Abstract
The blaSHV-12 β-lactamase gene is one of the most prevalent genes conferring resistance to extended-spectrum β-lactams in Enterobacteriaceae disseminating within and between reservoirs, mostly via plasmid-mediated horizontal gene transfer. Yet, studies regarding the biology of plasmids encoding blaSHV-12 are very limited. In this study, we revealed the emergence of IncX3 plasmids alongside IncI1α/γ in blaSHV-12 in animal-related Escherichia coli isolates. Four representative blaSHV-12-encoding IncX3 plasmids were selected for genome sequencing and further genetic and functional characterization. We report here the first complete sequences of IncX3 plasmids of animal origin and show that IncX3 plasmids exhibit remarkable synteny in their backbone, while the major differences lie in their blaSHV-12-flanking region. Our findings indicate that plasmids of this subgroup are conjugative and highly stable, while they exert no fitness cost on their bacterial host. These favourable features might have contributed to the emergence of IncX3 amongst SHV-12-producing E. coli in the Netherlands, highlighting the epidemic potential of these plasmids.
Introduction
The emerging IncX plasmid family consists of narrow host-range, self-transferable, iteron-containing plasmids with class A theta replication and sizes ranging approximately between 30 and 100 kb1–3. IncX plasmids have a highly syntenic backbone; yet based on phylogenetic analysis they can be assigned to six distinct subgroups, namely IncX1 to IncX62,4,5. Although it has been demonstrated that IncX plasmids occurred only infrequently among commensal and pathogenic E. coli isolates6, plasmids of this family encoding various resistance genes were recently described in Enterobacteriaceae originating from diverse sources and geographical areas2,4,7–11. Among this plasmid family, the IncX3 subgroup mediates the spread of genes encoding resistance for clinically relevant first-line (fluoroquinolones and extended-spectrum cephalosporins) and last-resort (carbapenems) antibiotics. IncX3 plasmids have been reported to encode qnrB79, qnrS12,9,12–15, blaCTX-M-311, blaSHV-129,16–19, blaKPC-220,21, blaKPC-322, blaNDM-118,19,23, blaNDM-424, blaNDM-525-28, blaNDM-715,26,29-33, blaNDM-1316, blaNDM-1734 and blaOXA-18112–14,35,36. Overall, these reports highlight the importance of this plasmid subgroup for the dissemination of antibiotic resistance genes within Enterobacteriaceae.
The blaSHV-12 gene ranks amongst the most predominant extended-spectrum β-lactamases within Enterobacteriaceae of diverse origins37. Plasmid-mediated horizontal gene transfer constitutes a key mechanism by which this gene disseminates among bacterial populations, therefore monitoring the spread of plasmids is essential to track the transmission of the blaSHV-12 gene between different reservoirs37. Several plasmid replicon types have been associated with the worldwide dissemination of blaSHV-12, including A/C, colE, F, HI2, I1α/γ, K, L/M, N, P, R, as well as the recently emerging X337. The few available data on the prevalence of blaSHV-12-encoding plasmids in the Netherlands, report IncHI2 plasmids in human Salmonella enterica isolates, IncN and IncF plasmids in human Escherichia coli37,38, as well as IncK plasmids in E. coli from poultry37.
To understand blaSHV-12 diffusion in the Netherlands, also in light of the emerging role of IncX3 plasmids worldwide, we investigated a collection of previously uncharacterized SHV-12-encoding E. coli isolates. We report here the plasmid epidemiology of SHV-12-producing E. coli from different reservoirs in The Netherlands, the first fully assembled and annotated sequence of three blaSHV-12 encoding-IncX3 plasmids of animal origin, and the genetic and functional characteristics of IncX3 plasmids from both human and animal origin.
Results
Plasmid epidemiology of blaSHV-12 and emergence of IncX3
Among the 129 blaSHV-12 encoding E. coli isolates included in this study (Table 1), 49.6% (n = 64) was isolated from food-producing animals, 41.1% (n = 53) from retail meat and 9.3% (n = 12) from humans. Plasmid typing revealed that blaSHV-12 was encoded by nine different plasmid families: I1α/γ (n = 86; 66.7%), X3 (n = 21; 16.3%), X1 (n = 6; 4.7%), F (n = 5; 3.9%), B/O (n = 4; 3.1%), K (n = 4; 3.1%), N (n = 1; 0.8%), colE (n = 1; 0.8%) and multi-replicon F-X1 (n = 1; 0.8%). A gradual decrease from 90.0% to 55.6% of IncI1α/γ in parallel with a significant increase from 0.0% to 24.1% (p = 0.041) of IncX3 was documented between 2011 and 2014 among food-producing animals and retail meat (Fig. 1). As a result, IncX3 plasmids were among the predominant rep-types encoding blaSHV-12 in the Netherlands from 2012 onwards (Fig. 1).
Table 1.
Characteristics of the 129 non-duplicate blaSHV-encoding E. coli isolates included in the study.
| Isolate | Date of isolation | Origin | ESBL gene(s)* | Inc/rep-type of blaSHV-12-encoding plasmid |
|---|---|---|---|---|
| 1954014 | 2009 | Livestock (pig) | bla SHV-12 | IncF |
| 35474 | 2009 | Livestock (poultry) | bla SHV-12 | IncF |
| 36289 | 2009 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 36278 | 2009 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 513768 | 2009 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 37318 | 2009 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 35078 | 2009 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 37881 | 2009 | Livestock (poultry) | blaCTX-M-1 (IncI1α/γ), blaCMY-2 (IncK), blaSHV-12 | IncX3 |
| 35659 | 2009 | Livestock (poultry) | blaCTX-M-1 (IncN), blaSHV-12 | IncI1α/γ |
| 36498 | 2010 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 36700 | 2010 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 36809 | 2010 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 66191451 | 2010 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 35568 | 2010 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 636942 | 2010 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 1026302 | 2010 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 1025601 | 2010 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 55833907 | 2011 | Livestock (pig) | bla SHV-12 | IncF |
| 55927588 | 2011 | Livestock (pig) | bla SHV-12 | IncI1α/γ |
| 55927758 | 2011 | Livestock (pig) | bla SHV-12 | IncI1α/γ |
| 37156 | 2011 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 36239 | 2011 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 55727422 | 2011 | Livestock (cattle) | bla SHV-12 | IncI1α/γ |
| 884 | 2011 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 1105 | 2011 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 1109 | 2011 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 984 | 2011 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 65268442 | 2012 | Livestock (pig) | bla SHV-12 | IncI1α/γ |
| 36788 | 2012 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 29062012-02 | 2012 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 55580200139 | 2012 | Livestock (poultry) | blaSHV-12, blaTEM-52c | IncK |
| 65094754 | 2012 | Livestock (cattle) | bla SHV-12 | IncI1α/γ |
| 36458 | 2013 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 35658 | 2013 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 1399001 | 2013 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 859 | 2014 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 219 | 2014 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 1041 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 900 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 570 | 2014 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 500 | 2014 | Livestock (poultry) | bla SHV-12 | IncB/O |
| 374 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 287 | 2014 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 73 ¥ | 2014 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 1424 | 2014 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 71 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 828 | 2014 | Livestock (poultry) | bla SHV-12 | IncX1 |
| 1003 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 11 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 240 | 2014 | Livestock (poultry) | bla SHV-12 | IncF-X1 |
| 386 ¥ | 2014 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 990 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 1341 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 1420 | 2014 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 118 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 139 | 2014 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 864 | 2014 | Livestock (poultry) | bla SHV-12 | IncX1 |
| 1206 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 876 | 2014 | Livestock (poultry) | bla SHV-12 | IncI1α/γ |
| 20 | 2014 | Livestock (poultry) | bla SHV-12 | IncX1 |
| 229 | 2014 | Livestock (poultry) | bla SHV-12 | IncX3 |
| 1096 | 2014 | Livestock (poultry) | bla SHV-12 | IncF |
| 1433 | 2014 | Livestock (pig) | bla SHV-12 | IncX3 |
| 1116 | 2014 | Livestock (cattle) | bla SHV-12 | IncI1α/γ |
| 69438407 | 2012 | Meat (poultry) | blaCTX-M-1 (IncI1α/γ), blaSHV-12 | IncX3 |
| 69606962 | 2012 | Meat (poultry) | bla SHV-12 | IncX3 |
| 76495084 | 2012 | Meat (beef) | bla SHV-12 | IncI1α/γ |
| 76495084 02 | 2012 | Meat (beef) | bla SHV-12 | IncI1α/γ |
| 69843204 | 2012 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 69210023 | 2012 | Meat (poultry) | bla SHV-12 | IncK |
| 69843409 | 2012 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 69438105 | 2012 | Meat (poultry) | bla SHV-12 | IncX3 |
| 69585604 | 2012 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 69064655 | 2012 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 69770576 | 2012 | Meat (poultry) | blaSHV-12, blaTEM-52c | IncI1α/γ |
| 69477895 | 2012 | Meat (poultry) | blaSHV-12, blaTEM-52c | IncI1α/γ |
| 69927807 | 2013 | Meat (poultry) | blaCMY-2 (IncK), blaSHV-12 | IncB/O |
| 698975250004 | 2013 | Meat (poultry) | bla SHV-12 | colE |
| 699561810004 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 699819760004 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 69986056 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 693784120004 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 693785440004 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 693562060004 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 699898610004 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 699229530004 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 698980410004 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 69960219 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 69960316 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 69960316 | 2013 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 69345093 | 2013 | Meat (pork) | bla SHV-12 | IncI1α/γ |
| 69799639 | 2013 | Meat (beef) | bla SHV-12 | IncI1α/γ |
| 699799710004 | 2013 | Meat (beef) | bla SHV-12 | IncI1α/γ |
| 694658380004 | 2013 | Meat (poultry) | blaSHV-12, blaSHV-2A, blaTEM-52c (IncI1α/γ) | IncI1α/γ |
| 699813050004 | 2013 | Meat (poultry) | blaSHV-12, blaTEM-52c (IncI1α/γ) | IncI1α/γ |
| 699900020004 | 2013 | Meat (poultry) | bla SHV-12 | IncK |
| 693503480004 | 2013 | Meat (poultry) | bla SHV-12 | IncX1 |
| 699953490004 | 2013 | Meat (poultry) | bla SHV-12 | IncX1 |
| 699081950004 | 2013 | Meat (poultry) | bla SHV-12 | IncX1 |
| 699952170004 | 2013 | Meat (poultry) | bla SHV-12 | IncX3 |
| 69960189 ¥ | 2013 | Meat (poultry) | bla SHV-12 | IncX3 |
| 79158224 | 2014 | Meat (beef) | bla SHV-12 | IncF |
| M14P0112 | 2014 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 79197637 | 2014 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 79059943 | 2014 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 79230383 | 2014 | Meat (poultry) | bla SHV-12 | IncB/O |
| 79696536 | 2014 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 79194778 | 2014 | Meat (pork) | bla SHV-12 | IncI1α/γ |
| 79195006 | 2014 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 79156655 | 2014 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 79207004 | 2014 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 79626295 | 2014 | Meat (poultry) | bla SHV-12 | IncB/O |
| 79207101 | 2014 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 79352292 | 2014 | Meat (poultry) | bla SHV-12 | IncK |
| 79771872 | 2014 | Meat (poultry) | bla SHV-12 | IncX3 |
| 79445126 | 2014 | Meat (poultry) | bla SHV-12 | IncI1α/γ |
| 79042587 | 2014 | Meat (poultry) | bla SHV-12 | IncX3 |
| 1190900169 ¥ | 2009 | Human (urine) | bla SHV-12 | IncX3 |
| 1190900881 | 2009 | Human (urine) | bla SHV-12 | IncI1α/γ |
| 1190900890 | 2009 | Human (urine) | bla SHV-12 | IncI1α/γ |
| 306 | 2014 | Human (faeces) | bla SHV-12 | IncI1α/γ |
| 1.1 | 2014 | Human (faeces) | bla SHV-12 | IncI1α/γ |
| 1.58 | 2014 | Human (faeces) | bla SHV-12 | IncI1α/γ |
| 2.12 | 2014 | Human (faeces) | bla SHV-12 | IncI1α/γ |
| 2.25 | 2014 | Human (faeces) | bla SHV-12 | IncI1α/γ |
| 2.48 | 2014 | Human (faeces) | bla SHV-12 | IncI1α/γ |
| 2.49 | 2014 | Human (faeces) | bla SHV-12 | IncI1α/γ |
| 2.52 | 2014 | Human (faeces) | bla SHV-12 | IncI1α/γ |
| 1.71 | 2014 | Human (faeces) | bla SHV-12 | IncN |
*When known the inc/rep-type of the plasmid encoding ESBL genes (excluding the blaSHV-12) is given in parenthesis.
¥In bold the 4 plasmids sequenced and functionally characterized.
Figure 1.
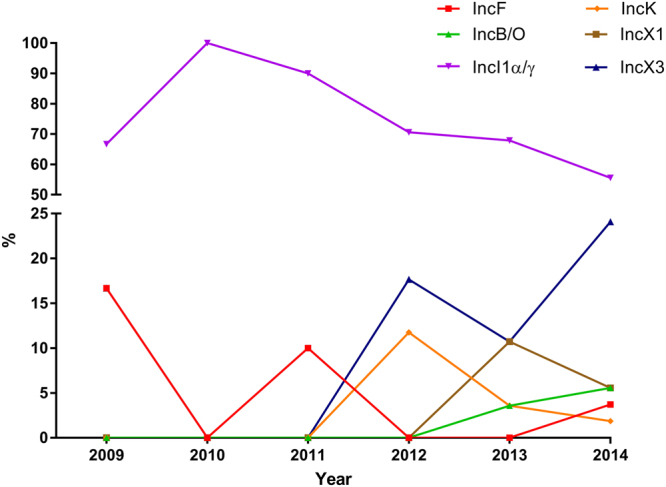
Prevalence of plasmid encoding blaSHV-12 in E. coli isolated between 2009 and 2014 in the Netherlands. Isolates were recovered from food-producing animals, retail meat and humans during national antimicrobial resistance monitoring programmes or national projects. Plasmids belonging to N, colE and F-X1 replicon types were recovered with prevalence of 0.85% and therefore were omitted from the figure.
To further study these emerging IncX3 plasmids, four of them (pEC-NRS18, pEC-393, pEC-125 and pEC-243) randomly selected from human-related E. coli ST69 and from diverse E. coli STs of animal origin (ST117, ST315 and ST410) were fully sequenced and functionally characterized in this study (Table 2).
Table 2.
IncX3 plasmids included in this study and their characteristics.
| Plasmid ID | Year | Host | Host source | Resistance gene(s) | Size (bp) | GC Content % | Open reading frames |
|---|---|---|---|---|---|---|---|
| pEC-NRS18 | 2009 | E. coli ST69/CC69 | Human UTI* | blaSHV-12, blaTEM-1, qnrS1 | 48,250 | 46.4 | 74 |
| pEC-393 | 2013 | E. coli ST410/CC23 | Turkey meat | bla SHV-12 | 43,506 | 46.8 | 65 |
| pEC-125 | 2014 | E. coli ST117 | Chicken faeces | blaSHV-12, qnrS1 | 46,338 | 46.4 | 73 |
| pEC-243 | 2014 | E. coli ST315/CC38 | Chicken faeces | blaSHV-12, qnrS1 | 46,338 | 46.4 | 73 |
*Urinary tract infection.
IncX3 plasmid backbone is highly syntenic and conserved
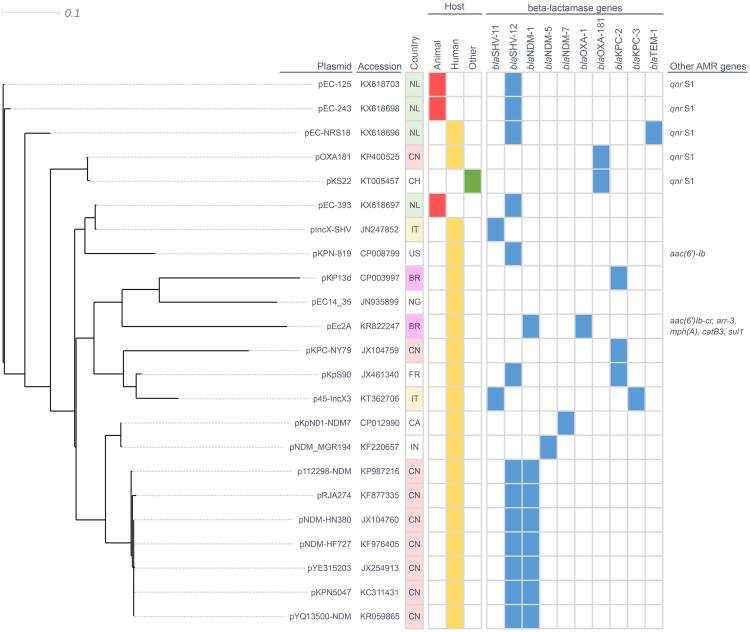
Comparison between whole sequences of the four IncX3 plasmids from this study and twenty IncX3 plasmids available in GenBank revealed a highly conserved plasmid backbone and their organization into a number of distinct clades (Fig. 2). Plasmids of animal origin pEC-125 and pEC-243 were closely clustered together (MUMi distance 0.018) and grouped with the human-derived plasmid pEC-NRS18 that showed distance from 0.085 (pEC-125) to 0.093 (pEC-243). The animal-derived plasmid pEC-393 (turkey meat) clustered with a Klebsiella pneumoniae-encoded pIncX-SHV from human source in Italy (MUMi distance 0.002). IncX3 plasmids recovered in the Netherlands clustered closely with pOXA181 (China) and pKS22 (Switzerland) encoding blaOXA-181 and qnrS1, respectively, with MUMi distances varying from 0.140 (pOXA181 with pEC-125) to 0.179 (pKS22with pEC-393).
Figure 2.
BioNJ MUMi distances phylogram of IncX3 plasmids. Plasmid sequences obtained here and those available in GenBank database were compared pair-wise and maximum unique matches converted to MUMi distances were hierarchically clustered and displayed as a phylogram using the BioNJ algorithm. GenBank accession number, antibiotic resistant gene content, country and source of isolation are indicated. NL: Netherlands, CN: China, CH: Switzerland, IT: Italy, US: United States, BR: Brazil, NG: Nigeria, FR: France, CA: Canada, IN: India and AMR: antimicrobial resistance.
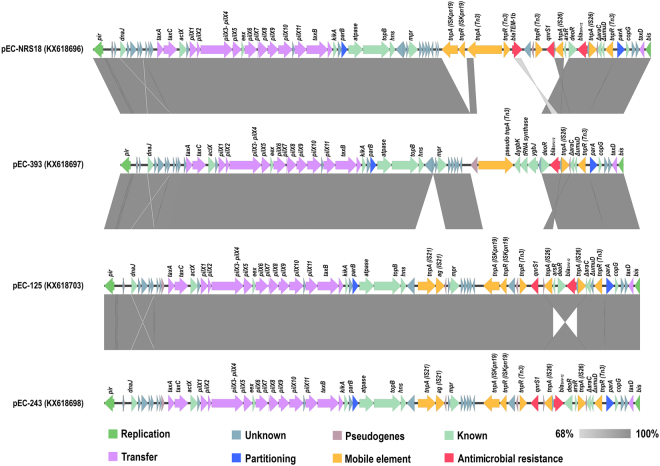
The four plasmids sequenced, assembled and fully annotated in this study had sizes varying from 43,506 (pEC-393) to 48,250 (pEC-NRS18) bp with an average GC content of 46.5% (Table 2). Similar to other IncX3 plasmids, they carried three putative origins of replication (oriV-α, oriV-β and oriV-γ), two origins of transfer (oriT-α and oriT-β), and approximately 6 iteron sequences. Nucleotide sequence analysis revealed 65 to 74 predicted open reading frames (Fig. 3). Comparative genomic of all four plasmids (Fig. 3) revealed high synteny among them, encoding genes for replication (pir: replication initiation protein and bis: replication accessory protein), partitioning (parAB), entry exclusion (eex), maintenance (topB and hns), transcriptional activation (actX), and conjugative transfer [pilX1-11 (type IV secretion system) and taxA-C]. In addition, a mosaic variable region containing resistance genes as well as intact and/or defective insertion sequences (i.e. IS21, ISKpn19, Tn3, and IS26) was identified in all four plasmids upstream of the partitioning gene parA (Fig. 3).
Figure 3.
Linear comparison in scale of IncX3 plasmids. The open reading frames identified in each sequence are represented with arrows, with the arrowhead indicating the direction of transcription. Their involvement in replication, partitioning, transfer, or antibiotic resistance, their association to mobile genetic elements, as well as other known or unknown functions and pseudogenes are colour-coded. Areas shaded in grey indicate nucleotide identity.
The variable region of pEC-NRS18 contained blaTEM-1 embedded in a Tn3 transposon, as well as genes blaSHV-12 and qnrS1 associated with the upstream presence of IS26 in the opposite and same orientation, respectively. Similarly, pEC-125 and pEC-243 contained both blaSHV-12 and qnrS1 genes, whereas pEC-393 encoded only blaSHV-12 associated to IS26. The genetic environment surrounding blaSHV-12 was characterized by two flanking copies of IS26 distributed in opposite orientation to form a composite IS26-IS26 transposon (3,633 bp); this structure was conserved in three of the four IncX3 plasmids (pEC-NRS18, pEC-125 and pEC-243; Fig. 3). BLAST analysis revealed that this composite transposon is 100% identical to previously described transposons located on plasmids of K. pneumoniae (IncFIB; GenBank accession no. CP019048.1) and Aeromonas veronii (IncA/C2; GenBank accession no. CP014775.1), as well as into the genome of Pseudomonas aeruginosa (GenBank accession no. GU592828.1). The flanking region of blaSHV-12 on pEC-393 showed a partial 2,484-bp overlap with corresponding regions of pEC-NRS18, pEC-125 and pEC-243 (Fig. 3) and encoded genes deoR, ygbJ and truncated ygbK also present on several K. pneumoniae chromosomes (GenBank accession no. CP000647, CP002910 and CP008831). This 4,783-bp region exhibits 100% identity to a fragment of a blaSHV-harboring IncR plasmid of K. pneumoniae (GenBank accession no. KF954150.1).
IncX3 plasmid transfer is temperature-dependent
Conjugation frequencies of the four IncX3 plasmids was determined and results are shown in Fig. 4. Transfer rates differed between solid matings at different temperatures. Geometric mean frequencies ranged below the detection limit (≤1 × 10−9, pEC-393) to 3.73 × 10−5 (pEC-NRS18) at 25 °C, from 6.36 × 10−6 (pEC-393) to 7.16 × 10−5 (pEC-243) at 30 °C, and from 1.33 × 10−6 (pEC-393) to 1.46 × 10−4 (pEC-NRS18) at 37 °C. The analysis showed a significant difference between conjugation frequencies at different temperatures (p = 0.027), mainly due to lower frequencies at 25 °C compared with frequencies at 30 °C. The difference in conjugation frequencies between different plasmids was nearly significant (p = 0.054). Comparisons of single plasmids at 30 °C and 37 °C indicated differences between the plasmids at 37 °C, with lower conjugation frequencies for animal-derived plasmids (pEC-125, pEC-243 and pEC-393), and higher frequencies for the human-derived plasmid pEC-NRS18. Overall, plasmid of animal origins seem to transfer better at 30 C, conversely to the human-derived plasmid (Fig. 4).
Figure 4.
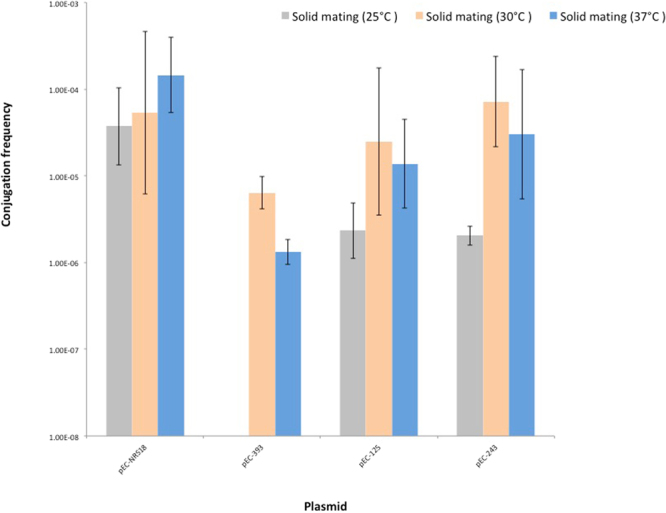
Conjugation frequencies of IncX3 plasmids. The reported values represent the average of three independent solid mating experiments (25 °C, 30 °C and 37 °C) and the error bars the standard deviation.
IncX3 plasmids exert no fitness cost to the bacterial host and are highly stable
The cost of IncX3 plasmid presence on the host cell fitness was assessed in the absence and in the presence of cefotaxime by comparing the exponential growth rate of E. coli DH10b with and without plasmid (Fig. 5). The exponential growth rates of E. coli DH10b harbouring each of the IncX3 plasmids singularly varied from 0.91 (95% CI 0.84–0.99; DH10b::pEC-125) to 1.22 (95% CI 1.13–1.32; DH10b::pEC-NRS18) in the absence of antibiotic selective pressure, and from 1.48 (95% CI 1.14–1.83; DH10b::pEC-NRS18) to 1.94 (95% CI 1.77–2.12; DH10b::pEC-243) in the presence of cefotaxime. In the absence of cefotaxime, DH10b::pEC-125 showed significantly lower (p = 0.04) and DH10b::pEC-NRS18 significantly higher (p < 0.001) exponential growth rates compared to E. coli DH10b control strain. Pairwise comparison of the relative growth rates for E. coli DH10b harbouring each of the IncX3 plasmids in the absence and presence of cefotaxime indicated significantly higher rates in the presence of selective pressure (p < 0.001) except for DH10b::pEC-NRS18.
Figure 5.
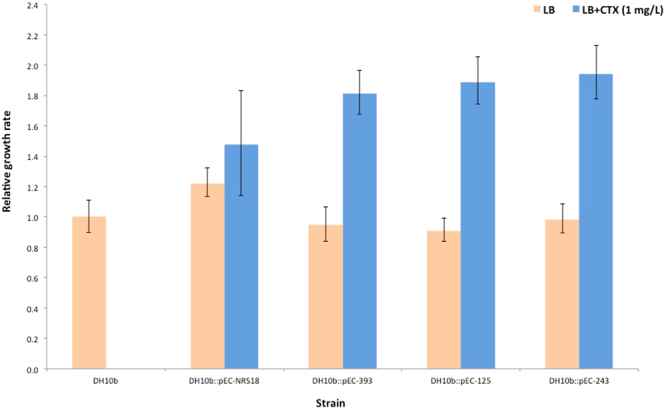
Relative exponential growth rates of IncX3-harbouring plasmid E. coli DH10b strains. All growth rates are set relative to plasmid-free E. coli DH10b. The reported values represent the average of three independent experiments and the error bars represent the 95% confidence interval for the ratio.
E. coli DH10b transformed strains were also propagated without positive antibiotic selective pressure as a measure of plasmid stability. After approximately 180 generations of growth, the percentage of plasmid-harbouring cells in each population (for all four plasmids singularly) was determined. All plasmids were found to be stably maintained in the E. coli population, ranging from 99.9% (95% CI 99.98–99.99) (pEC-NRS18) to 100% (95% CI 99.9–100) (pEC-393, pEC-125 and pEC-243) plasmid-harbouring cells per generation.
IncX3 plasmids do not contribute to bacterial pathogenicity
Annotation of the four IncX3 plasmids revealed the presence of a Type 4 Secretion System (pilX1-11) and several ORFs with unknown function that could potentially act as virulence effectors (Fig. 3). The Galleria mellonella in vivo infection model was employed to evaluate the impact of harbouring an IncX3 plasmid on bacterial pathogenicity. The LD50 value after 24 h was determined to be 107 CFU/larvae and survival curves were compared between the isogenic control E. coli DH10b strain and DH10b transformed strains harbouring each of the IncX3 plasmids (Fig. 6). All four transformed strains carrying IncX3 plasmids displayed comparable virulence to the plasmid-free control strain (mortality = 40–86%), with no significant difference in the 96 h survival curves (Fig. 6). In both control groups all larvae survived.
Figure 6.
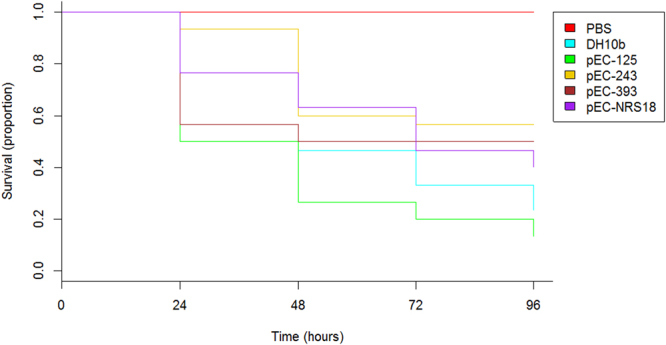
Impact of harbouring an IncX3 plasmid on E. coli DH10b strain pathogenicity. Kaplan-Meier plot of G. mellonella survival after injection with 107 CFU/larva of plasmid-free and IncX3-harbouring plasmid (pEC-NRS18, pEC-393, pEC-125 and pEC-243) E. coli DH10b strain is shown. Experiments were performed in triplicate and the plot represents the combined (additive) data from all experiments. No larval death was observed in control larvae injected with an equivalent volume of PBS.
Discussion
Our results confirmed that IncI1α/γ plasmids are the major facilitators of the blaSHV-12 diffusion in E. coli of human and animal origin and mirrored the global plasmid repertoire associated with blaSHV-1237,39. However, a gradual decrease in the prevalence of IncI1α/γ and a parallel increase in IncX3 plasmids encoding blaSHV-12 was documented mostly in animal-related commensal E. coli. Previously, the IncX3 plasmid subgroup was only incidentally associated with blaSHV-129,16–19 and/or qnrS12,9,12–15 among clinically recovered E. coli isolates, and very recently it was identified among poultry isolates in Germany40. IncX3 plasmids have been documented in other Enterobacteriaceae worldwide in association with multi-resistance, including to carbapenems18,20,22,24,26,34,36. Nevertheless, no association between IncX3 plasmids and other resistance genes (apart from blaSHV-12, blaTEM-1 and qnrS1) in Enterobacteriaceae was found in the Netherlands (data not shown).
The high degree of synteny and conservation in the backbone of IncX3 plasmids among E. coli isolates of both human and animal origin reflects the ecological success of this plasmid subgroup2. In addition to encoding genes essential for their maintenance and dissemination, IncX3 plasmids contained a variable region encoding resistance to clinically important antimicrobial agents (fluoroquinolones and/or extended spectrum cephalosporins). Our findings confirm the potential of this subgroup for accumulation of resistance genes via IS-mediated transposition, with the likely consequence of limiting effective treatment options for possible human infections9,13,15,16,18,21. As previously described41, the presence of IS26 linked to blaSHV-12 and other co-linear genes originating from the chromosome of K. pneumoniae was documented within the IncX3 variable region, confirming the hypothesis of IS26-mediated mobilization of a blaSHV ancestor gene from the chromosome of K. pneumoniae42. Although we cannot prove how blaSHV-12 was integrated into IncX3 plasmids, the documented ability of IS26 to participate in both replicative transposition and self-targeted transposition creating IS26-bounded transposons43 could have facilitated the formation of the composite blaSHV-12-encoding IS26-IS26 transposon seen in the majority of the IncX3 plasmids studied here. The presence of identical composite transposons, mostly on plasmids of diverse replicon types, indicates its mobilization ability preferentially onto plasmids rather than the chromosome, as previously suggested44. In addition to the contribution of IS26 to the mobilization on conjugative plasmids and the subsequent dissemination of blaSHV-12, it has been shown that IS26 supplies a promoter −35 box that can be coupled with a −10 box in the adjacent DNA45, possibly also contributing to the expression of this resistance gene.
IncX3 plasmids, as well as the archetypal R6K plasmid of the IncX family, have been investigated for their ability to conjugate13,15,16,18,19,46. We documented the temperature-dependent transfer of animal-derived IncX3 plasmids with frequencies higher at 30 °C, suggesting a more efficient transfer in the environment than in the animal gut. Similarly to other conjugative plasmids, IncX3 plasmids hold a gene encoding a H-NS-like protein2,47–51. Several studies demonstrated the inhibitory role of H-NS-like proteins on gene thermoregulation owing to their ability to polymerize along and bridge adjacent DNA regions at 37 °C and to derepress H-NS-regulated genes at lower temperatures47,48,52. The involvement of these proteins in the temperature-dependent conjugative transfer of IncHI1 plasmids suggests a similar role for IncX3 plasmids that can only be speculated here. The ability of IncX3 plasmids to replicate and be stably maintained in α-, β- and γ-Proteobacteria53, in combination with a higher conjugation frequency at 30 °C, underscore a potential mesophilic Proteobacteria reservoir of this plasmid subgroup.
In contrast with studies describing that plasmids impose fitness cost on their bacterial hosts54, growth kinetics obtained over 24 h showed no evidence of fitness cost on the bacterial E. coli host. It has been demonstrated that H-NS-like proteins are able to silence newly introduced foreign sequences (including plasmids), based on increased adenine and thymine (AT) content in comparison with the chromosome51,55,56. Taking into consideration the high AT content of the IncX3 plasmids, we hypothesize that the H-NS gene present on these plasmids allows them to invade bacterial hosts with a minimal impact on their fitness, ensuring the future competitiveness of the new plasmid-host combination even without the presence of antibiotic selective pressure. The significant IncX3 plasmid-mediated fitness enhancement of E. coli under antibiotic selective pressure highlights the ecological advantage and subsequent successful proliferation of these plasmids in antibiotic-rich reservoirs.
All IncX3 plasmids encoded the widespread partitioning system ParAB ensuring the correct inheritance of these plasmids to the daughter cells57. The observed high stability of IncX3 plasmids is potentially due to their high conjugation frequency and absence of a fitness burden, as well as to a low rate of segregational loss.
Our data show that IncX3 plasmids encode a type IV secretion system (T4SS, pilX1-11), typically used for the exchange of genetic material within bacteria, toxin secretion and the translocation of virulent effector proteins into eukaryotic host cells58,59. Yet, a virulence potential of IncX3-carrying E. coli DH10b was not observed, suggesting that T4SS does not play a role in the virulence of E. coli, at least in the G. mellonella model, conversely from other Gram-negative pathogens58.
In conclusion, we report the first genetic characterisation of IncX3 plasmids of animal origin, as well as the first functional analysis of human- and animal-derived plasmids of this subgroup, including their conjugation frequencies, stability, fitness cost and virulence potential. IncX3 plasmids are highly conserved, syntenic, conjugative and highly stable, while they exert no fitness cost on their bacterial host, independent of their origin. Although clonal expansion of E. coli strains could also play a role as suggested by our finding in The Netherlands of E. coli from the same clonal complex and carrying the same IncX3 plasmid (data not shown), the favourable plasmid functional features potentially contributed to their emergence amongst SHV-12-producing E. coli in the Netherlands, highlighting the epidemic potential of this plasmid subgroup.
Materials and Methods
Bacterial strains, transformants and plasmids
A total of 129 non duplicate blaSHV-12 encoding E. coli, consecutively recovered during national antimicrobial resistance monitoring programmes or national projects between 2009 and 2014 were included in the study60. Identification of the isolates was performed by MALDI-TOF Mass spectrometry (Brucker, Coventry, UK). Genes conferring an ESCR phenotype were sought by microarray analysis followed by PCR amplification and sequencing61. Plasmid location of blaSHV-12 was determined using a transformation-based approach. Briefly, plasmids encoding blaSHV-12 were extracted from the parental strain using a miniprep method and transformed into E. coli DH10b cells (Invitrogen, Van Allen Way, CA USA) by electroporation under the following conditions: 1.25 kV/cm, 200 Ω, 25 μFar, as previously described61. Transformants were selected on Luria-Bertani (LB) agar plates supplemented with cefotaxime (1 mg/L) and confirmed for the presence of blaSHV-12 gene. The presence of a single plasmid in the transformants was confirmed by S1-PFGE on the transformants, followed by Southern blot hybridization using DIG-labelled probe (DIG DNA Labeling and Detection Kit, Roche, Mannheim, Germany) targeting the blaSHV-12 gene, as previously described62. Plasmid typing was confirmed by PCR-based replicon typing (PBRT KIT, DIATHEVA, Fano, Italy) on the transformants. Host E. coli sequence type were assigned by MLST based on the allelic profiles of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA and recA)63.
Plasmid sequencing, assembly and analysis
Four genetically and epidemiologically unrelated IncX3 plasmids encoding blaSHV-12 were randomly selected for further analysis from E. coli isolates belonging to the single ST of human origin (ST69) and diverse STs of animal origin, including the predominant animal-related ST (ST117). The relevant characteristics of the selected plasmids are specified in Table 2. Plasmid DNA from transformants was isolated using the QIAfilter Plasmid Midi Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s recommendations. Deep sequencing of the plasmid genomes was performed using 300-bp paired-end sequencing libraries (Nextera TAG-mentation sequencing kits [Epicentre]) on an Illumina MiSeq sequencer. High-quality filtered reads were subsequently assembled de novo using SPAdes algorithm (SPAdes version 3.7.1) for Illumina-derived reads and then manually curated to close the gaps. Putative open reading frames (ORFs) were identified by RAST version 2.0 and manually curated when necessary64. BLASTP analyses of the putative ORFs against the NCBI non-redundant proteins (NR) database, Pfam, and Interpro scan were used to assess their putative functions by identification of structural features and motifs65,66. ResFinder (version 2.1), PlasmidFinder (version 1.3) and ISfinder were used to determine the presence of resistance genes, replicon types and insertion sequences, respectively67–69. Plasmid sequences were hierarchically clustered and displayed as a phenogram using the BioNJ algorithm, where the underlying distance matrix was calculated from the pairwise non-overlapping maximal unique matches (MUMs) using Nucmer version 3.0770,71. Relative pairwise distances were obtained by dividing the pairwise MUMs’ sum by the average genome size of the two paired genomes (MUMi genomic distance)72. BioNJ trees were generated from the MUMi distance matrix using SplitsTree473. BLAST analysis was used to assess sequence identity between the blaSHV-12-surrounding region and nucleotide sequences deposited to NCBI74.
Mating assays
Plasmid conjugation was assessed in solid mating assays at 25 °C, 30 °C and 37 °C conducted in triplicate. Chloramphenicol resistant (chlorR) E. coli MG1655::yfp was used as a recipient strain in 1:1 ratio with donor E. coli DH10b transformed strains carrying the different IncX3 plasmids, as previously described75. Overnight cultures of recipient and donor strains in mid-exponential phase were co-incubated (100 μl each) onto sterile nitrocellulose filters of 0.45 μm pore size (Schleicher and Schuell GmbH, Dassel, Germany) for 4 h at 25 °C, 30 °C and 37 °C. Transconjugants were selected on LB agar supplemented with chloramphenicol (32 mg/L) and cefotaxime (1 mg/L). Positive transconjugants were confirmed by PCR amplification for the resistance and yfp genes. Conjugation frequency was calculated as the number of transconjugants per donor cell, with the absence of transconjugants suggesting either non-conjugative plasmids or conjugation frequencies below the detection limit (≤1 × 10−9). For statistical analysis, conjugation frequencies were transformed to log10 values, the differences between the temperatures and plasmids were tested using a non-parametric Kruskal-Wallis test, and a p value < 0.05 was considered to be statistically significant. All analysis was performed using R and RStudio (version 1.0.143)76,77.
Fitness cost assays
Liquid cultures of E. coli DH10b transformed strains carrying different IncX3 plasmids were incubated overnight in 3 mL LB medium at 37 °C with 180 rpm shaking. Cultures were then diluted 100-fold into 3 mL of fresh pre-warmed LB medium with and without antibiotic (1 mg/L of cefotaxime) and incubated under the same conditions until mid-exponential phase (OD600 of ≈0.5). 200 µL of each culture were loaded in triplicate in wells of a 100-well honeycomb plate and incubated at 37 °C with shaking for 24 h. Growth rates were obtained by measuring optical density at 600 nm every 30 min by using a Bioscreen C Reader (Oy Growth Curves, Helsinki, Finland). Assays were performed in triplicate. Relative growth rates were calculated by dividing the generation time of each DH10b transformed strain by the generation time of the wild-type DH10b strain which was included in each individual assay78. Growth rates between strains were compared using the Wilcoxon rank sum test with a Bonferroni adjustment for multiple comparisons. All statistical analysis were performed in R studio (version 1.0.143)76.
Stability assays
E. coli DH10b transformants carrying different IncX3 plasmids were propagated in antibiotic-free LB medium at 37 °C with 180 rpm shaking for 10 days (∼180 generations) to determine their stability in an E. coli population. Cultures of each strain were daily diluted 1000-fold into 3 mL of fresh pre-warmed LB medium without antibiotics. On day 10, cultures were plated onto antibiotic-free LB agar and 100 randomly chosen colonies of each evolved line were replica-plated onto antibiotic-free and antibiotic-containing (1 mg/L of cefotaxime) LB agar plates. Plasmid presence was confirmed by colony PCR targeting the taxC gene of the IncX3 plasmids2. Colony growth on antibiotic-free but not on antibiotic-containing plates indicated the proportion of bacteria that lost the plasmid. Assays were performed in triplicate. The chance of E. coli DH10b keeping the plasmid was estimated for every plasmid using @Risk 6.3.1 (Palisade Corporation, Newfield, NY, USA), and the proportions of plasmid-harbouring colonies for each plasmid were compared.
Galleria mellonella survival assays
G. mellonella caterpillars in the final-instar larval stage were obtained in bulk from Livefood UK Limited (Rooks Bridge, Somerset, United Kingdom) and stored at 15 °C in the dark on wood shavings prior to use. Ten randomly chosen larvae weighing between 250 mg and 350 mg were employed for each group of an experiment. Strains included in the assay were grown overnight in LB broth and washed twice in sterile phosphate-buffered saline (PBS). The optimal bacterial inoculum was determined by injecting 10 larvae with 10 μl of bacterial suspensions containing 104 to 107 CFU/larva of organism in PBS. Bacterial inoculum concentration was determined by viable bacterial count on LB agar identifying the inoculum which killed 50% of larvae after 24 hours incubation at 37 °C (LD50). The optimal inoculum was then injected into the hemocoels of the caterpillars via a left proleg using 25-μl Hamilton syringes (Cole-Parmer, London, United Kingdom). Following injection, larvae were incubated in petri dishes lined with filter paper at 37 °C for 96 h and scored for survival by 2 independent observers daily. Larvae were considered dead when they displayed no movement in response to touch. Two control groups were used per experiment, including larvae that were inoculated with PBS to control for any lethal effects of the injection process and larvae that received no injection. All G. mellonella survival assays were performed in triplicate using different batches of larvae. Survival curves were plotted using the Kaplan-Meier method and differences in survival were calculated by the log-rank test using R studio (version 1.0.143)76.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Accession codes
The reported plasmid sequences are deposited in GenBank under the following accession numbers: KX618696 (pEC-NRS18), KX618697 (pEC-393), KX618698 (pEC-243) and KX618703 (pEC-125).
Acknowledgements
This work was supported by the Dutch Ministry of Economic Affairs through the 1Health4Food (1H4F) project under the ESBL Attribution (ESBLAT) consortium (project number: TKIAF-12067). We thank Nadine Handel and Benno ter Kuile for donation of the recipient strain E. coli MG1655::yfp and Frank Harders for technical assistance in plasmid genome sequencing.
Author Contributions
A.L. and D.M. designed the study. A.L., A.K. and J.B. acquired the data. A.L., J.v.G., A.B and D.C. performed the data analysis. A.L. prepared the first draft of the manuscript. All authors interpreted the data, read, contributed to, and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stalker DM, Helinski DR. DNA segments of the IncX plasmid R485 determining replication and incompatibility with plasmid R6K. Plasmid. 1985;14:245–254. doi: 10.1016/0147-619X(85)90008-3. [DOI] [PubMed] [Google Scholar]
- 2.Johnson TJ, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, et al. Decreased Fitness and Virulence in ST10 Escherichia coli Harboring blaNDM-5 and mcr-1 against a ST4981 Strain with blaNDM-5. Frontiers in cellular and infection microbiology. 2017;7:242. doi: 10.3389/fcimb.2017.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L, et al. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrobial agents and chemotherapy. 2013;57:269–276. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du H, et al. Genomic Characterization of Enterobacter cloacae Isolates from China That Coproduce KPC-3 and NDM-1 Carbapenemases. Antimicrobial agents and chemotherapy. 2016;60:2519–2523. doi: 10.1128/AAC.03053-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson TJ, et al. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Applied and environmental microbiology. 2007;73:1976–1983. doi: 10.1128/AEM.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kassis-Chikhani N, et al. Complete nucleotide sequence of the first KPC-2- and SHV-12-encoding IncX plasmid, pKpS90, from Klebsiella pneumoniae. Antimicrobial agents and chemotherapy. 2013;57:618–620. doi: 10.1128/AAC.01712-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du XX, et al. Genetic characteristics of blaNDM-1-positive plasmid in Citrobacter freundii isolate separated from a clinical infectious patient. Journal of medical microbiology. 2013;62:1332–1337. doi: 10.1099/jmm.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 9.Dobiasova H, Dolejska M. Prevalence and diversity of IncX plasmids carrying fluoroquinolone and beta-lactam resistance genes in Escherichia coli originating from diverse sources and geographical areas. The Journal of antimicrobial chemotherapy. 2016;71:2118–2124. doi: 10.1093/jac/dkw144. [DOI] [PubMed] [Google Scholar]
- 10.Doumith M, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. The Journal of antimicrobial chemotherapy. 2016;71:2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 11.Lo WU, et al. Highly conjugative IncX4 plasmids carrying blaCTX-M in Escherichia coli from humans and food animals. Journal of medical microbiology. 2014;63:835–840. doi: 10.1099/jmm.0.074021-0. [DOI] [PubMed] [Google Scholar]
- 12.Skalova, A. et al. Molecular Characterization of OXA-48-Like-Producing Enterobacteriaceae in the Czech Republic and Evidence for Horizontal Transfer of pOXA-48-Like Plasmids. Antimicrobial agents and chemotherapy61, 10.1128/AAC.01889-16 (2017). [DOI] [PMC free article] [PubMed]
- 13.Liu YB, et al. First Report of OXA-181-Producing Escherichia coli in China and Characterization of the Isolate Using Whole-Genome Sequencing. Antimicrobial agents and chemotherapy. 2015;59:5022–5025. doi: 10.1128/AAC.00442-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouedraogo AS, et al. First Description of IncX3 Plasmids Carrying blaOXA-181 in Escherichia coli Clinical Isolates in Burkina Faso. Antimicrobial agents and chemotherapy. 2016;60:3240–3242. doi: 10.1128/AAC.00147-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang LH, et al. Clinical isolates of uropathogenic Escherichia coli ST131 producing NDM-7 metallo-beta-lactamase in China. International journal of antimicrobial agents. 2016;48:41–45. doi: 10.1016/j.ijantimicag.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Lv J, et al. First Report of Complete Sequence of a blaNDM-13-Harboring Plasmid from an Escherichia coli ST5138 Clinical Isolate. Frontiers in cellular and infection microbiology. 2016;6:130. doi: 10.3389/fcimb.2016.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hargreaves ML, et al. Clonal Dissemination of Enterobacter cloacae Harboring blaKPC-3 in the Upper Midwestern United States. Antimicrobial agents and chemotherapy. 2015;59:7723–7734. doi: 10.1128/AAC.01291-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnevend A, et al. Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. Journal of medical microbiology. 2013;62:1044–1050. doi: 10.1099/jmm.0.059014-0. [DOI] [PubMed] [Google Scholar]
- 19.Feng Y, et al. Escherichia coli of sequence type 3835 carrying bla NDM-1, bla CTX-M-15, bla CMY-42 and bla SHV-12. Scientific reports. 2015;5:12275. doi: 10.1038/srep12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho PL, et al. Molecular Characterization of an Atypical IncX3 Plasmid pKPC-NY79 Carrying bla (KPC-2) in a Klebsiella pneumoniae. Current microbiology. 2013;67:493–498. doi: 10.1007/s00284-013-0398-2. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. O. et al. Outbreak of KPC-2-producing Enterobacteriaceae caused by clonal dissemination of Klebsiella pneumoniae ST307 carrying an IncX3-type plasmid harboring a truncated Tn4401a. Diagnostic microbiology and infectious disease, 10.1016/j.diagmicrobio.2016.12.012 (2016). [DOI] [PubMed]
- 22.Fortini D, et al. Double Copies of bla(KPC-3)::Tn4401a on an IncX3 Plasmid in Klebsiella pneumoniae Successful Clone ST512 from Italy. Antimicrobial agents and chemotherapy. 2016;60:646–649. doi: 10.1128/AAC.01886-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campos JC, et al. Characterization of Tn3000, a Transposon Responsible for bla(NDM-1) Dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrobial agents and chemotherapy. 2015;59:7387–7395. doi: 10.1128/AAC.01458-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espedido BA, Dimitrijovski B, van Hal SJ, Jensen SO. The use of whole-genome sequencing for molecular epidemiology and antimicrobial surveillance: identifying the role of IncX3 plasmids and the spread of bla(NDM-4)-like genes in the Enterobacteriaceae. J Clin Pathol. 2015;68:835–838. doi: 10.1136/jclinpath-2015-203044. [DOI] [PubMed] [Google Scholar]
- 25.Yaici L, et al. blaNDM-5-carrying IncX3 plasmid in Escherichia coli ST1284 isolated from raw milk collected in a dairy farm in Algeria. The Journal of antimicrobial chemotherapy. 2016;71:2671–2672. doi: 10.1093/jac/dkw160. [DOI] [PubMed] [Google Scholar]
- 26.Wailan AM, et al. Genomic Characteristics of NDM-Producing Enterobacteriaceae Isolates in Australia and Their bla(NDM) Genetic Contexts. Antimicrobial agents and chemotherapy. 2016;60:136–141. doi: 10.1128/AAC.01243-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishnaraju M, et al. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian journal of medical microbiology. 2015;33:30–38. doi: 10.4103/0255-0857.148373. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, et al. Widespread Dissemination of Carbapenem-Resistant Escherichia coli Sequence Type 167 Strains Harboring blaNDM-5 in Clinical Settings in China. Antimicrobial agents and chemotherapy. 2016;60:4364–4368. doi: 10.1128/AAC.00859-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch T, et al. Molecular Evolution of a Klebsiella pneumoniae ST278 Isolate Harboring blaNDM-7 and Involved in Nosocomial Transmission. The Journal of infectious diseases. 2016;214:798–806. doi: 10.1093/infdis/jiw240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moussounda M, et al. Emergence of blaNDM-7-Producing Enterobacteriaceae in Gabon, 2016. Emerging infectious diseases. 2017;23:356–358. doi: 10.3201/eid2302.161182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottig S, Hamprecht AG, Christ S, Kempf VA, Wichelhaus TA. Detection of NDM-7 in Germany, a new variant of the New Delhi metallo-beta-lactamase with increased carbapenemase activity. The Journal of antimicrobial chemotherapy. 2013;68:1737–1740. doi: 10.1093/jac/dkt088. [DOI] [PubMed] [Google Scholar]
- 32.Paul, D. et al. Occurrence of blaNDM-7 within IncX3-type plasmid of Escherichia coli from India. Journal of infection and chemotherapy: official journal of the Japan Society of Chemotherapy, 10.1016/j.jiac.2016.12.009 (2017). [DOI] [PubMed]
- 33.Pal, T. et al. Characterization of NDM-7 Carbapenemase-Producing Escherichia coli Isolates in the Arabian Peninsula. Microbial drug resistance, 10.1089/mdr.2016.0216 (2017). [DOI] [PubMed]
- 34.Liu, Z. et al. Plasmid-Mediated Novel blaNDM-17 Gene Encoding a Carbapenemase with Enhanced Activity in a ST48 Escherichia coli Strain. Antimicrobial agents and chemotherapy, 10.1128/AAC.02233-16 (2017). [DOI] [PMC free article] [PubMed]
- 35.Zurfluh, K., Poirel, L., Nordmann, P., Klumpp, J. & Stephan, R. First detection of Klebsiella variicola producing OXA-181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrob Resist In4, ARTN 3810.1186/s13756-015-0080-5 (2015). [DOI] [PMC free article] [PubMed]
- 36.Kieffer N, Nordmann P, Aires-de-Sousa M, Poirel L. High Prevalence of Carbapenemase-Producing Enterobacteriaceae among Hospitalized Children in Luanda, Angola. Antimicrobial agents and chemotherapy. 2016;60:6189–6192. doi: 10.1128/AAC.01201-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liakopoulos A, Mevius D, Ceccarelli D. A Review of SHV Extended-Spectrum beta-Lactamases: Neglected Yet Ubiquitous. Frontiers in microbiology. 2016;7:1374. doi: 10.3389/fmicb.2016.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dohmen W, et al. ESBL carriage in pig slaughterhouse workers is associated with occupational exposure. Epidemiology and infection. 2017;145:2003–2010. doi: 10.1017/S0950268817000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Accogli M, et al. IncI1 plasmids associated with the spread of CMY-2, CTX-M-1 and SHV-12 in Escherichia coli of animal and human origin. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2013;19:E238–240. doi: 10.1111/1469-0691.12128. [DOI] [PubMed] [Google Scholar]
- 40.Alonso CA, et al. Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. The Journal of antimicrobial chemotherapy. 2017;72:1589–1596. doi: 10.1093/jac/dkx024. [DOI] [PubMed] [Google Scholar]
- 41.Haeggman S, Lofdahl S, Burman LG. An allelic variant of the chromosomal gene for class A beta-lactamase K2, specific for Klebsiella pneumoniae, is the ancestor of SHV-1. Antimicrobial agents and chemotherapy. 1997;41:2705–2709. doi: 10.1128/aac.41.12.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ford PJ, Avison MB. Evolutionary mapping of the SHV beta-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosome. The Journal of antimicrobial chemotherapy. 2004;54:69–75. doi: 10.1093/jac/dkh251. [DOI] [PubMed] [Google Scholar]
- 43.Harmer, C. J. & Hall, R. M. IS26-Mediated Formation of Transposons Carrying Antibiotic Resistance Genes. mSphere1, 10.1128/mSphere.00038-16 (2016). [DOI] [PMC free article] [PubMed]
- 44.He S, et al. Insertion Sequence IS26 Reorganizes Plasmids in Clinically Isolated Multidrug-Resistant Bacteria by Replicative Transposition. mBio. 2015;6:e00762. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee KY, Hopkins JD, Syvanen M. Direct Involvement of Is26 in an Antibiotic-Resistance Operon. Journal of bacteriology. 1990;172:3229–3236. doi: 10.1128/jb.172.6.3229-3236.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nunez B, Avila P, de la Cruz F. Genes involved in conjugative DNA processing of plasmid R6K. Molecular microbiology. 1997;24:1157–1168. doi: 10.1046/j.1365-2958.1997.4111778.x. [DOI] [PubMed] [Google Scholar]
- 47.Dorman CJ. H-NS: A universal regulator for a dynamic genome. Nat Rev Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 48.Forns N, Banos RC, Balsalobre C, Juarez A, Madrid C. Temperature-dependent conjugative transfer of R27: Role of chromosome- and plasmid-encoded Hha and H-NS proteins. Journal of bacteriology. 2005;187:3950–3959. doi: 10.1128/JB.187.12.3950-3959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.More MI, Pohlman RF, Winans SC. Genes encoding the pKM101 conjugal mating pore are negatively regulated by the plasmid-encoded KorA and KorB proteins. Journal of bacteriology. 1996;178:4392–4399. doi: 10.1128/jb.178.15.4392-4399.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tietze E, Tschape H. Temperature-dependent expression of conjugation pili by IncM plasmid-harbouring bacteria: identification of plasmid-encoded regulatory functions. Journal of basic microbiology. 1994;34:105–116. doi: 10.1002/jobm.3620340206. [DOI] [PubMed] [Google Scholar]
- 51.Doyle M, et al. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- 52.White-Ziegler CA, Davis TR. Genome-Wide Identification of H-NS-Controlled, Temperature-Regulated Genes in Escherichia coli K-12. Journal of bacteriology. 2009;191:1106–1110. doi: 10.1128/JB.00599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grudniak AM, Kraczkiewicz-Dowjat A, Wolska KI, Wild J. Conjugal transfer of plasmid R6K gamma ori minireplicon derivatives from Escherichia coli to various genera of pathogenic bacteria. Current microbiology. 2007;55:549–553. doi: 10.1007/s00284-007-9032-5. [DOI] [PubMed] [Google Scholar]
- 54.Baltrus DA. Exploring the costs of horizontal gene transfer. Trends in ecology & evolution. 2013;28:489–495. doi: 10.1016/j.tree.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 55.Lucchini S, et al. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS pathogens. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Navarre WW, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 57.Volante A, Alonso JC. Molecular Anatomy of ParA-ParA and ParA-ParB Interactions during Plasmid Partitioning. The Journal of biological chemistry. 2015;290:18782–18795. doi: 10.1074/jbc.M115.649632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 60.Central Veterinary Institute of Wageningen University and Research Centre. Monitoring of Antimicrobial Resistance and Antibiotic Usage in Animals in the Netherlands in 2013 (MARAN 2014). (Central Veterinary Institute of Wageningen University and Research Centre, Lelystad, The Netherlands, 2014).
- 61.Liakopoulos A, et al. Extended-Spectrum Cephalosporin-Resistant Salmonella enterica serovar Heidelberg Strains, the Netherlands. Emerging infectious diseases. 2016;22:1257–1261. doi: 10.3201/eid2207.151377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Analytical biochemistry. 1995;226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 63.Wirth T, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Molecular microbiology. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finn RD, et al. Pfam: the protein families database. Nucleic acids research. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hunter S, et al. InterPro: the integrative protein signature database. Nucleic acids research. 2009;37:D211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemoth. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrobial agents and chemotherapy. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Molecular biology and evolution. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 71.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome biology. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deloger M, El Karoui M, Petit MA. A Genomic Distance Based on MUM Indicates Discontinuity between Most Bacterial Species and Genera. Journal of bacteriology. 2009;191:91–99. doi: 10.1128/JB.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular biology and evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 74.Johnson M, et al. NCBI BLAST: a better web interface. Nucleic acids research. 2008;36:W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 76.RStudio: Integrated Development for R (RStudio, Inc, Boston (MA), USA, 2016).
- 77.R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2008).
- 78.Knopp M, Andersson DI. Amelioration of the Fitness Costs of Antibiotic Resistance Due To Reduced Outer Membrane Permeability by Upregulation of Alternative Porins. Molecular biology and evolution. 2015;32:3252–3263. doi: 10.1093/molbev/msv195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).