Abstract
Brassinosteroids (BRs) play an essential role in plant growth, and BRI1-EMS suppressor 1 (BES1)/brassinazole-resistant 1 (BZR1) family transcription factors integrate a variety of plant signaling pathways. Despite the fact that BRs inhibit nodulation in leguminous plants, how BRs modulate rhizobia-host interactions and nodule morphogenesis is unknown. Here, we show that GmBEHL1, a soybean homolog of Arabidopsis BES1/BZR1 homolog 1 (BEH1), is an interacting partner of Nodule Number Control 1, a transcriptional repressor that mediates soybean nodulation. GmBEHL1 was highly expressed at the basal parts of emerging nodules, and its expression gradually expanded during nodule maturation. The overexpression and downregulation of GmBEHL1 inhibited and enhanced the number of nodules, respectively, in soybean. Intriguingly, alterations in GmBEHL1 expression repressed the expression of genes in the BR biosynthesis pathway, including homologs of Arabidopsis Constitutive Photomorphogenesis and Dwarf and Dwarf 4. We also detected an interaction between GmBEHL1 and GmBIN2, a putative BR-insensitive 2 (BIN2) homolog, in soybean. Moreover, BR treatment reduced the number, but increased the size, of soybean nodules. Our results reveal GmBEHL1 to be a potent gene that integrates BR signaling with nodulation signaling pathways to regulate symbiotic nodulation.
Introduction
Nitrogen is an essential element for plant growth and development. To obtain sufficient nitrogen, non-leguminous plants have evolved developmental plasticity of the root system that enables them to remodel their root architecture (i.e., lateral root formation) in response to fluctuating levels of nitrogen in the growth environment1. Given their high demand for nitrogen, leguminous plants have developed an additional form of root developmental plasticity (the formation of symbiotic root nodules) that allows plants to adapt to nitrogen-deficient conditions. Root nodules are formed from the cortical cells of a primary root according to a unique genetic program that allows a rhizobial infection to take place and de novo nitrogen-fixing organ formation to occur2,3. Thus, root nodules are capable of fixing atmospheric nitrogen to meet the demands of leguminous plants. Interestingly, indeterminate nodules in the roots of leguminous plants such as Medicago truncatula have a similar structure to lateral roots that includes functionally and developmentally different zones, including an apical meristematic zone, which allow the indeterminate nodules to continue to grow3. By contrast, the determinate nodules in roots of Lotus japonicus and Glycine max (soybean) are spherical, lateral organs with no apparent developmental zones3.
In recent decades, many studies have focused on the early stages of legume-rhizobial symbioses. Nodule development is trigged by a rhizobial infection, and both the initiation of rhizobial entry into roots and the onset of nodulation are dependent on the perception of nodulation factors (NFs) by LysM receptors (e.g., NF Perception [NFP] in M. truncatula, NF Receptors 1 and 5 [NFR1/5] in L. japonicus, and NFR1/5α in soybean), which activate a signaling cascade (the NF signaling pathway) that triggers nodule formation4–9. In M. truncatula, the key event following NFP binding is the accumulation of calcium within and around the nucleus of infected root hair cells mediated by Does Not Make Infections 1 and 2 (DMI2 and DMI1, respectively) and cyclic nucleotide-gated channels localized to the nuclear envelope10–12. Calcium oscillations are then decoded via a calmodulin 1 and calcium and calmodulin-dependent kinase-mediated network that is activated by the phosphorylation of a transcription factor (CYCLOPS) and its interacting partner (DMI3) in L. japonicus and M. truncatula, respectively13,14. CYCLOPS subsequently trans-activates Nodule Inception (NIN) and ERF Required for Nodulation1 (ERN1). NIN is a central regulator of nodulation, which targets early nodulation genes (ENODs) such as ENOD11 in the root epidermis and Cytokinin Receptor 1 (CRE1) in the cortex of M. truncatula roots to repress or activate their expression15. Several transcription factors belonging to the GRAS (e.g., Nodulation Signaling Pathway 1/2 [NSP1/2]); Nuclear factor Y (e.g., NF-YA1); Ethylene Response Factor ERF/APETALA2 (ERF/AP2); and NAM, ATAF1/2, and CUC2 domain protein families are also involved in nodulation16–19. Recently, it was shown that microRNAs, noncoding RNAs that are 20–22 nucleotides in length, are involved in regulating nodulation through the repression of their target genes20,21. In soybean, we identified miR172c as a key positive regulator of nodulation that promotes the cleavage of mRNAs encoding its target gene Nodule Number Control 1 (NNC1), which directly suppresses the transcription of ENOD40 and nodule development22. In addition, nodule number is controlled by the autoregulation of nodulation (AON) signaling pathway, which is initiated by the activation of CLE-RS1 and CLE-RS2 (CLE-Root Signal1/2) in rhizobia-infected roots during primordia formation; signaling is in turn perceived by specific receptors, including SUNN(Super Numerary Nodules)in M. truncatula, HAR1(Hypernodulation Aberrant Root Formation 1) in L. japonicus, and GmNARK (Nodule Autoregulation Recepter Kinase) in soybean23–25. Despite extensive progress, many questions concerning cell priming for nodule initiation, primordia formation, and nodule organogenesis remain unanswered.
It is well known that phytohormones are involved in nodule formation and development in legumes. Among them, auxin and cytokinin (CK) are the major hormones regulating root nodule development26,27; however, other phytohormones, including abscisic acid (ABA), strigolactones, gibberellic acid, ethylene, and jasmonic acid, and their interplay also participate in root nodule development28–32. Brassinosteroids (BRs) are a group of steroid hormones in plants that play crucial roles in shoot elongation, plant architecture, photomorphogenesis, and seed germination33. Because they are highly mobile, BRs can effectively regulate cell elongation and lateral organ development33. In Arabidopsis, many key components, including BR receptors, co-receptors, and their direct downstream components, have been identified and the core BR signaling pathway has been defined. BRASSINAZOLE-RESISTANT 1 (BZR1) and its homologs (BES1/BZR1-like genes [BEHs]) are key regulators of BR signaling that repress or activate the transcription of BR-responsive genes to mediate plant growth and development34,35. BZR1 and BEHs can directly repress the expression of genes involved in BR biosynthesis, thereby suppressing BR signaling through feedback regulation36. In legumes, the role of BRs in nodulation has been explored. In pea, the BR biosynthesis mutants lk (affecting 5α-reductase) and lkb (affecting sterol C-24 reductase), as well as the BR receptor mutant lka, all exhibit a reduced number of nodules37. Meanwhile, a genetic analysis of double mutants (lk and an AON-related mutant such as nark) revealed that BRs regulate nodule number in an AON-independent manner in pea plants38. In M. truncatula, a loss-of-function mutant of the BR receptor MtBRI1 (mtbri1) displayed a reduced number of nodules and a range of defects in symbiotic nitrogen fixation39. Interestingly, an earlier study indicated an opposing role for BRs in soybean nodulation; the exogenous application of brassinazole, an effective inhibitor of BR biosynthesis, resulted in an increase in nodule number in the soybean cultivar Enrei40. Despite limited data, there is no doubt that BRs mediate symbiotic nodulation in leguminous plants. Still, further molecular evidence is needed to address how BRs are involved in nodulation in different legumes.
In this study, we identified a soybean homolog of Arabidopsis BES1/BZR1 homolog (BEH1) as an NNC1-interacting protein and named it GmBEHL1. The knockdown and overexpression of GmBEHL1 resulted in increased and decreased numbers of nodules, respectively, in composite transgenic roots. Similar to BEH1, GmBEHL1 is localized to both the cytoplasm and nucleus, and it can bind directly to BR-responsive Responsive Elements and interact with GmBIN2, a soybean homolog of Arabidopsis BR-insensitive 2 (BIN2). Furthermore, we uncovered the diverse roles of BRs in determining the number and size of the nodules in soybean plants. Our results suggest that GmBEHL1 functions as a co-repressor to negatively regulate soybean nodulation; moreover, they reveal for the first time direct crosstalk between the NF and BR signaling pathways in soybean.
Results
GmBEHL1 is an NNC1-interacting protein and homolog of Arabidopsis BES1/BZR1
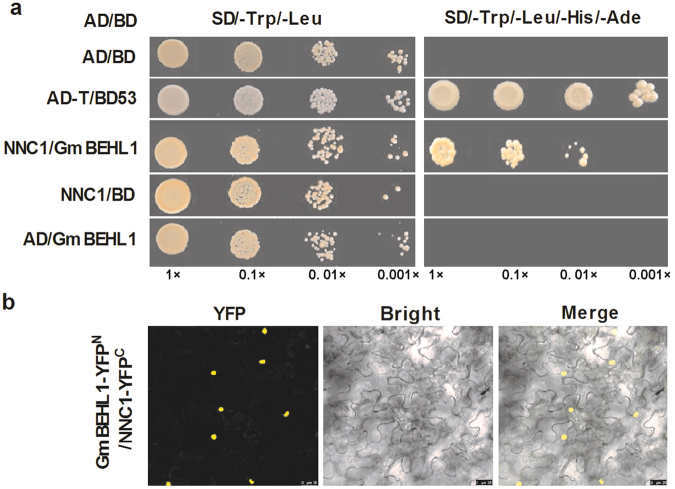
Because NNC1 is a key transcriptional repressor of ENOD40-1 that modulates soybean nodulation, we sought to identify its functional partners in order to uncover the regulatory mechanisms it mediates. To do this, we performed a yeast two-hybrid (Y2H) screen to isolate NNC1-interacting proteins. Interestingly, we found that one of the NNC1-interacting proteins (Glyma.01G178000) was an Arabidopsis BEH1-like protein41; therefore, it was named GmBEHL1. Based on this, we speculated that GmBEHL1 acts as a node in the NF and BR signaling pathways. To test this hypothesis, we first confirmed the NNC1-GmBEHL1 interaction using in vitro and in vivo protein-protein interaction assays. Our Y2H assay results showed that GmBEHL1 interacted directly with NNC1 (Fig. 1a). We next performed a bimolecular fluorescence complementation (BiFC) assay by co-expressing GmBEHL1-YFPN and NNC1-YFPC in Nicotiana benthamiana leaves. As shown in Fig. 1b, GmBEHL1 interacted strongly with NNC1 in the nucleus of each transformed cell, consistent with their transcriptional properties. These results indicate that GmBEHL1 may form a complex with NNC1 in rhizobia-inoculated soybean roots to mediate nodulation.
Figure 1.
NNC1 interacts directly with GmBEHL1. (a) Results of Y2H assay to detect interactions between NNC1 and GmBEHL1. Yeast cells co-transformed with the constructs pGADT7/pGBKT7-GmBEHL1, pGADT7-NNC1/pGBKT7-GmBEHL1, and pGADT7-NNC1/pGBKT7 were selected and grown on selective media lacking Leu, and Trp (SD/−2); surviving cells were subsequently transferred to selective media lacking Ade, His, Leu, and Trp (SD/−4) to test for protein-protein interactions. (b) Results of a BiFC assay to detect the interaction of NNC1 with GmBEHL1. GmBEHL1 and NNC1 were fused to the N-terminus of YFP (nYFP) and C-terminus of YFP (cYFP), respectively. Bars = 25 μm.
GmBEHL1 is a negative regulator of soybean nodulation
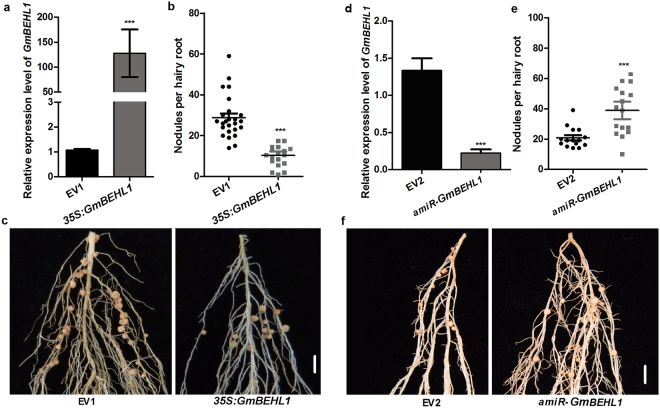
Our immediate question was whether GmBEHL1 plays a regulatory role in soybean nodulation. To address this, we performed a systemic phenotypic analysis of GmBEHL1 overexpression or knockout/knockdown roots using the hairy root transformation system. First, we made a construct containing CaMV35S promoter (35S):GmBEHL1 and obtained transformed roots overexpressing GmBEHL1 (GmBEHL1OX) for the evaluation of nodulation; this was confirmed by checking the Bar gene (Fig. S1) and qRT-PCR analysis (Fig. 2a). The effects of GmBEHL1 overexpression on the early and late stages of nodulation were evaluated at 6 and 28 days after inoculation (DAI) using Bradyrhizobium (B.) japonicum USDA110. The number of root hairs showing deformation was markedly decreased in GmBEHL1OX hairy roots at 6 DAI (Fig. S2). Transformed hairy roots overexpressing GmBEHL1 were also used to determine nodule numbers at 28 DAI. Intriguingly, the GmBEHL1OX roots produced significantly fewer nodules than the control roots did (Fig. 2b and c). The average number of nodules per vector control root was 27.3, whereas the average number of nodules per GmBEHL1OX root was only 10.9. Thus, the total number of nodules per GmBEHL1OX root was reduced by approximately 60.1%.
Figure 2.
Changes in GmBEHL1 expression affect nodulation. (a–c) GmBEHL1 overexpression suppresses nodulation. (a) qRT-PCR analysis of transgenic hairy roots expressing 35S:GmBEHL1. The relative expression levels of GmBEHL1 in hairy roots transformed with empty vector 1 (EV1) or 35S:GmBEHL1. The expression levels were normalized against the geometric mean of soybean GmELF1b for GmBEHL1. Student’s t-test was performed (***p < 0.001, n = 25). (b) Quantitative analysis of the nodule number per hairy root expressing EV1 and 35S:GmBEHL1 (n = 25). All values are the means ± SDs from more than three independent experiments. Asterisks represent statistically significant differences (Student’s t-test, ***p < 0.001). (c) Nodules of individual hairy roots expressing EV1 and 35S:GmBEHL1 at 28 DAI. Bar = 3 mm. (d–f) Knocking down GmBEHL1 promotes nodulation. (d) Expression analysis of transgenic hairy roots expressing empty vector 2 (EV2) and amiR-GmBEHL1. The expression levels of GmBEHL1 were normalized against the geometric mean of soybean GmELF1b. Student’s t-test was performed (n = 25, ***p < 0.001). (e) Quantitative data showing the nodules per hairy root for EV2 and the vector harboring amiR-GmBEHL1 (n = 25). The nodule numbers are the means ± SDs from more than three independent experiments. Asterisks represent statistically significant differences (Student’s t-test; ***p < 0.001). (f) Nodules of individual hairy roots expressing EV2 and amiR-GmBEHL1 at 28 DAI. Bar = 3 mm.
To investigate whether endogenous GmBEHL1 is required for nodulation, we created an amiR-GmBEHL1 construct to knock down the GmBEHL1 gene (Fig. 2d) and evaluated the effect of reduced GmBEHL1 expression on nodulation (Fig. 2e and f). As shown in Fig. 2e and f, amiR-GmBEHL1 transgenic roots with reduced expression of GmBEHL1 produced significantly more nodules compared with vector control roots. The average number of nodules per vector control root was only 21.2, whereas the average number of nodules per GmBEHL1-RNAi transgenic root was 39.1. Thus, the total number of nodules per GmBEHL1-RNAi transgenic root was increased by approximately 84.4%. Together, these data suggest that GmBEHL1 negatively regulates nodulation in soybean.
Changes in GmBEHL1 expression affect marker genes in the NF pathway
Given that altering the expression of GmBEHL1 dramatically affected the nodule number in soybean, we questioned whether GmBEHL1 regulates soybean nodulation through the NF and AON signaling pathways. To this end, we examined the expression pattern of a number of nodulation and AON marker genes during nodulation using roots in which GmBEHL1 was overexpressed or knocked down. The marker genes included ENOD40-1 and functional orthologs of LjNIN (Glyma.04G000600 [GmNINa]), LjNSP1 (Glyma.07G039400 [GmNSP1]), LjNSP2 (Glyma.04G251900 [GmNSP2]), and miR172c in the NF signaling pathway, and GmRIC1 in the AON signaling pathway. As shown in Fig. 3, the expression of ENOD40-1, GmNINa, GmNSP1, GmNSP2, and pre-miR172c in GmBEHL1OX roots was significantly reduced compared with that in empty vector control roots at 2 DAI, while GmRIC1 was upregulated (Fig. 3a). In contrast, the transcript levels of ENOD40-1, GmNIN, and pre-miR172c in amiR-GmBEHL1 hairy roots were markedly increased, although NSP1, NSP2, and GmRIC1 expression was not dramatically affected (Fig. 3b). Our results suggest that GmBEHL1 acts upstream of these symbiosis-related genes in controlling rhizobial infection and nodule development, and that GmBEHL1 participates in nodule number regulation via the AON signaling pathway.
Figure 3.
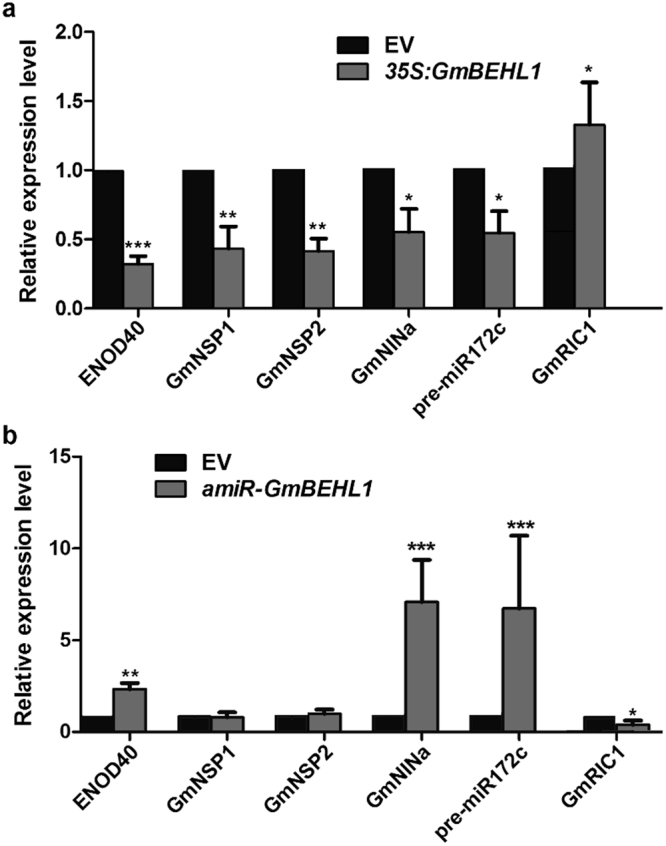
Alterations in GmBEHL1 expression affect the transcript levels of nodulation-related genes. (a) qRT-PCR analysis of ENOD40, GmNSP1, GmNSP2, GmNIN, pre-miR172c, and GmRIC1 in roots transformed with EV1 and 35S:GmBEHL1 at 2 DAI (n = 6). (b) qRT-PCR analysis of ENOD40, GmNSP1, GmNSP2, GmNIN, pre-miR172c, and GmRIC1 in roots transformed with EV2 and amiR-GmBEHL1 at 2 DAI (n = 6). We set all of the transcript profiles of the GmENOD40, GmNSP1, GmNSP2 and GmNINa in 2 DAI EV hairy roots as “1”. The transcript amounts in each sample were normalized to those of ELF1b. The expression levels shown are the means ± SDs from three replicates. Asterisks represent statistically significant differences compared to the empty control (Student’s t-test; *p < 0.05, **p < 0.01, and ***p < 0.001).
GmBEHL1 is expressed in multiple organs and is dynamically expressed during nodulation
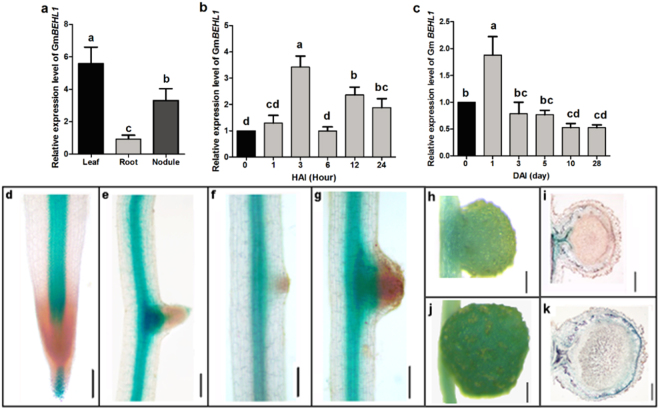
To validate the expression pattern of GmBEHL1, we performed a qRT-PCR analysis of samples collected from leaves, roots, and root nodules at 28 DAI. As expected, GmBEHL1 was expressed in all of the tested organs, though the transcript level of the gene was highest in leaves, intermediate in nodules, and lowest in roots (Fig. 4a). Next, we measured the expression of GmBEHL1 in roots at the early or late stages of nodulation. Within 24 h after inoculation (HAI), GmBEHL1 was rapidly upregulated and reached its peak at 3 HAI in the roots; further, GmBEHL1 expression was restored to its original level before another peak (Fig. 4b). Interestingly, the GmBEHL1 expression level started to decline at 3 DAI and dropped continously, exhibiting its lowest level when the nodules reached maturity (Fig. 4c). The dynamic pattern of GmBEHL1 expression during nodulation suggests diverse roles for the gene during nodulation in soybean.
Figure 4.
The expression pattern of GmBEHL1. (a–c) qRT-PCR analysis of GmBEHL1. (a) Seven-day-old seedlings were inoculated with B. japonicum strain USDA110; roots, leaves, and nodules harvested at 28 DAI were used for gene expression analyses (n = 15). (b) Inoculated roots were harvested at 0, 1, 3, 6, 12, and 24 HAI and the expression of GmBEHL1 at the early stage of nodulation was analyzed. (c) The GmBEHL1 expression pattern during nodulation was analyzed using root samples harvested at 0, 1, 3, 5, and 10 DAI (n = 15). GmELF1b was used as an endogenous control for gene expression. The expression levels are shown as the means ± SDs from three replicates. Means with different letters are significantly different (p < 0.05; Tukey’s test). (d–i) Histochemical localization of GUS activity in the transgenic hairy roots and at different stages of nodulation. (d and e) Expression pattern of GmBEHL1pro:GUS in primary (d) and lateral roots (e). (f–i) GmBEHL1pro:GUS activity at different stages of soybean nodulation. (f and g) GUS activity during nodule initiation (f) and primordia formation (g). (h,i) GUS staining of GmBEHL1pro:GUS in young nodule at 14 DAI (h) and in paraffin section of young nodule (i); (j,k) GUS staining of mature nodule at 28 DAI (j) and paraffin section of the stained mature nodule (k). Bars in (d–k) = 200 μm.
To further validate the transcriptional actitivty of the GmBEHL1 promoter, we made a construct harboring GmBEHL1pro:GUS and generated transformed hairy roots expressing GmBEHL1pro:GUS for a GUS assay. In uninfected hairy roots, GmBEHL1 was expressed at high levels in the root cap and mature regions; in sharp contrast, there was no visible expression of GmBEHL1 in the apical meristems of the primary and lateral roots (Fig. 4d and e). The expression pattern of GmBEHL1 in hairy roots inoculated with rhizobia was not significantly different from that in uninfected roots. However, increased expression of GmBEHL1 was observed in nodule primordia; moreover, when the nodules emerged, strong GmBEHL1 expression was observed only in the basal parts of the emerging and developing nodules (Fig. 4f–i). Interestingly, GUS staining of young and mature nodules showed gradual expansion of GmBEHL1 expression from the basal parts to the top of each nodule during nodule development. In addition, GmBEHL1 was expressed throughout the tissues of fully developed nodules, with the highest level of expression observed in the vascular bundles of the nodules (Fig. 4j and k). Our results confirm that GmBEHL1 participates in various processes during nodulation and root development.
GmBEHL1 is a nucleocytoplasmic protein with DNA-binding activity
Since GmBEHL1 is annotated as a BEH1-related protein, we conducted a phylogenetic analysis to verify the relationship between GmBEHL1 and its homolog in Arabidopsis. Our results confirmed that GmBEHL1 is the closest relative of BEH1 (Fig. S3a). GmBEHL1 shares high levels of amino acid sequence identity and structural similarities with Arabidopsis BEH1 (Fig. S3b). Both proteins have an N-terminal BES1_N domain, which is typical of BES1/BZR1 family proteins, and other domains, including a nuclear localization signal, P domain, and PEST domain36.
To assess the localization of GmBEHL1 in plant cells, we produced a construct harboring the 35S:GmBEHL1-GFP expression cassette and expressed the GmBEHL1-GFP fusion protein in N. benthamiana leaves. Confocal laser scanning microscopy revealed that GmBEHL1-GFP was localized in both the cytoplasm and nucleus (Fig. S3c), consistent with previous findings on the subcellular localization of Arabidopsis BZR1, implying that GmBEHL1 is a nucleocytoplasmic shuttling protein, like Arabidopsis BZR1 family proteins42.
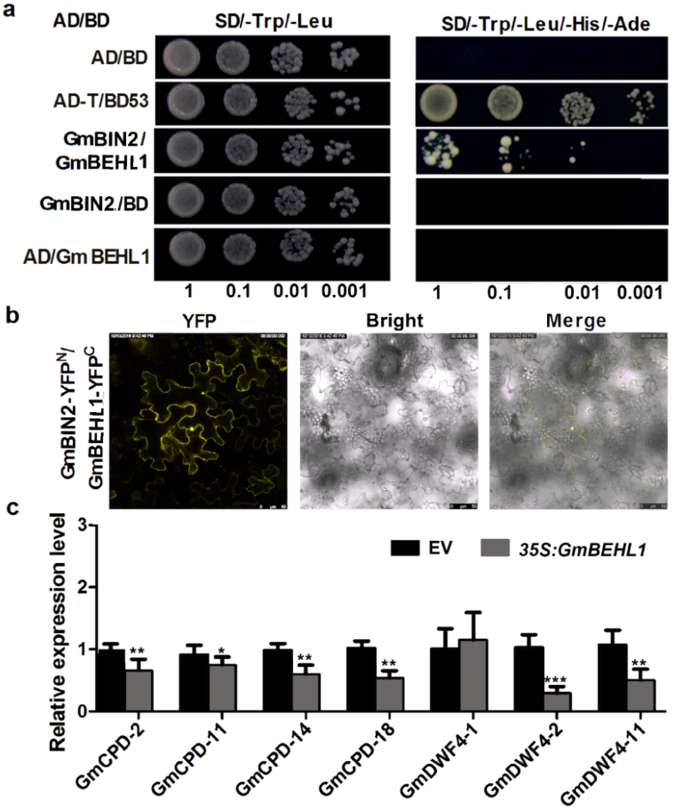
In Arabidopsis, the protein stability and subcellular localization of BZR1 are regulated by a BIN2-mediated interaction and phosphorylation43,44. To test whether GmBEHL1 exhibits these same features, we cloned the GmBIN2 (Glyma.13g228100) gene, which showed the highest level of protein sequence identity to Arabidopsis BIN2 (Fig. S4), and analyzed the interaction between GmBEHL1 and GmBIN2 by Y2H and BiFC approaches. As shown in Fig. 5a and b, GmBEHL1 exhibited a strong interaction with GmBIN2 in yeast cells and transformed N. benthamiana leaf cells, confirming that the subcellular localization of GmBEHL1 is regulated by the same mechanism as in Arabidopsis.
Figure 5.
GmBEHL1 encodes a functional ortholog of Arabidopsis BEH1. (a and b) GmBEHL1 interacts directly with GmBIN2. (a) Results of a Y2H assay to detect GmBEHL1 and GmBIN2L interactions. Yeast cells co-transformed with pGADT7/pGBKT7-GmBEHL1, pGADT7-GmBIN2/pGBKT7-GmBEHL1, and pGADT7-GmBIN2/pGBKT7 were grown on selective media lacking Leu and Trp (SD/−2) to check for transformation. The cells were subsequently grown on selective media lacking Ade, His, Leu, and Trp (SD/−4) to detect protein-protein interactions. (b) Results of a BiFC assay to detect the interaction of GmBEHL1 with GmBIN2. GmBIN2 and GmBEHL1 were fused to the N-terminus of YFP (nYFP) and C-terminus of YFP (cYFP), respectively. Bars = 25 μm. (c) qRT-PCR analysis of putative BR biosynthetic genes (GmCPDs and GmDWF4s) in roots transformed with EV1 and 35S:GmBEHL1 at 2 DAI (n = 6). The transcript amounts in each sample were normalized to those of ELF1b. The expression levels shown are the means ± SDs from three replicates. Asterisks represent statistically significant differences compared to empty vector (Student’s t-test; *p < 0.05, **p < 0.01, and ***p < 0.001).
Previous work showed that BZR1/BEH1 is a transcriptional repressor that contains a DNA-binding domain and which binds directly to the promoters of feedback-regulated BR biosynthetic genes36. To test whether GmBEHL1 has the same effect on BR biosynthetic genes in soybean, we assessed the expression patterns of the putative soybean orthologs of Arabidopsis Constitutive Photomorphogenesis and Dwarf (CPD) and Dwarf 4 (DWF4) (referred to as GmCPDs and GmDWF4s, respectively; Fig. S5) in GmBEHL1OX transgenic hairy roots. Our qRT-PCR results show that GmBEHL1 overexpression caused a significant reduction in the expression of most of the GmCPDs and GmDWF4s tested compared with vector control roots (Fig. 5c). Thus, GmBEHL1 may also repress BR biosynthetic genes via a negative transcriptional feedback loop.
Previous studies have shown that the N-terminal sequence of BEH1 can bind to a BRRE (5′-CGTG[T/C]G-3′) in the promoters of its downstream target genes to regulate their expression36. To test whether GmBEHL1 has DNA-binding activity through the same domain, we made a construct to express the N-terminal sequence of GmBEHL1. The purified, truncated version of GmBEHL1 was used in an electrophoretic mobility shift assay (EMSA) to analyze the binding activity of the peptide to a BRRE-containing probe. As shown in Fig. S6, the N-terminus of GmBEHL1 bound directly to the probe, confirming that (like BES1 and its homologs) GmBEHL1 was able to bind the cis-regulatory element through its N-terminal domain. Together, these data suggest that GmBEHL1 is a functional ortholog of BEH1.
Exogenous BRs reduced the nodule number but increased the nodule size in soybean
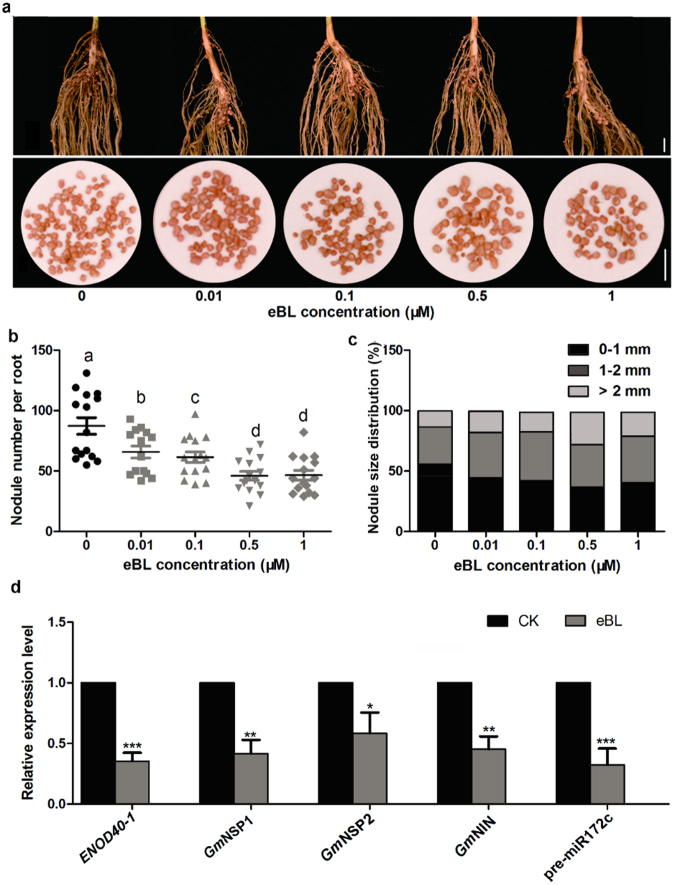
To assess the effects of BRs on soybean nodulation, we first examined the BR sensitivity of wild-type plants to 2, 4-epibrassinolide (eBL). Five-day-old plants were soaked in BD medium containing different concentrations of eBL (0, 0.01, 0.1, 0.5, and 1 μM) for 4 days, and then the hypocotyl lengths of the seedlings were measured. The hypocotyls of the young seedlings were sensitive to eBL; hypocotyl growth was significantly increased at 0.5 μM eBL (Fig. S7a and b). Since the epicotyls of the young seedlings were less sensitive to eBL, 9-day-old plants were treated with various concentrations of eBL. Elongated epicotyls were observed at eBL concentrations exceeding 0.1 μM (Fig. S7c and d). Since both the epicotyls and hypocotyls of young soybean seedlings showed the strongest response to eBL at a concentration of 0.5 μM (Fig. S7a–d) regardless of seedling age, 0.5 μM eBL was used in our subsequent rhizobial inoculation experiments.
Next, we simultaneously treated 9-day-old soybean plants with various concentrations of eBL and B. japonicum strain USDA110 for 3 days and then transplanted them to pots for further growth and nodulation. At 2 weeks after eBL treatment, a substantially reduced number of nodules was observed compared with eBL-untreated plants (Fig. 6a and b). Interestingly, the eBL-treated plants exhibited fewer and larger nodules as the eBL concentration increased, suggesting that BRs have inhibitory and promoting effects on nodule initiation and development, respectively (Fig. 6c). To explore the regulatory roles of BRs in soybean nodulation further, we analyzed the expression levels of several marker genes belonging to the NF signaling pathway in eBL-treated plants. As shown in Fig. 6d, the rhizobia-induced upregulation of ENOD40-1, GmNSP1, GmNSP2, GmNINa, and pre-miR172c (NF signaling pathway) was markedly repressed in plants treated with eBL compared with control plants. These results suggest that BRs interact antagonistically with the NF signaling pathway to regulate nodule formation in soybean.
Figure 6.
Effects of exogenous eBL on nodulation and the expression of nodulation-related genes. (a–c) Effects of exogenous eBL on soybean nodulation. Nine-day-old soybean seedlings were simultaneously treated with the indicated doses of eBL and B. japonicum USDA110 for 4 days; nodule number and size were then evaluated at 14 DAI. (a) The root system and nodulation phenotypes of the soybean seedlings (bars = 5 mm). (b) The average nodule number per treated plant. (c) A quantitative analysis of nodules of different sizes. (d) Results of the qRT-PCR analysis of ENOD40-1, GmNSP1, GmNSP2, GmNIN, and pre-miR172c expression in roots treated with 0.5 μM eBL (n = 12). The transcript amounts in each sample were normalized to those of GmELF1b. The expression levels shown are the means ± SDs from three replicates. Asterisks represent statistically significant differences compared to the control (Student’s t-test; *p < 0.05, **p < 0.01, and ***p < 0.001).
Discussion
Nodulation is a complex process involving two tightly coupled steps: rhizobial infection and nodule organogenesis. Successful establishment of a symbiosis between rhizobia and plants is precisely controlled by endogenous cues and environmental conditions. Although the molecules and mechanisms that participate in these processes are unclear, there is increasing evidence that phytohormones integrate relatively independent but closely related biological processes during legume nodulation. To date, it has been shown that most phytohormones mediate nodulation. It is conceivable that multiple phytohormones modulate nodule development antagonistically or synergistically in legumes. In recent decades, it has been reported that auxin, CK, ABA, and ethylene play crucial roles in legume nodulation, but it is unknown how the rest of the phytohormones (e.g., BRs) act during nodulation and how they interact to ensure successful nodulation. In this study, we found that BRs exert opposite effects on nodule number and size in soybean, and we identified GmBEHL1 as an NNC1-interacting protein that regulates soybean nodulation.
BES1/BRZ1 family proteins are central transcription factors in the BR signaling pathway that regulate plant responses to BRs by targeting many genes related to plant growth and stress tolerance41,45. It has been shown that BZR1 and BES1 (BZR2) function redundantly with their homologs BEH1-4 in the Arabidopsis BR signaling pathway42. Recently, an analysis of an Arabidopsis BZR1-like gene in soybean (GmBZL2) revealed that GmBZLs are highly conserved with Arabidopsis BZR1 in the BR signaling pathway46. Here, we provide further evidence that GmBEHL1 is an ortholog of Arabidopsis BEH1. GmBEHL1 shares a high level of amino acid sequence identity with Arabidopsis BEH1 and contains the typical BES1_domain of BES1/BZR1 family proteins. Further, GmBEHL1 is localized to both the nucleus and cytoplasm, implying that it can shuttle from the cytoplasm to the nuclues in response to BRs. Importantly, GmBEHL1 can bind directly to BRRE-containing DNA fragments and it is able to interact with GmBIN2, a homolog of Arabidopsis BIN2, a GSK3-like kinase. These data suggest that GmBEHL1 is a key regulator of the BR signaling pathway in soybean. Determining whether BRs induce GmBEHL1 dephosphorylation, and whether the dephosphorylated protein is localized to the nucleus, will provide convincing evidence for the role of GmBEHL1 in mediating BR signaling.
Importantly, our results show that GmBEHL1 mediates soybean nodulation. Firstly, GmBEHL1 interacts with NNC1. We previously demonstrated that NNC1 is the target of miR172c and that it negatively modulates soybean nodulation by directly repressing ENOD40 transcription22. The GmBEHL1-NNC1 interaction implies that GmBEHL1 participates in the regulation of nodulation as a co-repressor of NNC1. Secondly, GmBEHL1 exhibits a unique expression pattern during nodule primordia formation, nodule development, and nodule maturation, indicating multiple roles for the gene in symbiotic nodulation. Thirdly, our genetic data support the notion that GmBEHL1 negatively regulates nodulation through the classical NF signaling pathway because the alteration of GmBEHL1 affected the expression of several marker genes in the nodulation pathway. Finally, GmBEHL1 may affect the functionality of nitrogen-fixing nodules because GmBEHL1 was expressed in the nitrogen fixation zone of functioning nodules.
In Arabidopsis, BZR1 interacts with other transcription factors to co-repress target gene expression47. Since both NNC1 and GmBEHL1 are negative regulators of nodulation, it is possible that NNC1 and GmBEHL1 regulate soybean nodulation by acting as co-repressors of target genes. NNC1 is an AP2 transcription factor family member that represses ENOD40 transcription22. However, the 2-kb promoters of ENOD40 genes do not contain typical cis-regulatory BRREs (data not shown). Because both AP2 and BES1/BZR1 family transcription factors have many target genes in plant genomes, NNC1 and GmBEHL1 likely co-repress the transcription of other genes to mediate nodule organogenesis and nitrogen fixation in mature nodules. The fact that GmBEHL1 expression affects many marker genes, including GmNSPs and GmNINa, suggests that GmBEHL1 exerts its regulatory effect at multiple levels of soybean nodulation, beginning at the early stages of infection and nodule organogenesis. Thus, we also do not exclude the possibility that GmBEHL1 complexes with other transcriptional regulators to excert it functions during nodulation. The application of ChIP-seq technology will facilitate the identification of genes targeted by NNC1 and GmBEHL1 and help elucidate the novel molecular mechanism underlying the NNC1/GmBEHL1-mediated regulation of nodulation in soybean. Since BES1/BZR1 family proteins have redundant roles in various biological processes, it is conceivable that these proteins dynamically and coordinately regulate several processes involved in soybean nodulation.
Based on our data, it appears that NNC1 and GmBEHL1 mediate crosstalk between the nodulation and BR signaling pathways to orchestrate nodule organogenesis in soybean roots. BRs are important regulators of plant growth and development33; however, the roles of BRs in soybean nodulation remain unclear. Previous studies showed that BR treatment did not affect nodulation in the soybean cultivar Enrei40. In this study, we treated the soybean reference cultivar Williams 82 with various concentrations of eBL. Unexpectedly, exogenous treatment with eBL reduced the nodule number but enlarged the nodules in the plants. Our data show that BRs lower the nodule number in soybean by antagonistically regulating the NF signaling pathway (BR treatment repressed the expression of all positive regulators of the NF signaling pathway). However, we still do not know how BRs cause nodule enlargement. Given the role of BRs in cell division and proliferation, it is possible that BR treatment enhances cell cycle progression. Elucidation of the mechanism underlying the control of nodule size by BRs will further our understanding of how nodule organ size is maintained in legumes. We speculate that the different responses of cultivars Williams 82 and Enrei to BRs are largely due to the BR treatment protocols used; however, we cannot exclude the possible impact of their genetic backgrounds.
In summary, our data demonstrate that the BR signaling pathway plays diverse roles in soybean nodule organogenesis and nodule size regulation. We identified GmBEHL1 as a potential factor that mediates crosstalk between the BR and NF signaling pathways, possibly through an interaction with NNC1. To the best of our knowledge, this is the first study to establish a direct link between the nodulation pathway and BR signaling pathway in soybean. Our findings provide novel insight into the regulation of soybean nodulation by BRs and will facilitate the molecular breeding of new soybean varieties with improved symbiotic nitrogen fixation efficiency.
Methods
Plant and rhizobia growth, hairy root transformation, and B. japonicum inoculation
Soybean [G. max (L.) Merrill cv. Williams 82] was used to clone and analyze GmBEHL1. Soybean seedlings were cultured under 16 h/8 h light/dark conditions in a growth room at 25–26 °C and inoculated with B. japonicum strain USDA110 for nodulation pheonotype analyses as described previously22. Briefly, each young soybean seedling was inoculated with 30 ml of bacteria suspended in distilled water. For RNA extraction, plant materials were rinsed briefly in phosphate-buffered saline (pH 7.5) to remove vermiculite. All harvested marterials were then immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction. For soybean hairy root transformation, healthy and uniform soybean seeds were sterilized with chlorine gas for 12–14 h. The sterilized seeds were then germinated in B5 medium for 4 days under 16 h/8 h light/dark conditions in a growth chamber at 25–26 °C. Germinating seedlings were used for hairy root transformation as described previously with Agrobacterium rhizogenes strain K59948. For all nodulation assays, transgenic composite plants or soybean seedlings were transplanted to pots (8 × 8 cm) containing vermiculite and irrigated with a nitrogen-deficient solution as described elsewhere22. The plants were grown for 1 week (16 h of light at 25 °C and 50% relative humidity) to allow rooting and acclimation to the enviroment. The plants were then inoculated with a suspension of B. japonicum strain USDA110 (OD600 = 0.08). Nodule phenotypes, including nodule number and nodule size, were evaluated at 28 DAI.
Y2H assay
The full-length coding sequences of GmBEHL1 and NNC1 were amplified using the listed primers (Supplemental Table 1). Gateway PCR products were cloned into the vectors pGBKT7 and pGADT7 by BP and LR reactions. Y2H assays were done according to the Matchmaker GAL4 Two-Hybrid System (Clontech, Mountain View, CA). Yeast transformants were exhaustively selected on SD/-Ade/-His/-Leu/-Trp (SD/−4) medium. The constructs used to validate protein-protein interactions were cotransformed into Saccharomyces cerevisiae strain AH109. Suspended, transformed yeast (5 μl) were spread onto plates containing SD/-Ade/-His/-Leu/-Trp medium, and protein-protein interactions were judged based on the growth of the yeast after 2–3 days of incubation at 30 °C.
BiFC assays
The coding sequences of GmBEHL1 and NNC1 were cloned into the N-terminus and C-terminus of YFP through the Gateway reaction using the pDONOR vector system (Invitrogen, Carlsbad, CA), respectively. The primers used are listed in Supplemental Table 1. The resulting constructs were transformed into Agrobacterium strain GV3101 for transient expression of the proteins in N. benthamiana leaf cells22. The N. benthamiana plants were cultured for at least 36 h; YFP fluorescence was observed using a Leica Microsystems confocal laser scanning microscope (Wetzlar, Germany).
DNA extraction from and PCR-based analysis of the transgenic roots
DNA from the hairy roots of transgenic composite plants or soybean seedlings was extracted as described previously49 and used for detection of the Bar gene by PCR using the primers listed in Supplemental Table 1.
RNA extraction and quantitative PCR analysis
Total RNAs were extracted from transgenic hairy roots or soybean seedlings using TRIpure Reagent (Aidlab Biotechnologies Ltd., Beijing, China). The RNA samples were then treated with gDNA Wiper Mix (Vazyme Biotech Co., Ltd., Nanjing, China) to remove contaminating genomic DNA. cDNA strands were synthesized from the RNAs using a FastQuant RT Kit (Vazyme Biotech Co., Ltd.). qRT-PCR was done using SuperReal PreMix Plus (Vazyme Biotech Co., Ltd.) with gene-specific primers (Table S1). GmELF1b was used as an internal control.
Vector construction
For the GmBEHL1 promoter:GUS reporter construct, the putative promoter region (2,000 bp) of GmBEHL1 was amplified from cv. Williams 82 genomic DNA and cloned into a T-vector by the BP reaction for sequencing. Positive plasmids (T-vector containing the GmBEHL1 promoter sequence) were used to generate the construct pCAMBIA1391-GmBEHL1pro:GUS through the LR reaction. For the 35S:GmBEHL1 construct and the constructs for the Y2H (BD-GmBEHL1) and BiFC (GmBEHL1-YFPN) assays, the coding DNA sequence of GmBEHL1 was amplified and cloned into pDORNOR207 by the BP reaction for sequencing, and positive plasmids (pDORNOR207 with the GmBEHL1 coding DNA sequence) were used to generate the constructs by the LR reaction. The primers used for plasmid construction are listed in Table S1.
Histochemical analysis of GmBEHL1 transcription
Composite transgenic plants expressing GmBEHL1pro:GUS were generated through A. rhizogenes-mediated hairy root transformation. The transformed hairy roots of the composite seedlings were stained with X-Gluc to test for β-glucuronidase activity before and after inoculation with B. japonicum strain USDA110 at the specified time points.
EMSAs
EMSAs were performed as described previously22 using a Light Shift Chemiluminescent EMSA Kit (Pierce, Rockford, IL) according to the manufacturer’s protocol. Briefly, MBP-tagged nGmBEHL1 (amino acids 10–91) was expressed in Escherichia coli BL21 cells. The probe-binding activity of the protein was analyzed using oligonucleotides labeled with biotin at the 5′ end (Invitrogen). As competition, 200-fold unlabeled probe was added to the reactions.
Brassinolide treatment
To examine the response of soybean to BR treatment, 5-day-old seedlings germinated and grown in vermiculite were harvested for root treatment with BD media containing different concentrations (0, 0.01, 0.1, 0.5, and 1 μM) of eBL (Realtimes Beijing Technology Co., Ltd., Beijing, China). The lengths of the hypocotyls were measured at 4 days after treatment. To confirm the BR response of Williams 82, 9-day-old seedlings were treated with eBL and the epicotyl lengths were measured as described above. For the nodulation assay, 9-day-old seedlings treated simultaneously with eBL and a rhizobia inoculum for 4 days were transferred to vermiculite for further growth, and the number of nodules was evaluated 2 weeks after transplanting. For the expression analysis of marker genes in the NF signaling pathway, 5-day-old seedlings germinated in 50-ml centrifuge tubes containing vermiculite were watered with 0.5 μM eBL and the roots were collected for RNA extraction and qRT-PCR analysis.
Statistical analysis
Data analysis was done using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA). The means and standard deviations (SDs) of the data were calculated. Student’s t-test and an analysis of variance were applied to generate p-values. Student-Newman-Kuels tests were conducted when statistically significant differences existed.
Electronic supplementary material
Acknowledgements
The authors thank Xingke Zhang for providing technical assistance. This research was supported by the National Key Research and Development Program of China (2016YFA0500503), the National Natural Science Foundation of China (31730066 and 31230050), the Ministry of Agriculture of the People’s Republic of China (2014ZX0800929B), and the Huazhong Agricultural University Scientific and Technological Self-innovation Foundation (2015RC014). The authors declare no conflict of interest.
Author Contributions
L.W. and X.L. conceived the study and designed the experiments; L.W. and Q.Y. performed the experiments; L.W., Q.Y. and X.L. analyzed the data; and L.W. and X.L. wrote the manuscript with input from all of the authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Qiqi Yan and Lixiang Wang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25910-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.De Smet I. Lateral root initiation: one step at a time. New Phytol. 2012;193(4):867–873. doi: 10.1111/j.1469-8137.2011.03996.x. [DOI] [PubMed] [Google Scholar]
- 2.Oldroyd GE, Robatzek S. The broad spectrum of plant associations with other organisms. Curr. Opin. Plant Biol. 2011;14(4):347–350. doi: 10.1016/j.pbi.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Oldroyd GD, Downie JM. Coordinating nodule morphogenesis with rhizobial infection in legumes. Ann. Rev. Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 4.Madsen EB, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 5.Radutoiu S, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425(6958):585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 6.Arrighi JF, et al. The Medicago truncatula LysM motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142(1):265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Indrasumunar A, et al. Inactivation of duplicated Nod Factor Receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.) Plant Cell Physiol. 2010;51(2):201–214. doi: 10.1093/pcp/pcp178. [DOI] [PubMed] [Google Scholar]
- 8.Indrasumunar A, et al. Nodulation Factor Receptor kinase 1 alpha controls nodule organ number in soybean (Glycine max L. Merr) Plant J. 2011;65(1):39–50. doi: 10.1111/j.1365-313X.2010.04398.x. [DOI] [PubMed] [Google Scholar]
- 9.Kelly S, Radutoiu S, Stougaard J. Legume LysM receptors mediate symbiotic and pathogenic signalling. Curr. Opin. Plant Biol. 2017;39:152–158. doi: 10.1016/j.pbi.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Stracke S, et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417(6892):959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- 11.Ané JM, et al. Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science. 2004;303(5662):1364–1367. doi: 10.1126/science.1092986. [DOI] [PubMed] [Google Scholar]
- 12.Charpentier M, et al. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science. 2016;352(6289):1102–1105. doi: 10.1126/science.aae0109. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, et al. Recruitment of novel calcium-binding proteins for root nodule symbiosis in Medicago truncatula. Plant Physiol. 2006;141(1):167–177. doi: 10.1104/pp.106.076711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh S, Katzer K, Lambert J, Cerri M, Parniske M. Cyclops, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe. 2014;15(2):139–152. doi: 10.1016/j.chom.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez-Rizzo S, Crespi M, Frugier F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell. 2006;18(10):2680–2693. doi: 10.1105/tpc.106.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smit P, et al. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308(5729):1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- 17.Murakami Y, et al. Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 2006;13(6):255–265. doi: 10.1093/dnares/dsl017. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch S, et al. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell. 2009;21(2):545–557. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerri MR, et al. Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 2012;160(4):2155–2172. doi: 10.1104/pp.112.203190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauressergues D, et al. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 2012;72(3):512–522. doi: 10.1111/j.1365-313X.2012.05099.x. [DOI] [PubMed] [Google Scholar]
- 21.Lelandais-Brière C, Moreau J, Hartmann C, Crespi M. Noncoding RNAs, emerging regulators in root endosymbiosis. MPMI. 2016;29(3):170–180. doi: 10.1094/MPMI-10-15-0240-FI. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell. 2014;26(12):4782–4801. doi: 10.1105/tpc.114.131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim C-W, Lee Y-W, Hwang C-H. Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant Cell Physiol. 2011;52(9):1613–1627. doi: 10.1093/pcp/pcr091. [DOI] [PubMed] [Google Scholar]
- 24.Reid DE, Ferguson BJ, Gresshoff PM. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. MPMI. 2011;24(5):606–618. doi: 10.1094/MPMI-09-10-0207. [DOI] [PubMed] [Google Scholar]
- 25.Krusell L, et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature. 2002;420(6914):422–426. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- 26.Overvoorde P, Fukaki H, Beeckman T. Auxin control of root development. CSH Perspect. Biol. 2010;2(6):a001537. doi: 10.1101/cshperspect.a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saini S, Sharma I, Kaur N, Pati PK. Auxin: a master regulator in plant root development. Plant Cell Rep. 2013;32(6):741–757. doi: 10.1007/s00299-013-1430-5. [DOI] [PubMed] [Google Scholar]
- 28.Su Y-H, Liu Y-B, Zhang X-S. Auxin-cytokinin interaction regulates meristem development. Mol. Plant. 2011;4(4):616–625. doi: 10.1093/mp/ssr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng X, Ruyter-Spira C, Bouwmeester H. The interaction between strigolactones and other plant hormones in the regulation of plant development. Front. Plant Sci. 2013;4:199. doi: 10.3389/fpls.2013.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rehman NU, Ali M, Ahmad MZ, Liang G, Zhao J. Strigolactones promote rhizobia interaction and increase nodulation in soybean (Glycine max) Microb. Pathogen. 2018;114:420–430. doi: 10.1016/j.micpath.2017.11.049. [DOI] [PubMed] [Google Scholar]
- 31.Bensmihen S. Hormonal control of lateral root and nodule development in legumes. Plants. 2015;4(3):523–547. doi: 10.3390/plants4030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferguson BJ, Mathesius U. Phytohormone regulation of legume-rhizobia interactions. J. Chem. Ecol. 2014;40(7):770–790. doi: 10.1007/s10886-014-0472-7. [DOI] [PubMed] [Google Scholar]
- 33.Chaiwanon J, Wang W, Zhu J-Y, Oh E, Wang Z-Y. Information integration and communication in plant growth regulation. Cell. 2016;164(6):1257–1268. doi: 10.1016/j.cell.2016.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 2010;19(5):765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65(4):634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 36.He J-X, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307(5715):1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferguson BJ, Ross JJ, Reid JB. Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 2005;138(4):2396–2405. doi: 10.1104/pp.105.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foo E, Ferguson BJ, Reid JB. The potential roles of strigolactones and brassinosteroids in the autoregulation of nodulation pathway. Ann. Bot. 2014;113(6):1037–1045. doi: 10.1093/aob/mcu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng X, et al. Functional characterization of brassinosteroid receptor mtbri1 in Medicago truncatula. Sci. Rep. 2017;7(1):9327. doi: 10.1038/s41598-017-09297-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terakado J, et al. Systemic effect of a brassinosteroid on root nodule formation in soybean as revealed by the application of brassinolide and brassinazole. Soil Sci. Plant Nutr. 2005;51(3):389–395. doi: 10.1111/j.1747-0765.2005.tb00044.x. [DOI] [Google Scholar]
- 41.Vert G, Chory J. Downstream nuclear events in brassinosteroid signaling. Nature. 2006;441(7089):96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 42.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120(2):249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 43.Ryu H, et al. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19(9):2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109(2):181–191. doi: 10.1016/S0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 45.Oh E, Zhu J-Y, Wang Z-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012;14(8):802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. Functional characterization of GmBZL2 (AtBZR1 like gene) reveals the conserved BR signaling regulation in Glycine max. Sci. Rep. 2016;6:31134. doi: 10.1038/srep31134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, et al. Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell. 2017;29(6):1425–1439. doi: 10.1105/tpc.17.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kereszt A, et al. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007;2(4):948–952. doi: 10.1038/nprot.2007.141. [DOI] [PubMed] [Google Scholar]
- 49.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.