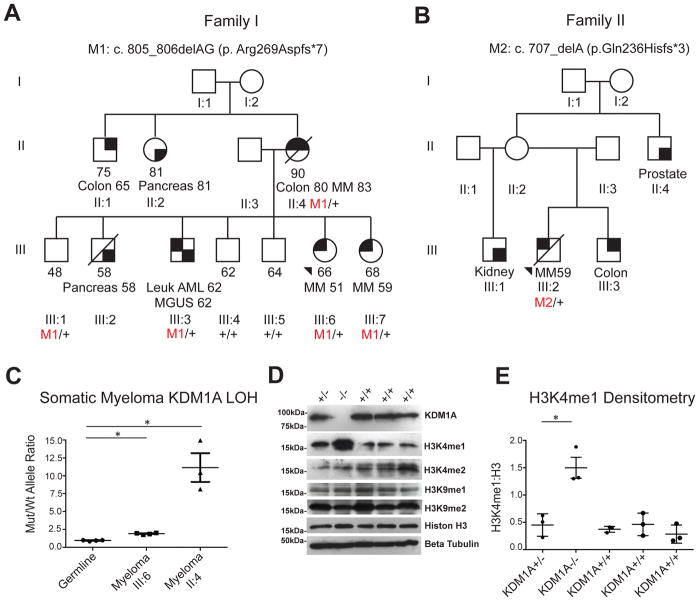
Figure 1. Identification of germline KDM1A mutations in familial and early onset multiple myeloma patients.
A. Pedigree of familial myeloma kindred carrying germline KDM1A c.805_806delAG (p.Arg269Aspfs*7) mutation.
B. Pedigree of early onset myeloma proband carrying KDM1A c.707delA (p.Gln236Hisfs*3) mutation. Last known ages, age of death if applicable, affected status with different malignancies, and ages at diagnosis are indicated. MM, multiple myeloma. AML, acute myeloid leukemia. MGUS, monoclonal gammopathy of uncertain significance. Colon, colon cancer. Pancreas, pancreatic cancer. Prostate, prostate cancer. +, wild-type. M1 and M2, mutations. For kindred shown in B, only the proband had a biospecimen available for testing. For this kindred, ages of diagnosis for colon, kidney and prostate cancers in family members could not be rigorously confirmed and so are not included. C. Somatic loss of heterozygosity (LOH) in germline and myeloma cells from probands III:6 and II:4 in family carrying KDM1A c.805_806delAG (p.Arg269Aspfs*7) using digital droplet PCR (ddPCR). Ratio of KDM1A c.805_806delAG (p.Arg269Aspfs*7) mutant:wild-type allele for germline and myeloma cells are indicated with mean. * P< 0.01 by Mann-Whitney test, LOH, loss of heterozygosity. N=3 and 4 respectively. Mean and s.e.m. are shown. D, E. Western and densitometry analysis showing KDM1A null (−/−) LCLs (generated from family member III:6 from the kindred shown in Figure 1A) have undetectable KDM1A and increased H3K4 mono-methylation (H3K4me1):H3 ratio. H3 di-methylated isoform (H3K4me2), H3 and tubulin are shown for comparison. Heterozygote and compound heterozygote LCLs were made from Family 1 III:6 (lanes 1 and 2) and wild type LCLs were made from Family 1 II:3, III:4 and III:5 (lanes 3–5). Densitometry represents 3 independent measurements from each of the LCL cell lines. Mean and s.e.m. are shown. * P= 0.015 by two-tailed student t test. LCL, B lymphoblastoid cell line.
Sanger Sequencing
For the KDM1A-Sanger sequencing EA validation cohort, all coding regions of KDM1A (NM_015013) were amplified using primers available upon request. PCR amplicons were sequenced in the Applied Biosystems 3730 DNA (Applied Biosystems, Waltham, MA) Analyzer and analyzed using Mutation Surveyor (SoftGenetics, State College, PA). KDM1A mutations in probands and indicated family members (Figure 1A) were verified by Sanger sequencing.
Cell Culture
Cell culture methods used are described in Supplementary Data S1.
KDM1A Knockout by CRISPR/Cas9
To generate bi-allelic KDM1A truncating mutations in lymphoblastoid (LCL) cells that carry the KDM1A c.805_806delAG (p.Arg269Aspfs*7) heterozygous mutation, CRISPR/Cas9 gene editing sgRNA sequences were designed to target genomic sites near KDM1A c.805_806delAG (p.Arg269Aspfs*7). pX458_sgRNA (5′-CACCGTCAACTTC-GGCATCTATAAG-3′, 5′-AAACCTTATAGATGCCGAAGTTGAC-3′) was transfected into LCLs (Amax Nucleofector II device). GFP-expressing clones were seeded as single cell clones in 96-wells plates using flow cytometry and screened for mutations by PCR amplification, target sequencing and western blot.
Western Blot Analysis and Antibodies
LCL cells were harvested in lysis buffer containing complete Protease inhibitors [Roche, 4693159001]). Whole-cell extracts were subjected to SDS-PAGE. Antibodies used include anti-H3K4me1 (Abcam ab8895, 1:500 dilution), anti-H3K4me2 (Abcam ab32356, 1:500 dilution), anti-H3K9me1 (Abcam ab9045, 1:500 dilution), anti-H3K9me2 (Abcam ab1220, 1:500 dilution), anti-Histone H3 (Abcam ab1791, 1:1000 dilution), anti-γ-tubulin (Sigma T5192), and anti-KDM1A (Santa Cruz sc-398794, 1:200 dilution).
Digital Droplet PCR
A Biorad assay specific for detection of the KDM1A frameshift mutation was designed (Forward Primer: 5′ CGTCATGGTCTTATCAACT3′; Reverse primer: 5′ AGGAGG-TCCTTACTTGGT3′; Wt probe: 5′CGGCATCTATAagAGGATAAAACC3′-; mutation probe:5′ CGGCATCTATAAGGATAAAACC3′. Cycling conditions were optimized for annealing/extension temperature and separation of positive from empty droplets on a QX200 ddPCR system (Biorad). Technical duplicates were run. PCR reactions contain primers/probes, DNA and digital PCR Supermix for probes (no dUTP). Reactions were partitioned into median ~16,000 droplets/well using QX200 droplet generator. Emulsified PCRs were run using cycling conditions (95°C 10′;40 cycles of 94°C 30″ 55°C 1′, 98°C 10′, 4°C hold). Plates were read and analyzed with QuantaSoft software to assess the number of droplets positive for mutant DNA, wild-type (wt) DNA, both, or neither. Assay threshold sensitivity was set at 2 mutant droplets.
Mutation Burden Analysis
Exome data from 879 sporadic Multiple Myeloma patients (dbGaP phs000748 and phs000348) and 2389 control subjects (phs000179, phs000276, phs000403, phs000687 and phs000806) were simultaneously processed and variants jointly called. Exomes from cases/controls were required to have ≥75% of the 33.27Mbps of CCDS with >10-fold coverage and ≤3% contamination using VerifyBamID(14). Exomes from cryptically related individuals were removed using KING algorithm(15). Principal component analysis (PCA) was performed using SMARTPCA(16) to identify European ancestry (EA). EA exomes meeting these quality control metrics were selected (733 sporadic Multiple Myeloma patients and 1,480 controls). We compared the sum of (a) predicted deleterious mutation or (b) neutral synonymous coding variant qualifying carriers for each gene in MM cases and controls using Fishers exact test (Statistical function scipy.stats.fisher_exact in SciPy v0.14.0.). Qualifying variant criteria includes (1) a quality score (QUAL) ≥50, (2) a genotype quality (GQ) score ≥20, (3) a quality by depth (QD) score ≥2, (4) a mapping quality (MQ) score ≥40, (5) a read position rank sum (RPRS) score ≥-3, (6) mapping quality rank sum (MQRS) score ≥-10, (7) coding variants with mean allele frequency (MAF) ≤ 0.05% in all three databases of the 1000 Genomes Project, ESP, and ExAC. For rare synonymous variant burden testing, synonymous coding variants meeting criteria (1)-(7) were used. For mutation burden testing, loss-of-function (frameshift insertion or deletion, stop-gain, splice-site change) and missense variants with Phred CADD (Combined Annotation Dependent Depletion) score ≥15 meeting criteria (1)-(7) were used. For association testing, common variants were required to meet criteria (1)-(6), have MAF ≥5%, and be in Hardy-Weinberg equilibrium in cases, controls, and the combined group (p>0.001 for each). PCA, rare synonymous, predicted mutation gene burden and common variant association lambda statistics were used to confirm case-control matching and lack of notable population stratification (Supplementary Figure S1 and Figure 2A–D).
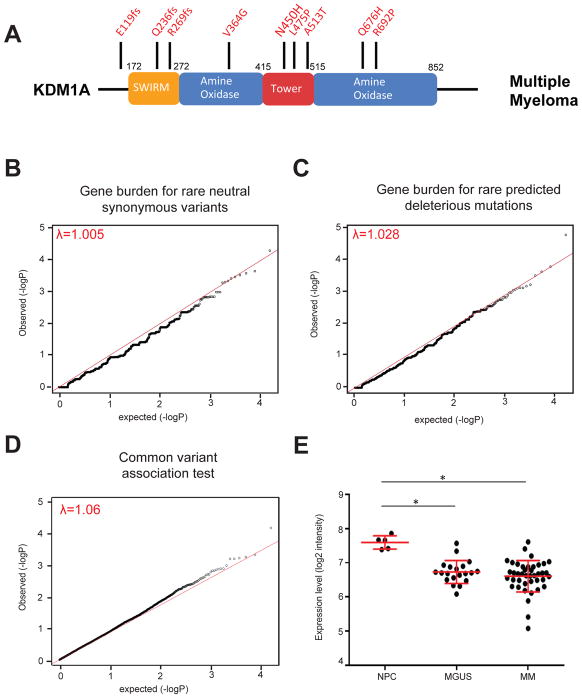
Figure 2. KDM1A germline mutations in multiple myeloma patients.
A. Schematic of KDM1A protein. Germline mutations from Multiple Myeloma patients are shown on top. B–C. Quantile-quantile plots of gene-based burden tests for rare neutral synonymous variants (B) and rare predicted deleterious mutations (C). D. Quantile-quantile plot of MM case and control common variants. Lambda statistics are shown. E. KDM1A mRNA levels in normal plasma cells (NPC), MGUS and MM cells. *, p=7.45 × 10−6 (NPC-MGUS) and 1.70 × 10−5 (NPC-MM) respectively, moderated t-test. Mean and SEM bars are indicated.
Multiple Myeloma Organoids
Peg-Mal (polyethylene glycol-malate) (AA Peptides) hydrogels with REDV (AA Peptides) peptidic ligands were impregnated with 30,000 U266 myeloma cells and 5,000 HS5 non-proliferating (mitomycin-treated) stromal cells, or 30,000 mono-nucleated cells from a primary bone marrow biopsy sample as previously described(17) with the addition of VLA-4/Integrin α4β1 ligand fibronectin(18). After gelation, hydrogel 3D organoids were rehydrated with media and cultured as described(17, 19). A detailed protocol for MM organoid culture is available upon request. U266 organoids untreated or treated with 1 μM LSD1 inhibitor (GSK-LSD1) were followed by microscopy. Cells were harvested and counted by measuring ATP content/well (CellTiter Glo, Promega) at days 0, 1, 3, 5, and 7. 3–5 replications were used per organoid datapoint. For primary bone marrow biopsy sample, after 6 days in culture 2 organoids per experimental condition were mechanically digested and single cells assessed for plasma cells markers CD138 and p63 using flow cytometry, and growth assessed at the same time point for 3 organoid datapoints per treatment condition using CellTiter Glo. To assess proliferation in a bone marrow sample of a MM patient in remission, mononuclear cells were pre-loaded with CellTrace Violet (ThermoFisher).
RNAseq Analysis
For mouse plasma cells, polyadenylated single-read RNAseq was performed using standard Illumina Truseq protocols and run at 3/lane on HiSeq 2500. Reads were trimmed of adapters with Flexbar then aligned to mm10 mouse genome(20). Uniquely mapped reads were used for read counts. Raw read counts were processed in R with DESeq2(21) and accessioned as NCBI GEO GSE85596. Gene Set Enrichment Analysis (GSEA) scores were calculated as the negative log10 of the False Discovery Rate (FDR) multiplied by the sign of the log2 fold change (log FC). Scores were run against pre-ranked GSEA Hallmarks(22) (weighted score type, 1000 permutations) or Boylan Multiple Myeloma gene sets(23). Differentially expressed genes were graphed as a volcano plot in R.
For analysis of MMRF CoMMpass RNAseq MM transcriptomes from KDM1A mutation carriers and wt MM patients, raw counts[19] were analyzed in R with DESeq2[20]. MM RNAseq data was available from 4 KDM1A mutation carriers (MMRF_1201, 1730, 2293 and 2068). RNAseq data from these 4 tumors were compared against 4 randomly picked MM tumors from the 799 non-mutation carriers and run for 1000 iterations. Significantly up- or down-regulated genes obtained from 1000 permutation analysis outputs (P-adjusted<0.05) were compared against mouse GSK-LSD1 treated plasma cells. Outputs with >5% frequency were input for pathway enrichment in ClueGO[23], using GO Biological Process, KEGG, REACTOME and Wikipathways ontologies. Two-sided hypergeometric test was used to calculate P-values, followed by Bonferroni correction. Pathways with adjusted P-value <0.05 were considered significant. The complete lists of genes and pathways are given in Supplementary Table S3.
For analysis of hypodiploid ALL gene expression data(24), we compared samples with truncating KDM1A mutations (SJHYPO021 and SJHYPO032) to 50 KDM1A wt hypodiploid ALL tumors and used GSEA (n=1000 permutations) to test for enrichment of HoxA9 and LSK gene sets (25).
Mouse Immunization and GSK-LSD1 Treatment
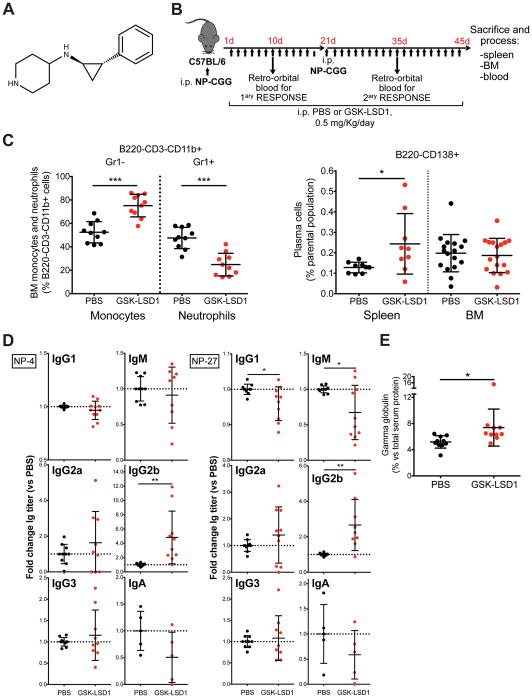
C57BL/6 male mice age 11 weeks (n=18) were immunized intraperitoneally with 100 μg NP-CGG in alum on day 0 and day 21. On day 1, intraperitoneal treatment with either vehicle (PBS) or GSK-LSD1 (Cayman Chemical #16439) (0.5 mg/Kg//day) started and continued until day 45. On day 10 and 35, mice were bled. On day 45, mice were sacrificed, heart blood, femurs and spleen collected and analyzed.
Cell Sorting and FACS
Mouse spleens were processed into single cell suspensions, pooled in groups of 3–4 and washed before resuspending Ficoll-isolated mononuclear cells in PBS containing 0.5% BSA and antibodies B220-PECy7 (eBioscience, #25-0452-82) and CD138-APC (BD Pharmigen #558626). After one wash, cells were resuspended at 20 million/mL and DAPI (1 μg/mL) added to exclude dead cells. Cells collected were B220+ CD138- (B cells), and CD138+ (plasma cells), and were >95% pure. Bone marrow extracted from mouse femurs and spleen mononuclear cells were also analyzed by FACS with B220, CD138, CD3, CD11b and Gr1 antibodies.
NP-immunoglobulin ELISA and Serum Electrophoresis
Day 35 retro-orbital blood sera were serially diluted and incubated overnight in 96-well plates pre-coated with NP (4)-CGG or NP (27)-CGG (Biosearch, #N-5055A and N-5055C) and pre-blocked with PBS with 1% BSA. After three washes with 0.05% Tween-20 PBS, HRP-conjugated antibodies against IgG1, IgG2a, IgG2b, IgG3, IgM or IgA (SouthernBiotech, #5300-05) were used to detect NP-specific antibodies. ABTS (SouthernBiotech, #0404-01) was used as peroxidase substrate and the reaction stopped with 1% SDS. Absorbance of the product was measured at 410 nm.
Serum electrophoresis (Helena Laboratories) in agarose was performed with day 45 heart blood sera. Densitometry-based quantification of serum protein bands was expressed as % of total protein in samples quantified in a Roche/Hitachi Modular P Chemistry Analyzer.
Study Approval
Patients were consented under IRB approved protocols in accord with CIOMS at Weill-Cornell Medicine, Memorial Sloan-Kettering, Creighton University, University of Utah, Mayo Clinic, Dana Farber Cancer Institute and University of Lyon. All mouse experiments were performed under IACUC approved protocols at Weill Cornell.
Statistics
Statistical tests used are described in Supplementary Data S1.
Internet Resources Cited
Internet resources cited are listed in Supplementary Data S1.
RESULTS
Germline truncating KDM1A mutations in familial multiple myeloma
Germline exome sequencing of 50 familial MM kindreds (Supplementary Table S1) revealed an EA family carrying an N-terminal truncating KDM1A mutation (c.805_806delAG [p.Arg269Aspfs*7]) (Figure 1A) absent in ClinVar or Exome Aggregation Consortium (ExAc) databases. This was the only kindred with a qualifying truncating mutation (Supplementary Table S2). Therefore we focused follow-up on this family. Sanger sequencing identified five relatives carrying this KDM1A mutation. Three were MM affected (II:4, III:6 and III:7), one (III:3) with both acute myelogenous leukemia (AML) and MGUS, and one unaffected at age 48 (III:1) (Figure 1A). Three additional family members were diagnosed with pancreatic (II:2, III:2) and colon cancer (II:1).
KDM1A is composed of three major domains: SWIRM and Tower, which mediate protein-protein interactions, and a C-terminal amine oxidase (AO) domain(26). The truncating KDM1A c.805_806delAG (p.Arg269Aspfs*7) mutation eliminates the AO and Tower domains. Western blot of an LCL derived from patient III:6 showed normal levels of full length KDM1A and no truncated protein. RT-PCR and Sanger sequencing of cDNA revealed the absence of KDM1A mutant transcripts.
Identification of germline truncating KDM1A mutations in early onset multiple myeloma
We next sequenced KDM1A exons in 400 additional EA MM early-onset (<age 60) or familial myeloma probands. This identified another EA patient with early-onset MM (age 59) and a truncating KDM1A mutation (KDM1A c.707delA [p.Gln236Hisfs*3]) in the SWIRM domain (Figure 1B). No other family members were diagnosed with MM/MGUS. One sibling was affected with colorectal cancer, a half-sibling with renal cell cancer and one uncle with prostate cancer.
Combined Annotation Dependent Depletion (CADD) scoring is a widely used computational tool to predict whether germline variants are benign or deleterious(13). The higher a CADD score, the higher the likelihood that a genetic variant is deleterious. Using CADD ≥15 threshold we identified two predicted rare deleterious KDM1A missense mutations in the Tower and AO domains, (KDM1A c.1424T>C [p.Leu475Pro] and c.2003G>C [p.Arg692Pro]) in MM patients with first degree relatives also affected with MM/MGUS. Collectively, these data confirm the presence of rare truncating and predicted deleterious missense KDM1A mutations in familial and early onset MM patients.
To understand the prevalence of KDM1A truncating mutations in controls, we examined the ExAC database. Including all ethnicities only 9 predicted truncating and splice-site loss-of-function (LoF) variants were identified, carried by 10 individuals (10/60,694) (LoF constraint metric pLI=0.99). Seven of these are of European ancestry. All KDM1A truncating mutations had mean allele frequencies <1.6 × 10−5 and no homozygotes were identified. Joint analysis of LoF mutations in familial and early onset MM cases (2/450) and ExAC EA participants as controls (7/36,664), showed enrichment of KDM1A LoF mutations among familial and early onset myeloma patients (p=0.005, Fisher exact test; relative risk 23.28, 95% confidence interval 4.85 to 111.75). Overall, despite possible small mismatch in mean age and the precise definitions of our MM probands and ExAC database assignments of European ancestry, our data provide evidence of KDM1A LoF mutation enrichment in familial and early-onset MM.
Digital Droplet PCR Analysis of germline KDM1A mutant MM cells
Myeloma cells from bone marrow biopsies were available for two members (II:4 and III:6) of the kindred carrying the KDM1A c.805_806delAG (p.Arg269Aspfs*7) mutation. Digital droplet PCR (ddPCR) was performed on DNA isolated from MM cells and normal bone marrow. The KDM1A mutation was enriched in MM cells from deletion of the wt allele compared to germline bone marrow cells carrying the heterozygous germline mutation in both family members tested (both P<0. 01, Mann-Whitney test (Figure 1C). Next, we generated LCLs from members of this kindred carrying KDM1A heterozygous or wt genotypes. We used CRISPR/Cas9 gene editing technology to introduce a second hit on the wt allele and an LCL cell line was derived carrying bi-allelic KDM1A truncating mutations [(c.805_806delAG (p.Arg269Aspfs*7) and c.801_802insT (p.Lys268*)]. Western blot analysis of KDM1A bi-allelic (−/−), heterozygous (+/−) and wt (+/+)LCLs revealed that heterozygous and wt LCLs had similar KDM1A protein levels, but that bi-allelic mutant LCLs had no detectable KDM1A protein (Figure 1D). Accordingly, KDM1A −/− LCLs have significantly higher overall cellular levels of H3K4me1 whereas total H3K4me2, H3K9me1 and H3K9me2 levels were not significantly changed (Figure 1D, 1E).
Increased KDM1A mutations in sporadic MM patients
We performed gene burden tests using jointly called germline exomes from EA MM patients unselected for family history and EA controls (Figure 2A). Exomes were matched for quality metrics as before (27, 28), and KING algorithm used to exclude individuals with cryptic relatedness(15). PCA showed no apparent population stratification between EA MM cases/controls (Supplementary Figure S1). The distribution of neutral synonymous coding variants showed no significant difference in the number of genes carrying neutral synonymous coding variants in MM cases (N=733; yellow, average 17.55±0.26/gene) and controls (N=1480; blue, average 17.52±0.18/gene) (Mann-Whitney U test, p=0.91) (Supplementary Figure S1). We used Fisher’s exact test (28, 29) to generate quantile-quantile plots for neutral synonymous coding variant and predicted deleterious coding mutation gene burden tests. We also used Fisher’s exact test to generate a quantile-quantile plot comparing MM case and control common variants (MAF≥5%). The genomic inflation factor (λ) statistic confirmed appropriate matching between MM cases and controls (λ=1.005, 1.028 and 1.06 respectively) (Figure 2B–D). This showed significant enrichment of KDM1A predicted deleterious missense and LoF/truncating mutations (p=1.27× 10−3) in MM patients compared to controls (Table 1).
Table 1.
Predicted deleterious KMD1A mutation frequencies in Multiple Myeloma probands and control subjects
| Probands | Deleterious | Benign | Total | Mutation Burden Test (Fisher’s exact test) |
|---|---|---|---|---|
| Multiple Myeloma | 9 (1.23%) | 724 (98.77%) | 733 | p value= 1.27×10−3 |
| Control Subjects | 2 (0.14%) | 1478 (99.86%) | 1480 | |
| Total | 11 | 2202 | 2213 |
Among MM patients, we detected another early-onset (56 year old) MM patient with an exon 2 splice-donor LoF/truncating mutation KDM1A c.517+1G>A, (p.Glu119Trpfs*15), but no LoF/truncating mutation in controls. The myeloma tumor from this patient carried a somatic deletion encompassing the entire KDM1A locus (chr1:830,800-120,666,600). Additionally, screening for predicted deleterious KDM1A missense mutations with CADD scores ≥15 identified 8 MM patients with heterozygous predicted deleterious missense mutations and 2 non-MM control subjects (1.23% LoF/missense mutations in MM patients and 0.14% LoF/missense mutations in controls; Relative Risk 9.08 95% confidence interval 1.97 to 41.95) (Table 1). The rate of predicted KDM1A LoF and missense mutations in EA controls is similar to the ExAc database (0.22%; 81/36,664). However, none of the 7 tumors from MM patients carrying KDM1A missense mutations whose tumor somatic mutation data were available demonstrated LOH(30).
We analyzed published transcriptome databases containing gene expression from normal plasma cells (PC), MGUS and MM cells and found that KDM1A mRNA levels are significantly lower in MGUS or MM cells vs. PC (7.45×10−6 and 1.7×10−05 respectively, moderated t-test) (Figure 2E)(31). Overall, these data support a broader role for KDM1A inactivation in sporadic MGUS and MM, in addition to its role in familial and early-onset multiple myeloma. Because they have significantly lower KDM1A levels compared with normal PC, MGUS and MM cells may be particularly vulnerable to epigenetic dysregulation from KDM1A haploinsufficiency or loss of function mutations.
KDM1A pharmacological inhibition promotes expansion of MM and plasma cells
To test a role of KDM1A in post-germinal center B cell differentiation we conducted antigen-driven B cell differentiation in vivo experiments. To induce differentiation of B cells into PC, we challenged immunologically mature C57BL/6 mice with T-cell dependent antigens on day 1 and boosted at day 21. In one group of mice we used sheep red blood cells (SRBC) as antigen and in another group NP-chicken gamma globulin (NP-CGG), which allows quantification of total and high-affinity immunoglobulins. KDM1A was inhibited with GSK-LSD1, an irreversible KDM1A inhibitor (Figure 3A) with an IC50 of 16 nM that shows >1000X selectivity for KDM1A over closely related FAD utilizing enzymes including LSD2, MAOA and B. Mice were randomized to receive vehicle (PBS) or GSK-LSD1 0.5 mg/kg daily by intraperitoneal injection (Figure 3B). Mice were monitored for primary (day 10) and secondary (day 35) antibody immune response, and sacrificed (day 45).
Figure 3. KDM1A pharmacologic inhibition in mice promotes secondary immune response, immunoglobulin class switching and serum paraprotein.
A. Chemical structure of KDM1A inhibitor GSK-LSD1. B. Schedule of administration to mice and immune response elicited by antigen NP-CGG. C. Kdm1a inhibition recapitulates Kdm1a−/− mouse phenotype, with increased monocytes and reduced neutrophils in bone marrow (BM) (left), and causes increased splenic PC in the secondary immune response (right panel). At least n=9 per condition. * p<0.05, *** p<0.001, Mann-Whitney test. Mean and s.e.m. are shown. D. Kdm1a inhibition promotes NP-specific IgG2b antibodies production (high affinity antibodies, which detect the less haptenated antigen NP-4, and total antibodies, which detect the highly haptenated antigen NP-27). At least n=5 per condition. * p<0.05, ** p<0.01, Mann-Whitney test. Mean and s.e.m. are shown. E. Gamma globulin fraction in serum electrophoresis of same mice at day 45. n=10 per condition. * p<0.05, Mann-Whitney test. Mean and s.e.m. are shown.
Phenocopying the Kdm1a knockout mouse models(25, 32), GSK-LSD1 significantly decreased neutrophils and increased monocytes in bone marrow (Figure 3C). Antigen-challenged mice treated with GSK-LSD1 showed increased splenic PCs (Figure 3C). Analysis of serum total and high affinity anti-NP antibody production revealed that KDM1A inhibition promoted secondary immune response to the T-cell dependent antigen NP-CGG (Figure 3D). KDM1A inhibition also promoted IgM to IgG2b isotype class-switching, a process where B cells activated upon antigen encounter are co-stimulated by T helper cells (Figure 3D). The increase in immunoglobulin production could be detected by serum electrophoresis as an increased gamma globulin fraction (Figure 3E). These findings are consonant with a model whereby KDM1A activity is required to control for abnormal generation and/or expansion of PC.
KDM1A inhibition promotes in vivo plasma cell upregulation of MYC transcriptional targets
To characterize the transcriptional consequences of KDM1A inhibition in PC, we performed RNA-sequencing of differentially expressed genes (DEG) in sorted PC (CD138+) from GSK-LSD1 treated and control mice. To functionally categorize these genes we conducted pathway analysis using GSEA (33) and identified “MYC targets (v1)” as a significant pathway associated with up-regulated genes (Figure 4A, p-value=0.0, FDR 0.0).
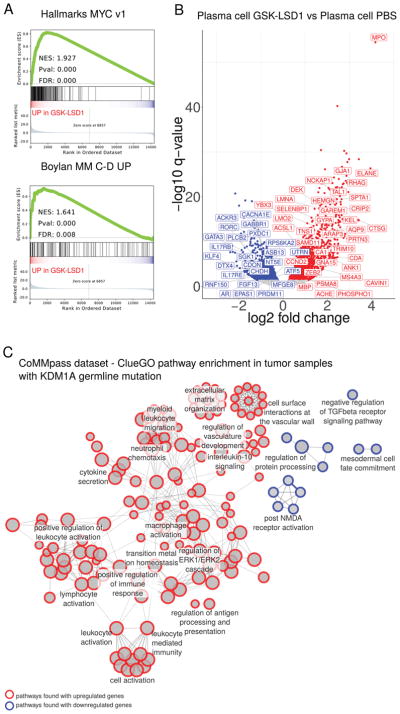
Figure 4. KDM1A inhibition in mouse plasma cells and germline KDM1A mutations in MM patients alters gene expression.
A. Gene set enrichment analysis (GSEA) of differentially expressed genes in plasma cells from mice treated with GSK-LSD1 in MYC v1 MSigDB Hallmark (top) and Bcl-xL/Myc transgenic mouse MM (bottom) gene sets. FDR, False Discovery Rate. NES, normalized enrichment score. B. Volcano plot of differentially expressed genes in sorted plasma cells from mice treated with GSK-LSD1. Labels (human nomenclature) indicate genes differentially regulated in common for both GSK-LSD1 treated mouse plasma cells and MM tumors from KDM1A germline predicted mutation carriers. C. ClueGO analysis pathways enriched in differentially expressed genes comparing MM tumors from KDM1A germline mutation carriers to those from wild-type patients. Pathways with adjusted P-value <0.05 are shown. Circles represent pathways. Size represents the percentage of genes shared with a pathway. Red, upregulated gene pathways. Blue, downregulated gene pathways. Edges (lines) indicate shared genes between pathways.
To evaluate mouse genes dysregulated by Kdm1a inhibition in PC with development of MM, we compared DEG in PC from GSK-LSD1 treated vs. control mice with Bcl-xL/Myc transgenic mouse MM cells(23). GSEA showed enrichment of genes upregulated in Bcl-xL/Myc transgenic mouse MM cells in GSK-LSD1 treated PC DEG (NES 1.64, P value 0.0, FDR 0.008) (Figure 4A), including Myc-driven MM oncogenes Ccnd1 and Ccnd2 (Supplementary Table S3) (34–36). Thus, Kdm1a pharmacological inhibition enriched for upregulated MYC transcriptional targets including Ccnd1 and Ccnd2 as potential drivers of PC proliferation.
Increased CCND2 and alterations in other pathways in MM cells from KDM1A mutation carriers
To determine the impact of germline KDM1A mutations on patient MM tumors, we compared MM tumor transcriptomes from patients with KDM1A predicted deleterious mutations to those from KDM1A wild-type patients. First, we evaluated shared DEG common to both (a) GSK-LSD1 treated PC and (b) MM cells from KDM1A mutation carriers vs wt patients. Upregulated shared DEG included CCND2 (Figure 4B). Among down-regulated shared DEG were the transcription factor KLF4 that has tumor suppressor functions in MM(37) and PRDM11, a tumor suppressor in MYC-driven B lymphomagenesis(38) (Figure 4B). Thus, CCND2, KLF4 and PRDM11 are potential transcriptional targets downstream of KDM1A whose dysregulation promotes MGUS and MM in patients.
We then analyzed DEG in MM tumors from KDM1A mutation carriers compared to wt patients by ClueGO pathway analysis(39) (Figure 4C). Pathways related to myelomagenesis enriched in upregulated genes include Immunoglobulin Production, Antigen Processing and Presentation(40), IL-10 signaling(41), ERK Signaling Cascade(42), Vasculature Development(43) and Extracellular Matrix Organization(44), while in downregulated genes the TGFβ signaling pathway was enriched (Figure 4C). This included SMAD1, which suppresses MYC-dependent transcription in MM cells(45). Altogether, these data reveal enrichment in pathways associated with both intrinsic MM pathogenesis and MM-bone marrow microenvironment interactions in KDM1A mutation carriers.
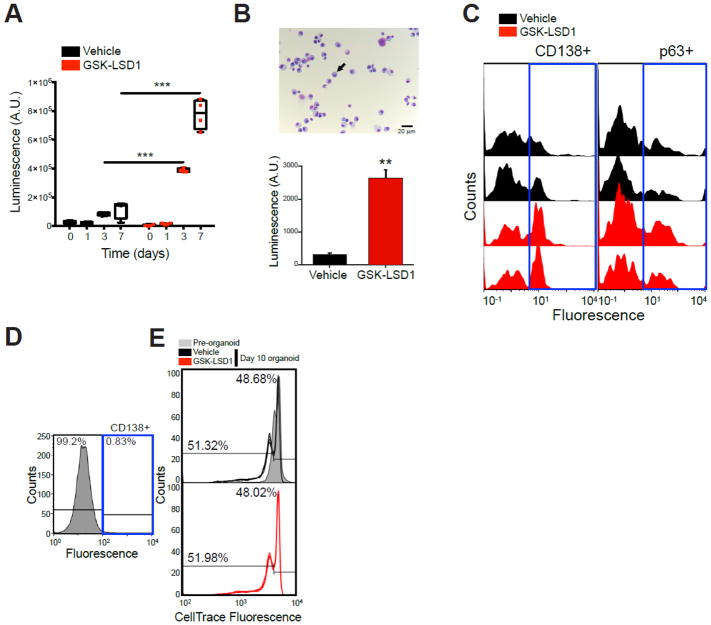
To directly test whether KDM1A affects myeloma cell proliferation, we treated human myeloma cells with GSK-LSD1 in 3D organoids with extracellular matrix-like biophysical properties that resemble the microenvironment of lymphoid tissues(17, 19). When U266 MM cell line (Figure 5A) or primary sporadic MM patient cell (Figure 5B, C) organoids were treated with GSK-LSD1, KDM1A inhibition significantly stimulated MM cell number (p<0.001 and <0.01, t-test). In contrast, GSK-LSD1 treatment did not affect proliferation of non-MM CD138neg bone marrow cells as controls (Figure 5D, E). These data support roles for KDM1A regulating PC differentiation to prevent MGUS and MM development, and also to promote MM proliferation.
Figure 5. KDM1A inhibition increases multiple myeloma cell line and primary patient culture cell proliferation in 3D cultures.
A. 3D organoids containing an inert matrix presenting ligands for integrin α4β1 and U266 MM cells were treated with vehicle or 1 μM GSK-LSD1 for the indicated time points. Cell number was determined by contrast microscopy (not shown) and ATP content (CellTiter Glo). Experiment performed with 3–5 replicates. ***, p<0.01 Mann-Whitney Test. B. Bone marrow aspirate from an untreated MM patient presented 90 % plasma cells (black arrow) after 3 days of culture in flask (top panel). 3D organoids containing an inert matrix presenting ligands for integrin α4β1 and the 3-day cultured bone marrow aspirate were treated with vehicle or GSK-LSD1 1 uM once at day 1. After 6 days of culture, total cell number was measured by ATP content (bottom panel). **, p<0.05 Mann-Whitney Test. C. Myeloma cell count from the same organoids in (B) as measured by FACS analysis of CD138+ and p63+ cells with FITC-conjugated antibodies. Left part of histograms corresponds to matrix debris and other non-PC cells. D. Bone marrow aspirate from a MM patient in remission after 3 days of culture in flask. e. Bone marrow sample from (D) was loaded with the proliferation dye CellTrace Violet and cultured in 3D organoids for 10 days. Proliferation was assessed as a function of fluorescence decrease when compared to pre-organoid fluorescence.
DISCUSSION
MGUS and MM risk is increased in relatives of affected individuals (2). We used whole exome sequencing of familial myeloma kindreds, mutation co-segregation in family members, somatic tumor studies, CRISPR induced somatic second hit mutations, targeted sequencing of additional familial and early-onset MM patients, mutation burden analysis in MM patients unselected for family history, MM tumor transcriptome studies, and functional studies of a KDM1A inhibitor on human MM cells and mouse PC that promoted PC expansion, increased immunoglobulin production and MM cell proliferation to nominate KDM1A as the first autosomal dominant MM predisposition gene. Our data are consistent with germline KDM1A mutations predisposing to familial MM, having a prevalence of ~1.23% in MM patients unselected for family history and conferring ~9-fold increased risk of developing MM.
Whole body knockout or shRNA knockdown of mouse Kdm1a is embryonic lethal (25, 32). In hematopoietic cells, Kdm1a knockout/knockdown causes immature HSPC blasts, LSK progenitors and monocytes to proliferate while LT-HSC, granulocytes, megakaryocytes and erythrocytes are reduced(25, 32). The early lethality of Kdm1a mutant mouse models has limited studies of B lymphocytes, which continue to be produced despite absent Kdm1a(25, 32). Therefore, the roles of KDM1A in B lymphocytes are poorly understood.
Previously, chronic exposure to lysolipid and dioxin antigens were proposed to drive the transition from PC to MGUS(46, 47). In mice, Kdm1A pharmacological inhibition upregulates PC MYC proto-oncogene targets including Ccnd1 and Ccnd2, increases PC numbers, high-affinity secondary immune response to T-cell dependent antigens, immunoglobulin production and causes the appearance of serum paraprotein (Figure 3C). Based on similarities between these PC and Bcl-xL/Myc driven mouse MM cells, this transcriptional dysregulation likely predisposes to additional myelomagenesis driving events. Furthermore, analysis of tumor transcriptomes from KDM1A mutant patients is consonant with selected alterations persisting in MM cells, such as increased CCND2, reduced KLF4 and PRDM11(37, 38). In line with MM organoid findings, tumor transcriptomes from KDM1A mutant patients are enriched in pathways associated with both intrinsic MM pathogenesis and extrinsic MM-bone marrow microenvironment interactions. These data are consistent with KDM1A inhibition/mutation acting at distinct steps to both promote PC immune response and MM proliferation by dysregulating both cell-intrinsic (including MYC driven) and extrinsic microenvironment signaling pathways.
We identified both KDM1A germline truncating and missense mutation carriers (Figure 2). Somatic MM tumor analyses performed with two members of our index family carrying a KDM1A truncating mutation (Figure 1) showed evidence of possible LOH. However, this finding may be by chance, as chromosome 1p deletion is frequent in myeloma(1, 42). Of seven additional KDM1A predicted germline mutation carriers with somatic MM mutation data, only one MM patient’s tumor showed somatic LOH The other 6 MM patients without tumor LOH all carried germline missense mutations. KDM1A is a component of the NuRD and CoRest chromatin remodeling complexes, and participates in multiple protein-protein interactions with distinct histone demethylase activities (10–12). It is possible that KDM1A missense mutations act through a dominant negative mechanism in PC NuRD or CoRest complexes to promote MGUS/MM predisposition. Alternatively, because MGUS and MM cells both have significantly lower KDM1A levels compared with normal PC (Figure 2E), they may be particularly vulnerable to epigenetic dysregulation from KDM1A hypomorphic missense mutations and haploinsufficiency.
Scales et al recently reported an MM exome mutation burden test study in 513 United Kingdom northern EA MM cases from two clinical trials (48). While no individual gene reached genomewide significance in this study, the authors nominated KIF18A as a promising MM predisposition candidate. However, in our EA MM mutation burden study, we did not see an association between KIF18A and MM (Fishers exact test, p=0.787). Furthermore, the study of Scales et al did not nominate KDM1A as an MM predisposition gene. A likely reason for these different findings is that Scales et al used variant MAF filter <1% while our study used MAF ≤0.05% to define rare candidate mutations. Thus, each study used different inputs for mutation burden testing. Additional potential reasons include methodological differences in variant annotation and mutation burden testing pipelines, possible differences in European ancestry population substructure (American vs UK MM patients) and different rates of clinical covariates for MM patients and their tumor molecular sub-types.
Bolli et al performed exome sequencing of a large, multiplex MM/MGUS kindred(49). The authors nominated EP300 and PDPK1, but not KDM1A, as candidate MGUS/MM predisposition genes. However, in contrast to the KDM1A mutation carrying kindreds we identified (Figure 1A, 1B), this family did not have non-MM/MGUS cancers. Thus, additional MGUS/MM specific high-risk predisposition genes may exist.
Recently, recurrent somatic KDM1A mutations were identified in chronic myeloid leukemia (CML)(50). We also identified one patient with a germline truncating KDM1A mutation diagnosed with MGUS and AML (Figure 1A). In mice, Kdm1a knockout causes monocytosis(25, 32). However, KDM1A inhibition was found to induce terminal differentiation of AML cell lines and several primary AML cultures(51). Based on this, KDM1A small molecule inhibitors are used in clinical trials for AML and other conditions. Our study of germline KDM1A mutations supports individuals being treated with KDM1A inhibitors be monitored for MGUS/MM.
We also surveyed germline data from pediatric hypodiploid ALL patients (24). This revealed a germline KDM1A truncating mutation c.1368_1371delAGAA (p.Lys456Asnfs*12) in a near-haploid hypodiploid ALL patient whose tumor demonstrated LOH and another near-haploid hypodiploid ALL patient whose tumor carried a somatic KDM1A truncating mutation c.1249C>T (p.Gln417*, MAF 83%). Mouse Kdm1a−/− LSK hematopoietic stem-progenitor cells have upregulated HOXA9 and LSK leukemia stem cell signatures(25). Re-analysis of KDM1A mutant and wt hypodiploid ALL tumor transcriptomes revealed that KDM1A mutations enriched for these same leukemia stem cell signatures(25) (Supplementary Figure S2). Additionally, query of germline sequence data from The Cancer Genome Atlas (TCGA) revealed germline truncating and splice-site KDM1A mutations in 3 patients with glioblastoma multiforme (KDM1A c.2371C>T [p.Gln791*]), glioma and ovarian serous cystadenocarcinoma (both KDM1A c.1476+1_1476+2delGT [p.Met448_Lys492del]). Thus, our data are consistent with KDM1A mutations contributing to predisposition of other tumors in addition to MM.
Supplementary Material
Acknowledgments
We acknowledge funding from R01 CA167824, R01 CA13464, R01 CA178765, T15 LM007124, R21 CA152336, LLS 6067-09, NCI P30 CA42014, HHSN261201000026C, the V Foundation (V2015-003), the Weill-Cornell Program in Mendelian Genetics, the Utah Genome Project, Huntsman Cancer Institute, Utah Population Database (UPDB), the Utah Cancer Registry (UCR), Icahn School of Medicine at Mount Sinai Office of Research Infrastructure of the National Institutes of Health award number S10OD018522 and a generous donation from Matthew Bell.
Footnotes
Conflicts of Interest
There are no potential conflicts of interest related to this study for all authors.
References
- 1.Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364(11):1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Schinasi L, Brown E, Camp N, Wang S, Hofmann J, Chiu B, Miligi L, Beane Freeman L, de Sanjose S, Bernstein L, et al. Multiple myeloma and family history of lymphohaematopoietic cancers: Results from the International Multiple Myeloma Consortium. Br J Haematol. 2016;175:87–101. doi: 10.1111/bjh.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chubb D, Weinhold N, Broderick P, Chen B, Johnson DC, Forsti A, Vijayakrishnan J, Migliorini G, Dobbins SE, Holroyd A, et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13. 1 influences multiple myeloma risk. Nat Genet. 2013;45(10):1221–1225. doi: 10.1038/ng.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell JS, Li N, Weinhold N, Forsti A, Ali M, van Duin M, Thorleifsson G, Johnson DC, Chen B, Halvarsson BM, et al. Genome-wide association study identifies multiple susceptibility loci for multiple myeloma. Nat Commun. 2016;7:12050. doi: 10.1038/ncomms12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rand KA, Song C, Dean E, Serie DJ, Curtin K, Sheng X, Hu D, Huff CA, Bernal-Mizrachi L, Tomasson MH, et al. A Meta-analysis of Multiple Myeloma Risk Regions in African and European Ancestry Populations Identifies Putatively Functional Loci. Cancer Epidemiol Biomarkers Prev. 2016;25(12):1609–1618. doi: 10.1158/1055-9965.EPI-15-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swaminathan B, Thorleifsson G, Joud M, Ali M, Johnsson E, Ajore R, Sulem P, Halvarsson BM, Eyjolfsson G, Haraldsdottir V, et al. Variants in ELL2 influencing immunoglobulin levels associate with multiple myeloma. Nat Commun. 2015;6:7213. doi: 10.1038/ncomms8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halvarsson B-M, Wihlborg Anna-Karin, Ali Mina, Lemonakis Konstantinos, Johnsson Ellinor, Niroula Abhishek, Cibulskis Carrie, Weinhold Niels, Forsti Asta, Alici Evren, Langer Christian, Pfreundschuh Michael, Goldschmidt Hartmut, Mellqvist Ulf-Henrik, Turesson Ingemar, Waage Anders, Hemminki Kari, Golub Todd, Nahi Hareth, Gullberg Urban, Hansson Markus, Nilsson Bjorn. Direct evidence for a polygenic etiology in familial multiple myeloma. Blood Advances. 2017;1(10):619–623. doi: 10.1182/bloodadvances.2016003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinhold N, Meissner T, Johnson D, Seckinger A, Moreaux J, Försti A, Chen B, Nickel J, Chubb D, Rawstron A, et al. The 7p15. 3 (rs4487645) association for multiple myeloma shows strong allele-specific regulation of the MYC-interacting gene CDCA7L in malignant plasma cells. Haematologica. 2015;100(3):110–113. doi: 10.3324/haematol.2014.118786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19(6):857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437(7057):432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 12.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437(7057):436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 13.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jun G, Flickinger M, Hetrick KN, Romm JM, Doheny KF, Abecasis GR, Boehnke M, Kang HM. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91(5):839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5(9):1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian YF, Ahn H, Schneider RS, Yang SN, Roman-Gonzalez L, Melnick AM, Cerchietti L, Singh A. Integrin-specific hydrogels as adaptable tumor organoids for malignant B and T cells. Biomaterials. 2015;73:110–119. doi: 10.1016/j.biomaterials.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiziltepe T, Ashley JD, Stefanick JF, Qi YM, Alves NJ, Handlogten MW, Suckow MA, Navari RM, Bilgicer B. Rationally engineered nanoparticles target multiple myeloma cells, overcome cell-adhesion-mediated drug resistance, and show enhanced efficacy in vivo. Blood Cancer J. 2012;2(4):e64. doi: 10.1038/bcj.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Puente P, Muz B, Gilson RC, Azab F, Luderer M, King J, Achilefu S, Vij R, Azab AK. 3D tissue-engineered bone marrow as a novel model to study pathophysiology and drug resistance in multiple myeloma. Biomaterials. 2015;73:70–84. doi: 10.1016/j.biomaterials.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boylan KL, Gosse MA, Staggs SE, Janz S, Grindle S, Kansas GS, Van Ness BG. A transgenic mouse model of plasma cell malignancy shows phenotypic, cytogenetic, and gene expression heterogeneity similar to human multiple myeloma. Cancer Res. 2007;67(9):4069–4078. doi: 10.1158/0008-5472.CAN-06-3699. [DOI] [PubMed] [Google Scholar]
- 24.Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, Payne-Turner D, Churchman M, Andersson A, Chen SC, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45(3):242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerenyi MA, Shao Z, Hsu YJ, Guo G, Luc S, O’Brien K, Fujiwara Y, Peng C, Nguyen M, Orkin SH. Histone demethylase Lsd1 represses hematopoietic stem and progenitor cell signatures during blood cell maturation. Elife. 2013;2:e00633. doi: 10.7554/eLife.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudolph T, Beuch S, Reuter G. Lysine-specific histone demethylase LSD1 and the dynamic control of chromatin. Biol Chem. 2013;394(8):1019–1028. doi: 10.1515/hsz-2013-0119. [DOI] [PubMed] [Google Scholar]
- 27.Linderman MD, Brandt T, Edelmann L, Jabado O, Kasai Y, Kornreich R, Mahajan M, Shah H, Kasarskis A, Schadt EE. Analytical validation of whole exome and whole genome sequencing for clinical applications. BMC Med Genomics. 2014;7:20. doi: 10.1186/1755-8794-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrovski S, Todd JL, Durheim MT, Wang Q, Chien JW, Kelly FL, Frankel C, Mebane CM, Ren Z, Bridgers J, et al. An Exome Sequencing Study to Assess the Role of Rare Genetic Variation in Pulmonary Fibrosis. Am J Respir Crit Care Med. 2017;196(1):82–93. doi: 10.1164/rccm.201610-2088OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji X, Kember RL, Brown CD, Bucan M. Increased burden of deleterious variants in essential genes in autism spectrum disorder. Proc Natl Acad Sci U S A. 2016;113(52):15054–15059. doi: 10.1073/pnas.1613195113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller A, Asmann Y, Cattaneo L, Braggio E, Keats J, Auclair D, Lonial S, Network MC, Russell SJ, Stewart AK. High somatic mutation and neoantigen burden are correlated with decreased progression-free survival in multiple myeloma. Blood Cancer J. 2017;7(9):e612. doi: 10.1038/bcj.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Corral L, Corchete LA, Sarasquete ME, Mateos MV, Garcia-Sanz R, Ferminan E, Lahuerta JJ, Blade J, Oriol A, Teruel AI, et al. Transcriptome analysis reveals molecular profiles associated with evolving steps of monoclonal gammopathies. Haematologica. 2014;99(8):1365–1372. doi: 10.3324/haematol.2013.087809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprussel A, Schulte JH, Weber S, Necke M, Handschke K, Thor T, Pajtler KW, Schramm A, Konig K, Diehl L, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26(9):2039–2051. doi: 10.1038/leu.2012.157. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18(19):5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagde A, Rajabi H, Bouillez A, Alam M, Gali R, Bailey S, Tai YT, Hideshima T, Anderson K, Avigan D, et al. MUC1-C drives MYC in multiple myeloma. Blood. 2016;127(21):2587–2597. doi: 10.1182/blood-2015-07-659151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah V, Sherborne AL, Walker BA, Johnson DC, Boyle EM, Ellis S, Begum DB, Proszek PZ, Jones JR, Pawlyn C, et al. Prediction of outcome in newly diagnosed myeloma: a meta-analysis of the molecular profiles of 1905 trial patients. Leukemia. 2017 doi: 10.1038/leu.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoenhals M, Kassambara A, Veyrune JL, Moreaux J, Goldschmidt H, Hose D, Klein B. Kruppel-like factor 4 blocks tumor cell proliferation and promotes drug resistance in multiple myeloma. Haematologica. 2013;98(9):1442–1449. doi: 10.3324/haematol.2012.066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fog CK, Asmar F, Come C, Jensen KT, Johansen JV, Kheir TB, Jacobsen L, Friis C, Louw A, Rosgaard L, et al. Loss of PRDM11 promotes MYC-driven lymphomagenesis. Blood. 2015;125(8):1272–1281. doi: 10.1182/blood-2014-03-560805. [DOI] [PubMed] [Google Scholar]
- 39.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tai YT, Anderson KC. Targeting B-cell maturation antigen in multiple myeloma. Immunotherapy. 2015;7(11):1187–1199. doi: 10.2217/imt.15.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Wang L, Chi PD, Wang WD, Chen XQ, Geng QR, Xia ZJ, Lu Y. High level of interleukin-10 in serum predicts poor prognosis in multiple myeloma. Br J Cancer. 2016;114(4):463–468. doi: 10.1038/bjc.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, Sougnez C, Knoechel B, Gould J, Saksena G, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menu E, Kooijman R, Van Valckenborgh E, Asosingh K, Bakkus M, Van Camp B, Vanderkerken K. Specific roles for the PI3K and the MEK-ERK pathway in IGF-1-stimulated chemotaxis, VEGF secretion and proliferation of multiple myeloma cells: study in the 5T33MM model. Br J Cancer. 2004;90(5):1076–1083. doi: 10.1038/sj.bjc.6601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shay G, Hazlehurst L, Lynch CC. Dissecting the multiple myeloma-bone microenvironment reveals new therapeutic opportunities. J Mol Med (Berl) 2016;94(1):21–35. doi: 10.1007/s00109-015-1345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Y, Saga K, Miyamoto Y, Kaneda Y. Cytoplasmic calcium increase via fusion with inactivated Sendai virus induces apoptosis in human multiple myeloma cells by downregulation of c-Myc oncogene. Oncotarget. 2016;7(24):36034–36048. doi: 10.18632/oncotarget.9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nair S, Branagan AR, Liu J, Boddupalli CS, Mistry PK, Dhodapkar MV. Clonal Immunoglobulin against Lysolipids in the Origin of Myeloma. N Engl J Med. 2016;374(6):555–561. doi: 10.1056/NEJMoa1508808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landgren O, Shim YK, Michalek J, Costello R, Burton D, Ketchum N, Calvo KR, Caporaso N, Raveche E, Middleton D, et al. Agent Orange Exposure and Monoclonal Gammopathy of Undetermined Significance: An Operation Ranch Hand Veteran Cohort Study. JAMA Oncol. 2015;1(8):1061–1068. doi: 10.1001/jamaoncol.2015.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scales M, Chubb D, Dobbins SE, Johnson DC, Li N, Sternberg MJ, Weinhold N, Stein C, Jackson G, Davies FE, et al. Search for rare protein altering variants influencing susceptibility to multiple myeloma. Oncotarget. 2017;8(22):36203–36210. doi: 10.18632/oncotarget.15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolli N, Barcella M, Salvi E, D’Avila F, Vendramin A, De Philippis C, Munshi NC, Avet-Loiseau H, Campbell PJ, Mussetti A, et al. Next-generation sequencing of a family with a high penetrance of monoclonal gammopathies for the identification of candidate risk alleles. Cancer. 2017;123(19):3701–3708. doi: 10.1002/cncr.30777. [DOI] [PubMed] [Google Scholar]
- 50.Togasaki E, Takeda J, Yoshida K, Shiozawa Y, Takeuchi M, Oshima M, Saraya A, Iwama A, Yokote K, Sakaida E, et al. Frequent somatic mutations in epigenetic regulators in newly diagnosed chronic myeloid leukemia. Blood Cancer J. 2017;7(4):e559. doi: 10.1038/bcj.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris WJHX, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, Miller CJ, Ogilvie DJ, Somervaille TC. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.