Abstract
Overexpression of Cyclin E has been seen in many types of cancers. However, the underlying mechanism remains enigmatic. Herein, we identified ubiquitin-specific peptidase 27 (USP27) as a Cyclin E interactor. We found that USP27 promoted Cyclin E stability by negatively regulating its ubiquitination. In addition, suppression of USP27 expression resulted in the inhibition of the growth, migration, and invasion of hepatocellular carcinoma. Furthermore, we detected a positive correlation between USP27 and Cyclin E expression in hepatocellular carcinoma tissues. Finally, we found that USP27 expression is inhibited by 5-fluorouracil (5-FU) treatment and USP27 depletion sensitizes Hep3B cells to 5-FU-induced apoptosis. USP27-mediated Cyclin E stabilization is involved in tumorigenesis, suggesting that targeting USP27 may represent a new therapeutic strategy to treat cancers with aberrant overexpression of Cyclin E protein.
Introduction
Cell cycle are controlled by cyclin-dependent kinases (CDKs), which form a complex with a specific cyclin that periodically activates and directs its kinase activity toward specific substrates with temporal and spatial selectivity [1, 2]. Different cyclins promote distinct cell cycle events [3]. Cyclin E, one of the cyclin family members, forms a complex with CDK2 to function at G1/S transition [4, 5]. Cyclin E-Cdk2 activity is lowest in quiescent cells and highest in G1–S cells. This periodicity is caused by many factors including transcriptional and proteolysis control of Cyclin E abundance, regulation of CDK2 activity by inhibitory and activating phosphorylations, and the binding of Cip/Kip CDK inhibitors [6]. These multiple layers of control ensure that the activity of Cyclin E/CDK2 is finely tuned during cell cycle progression [3]. In contrast, Cyclin E is frequently dysregulated in tumor cells and this may contribute to the development of various types of cancers including adrenocortical tumors and carcinomas (breast, lung, cervix, endometrium, and gastrointestinal tract), lymphoma, leukemia, and sarcomas [7–18]. In addition, overexpression of Cyclin E causes aneuploidy [19], genomic instability [20], or drives the cell cycle progression by curtailing the G1 to S transition, and thus accelerates cell proliferation and oncogenesis [5, 21–27].
Ubiquitination and deubiquitination are important mechanisms for regulating cell cycle progression. A number of components of the cell cycle apparatus, including various cyclins and inhibitors of CDK, are modified by ubiquitination [28]. Ubiquitin-specific peptidase 27 (USP27) belongs to a family of over 70 deubiquitinases (DUB) in human and mammals. It contains an N terminus and an ubiquitin-specific peptidase domain at its C terminus. USP27 is required for normal cell proliferation and its depletion suppresses tumor growth [29]. How USP27 regulates cell proliferation and tumorigenesis remains unclear. We previously depicted a molecular mechanism by which USP22 regulates cell cycle progression by affecting CCNB1 stability [30]. Whether USP27 affects cell proliferation and tumorigenesis through cyclin regulation needs investigation.
Herein, we demonstrate that USP27 regulates Cyclin E abundance to accelerate cell cycle progression, cell proliferation, and tumor cell growth as well. In support of this notion, we found that USP27 and Cyclin E expression are positively correlated in human hepatocellular carcinoma.
Results
Library screening identified USP27 as Cyclin E interactor
To determine the molecular mechanisms underlying the aberrant protein expression of the cell cycle protein Cyclin E in tumorigenesis, we used a DUB library to screen Cyclin E-interacting protein, which might be responsible for its dysregulation. Briefly, we transfected each of Flag-tagged DUB (USP1, 2, 3, 4, 7, 8, 10, 12, 14, 15, 17, 18, 19, 21, 22, 25, 26, 27, 28, 29, 30, 33, 36, 37, 38, 39, 42, 44, 46, 47, 51, OTUB1, and A20) with myc-tagged Cyclin E or Flag-tagged Cyclin E with myc-tagged DUB (USP5, 11, 16, COPS5, COPS6, BRCC3, UCHL1, UCHL3, UCHL5, JOSD1, JOSD2, CYLD, ATXN3, ATXN3L, OTUB1, OTUB2, OTUD6A, STAMBP, STAMBPL1, DUB3, EIF3S3,MPND, PSMD7, PSMD14, PARP11, BAP1, EIF3S5, MYSM1, and ZRANB1) constructs into 293T cells, and 48 h later we test the interactions between Cyclin E and each DUB by co-immunoprecipitation (Co-IP) and western blotting (WB). As seen in Fig. 1a, USP27 was identified as Cyclin E interaction partner among 60 DUBs (other DUBs not shown). To further confirm that the interaction between Cyclin E and USP27 was specific, we transfected them alone or together into HEK293T cells and evaluated their interaction by immunoprecipitation and WB. As shown in Fig. 1b, USP27 could be pulled down by Cyclin E, suggesting that USP27 are true Cyclin E interaction partner. In addition, we examined whether endogenous Cyclin E interacts with USP27 in the Hep3B cells. As expected, Cyclin E was detected in the anti-USP27 but not normal rabbit IgG immunoprecipitates from the cell lysate (Fig. 1c).
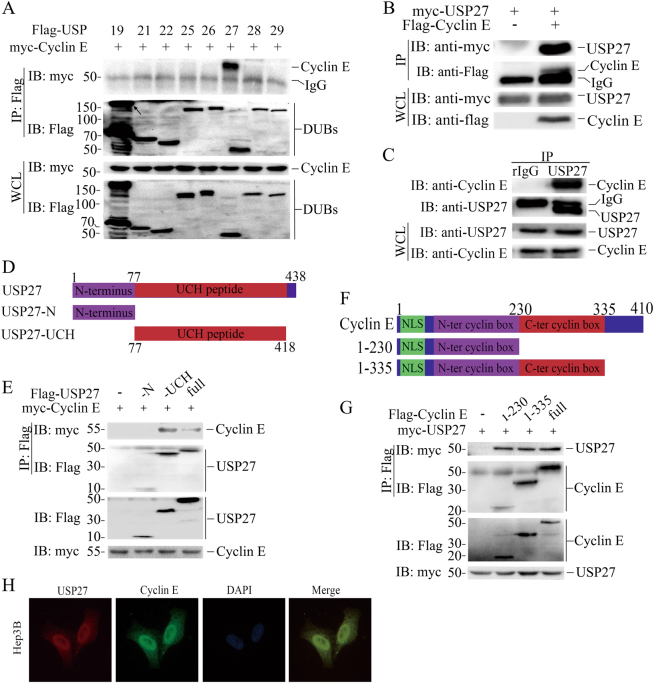
Fig. 1.
USP27 interacts and colocalizes with Cyclin E. a Identification of Cyclin E interaction partner. The interactions between Cyclin E and DUBs (a library containing about 60 deubiquitinases) were tested in 293T cells. 293T cells were transfected with Cyclin E and each of DUBs and immunoprecipitation and immunoblot were performed with indicated antibodies. b USP27 plasmid was transfected alone or with Cyclin E into HEK293T cells and their interaction was determined by immunoprecipitation and immunoblot. c The endogenous interaction of USP27 and Cyclin E was tested in Hep3B cells. d Schematic picture of USP27 and its mutants. e Cyclin E interactions with USP27 and its mutants were tested as indicated. f Cyclin E and its domains. NLS, nuclear localization signal. g The interactions of Cyclin E and its mutants with USP27 protein were tested. h Localization of Cyclin E and USP27 in Hep3B cells. DAPI was used to stain the DNA. DAPI, 4, 6-diamidino-2-phenylindole
Next, we mapped the regions of USP27 that mediate its interaction with Cyclin E by generating truncated mutants. USP27 protein carries an N terminus and C-terminal UCH domain (Fig. 1d). Co-IP and WB assay uncovered that the C-terminal UCH domain of USP27 is required for its interaction with Cyclin E, as deletion of this region completely abolished its interaction with Cyclin E. In contrast, expression of the USP27 C-terminal 77–438 fragment alone is sufficient to pull Cyclin E protein down (Fig. 1e). Similarly, truncated mutation analyses showed that the N terminus of Cyclin E is required for its interaction with USP27 (Fig. 1f,g). Furthermore, we detected a colocalization of USP27 and Cyclin E in Hep3B cells (Fig. 1h). Taken together, these data suggested that USP27 interacts and colocalizes with Cyclin E.
USP27 negatively regulates Cyclin E ubiquitination
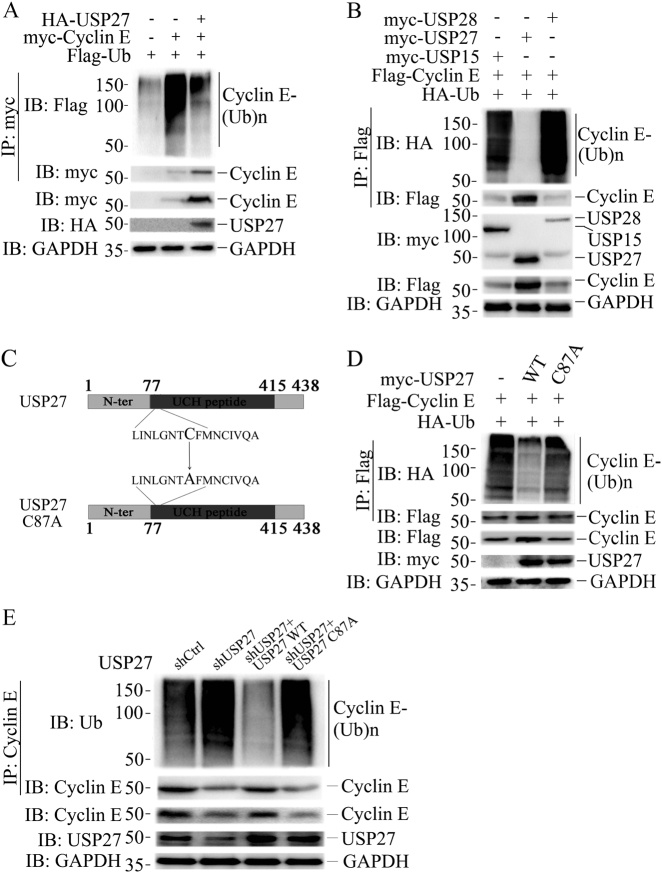
As USP27 is a DUB that might protect its substrate from proteasome-mediated degradation, we determined whether this DUB removes ubiquitin from Cyclin E. Cyclin E and ubiquitin were transfected into 293T cells without or with USP27 addition. As shown in Fig. 2a, in the presence of USP27 expression, the ubiquitination of Cyclin E protein was largely inhibited. USP27-mediated Cyclin E deubiquitination is highly specific, as co-expression of USP15, which was recently shown to be a PRP31-specific DUB [31], and USP28, did not have any effect on Cyclin E ubiquitination (lane 1 and 3 in Fig. 2b and Figure S1E). Next, we generated a DUB-inactive mutant of USP27 [29] (Fig. 2c) and assessed the effect of the mutation on Cyclin E ubiquitination. As seen in Fig. 2d, this mutant failed to inhibit the ubiquitination of Cyclin E protein, implying that the DUB activity is required for USP27 to remove ubiquitin from Cyclin E. Finally, we examined the endogenous ubiquitination of Cyclin E in Hep3B cells. As shown in Fig. 2e, knockdown of USP27 expression resulted in elevated Cyclin E ubiquitination compared with nonspecific short hairpin RNA (shRNA) control. Importantly, reintroducting wildtype but not the CA mutant form of USP27 could rescue Cyclin E from ubiquitination. Collectively, these data suggested that USP27 is a Cyclin E DUB.
Fig. 2.
USP27 inhibits Cyclin E ubiquitination. a Flag-Ubiquitin, HA-USP27, and myc-Cyclin E plasmids were transfected alone or cotransfected into HEK293T cells and the ubiquitination of Cyclin E was detected by immunoprecipitation and western blotting. The expression of Cyclin E and USP27 were confirmed. b Flag-Ubiquitin and myc-Cyclin E expression plasmids were cotransfected with USP15, USP27, or USP28 into HEK293T cells and Cyclin E ubiquitination was performed as in a. c Schematic diagram of USP27 and its mutants. The cysteine residue in position 87 in the UCH domains was replaced by alanine a. d The effects of USP27 and its mutants on Cyclin E ubiquitination were analyzed as in a. e The ubiquitination of endogenous Cyclin E in Hep3B cells was analyzed by immunoprecipitation and western blotting. GAPDH are used as loading control
USP27 stabilizes Cyclin E
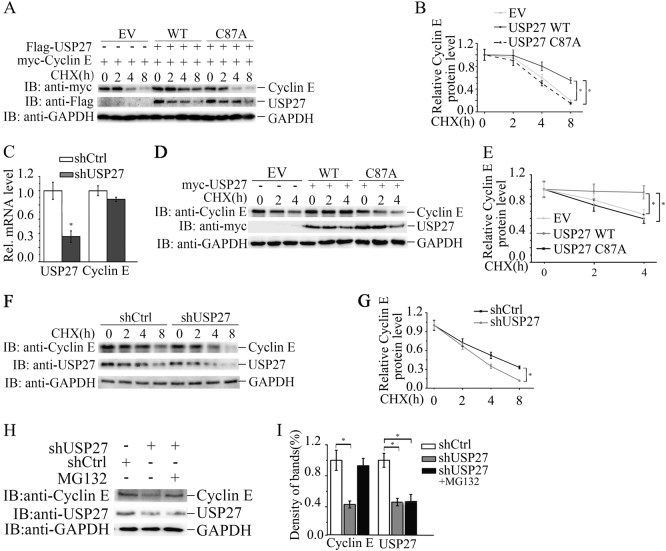
The observation that USP27 interacts with and deubiquitinates Cyclin E led us to think that USP27 might involve in cell cycle progression by regulating Cyclin E stability. Indeed, USP27 but not its mutant markedly extended the half-life of Cyclin E as seen in Hep3B cells (Fig. 3a, b). In addition, the regulation of USP27 on Cyclin E stability might occur at the posttranscriptional level, as we did not see mRNA change of Cyclin E in USP27 knockdown Hep3B cells (Fig. 3c). Then we determined whether the DUB activity is required for USP27 to regulate Cyclin E stability. As shown in Fig. 3d, e, catalytically inactive USP27/CA mutant failed to protect Cyclin E from degradation, implying that the DUB activity is required for USP27-mediated Cyclin E stabilization. To further confirm our notion that USP27 regulates Cyclin E stability, we knocked down USP27 expression and found that the endogenous Cyclin E protein levels decreased more rapidly than control (Fig. 3f, g). Finally, we determined whether USP27-mediated Cyclin E stability is through proteasome. As seen from Fig. 3h, i, MG132 (proteasome inhibitor) treatment could rescue the USP27 knockdown-mediated degradation of Cyclin E protein. In conclusion, our data suggested that USP27 regulates Cyclin E stability.
Fig. 3.
USP27 promotes Cyclin E abundance. a, b 293T cells were transfected with indicated plasmids and two days later were treated with 10 mg/ml cycloheximide (CHX) for 2 h and harvested. The expression of Cyclin E, USP27, and GAPDH were analyzed by western blotting a and quantified b. c The mRNA levels of Cyclin E and USP27 were determined by real-time PCR. The error bar represents the SEM of triplicate experiments. d, e Cyclin E protein stability was examined by cotransfection of USP27 or its mutant and quantified. f, g Hep3B cells were transfected with control shRNA or with shRNA specifically against USP27. Cyclin E stability was analyzed f and quantified g. h, i USP27 knockdown or control plasmids were transfected into Hep3B cells and cell were treated with the proteasome inhibitor MG132 2 h before collecting. The expression of Cyclin E and USP27 were determined and quantified
USP27 regulates cell cycle progression and cell proliferation
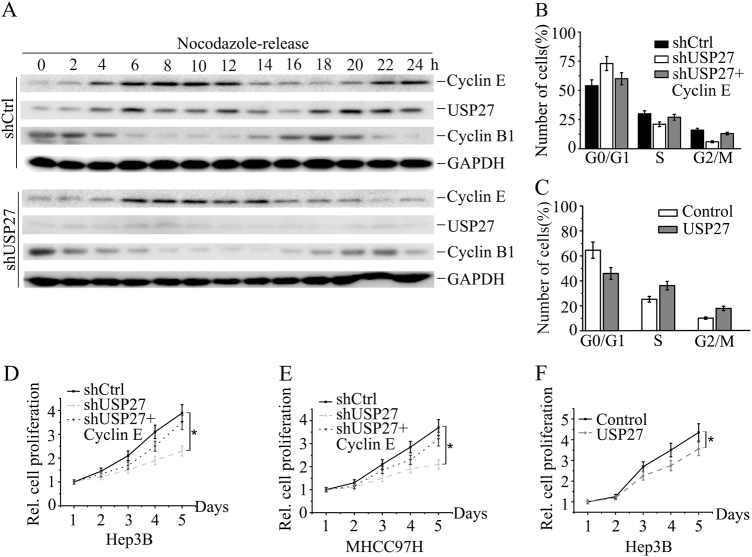
Cyclin E overexpression has been reported to promote cell proliferation by passing Rb-imposed G1 control on rate limiting [32]. Our discovery that USP27 mediated Cyclin E stabilization implied that USP27 might modulate cell cycle and cell proliferation through Cyclin E regulation. We first examined whether USP27 protein expression is oscillated during cell cycle progression. As shown in Fig. 4a, USP27 expression was oscillated during 24 h. Notably, the oscillation of USP27 expression was similar to Cyclin E, implying a correlation between two proteins during cell cycle progression. In agreement with above results, knocking down USP27 expression, the cell cycle phase was delayed about 2–4 h as judged by the expression level of Cyclin E and Cyclin B1 as well as the results from the flow cytometry analysis (Fig. 4a and S2A–C). The corresponding relative mRNA levels were determined by quantitative reverse-transcriptase PCR (qRT-PCR) (Figure S2D). It seemed that USP27 was also transcriptionally regulated in the cell cycle progression (Figure S2D). Interestingly, USP27 expression might also be regulated by Fbxw7, a well-known E3 ubiquitin ligase of Cyclin E, which interacts with and degrades USP27 (Figure S3A–C). Next, we investigated whether USP27 regulates cell cycle progression. We found that USP27 knockdown suppresses cell cycle progression by increasing the percentage of cells in G0/G1 phase (Fig. 4b, S1A, S2E, and S2F), which can be reversed by the addition of Cyclin E expression (Fig. 4b). In contrast, USP27 overexpression facilitates cell cycle progression by increasing the percentage of cells in G2/M phase but also by decreasing the percentage of cells in G0/G1 phase (Fig. 4c, S1B, and S2G). To further support our notion that USP27 promotes cell cycle progression, we detected a statistically significant reduction in the growth of Hep3B (Fig. 4d and S2H) and MHCC97H cells (Fig. 4e, S1C, and S2I) compared with that of control cells with single stable knockdown of USP27. Notably, reintroducing Cyclin E expression into USP27-depleted hepatoma cells (shUSP27 + Cyclin E) reversed the growth-retarding effect caused by single USP27 knockdown (Fig. 4d, e). In accordance with this, ectopic expression of USP27 leads to an increase in cell proliferation (Fig. 4f). Taken together, USP27 is involved in cell cycle progression and cell proliferation through Cyclin E regualtion.
Fig. 4.
USP27 regulates cell cycle progression and cell proliferation. a Hep3B cells stably expressing control or USP27 knockdown plasmids were synchronized at G2/M stage by treatment of nocodazole (50 ng/ml) twice for 20 h separately and then released into fresh medium and analyzed by western blot. b, c The effects of USP27 knockdown, USP27 knockdown plus Cyclin E overexpression b or single overexpression of USP27 c together with their respective controls on Hep3B cell cycle progression were measured by flow cytometry analysis. d–f The growth of Hep3B d, f or MHCC97H e cells stably expressing shUSP27, shUSP27 + Cyclin E, USP27, or their corresponding control vector was examined
Depletion of USP27 inhibits tumor growth
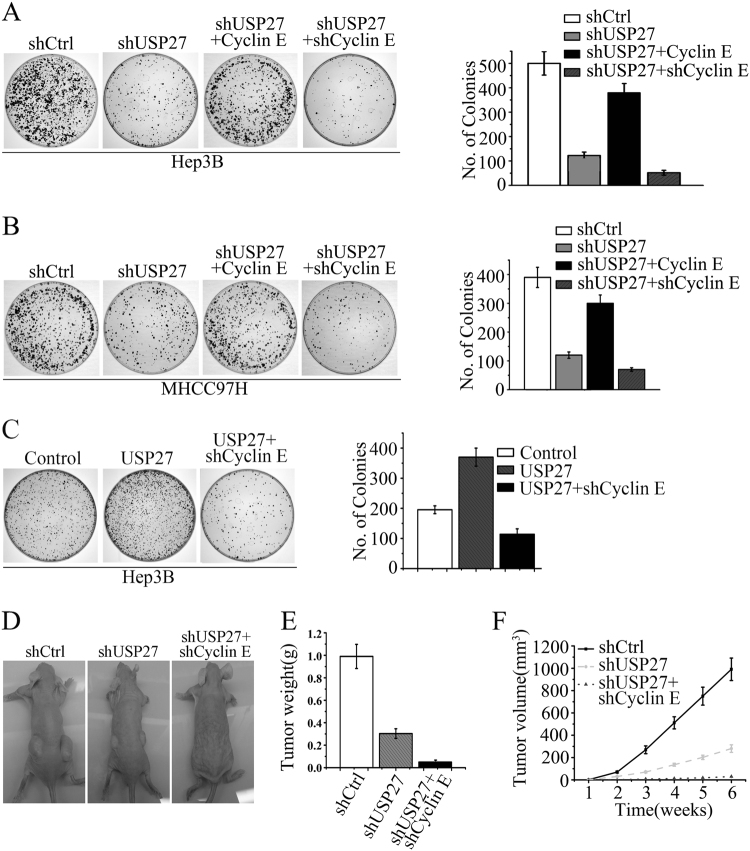
The observation that USP27 promotes cell cycle progression and cell proliferation prompted us to imagine that depletion of USP27 expression might suppress tumor progression. To test this hypothesis, we performed colony formation assay to determine whether the tumor growth is affected by suppression of USP27 expression. As indicated in Fig. 5a, b and S4, a soft agar assay revealed that USP27 knockdown dramatically inhibited the colony formation of Hep3B and MHCC97H cells. Reintroducing Cyclin E into USP27-depleted cells restored the malignant phenotype, while further suppression of Cyclin E rescued the cells from transformation phenotype. Conversely, USP27 overexpression got more colonies compared with the vector control (Fig. 5c). As expected, this effect was reversed by knockdown of Cyclin E expression in USP27-overexpressing cells (Fig. 5c). Next, we used a xenograft model to assess the effect of USP27 suppression on tumor growth. USP27 knockdown significantly inhibited the tumor growth in nude mice (Fig. 5d). Both the tumor weight and tumor volume were remarkably shrinked in mice that had been inoculated with USP27-knockdown Hep3B cells (Fig. 5e, f). Importantly, double knockdown of USP27 and Cyclin E expression rescued the mice from malignant phenotype (Fig. 5d, third panel). Collectively, these findings suggested that depletion of USP27 inhibited the tumor growth.
Fig. 5.
Suppression of USP27 expression delayed tumor growth. a Hep3B cells stably expressing control shRNA, shRNA against USP27 (shUSP27), Cyclin E overexpression plasmids plus shUSP27 (shUSP27 + Cyclin E), or double knockdown plasmids of Cyclin E and USP27 (shUSP27 + shCyclin E) were seeded in soft agar and cultured for 3–4 weeks. Colonies were visualized by light microscopy and quantified. b MHCC97H cells stably expressing indicated plasmids were seeded in soft agar and the numbers of colonies were quantified as in a. c Hep3B cells stably expressing indicated plasmids were seeded in soft agar and the numbers of colonies were quantified as in a. d–f Hep3B cells stably expressing control, single knockdown of USP27 or double knockdown of USP27 and Cyclin E plasmids were inoculated subcutaneously into nude mice. Six weeks after injection, mice were killed, photographed, and weighed. Tumor size was measured every week
Down regulation of USP27 suppresses hepatocellular migration and metastasis
To further explore the pathological roles of USP27 in HCC cancer progression, we examined HCC cell migration and invasion by knockdown or overexpression of USP27. As shown in Figure S5A–D, depletion of USP27 expression significantly decreased cell migration compared with the control in Hep3B (Figure S5A and S5B) and MHCC97H (Figure S5C and S5D) cells using wound-healing assay. Consistent with this notion, knockdown of USP27 markedly decreased the invasion of Hep3B and MHCC97H cells in transwell matrigel invasion assays (Figure S5E and S5F). Conversely, overexpression of USP27 evidently enhanced cell invasion (Figure S5G). Taken together, these results suggested that USP27 might promote hepatoma metastasis.
USP27 and Cyclin E expression are upregulated in hepatocellular carcinoma
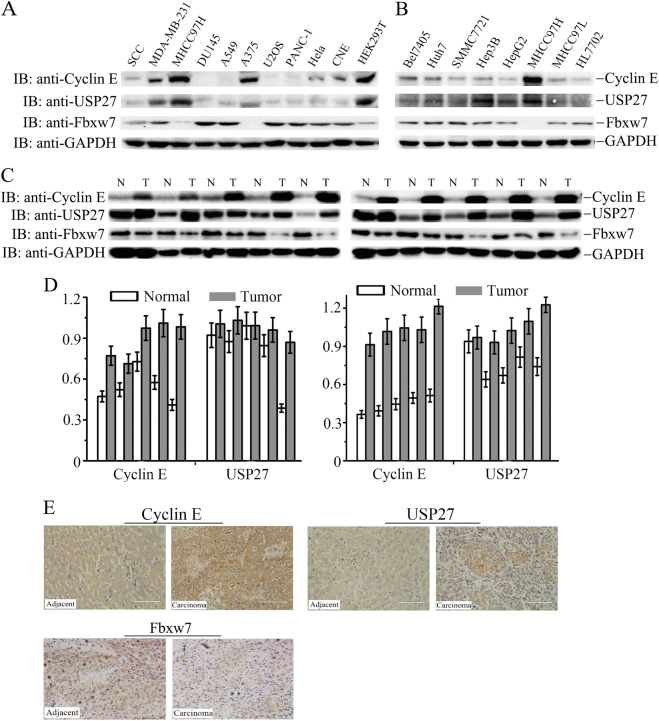
The observation that USP27 expression promotes the colony formation of Hep3B cells and tumor growth prompted us to think that USP27 might be upregulated in hepatocellular carcinoma. We first examined the expression of USP27 and Cyclin E in various types of cancer cell lines. As seen in Fig. 6a, USP27 and Cyclin E highly expressed in MDA-MB-231 (breast cancer), MHCC97H (hepatoma), A375 (melanoma), and HEK293T (kidney) cells, whereas the upstream E3 ubiquitin ligase of Cyclin E, Fbxw7, showed a reverse expression pattern. Next, we evaluated their expression in different hepatoma cell lines and found that they expressed in all of these cells (Fig. 6b). In fact, WB analysis detected dramatically higher protein expression of USP27 and Cyclin E in hepatoma tissues (in 8 of 10 patients) compared with those in adjacent normal control tissues (Fig. 6c, d). Consistent with the correlated upregulation of both proteins, USP27 and Cyclin E were co-localized in hepatoma tissues as revealed by immunohistochemical staining, whereas Fbxw7 showed an opposite expression pattern (Fig. 6e). Notably, a strong correlation between USP27 and Cyclin E protein expression was found, indicating that the USP22-Cyclin E interaction is associated with human hepatocellular cancer development.
Fig. 6.
USP27 and Cyclin E expression are upregulated in hepatocellular carcinoma cells and tissues. a Western blot analysis of Cyclin E, USP27, and Fbxw7 expression in various kinds of cancer cell lines. GAPDH was used as loading control. b Cyclin E, USP27, and Fbxw7 expression in various kinds of hepatocellular carcinoma cell lines were analyzed. c, d The expression of USP27, Cyclin E, and Fbxw7 were measured in HCC and normal tissue c and quantified d. e Representative images of immunohistochemical staining of USP27, Cyclin E, and Fbxw7. Scale bars, 100 μm
Anti-cancer drug 5-FU regulates cell cycle progression and inhibits hepatocellular proliferation by downregulating USP27 expression
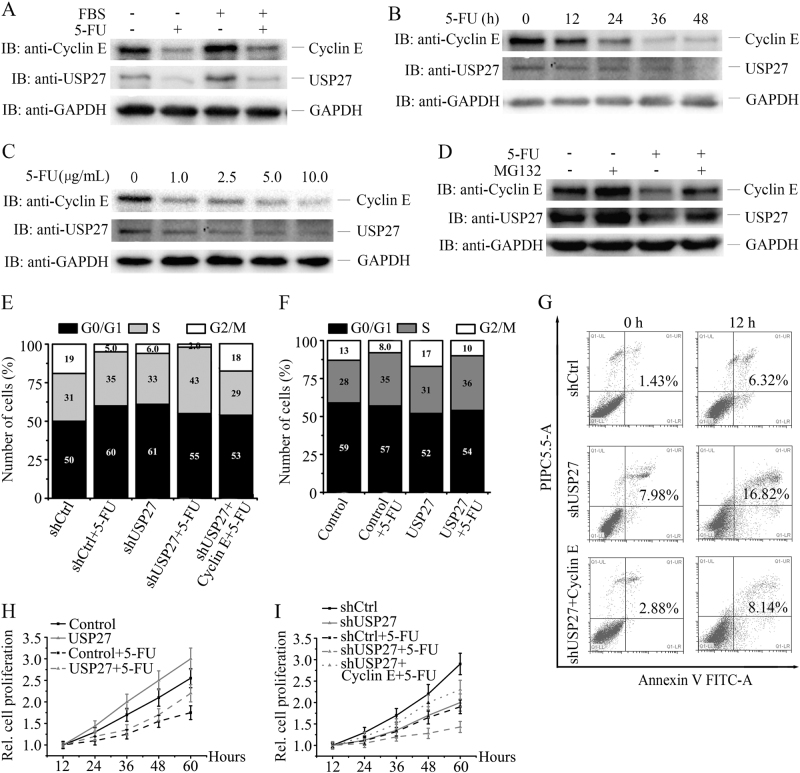
To evaluate the therapeutic potential of the USP27-Cyclin E interaction targeting in cancer treatment, we treated Hep3B cells with or without serum starvation. As shown in Fig. 7a, USP27 and Cyclin E expression were downregulated by 5-fluorouracil (5-FU) treatment. The protein expression of USP27 decreased in a dose-dependent manner (Fig. 7b) or changed with time increasing following 5-FU treatment (Fig. 7c). This downregulation of USP27 expression can be rescued by MG132 treatment, suggesting that the regulation might occur at the posttranscriptional level (Fig. 7d). In agreement with this notion, we did not see mRNA change of USP27 following 5-FU treatment (data not shown). Next, we examined the effects of USP27 overexpression or knockdown on cell cycle progression, apoptosis, and cell proliferation with or without 5-FU treatment. As shown in Fig. 7e, USP27 knockdown suppresses cell cycle progression by increasing the percentage of cells in G1/S phase but decreasing the percentage of cells in G2/M phase, similar to 5-FU treatment. In contrast, USP27 overexpression facilitates cell cycle progression by increasing the percentage of cells in G2/M phase but also by decreasing the percentage of cells in G0/G1 phase (Fig. 7f). Notably, knockdown of USP27 expression together with 5-FU treatment greatly reduced the percentage of cells in G2/M phase, whereas introducing Cyclin E to the USP27 knockdown cells (shUSP27 + Cyclin E) could increase the percentage (Fig. 7e). Consistent with this, USP27 knockdown sensitized Hep3B cells to apoptosis and Cyclin E overexpression plus USP27 knockdown (shUSP27 + Cyclin E) rescued cells from apoptosis, suggesting a therapeutic potential of USP27 inhibition for cancer treatment. Furthermore, cell proliferation was also remarkably reduced in the combination of USP27 knockdown and 5-FU treatment, whereas overexpression of USP27 (Fig. 7h) or addition of Cyclin E in USP27 knockdown cells could raise cell proliferation (Fig. 7i). Collectively, these results implied that USP27 might be targeted in cancer therapy for hepatocellular carcinoma patients (Fig. 8).
Fig. 7.
5-FU blocks cell cycle progression and inhibits hepatocellular proliferation by downregulating USP27 expression. a Hep3B cells were treated with 5-FU (5 µg/ml). The expression of USP27 and Cyclin E were measured. b Hep3B cells were treated with 5-FU (5 µg/ml) at indicated time. USP27 and Cyclin E expression were measured by western blotting. c Hep3B cells were treated with 5-FU (5 µg/ml) at different concentrations as indicated for 12 h. USP27 and Cyclin E expression were examined by western blotting. d Hep3B cells were not treated or treated with MG132 or 5-FU (5 µg/ml) or together. USP27 and Cyclin E expression were measured by western blotting. e, f The cell cycle stage of Hep3B cells stably expressing indicated plasmids with or without 5-FU treatment was determined by PI staining and flow cytometry analysis. g Hep3B cells stably expressing USP27 knockdown constructs, USP27 knockdown plus Cyclin E overexpression plasmids, or control vectors were treated with 5-FU for 12 h, and apoptosis was evaluated by Annexin V-FITC and PI staining followed by flow cytometry analysis. h, i Hep3B cells stably overexpressing USP27 or its knockdown constructs or indicated combination of constructs together with their respective controls were treated or not treated with 5-FU for indicated time. The growth of Hep3B cells were evaluated as in Fig. 4d
Fig. 8.
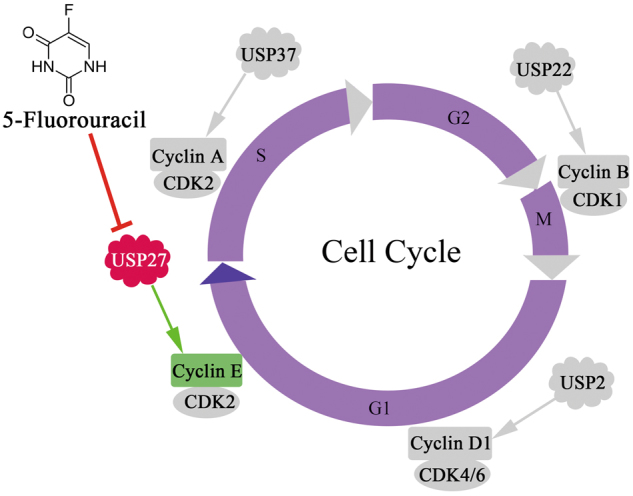
Working model of Cyclin E regulation by USP27. Cyclin E abundance is positively regulated by USP27, which expression may be downregulated by 5-fluorouracil
Discussion
Our discovery demonstrates that USP27 is a Cyclin E DUB. This conclusion is supported by the following observations. First, USP27 interacts with and colocalizes with Cyclin E. Second, USP27 negatively regulates Cyclin E ubiquitination. Third, USP27 positively regulates Cyclin E protein level. Fourth, USP27 and Cyclin E expression are oscillated and show a similar pattern during cell cycle progression. Finally, both USP27 and Cyclin E protein expression are elevated and positively correlated in human hepatocellular carcinomas. Collectively, our study implied that USP27 functions as a Cyclin E DUB and might be involved in hepatocellular carcinogenesis.
Cyclin E abundance is periodically changed and tightly controlled during cell cycle progression. Its abundance is determined by control of transcription and proteolysis. Transcriptional activity of the Cyclin E genes is primed during G1 phase and depends on mitogenic input, which is integrated through Myc and E2F activities [32, 33]. Cyclin E is able to stimulate its own transcription by an E2F-involved mechanism [33]. Its periodicity and turnover is mainly determined by ubiquitin-mediated proteolysis [34]. Cyclin E abundance is controlled by at least two distinct ubiquitin-proteasome pathways: BCR (BTB-Cul-3-Rbx1) pathway [35] and the SCFFbxw7 pathway [36–38]. Our findings demonstrated that USP27-mediated Cyclin E deubiquitination promoted its stability, thus adding another layer of control to the complexity of the cellular network that regulates Cyclin E transcription, proteolysis, and activity. Whether and how USP27 and Fbxw7 cooperatively regulate Cyclin E abundance deserves investigation. In our study, USP27 protein is also oscillated during cell cycle progression (Fig. 4a). The question how USP27 is regulated during cell cycle progression remains to be answered. It is possible that Fbxw7 regulates USP27 protein stability in the cell cycle progression.
How is Cyclin E overexpression involved in tumorigenesis? One possibility is when Cyclin E overexpressed, cell cycle progression passes Rb-imposed G1 control and becomes partially independent of the rate-restricting mitogens [34]. The other possibility is that Cyclin E overexpression may cause aneuploidy [19] or genomic instability [20]. Our data showed that USP27 promotes cell proliferation and migration. Whether USP27 cause aneuploidy or genomic instability through stabilizing Cyclin E need further investigation. How does USP27 promote migration and metastasis? Is Cyclin E dependent or independent? Although Cyclin E has been reported to involve in metastasis [15], how USP27 promotes migration and metastasis also need further detailed exploration.
In addition to USP27, USP2, USP22, and USP37 have been involved in cell cycle regulation by acting as DUB for certain cyclin protein [30, 39, 40]. USP2 was previous identified as a DUB specific for cyclin D1 by in vitro screening of over than 70 DUBs, presumably to modulate the G1–S transition [39]. USP22 is possibly responsible for CCNB1 protein elevation during colon cancer progression [30]. USP37 is suggested to promote S-phase entry by acting as cyclin A DUB [40]. Therefore, different cyclin protein is possibly regulated by its specific DUB during cell cycle progression (Fig. 8).
As Cyclin E is frequently deregulated and may contribute to the development of various types of cancers [7–18] as well as the downstream effector of multiple cancer-related processes, Cyclin E-Cdk2 is an attractive target for therapy. However, CDKs and cyclins are required for the cell cycle progression of normal and cancer cells. In our study, we found that both USP27 and Cyclin E are downregulated by 5-FU treatment in a dose-dependent manner. In addition, USP27 depletion sensitizes Hep3B cells to 5-FU-induced apoptosis. Thus, selective USP27 inhibition by chemical drugs may provide therapeutic benefit for certain human neoplasias where USP27/Cyclin E is overexpressed.
Materials and methods
Nude mice
Animal experiments were conducted under the guidance of Animal Management Regulations in Chongqing University.
Cells, plasmids, and antibodies
Hep3B cells were purchased from Procell Company (Wuhan, China), and MHCC97H was from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). USP27, Cyclin E, and their mutants were cloned into pCMV-myc or pCMV-Flag2.0 (Addgene Company).
To knockdown USP27, lentiviral vector [41] was obtained from GeneCopoeia (Rockville, MD) and packaging was performed in 293T cells. Nonspecific shRNA was used as a control. The sequences were shown as follows: shCtrl forward: 5′-GATCCGTTCTCCGAACGTGTCACGTAATTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTC-3′, Reverse: 5′-AATTGAAAAAATTCTCCGAACGTGTCACGTAATCTCTTGAATTACGTGACACGTTCGGAGAACG-3′, shUSP27-1 forward: 5ʹ-CCGGGACGCCGTTTATGGCCTCAAGTAAACTCGAGTTTACTTGAGGCCATAAACGGCGTCTTTTT-3ʹ, Reverse: 5ʹ-AATTAAAAAGACGCCGTTTATGGCCTCAAGTAAACTCGAGTTTACTTGAGGCCATAAACGGCGTC-3ʹ. shUSP27-2 forward: 5′-CCGGCACTGGTACATATCCTATATTCTCGAGAATATAGGATATGTACCAGTGTTTTTG-3′, Reverse:5′-AATTCAAAAACACTGGTACATATCCTATATTCTCGAGAATATAGGATATGTACCAGTG-3′.
Antibodies used in the study included anti-USP27 (Abcam-ab130642), anti-Cyclin E (Cell Signaling-Danvers-4129), anti-CCNB1 (Santa Cruz, sc-4073), anti-Flag (Sigma-Aldrich, F1804), anti-Myc (Santa Cruz, sc-40), anti-HA (Santa Cruz, sc-805), anti-Ub (Santa Cruz, sc-9133), and anti-FBXW7 (BBI Life Science, D122081).
WB and immunoprecipitation
WB analysis and immunoprecipitation were conducted as previously depicted [30]. If necessary, membranes were stripped using stripping buffer (Bio-Rad, Hercules, CA, USA) and reprobed with appropriate antibodies.
Immunofluorescence staining
Hep3B cells were grown on chamber slides and then transfected with corresponding plasmids. Cells were washed three times with phosphate-buffered saline (PBS), and fixed with 4% paraformaldehyde, permeabilized with PBS containing 0.1% Triton X-100 at room temperature, and blocked with 0.8% bovine serum albumin. Immunofluorescence staining was performed using the primary antibodies depicted in figure legend.
Ubiquitination analysis
Cells were lysed with high concentration of SDS lysis buffer, and immunoprecipitation with appropriate antibodies was conducted as described previously [41].
RNA extraction and real-time PCR analysis
Total RNA was extracted using Trizol reagent (Invitrogen) as described [21]. qRT-PCR was conducted according to the manufacture’s protocol (Clontech). β-Actin was used to normalize sample. Primers were listed as following. Cyclin E forward: 5ʹ-AAGGAGCGGGACACCATGA-3ʹ, Reverse: 5ʹ-ACGGTCACGTTTGCCTTCC-3ʹ; USP27 forward: 5ʹ-TCACCCACACGCCGATACT-3ʹ, Reverse: 5ʹ-CCAGACACAACTCGGGACTC-3ʹ; β-actin forward: 5ʹ-CATGTACGTTGCTATCCAGGC-3ʹ, Reverse: 5ʹ-CTCCTTAATGTCACGCACGAT-3ʹ.
Cell cycle and apoptosis analysis
Cells were seeded into six-well plate before analysis, then collected, fixed in 70% ethanol, and incubated at − 20 °C overnight. Cells were treated with 10 mg/ml RNase (Promega) for 30 min and stained with 5 mg/ml of propidium iodide (Promega), and then subjected to cell cycle analysis by a flow cytometry (Becton Dickinson, CA) [42].
Cell proliferation and colony-formation assay
Cell proliferation and colony-formation assay were conducted as previously described. Colonies with more than 50 cells were counted [42].
Wound-healing assay
A single scratch wound was created by dragging a 10 μl plastic pipette tip across the cell surface. The wound closure was measured using Image J software over 24 h. The experiments were repeated at least three times.
Statistical analysis
The statistical analyses of the experiment data was performed by using a two-tailed Student’s paired T-test and one-way ANOVA. Data were expressed as mean ± SEM of at least three independent experiments and significance was considered at either P-value ≤ 0.05.
Electronic supplementary material
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant Number 31571454 to Z. Lin), the Fundamental Research Funds for the Central Universities (Project Number 106112017CDJQJ298833 to Z. Lin), and the starting funds of ‘100 talent scholar program’ from Chongqing University.
Author contributions
L.D., L.Y., C.B., H.L. and L.L. performed the study. L.S. and Z.L. performed data analysis. Z.L. wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41388-018-0137-z) contains supplementary material, which is available to authorized users.
References
- 1.Ear PH, Booth MJ, Abd-Rabbo D, Kowarzyk Moreno J, Hall C, Chen D, et al. Dissection of Cdk1-cyclin complexes in vivo. Proc Natl Acad Sci USA. 2013;110:15716–21. doi: 10.1073/pnas.1305420110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–60. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 3.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–8. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 4.Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P. CDK substrate phosphorylation and ordering the cell cycle. Cell. 2016;167:1750–61. doi: 10.1016/j.cell.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–24. doi: 10.1128/MCB.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–63. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 7.Wolowiec D, Mekki Y, Ffrench P, Manel AM, Bertrand Y, Rimokh R, et al. Differential expression of cell proliferation regulatory proteins in B- and T-lineage acute lymphoblastic leukaemias. Br J Haematol. 1996;95:518–23. doi: 10.1046/j.1365-2141.1996.d01-1930.x. [DOI] [PubMed] [Google Scholar]
- 8.Yasui W, Akama Y, Kuniyasu H, Yokozaki H, Semba S, Shimamoto F, et al. Expression of cyclin E in human gastric adenomas and adenocarcinomas: correlation with proliferative activity and p53 status. J Exp Ther Oncol. 1996;1:88–94. [PubMed] [Google Scholar]
- 9.Yasui W, Kuniyasu H, Yokozaki H, Semba S, Shimamoto F, Tahara E. Expression of cyclin E in colorectal adenomas and adenocarcinomas: correlation with expression of Ki-67 antigen and p53 protein. Virchows Arch: Int J Pathol. 1996;429:13–19. doi: 10.1007/BF00196815. [DOI] [PubMed] [Google Scholar]
- 10.Iida H, Towatari M, Tanimoto M, Morishita Y, Kodera Y, Saito H. Overexpression of cyclin E in acute myelogenous leukemia. Blood. 1997;90:3707–13. [PubMed] [Google Scholar]
- 11.Molendini L, Benassi MS, Magagnoli G, Merli M, Sollazzo MR, Ragazzini P, et al. Prognostic significance of cyclin expression in human osteosarcoma. Int J Oncol. 1998;12:1007–11. [PubMed] [Google Scholar]
- 12.Dong Y, Sui L, Tai Y, Sugimoto K, Hirao T, Tokuda M. Prognostic significance of cyclin E overexpression in laryngeal squamous cell carcinomas. Clin Cancer Res. 2000;6:4253–8. [PubMed] [Google Scholar]
- 13.Fukuse T, Hirata T, Naiki H, Hitomi S, Wada H. Prognostic significance of cyclin E overexpression in resected non-small cell lung cancer. Cancer Res. 2000;60:242–4. [PubMed] [Google Scholar]
- 14.Erlanson M, Landberg G. Prognostic implications of p27 and cyclin E protein contents in malignant lymphomas. Leuk Lymph. 2001;40:461–70. doi: 10.3109/10428190109097645. [DOI] [PubMed] [Google Scholar]
- 15.Muller-Tidow C, Metzger R, Kugler K, Diederichs S, Idos G, Thomas M, et al. Cyclin E is the only cyclin-dependent kinase 2-associated cyclin that predicts metastasis and survival in early stage non-small cell lung cancer. Cancer Res. 2001;61:647–53. [PubMed] [Google Scholar]
- 16.Schraml P, Bucher C, Bissig H, Nocito A, Haas P, Wilber K, et al. Cyclin E overexpression and amplification in human tumours. J Pathol. 2003;200:375–82. doi: 10.1002/path.1356. [DOI] [PubMed] [Google Scholar]
- 17.Tissier F, Louvel A, Grabar S, Hagnere AM, Bertherat J, Vacher-Lavenu MC, et al. Cyclin E correlates with malignancy and adverse prognosis in adrenocortical tumors. Eur J Endocrinol. 2004;150:809–17. doi: 10.1530/eje.0.1500809. [DOI] [PubMed] [Google Scholar]
- 18.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–86. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 19.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 20.Minella AC, Swanger J, Bryant E, Welcker M, Hwang H, Clurman BE. p53 and p21 form an inducible barrier that protects cells against cyclin E-cdk2 deregulation. Curr Biol. 2002;12:1817–27. doi: 10.1016/S0960-9822(02)01225-3. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez I, Dynlacht BD. New insights into cyclins, CDKs, and cell cycle control. Semin Cell Dev Biol. 2005;16:311–21. doi: 10.1016/j.semcdb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Resnitzky D, Hengst L, Reed SI. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol Cell Biol. 1995;15:4347–52. doi: 10.1128/MCB.15.8.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keyomarsi K, Pardee AB. Redundant cyclin overexpression and gene amplification in breast cancer cells. Proc Natl Acad Sci USA. 1993;90:1112–6. doi: 10.1073/pnas.90.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keyomarsi K, Conte D, Jr., Toyofuku W, Fox MP. Deregulation of cyclin E in breast cancer. Oncogene. 1995;11:941–50. [PubMed] [Google Scholar]
- 25.Berglund P, Stighall M, Jirstrom K, Borgquist S, Sjolander A, Hedenfalk I, et al. Cyclin E overexpression obstructs infiltrative behavior in breast cancer: a novel role reflected in the growth pattern of medullary breast cancers. Cancer Res. 2005;65:9727–34. doi: 10.1158/0008-5472.CAN-04-3984. [DOI] [PubMed] [Google Scholar]
- 26.Keyomarsi K, Tucker SL, Buchholz TA, Callister M, Ding Y, Hortobagyi GN, et al. Cyclin E and survival in patients with breast cancer. N Eng J Med. 2002;347:1566–75. doi: 10.1056/NEJMoa021153. [DOI] [PubMed] [Google Scholar]
- 27.Luhtala S, Staff S, Tanner M, Isola J. Cyclin E amplification, over-expression, and relapse-free survival in HER-2-positive primary breast cancer. Tumour Biol. 2016;37:9813–23. doi: 10.1007/s13277-016-4870-z. [DOI] [PubMed] [Google Scholar]
- 28.Wickliffe K, Williamson A, Jin L, Rape M. The multiple layers of ubiquitin-dependent cell cycle control. Chem Rev. 2009;109:1537–48. doi: 10.1021/cr800414e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atanassov BS, Mohan RD, Lan X, Kuang X, Lu Y, Lin K, et al. ATXN7L3 and ENY2 coordinate activity of multiple H2B deubiquitinases important for cellular proliferation and tumor growth. Mol Cell. 2016;62:558–71. doi: 10.1016/j.molcel.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Z, Tan C, Qiu Q, Kong S, Yang H, Zhao F, et al. Ubiquitin-specific protease 22 is a deubiquitinase of CCNB1. Cell Discov. 2015;1:15028. doi: 10.1038/celldisc.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das T, Park JK, Park J, Kim E, Rape M, Kim EE, et al. USP15 regulates dynamic protein-protein interactions of the spliceosome through deubiquitination of PRP31. Nucleic Acids Res. 2017;45:5010–1. doi: 10.1093/nar/gkx486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santoni-Rugiu E, Falck J, Mailand N, Bartek J, Lukas J. Involvement of Myc activity in a G(1)/S-promoting mechanism parallel to the pRb/E2F pathway. Mol Cell Biol. 2000;20:3497–509. doi: 10.1128/MCB.20.10.3497-3509.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng Y, Eaton EN, Picon M, Roberts JM, Lundberg AS, Gifford A, et al. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–80. [PubMed] [Google Scholar]
- 34.Siu KT, Rosner MR, Minella AC. An integrated view of cyclin E function and regulation. Cell Cycle. 2012;11:57–64. doi: 10.4161/cc.11.1.18775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13:2375–87. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–22. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 37.Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413:311–6. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- 38.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–7. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 39.Shan J, Zhao W, Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell. 2009;36:469–76. doi: 10.1016/j.molcel.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, et al. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell. 2011;42:511–23. doi: 10.1016/j.molcel.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 41.Lin Z, Yang H, Kong Q, Li J, Lee SM, Gao B, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell. 2012;46:484–94. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 42.Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q, et al. USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep. 2013;5:1639–49. doi: 10.1016/j.celrep.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.