Abstract
Inspired by the high reactivity and specificity of thiyl radicals toward alkenes, we have developed a new charge derivatization method to enable fast and quantitative analysis of sterols via electrospray ionization-mass spectrometry (ESI-MS). Thioglycolic acid (TGA), a commercially available compound, has been established as a highly efficient tagging reagent. Initiated from photochemical reactions, the thiyl radical derived from TGA abstracts an allylic hydrogen in the B ring of sterols, forming a radical intermediate which rapidly recombines with a second thiyl radical to produce the final tagged product. Because of the incorporation of a carboxylic acid group, TGA tagging not only improves the limit of detection (sub-nM) for sterols but also facilitates their quantitation via characteristic 44 Da neutral loss scan. This radical based derivatization is fast (1 min) and efficient (>90% yield) when conducted in a flow microreactor. The analytical utility of thiyl radical charge tagging method has been demonstrated by quantifying sterols from human plasma and vegetable oil.
Graphical Abstract

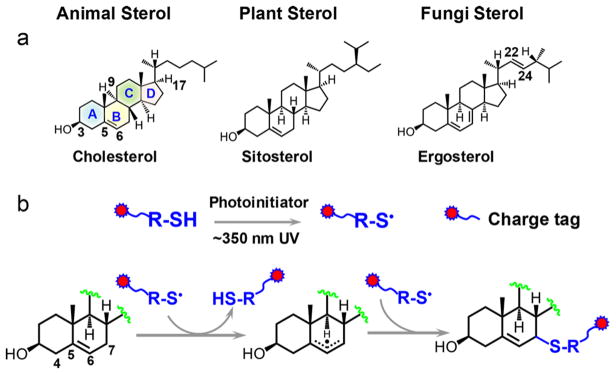
Sterols are naturally occurring and essential lipid molecules for animals (zoosterols), plants (phytosterols), and fungi. They commonly contain an unsaturated four-ring core structure (A, B, C, D rings), a 3β-hydroxyl (OH) group, and an aliphatic side chain attached to C17 (demonstrative structures shown in Scheme 1a). The major biological functions of sterols include regulating membrane fluidity and serving as precursors of steroids, vitamin D, hormones, and many other important mediator molecules.1 Gas chromatography–mass spectrometry (GC–MS) and liquid chromatography–mass spectrometry (LC–MS) are the primary methods for the analysis of sterols from complex mixtures.2,3 Since GC–MS suffers from lower sensitivity and throughput,4 more recent development focuses on enhancing separation and throughput using high-performance liquid chromatography (HPLC)–MS platform, which typically employs an electrospray ionization (ESI)-MS interface.5 Given the nonpolar nature of sterols, charge derivatization is a necessary and effective strategy to enhance the response of sterols via ESI. Established derivatization methods either target the hydroxyl function group via forming an ester bond with a charge tag (e.g., sulfate,6 phosphonium,7 picolinyl esters8) or convert the hydroxyl to ketone first and then use carbonyl chemistry to link a charge tag to sterols (e.g., forming Girard P hydrazones).9,10 These methods significantly improved sterol analysis via ESI with the limit of detection (LOD) achieved at nanomolar to sub-nanomolar.6–10 The derivatization reactions typically require several hours or even days to accomplish. Several derivatization reagents need to be synthesized in-house, further curbing wide accessibility to these methods.
Scheme 1.
(a) Chemical Structures of Representative Sterols in Animals, Plants, and Fungi and (b) Proposed Reaction Pathways for Thiyl Radical-Based Charge Tagging of Sterolsa
aTagging can happen at C7 and C5 (structure not shown).
Radical reactions, due to their fast reaction kinetics, are attractive candidates for chemical derivatization. Although only marginally explored, some notable examples of derivatization via radical chemistry include protein surface mapping via radical modification11,12 and carbon–carbon double location determination in lipids via the Paternò–Büchi reactions.13,14 Thiyl radicals are known to exhibit high reactivity toward alkenes, which has been harnessed as a type of “click chemistry” for forming a carbon–sulfur bond.15,16 Inspired by the reactions of thiyl radicals toward alkenes, we aim to tailor thiyl radical chemistry into a highly efficient charge derivatization strategy to enhance the analysis of nonpolar molecules, which are poorly detected by ESI-MS. In this report, we choose sterols as the first-time demonstration of this concept.
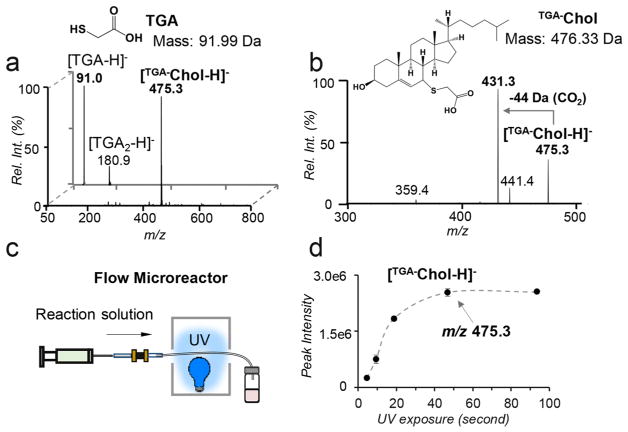
The C5–C6 double bond in the B ring of sterols makes them susceptible to thiyl radical tagging (Scheme 1b). In the survey studies, cholesterol was used as a model compound to react with a series of thiol reagents each consisting of a readily charged functional group via ESI (cysteamine, thioglycolic acid (TGA), and sodium 2-mercaptoethanesulfonate). On the basis of the reaction conditions reported in thiolene coupling,17 the thiol reagent (100 mM) and cholesterol (5 μM) were codissolved in dimethylformamide (DMF) with 1 mM 2,2-dimethoxy-2-phenylacetophenone (DMPA) added as a photo-initiator to effect thiyl radical formation. The reaction mixture was placed in a borosilicate glass vial and irradiated by a low-pressure mercury lamp with emission centered at 351 nm. After the completion of the reaction, 1 mL of sodium hydroxide (NaOH) solution (0.1 M) was added to the reaction mixture and the reaction product was extracted with ethyl acetate. The above procedure effectively removed remaining high concentration TGA, which could cause signal suppression upon subsequent nanoESI-MS analysis. The detailed experimental procedure can be found in the Supporting Information. Figure 1a compares nanoESI MS spectra of cholesterol before and after TGA tagging in the negative ion mode. Before reaction, only deprotonated TGA, [TGA – H]– (m/z 91.0) and a small amount of its oxidized product, [O-TGA2 – H]– (m/z 180.9), were observed. As a big contrast, the postreaction spectrum is dominated by a single peak at m/z 457.3 (front panel of Figure 1a). Accurate mass measurement proved that this product (m/z 457.3253) had a mass increase of 89.9776 Da relative to cholesterol, corresponding to a net addition of C2H2O2S, the elemental composition of deprotonated TGA (Figure S1a, Supporting Information). Collision-induced dissociation (CID) of the product ([TGA–Chol – H]–) produced a prominent loss of 44 Da (CO2) from the carboxylic acid group of the TGA moiety (Figure 1b). The above data all supported that TGA addition to cholesterol was successful. The reaction progress, monitored via the ion intensity of [TGA–Chol – H]–, showed a rapid increase in the first 15 min and a plateau afterward (Figure S2, Supporting Information). Quantitative analysis of cholesterol via GC–MS showed that the derivatization yield was above 90% after 20 min UV exposure (Figure S3, Supporting Information). TGA outperformed the rest thiol reagents tested in this study in terms of speed of reaction, product yield, and the easiness of sample cleanup for MS analysis; therefore, it was chosen for further method development.
Figure 1.
(a) Negative ion mode nanoESI MS spectra of cholesterol (5 μM) and TGA before reaction (back panel) and after reaction (front panel). TGA-tagged cholesterol appears at m/z 475.3 ([TGA–Chol-H]–). (b) MS2 CID of [TGA–Chol-H]–. (c) Schematic representation of a flow microreactor. (d) Plot of the peak intensity of [TGA–Chol-H]– as a function of UV exposure time using the flow microreactor setup.
Compared to bulk reactions, photochemical reactions in flow microreactors (micrometer (μm)-size inner dimensions) have been demonstrated to proceed more rapidly due to higher photon absorption efficiency.18 To further speed up TGA derivatization and make this reaction compatible with small sample size handling which is typically encountered in bioanalysis, a flow microreactor employing a fused silica capillary (100 μm i.d., 375 μm o.d.) has been developed (Figure 1c). The reaction kinetic curve was obtained by monitoring the ion intensity of [TGA–Chol – H]– (Figure 1d). Clearly, the reaction proceeded to a steady state within 50 s. GC–MS analysis of the remaining cholesterol suggested 94% conversion yield after 1 min UV irradiation, which was comparable to that obtained from the 20 min bulk reaction. The vastly accelerated reaction rate in the flow microreactor can be attributed to the large surface area to volume ratio of the microreactor, allowing maximal light transmission and thus significantly improving irradiation efficiency of the reaction mixture.
Initially, we expected to detect the thiolene coupling product, resulting from thiyl radical addition to the C5–C6 double bond followed by H atom abstraction from another thiol. This product should appear at m/z 477.3; however, the observed product (peak at m/z 475.3) is two Da less than the thiolene coupling product, suggesting the preservation of the C═C. This result can be rationalized from the reverse nature of thiyl radical addition to a C═C and the structure of cholesterol. The rate of thiyl radical addition to a ring C═C is several orders of magnitude smaller than the product dissociation rate,19 leading to ineffective thiolene coupling. This explains why the thiolene coupling product was not observed for cholesterol. On the other hand, although alkyl thiyl radicals cannot abstract alkyl hydrogen due to the relatively small S–H bond dissociation energies (BDEs, ~87 kcal/mol),20 they can abstract the allylic hydrogen at the C7 position (C7–H BDE = 83.2 kcal/mol).21 We hypothesize that the TGA thiyl radical abstracts the C7–H, forming a delocalized three-carbon allylic radical intermediate, which recombines with another thiyl radical at either C5 or C7 position, leading to the observed TGA-tagged cholesterol. It is worth noting that TGA thiyl radical is unlikely to abstract C4–H, the BDE of which is 89.0 kcal/mol,21 higher than the BDE of the alkyl thiol reagent. The proposed reaction pathway (Scheme 1b), however, needs to be further verified, e.g., using C7–D cholesterol, to provide more evidence on the C7–H abstraction process.
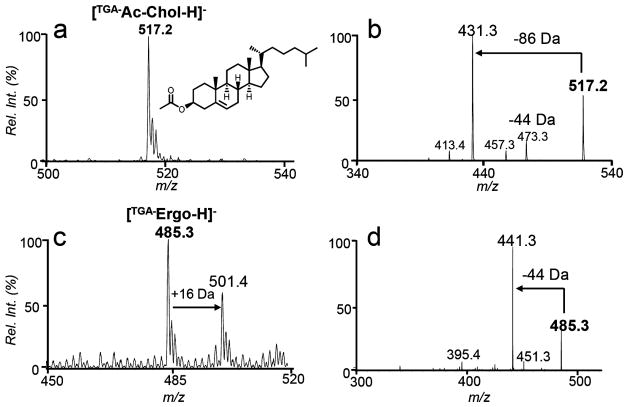
Established derivatization techniques target the free –OH functionality of sterols and thus have a limitation in analyzing the esterified form of sterols. As a contrast, the TGA tagging method can be readily applied to cholesterol ester (CE) analysis. Figure 2a shows the post-tagging spectrum using cholesterol acetate ([TGA–Ac-Chol – H]–, m/z 517.2) as a demonstration. MS2 CID of the tagged product (Figure 2b) produced 44 Da loss and 86 Da loss. The latter fragment resulted from sequential losses of CO2 (from TGA) and CH2CO (from the acetyl moiety).
Figure 2.
Negative ion mode nanoESI-MS of TGA derivatized (a) cholesterol acetate (1 μM, m/z 517.2) and (b) MS2 CID of [TGA–Ac-Chol – H]–, (c) ergosterol (1 μM, m/z 485.3), and (d) MS2 CID of [TGA–Ergo – H]–.
Ergosterol serves as a structural component of fungi cell membrane and thus it is a frequently used target for developing antifungal drugs.22 Ergosterol consists of three C═Cs, the diene structure in the B ring and a C22–C23 double bond in the aliphatic chain (structure shown in Scheme 1a). Since the aliphatic allylic hydrogen has a higher BDE than that of C9–H (about 10 kcal/mol higher),20 we expect that TGA tagging would be more competitive at the B ring. Indeed, only one TGA tagging product (the peak at m/z 485.3, Figure 2c) was observed from the reaction of TGA and ergosterol for an extended reaction period and no competing thiolene coupling product was observed (should appear at m/z 487.5, if formed). MS2 CID of tagged product produced abundant 44 Da loss (Figure 2d), characteristic to the TGA tag. Because of the presence of diene in the B ring, minor sequential oxidation of the TGA-tagged ergosterol was detected (the peak at m/z 501.4 in Figure 2c).
We further applied the TGA tagging to several other frequently encountered sterols from mammalian cells and plants (e.g., 7-dehydrocholesterol, Figure S4, Supporting Information). We found that single TGA tagging could be accomplished within 1 min for different sterol standards consisting at least one C═C in the B ring, likely resulting from the overwhelming excess of TGA (100 mM) relative to sterols (μM or lower) in the reaction system. TGA tagging, however, was not successful for steroids having a conjugated enone structure (e.g., progesterone) due to competitive photo-chemical rearrangement reactions.23 These results demonstrate that thiyl radical tagging of sterols exhibits selectivity to unsaturated B ring, which is an attractive feature for mixture analysis.
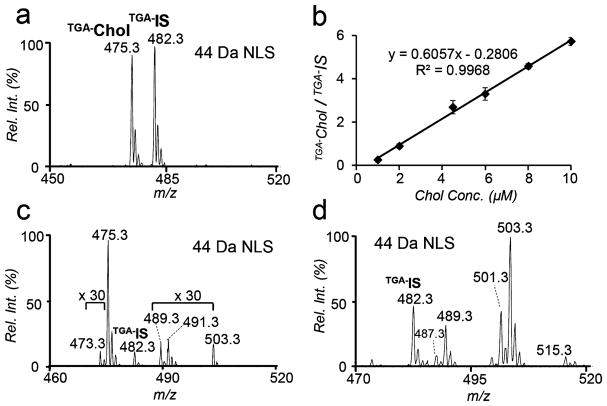
Tandem mass spectrometry (MS/MS) analysis of TGA-tagged sterols all showed abundant loss of CO2, due to the presence of deprotonated carboxylic acid moiety. On the basis of this characteristic fragmentation channel, 44 Da neutral loss scan (NLS) was evaluated for the quantitation of TGA-tagged sterols. Figure 3a shows a representative 44 Da NLS spectrum of 1 μM cholesterol with 5 μM cholesterol-d7 (Chol-d7) added as internal standard (IS). The calibration curve obtained by plotting the peak area ratios of [TGA–Chol – H]– and [TGA–IS – H]– against cholesterol concentration showed good linear regression (R2 = 0.9968). LOD was achieved at 0.5 nM (S/N ratio > 3, Figure S5, Supporting Information), comparable to those reported by other charge derivatization approaches, such as sulfation (0.2 nM),6 phosphonium labeling (0.05 nM),7 N-alkylpyridinium quaternization (54 nM),24 and Girard P reagent (0.1 nM).10 Most importantly, the whole process of TGA tagging and subsequent MS analysis is significantly shortened to less than 2 min per run. Although CO2 loss is a facile fragmentation channel for unsaturated fatty acids,25 their potential interference for sterol analysis via 44 Da NLS should be limited due to their appearance at lower m/z range than the commonly observed sterols.
Figure 3.
(a) 44 Da NLS of TGA tagged cholesterol (1 μM) and cholesterol-d7 (Chol-d7, IS, 5 μM). (b) Calibration curve obtained for cholesterol based on 44 Da NLS. Error bars stand for standard deviation (n = 3). 44 Da NLS profile of TGA tagged sterols from (c) 20 μL human plasma and (d) soybean oil with 5 μM Chol-d7 added as IS.
Because of the selectivity and sensitivity of TGA tagging toward sterols, this method is well suited for small sample volume analysis without a prior lipid extraction. As a demonstration, 20 μL of human plasma was dissolved in DMF at a final volume of 200 μL for TGA derivatization (1 min UV irradiation). Figure 3c shows the 44 Da NLS spectrum. Besides the TGA-tagged cholesterol peak (m/z 475.4), less abundant sterols such as dehydrocholesterol (m/z 473.4; intact mass, 384.6 Da), campesterol (m/z 489.3; intact mass, 400.37 Da), and sitosterol (m/z 503.3; intact mass, 414.4 Da) were also detected. The peak at m/z 491.4 corresponds to hydroxycholesterols (intact mass, 402.6 Da), which have been reported to be a mixture of multiple structural isomers in human plasma (Tables S1–S3, Supporting Information) and they typically exist in the sub-micromolar to nanomolar concentration range.26 Using Chol-d7 as the IS, quantitation of cholesterol in plasma was performed for cholesterol (procedure shown in the Supporting Information). The concentration of free cholesterol was found to be 1.4 ± 0.3 μmol/mL, while the concentrations of total and esterified cholesterol were 3.1 ± 0.1 μmol/mL and 2.0 ± 0.1 μmol/mL, respectively. These values fall within the range of reported cholesterol levels from human plasma using GC/LC–MS methods.26 Phytosterols typically exist at low concentrations (ppm) in vegetable oil and their quantitation demands laborious sample processing. To test the applicability the TGA tagging method, we performed quantitative analysis of phytosterols in soybean oil purchased from a local grocery store (detailed procedures shown in the Supporting Information). A variety of TGA derived sterols including brassicasterol (m/z 487.3), campesterol (m/z 489.3), stigmasterol (m/z 501.3), β-sitosterol (m/z 503.3), and cycloartenol (m/z 515.3) were clearly observed from 44 Da NLS spectrum (Figure 3d, 5 μM Chol-d7 added as IS). Table 1 summarizes the quantitative information on the free, esterified, and total phytosterols, expressed in mg/100 g of soybean oil. A significant portion of the sterols existed in their free form rather than in esterified form. β-Sitosterol was found to be the most abundant species in all sterol fraction (both free and esterified form) followed by stigmasterol and campesterol.
Table 1.
Free, Esterified, and Total (Free and Esterified) Sterols from Commercially Bought Soybean Oila
| sterol | MW [Da] | detection m/z | concentration [mg/100 g] | ||
|---|---|---|---|---|---|
| free | esterified | total | |||
| brassicasterol | 398.67 | 487.3 | 0.52 ± 0.07 | 0.41 ± 0.01 | 1.4 ± 0.1 |
| campesterol | 400.69 | 489.3 | 64 ± 5 | 17 ± 0.3 | 75 ± 4 |
| stigmasterol | 412.70 | 501.3 | 69 ± 8 | 23 ± 0.3 | 84 ± 5 |
| β-sitosterol | 414.70 | 503.3 | 135 ± 1 | 67 ± 1 | 190 ± 6 |
Chol-d7 (5 μM) was used as IS.
In summary, a rapid and simple charge derivatization method via thiyl radical tagging was developed for quantitative analysis of sterols from complex mixtures. The reaction allows linking the charged thiol group to the allylic position of the double bond in the B ring of a sterol. Thioglycolic acid (TGA), a commercially available compound, stands out as a highly efficient reagent. The carboxylic acid group in TGA allows tagged sterols to be detected by negative ion mode nanoESI-MS with high sensitivity. The deployment of a flow microreactor setup for this photochemical reaction further shortens the reaction time to less than 1 min with a yield higher than 90%, a significant improvement over conventional tagging methods which require several hours or even days to accomplish. Taking advantage of the dominant CO2 loss from CID of TGA-tagged sterols, 44 Da NLS has been established as a sensitive and quantitative method for sterol analysis from mixtures (e.g., total, free, and esterified sterols in human plasma and vegetable oil). Because of the fast reaction stemmed from radical chemistry, the TGA derivatization method shows potential to be incorporated into high-throughput workflows for applications in lipidomics and biomedical research. In this study, we noticed that structural isomers of several oxysterols cannot be distinguished using thiyl radical tagging. The same issue also exists for other conventional charge derivatization methods and has been resolved by adding an LC separation component in the workflow. In future studies, we plan to couple thiyl radical tagging with HPLC–MS/MS to further expand its capability in mixture analysis.
Supplementary Material
Acknowledgments
Financial support from NIHGM Grant R01GM118484 is greatly appreciated.
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.7b04080.
Details for chemicals and materials, lipid extraction, derivatization procedures of sterols in bulk and in the photochemical microreactor, and analysis of sterols in human plasma and vegetable oil (PDF)
References
- 1.Bloch K. New Compr Biochem. 1991;20:363–381. [Google Scholar]
- 2.McDonald JG, Smith DD, Stiles AR, Russell DW. J Lipid Res. 2012;53:1399. doi: 10.1194/jlr.D022285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy RC. Tandem Mass Spectrometry of Lipids Molecular Analysis of Complex Lipids. Royal Society of Chemistry; Cambridge, U.K: 2014. [Google Scholar]
- 4.Hübschmann H-J. Handbook of GC/MS. Wiley-VCH Verlag GmbH & Co. KGaA; 2008. pp. 7–292. [Google Scholar]
- 5.Liebisch G, Binder M, Schifferer R, Langmann T, Schulz B, Schmitz G. Biochim Biophys Acta, Mol Cell Biol Lipids. 2006;1761:121–128. doi: 10.1016/j.bbalip.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Chatman K, Hollenbeck T, Hagey L, Vallee M, Purdy R, Weiss F, Siuzdak G. Anal Chem. 1999;71:2358–2363. doi: 10.1021/ac9806411. [DOI] [PubMed] [Google Scholar]
- 7.Woo HK, Go EP, Hoang L, Trauger SA, Bowen B, Siuzdak G, Northen TR. Rapid Commun Mass Spectrom. 2009;23:1849–1855. doi: 10.1002/rcm.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honda A, Miyazaki T, Ikegami T, Iwamoto J, Yamashita K, Numazawa M, Matsuzaki Y. J Steroid Biochem Mol Biol. 2010;121:556–564. doi: 10.1016/j.jsbmb.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths W, Wang Y, Alvelius G, Liu S, Bodin K, Sjövall J. J Am Soc Mass Spectrom. 2006;17:341–362. doi: 10.1016/j.jasms.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Crick PJ, William Bentley T, Abdel-Khalik J, Matthews I, Clayton PT, Morris AA, Bigger BW, Zerbinati C, Tritapepe L, Iuliano L, Wang Y, Griffiths WJ. Clin Chem. 2015;61:400–411. doi: 10.1373/clinchem.2014.231332. [DOI] [PubMed] [Google Scholar]
- 11.Hambly DM, Gross ML. J Am Soc Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Rempel DL, Gau BC, Gross ML. J Am Chem Soc. 2012;134:18724–18731. doi: 10.1021/ja307606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma X, Xia Y. Angew Chem, Int Ed. 2014;53:2592–2596. doi: 10.1002/anie.201310699. [DOI] [PubMed] [Google Scholar]
- 14.Ma X, Chong L, Tian R, Shi R, Hu TY, Ouyang Z, Xia Y. Proc Natl Acad Sci U S A. 2016;113:2573–2578. doi: 10.1073/pnas.1523356113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tasdelen MA, Yagci Y. Angew Chem, Int Ed. 2013;52:5930–5938. doi: 10.1002/anie.201208741. [DOI] [PubMed] [Google Scholar]
- 16.Dénès F, Pichowicz M, Povie G, Renaud P. Chem Rev. 2014;114:2587–2693. doi: 10.1021/cr400441m. [DOI] [PubMed] [Google Scholar]
- 17.Jackson JA, Turner JD, Rentoul L, Faulkner H, Behnke JM, Hoyle M, Grencis RK, Else KJ, Kamgno J, Bradley JE, Boussinesq M. Int J Parasitol. 2004;34:1237–1244. doi: 10.1016/j.ijpara.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Su Y, Straathof NJW, Hessel V, Noël T. Chem - Eur J. 2014;20:10562–10589. doi: 10.1002/chem.201400283. [DOI] [PubMed] [Google Scholar]
- 19.Chatgilialoglu C, Ferreri C. Acc Chem Res. 2005;38:441–448. doi: 10.1021/ar0400847. [DOI] [PubMed] [Google Scholar]
- 20.Luo Y-R. Handbook of Bond Dissociation Energies in Organic Compounds. CRC Press; Boca Raton, FL: 2003. [Google Scholar]
- 21.Zielinski ZA, Pratt DA. J Am Chem Soc. 2016;138:6932–6935. doi: 10.1021/jacs.6b03344. [DOI] [PubMed] [Google Scholar]
- 22.Muller C, Binder U, Bracher F, Giera M. Nat Protoc. 2017;12:947–963. doi: 10.1038/nprot.2017.005. [DOI] [PubMed] [Google Scholar]
- 23.Asher JDM, Sim GA. J Chem Soc. 1965;0:1584–1594. [Google Scholar]
- 24.Wang H, Wang H, Zhang L, Zhang J, Leng J, Cai T, Guo Y. J Mass Spectrom. 2013;48:1101–1108. doi: 10.1002/jms.3265. [DOI] [PubMed] [Google Scholar]
- 25.Yang K, Zhao Z, Gross RW, Han X. Anal Chem. 2011;83:4243–4250. doi: 10.1021/ac2006119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CR, Guan Z, Laird GM, Six DA, Russell DW, et al. J Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.