Abstract
Bacteria commonly live in dense and genetically diverse communities associated with surfaces. In these communities, competition for resources and space is intense, and yet we understand little of how this affects the spread of antibiotic-resistant strains. Here, we study interactions between antibiotic-resistant and susceptible strains using in vitro competition experiments in the opportunistic pathogen Pseudomonas aeruginosa and in silico simulations. Selection for intracellular resistance to streptomycin is very strong in colonies, such that resistance is favoured at very low antibiotic doses. In contrast, selection for extracellular resistance to carbenicillin is weak in colonies, and high doses of antibiotic are required to select for resistance. Manipulating the density and spatial structure of colonies reveals that this difference is partly explained by the fact that the local degradation of carbenicillin by β-lactamase-secreting cells protects neighbouring sensitive cells from carbenicillin. In addition, we discover a second unexpected effect: the inducible elongation of cells in response to carbenicillin allows sensitive cells to better compete for the rapidly growing colony edge. These combined effects mean that antibiotic treatment can select against antibiotic-resistant strains, raising the possibility of treatment regimes that suppress sensitive strains while limiting the rise of antibiotic resistance. We argue that the detailed study of bacterial interactions will be fundamental to understanding and overcoming antibiotic resistance.
Introduction
Antibiotic resistance is a major concern that is threatening our ability to treat the most common of bacterial infections [1]. As well as attempting to find new drugs, we need to better understand the processes that promote, or inhibit, the spread of antibiotic-resistant strains [2]. There is a large literature on the evolution of antibiotic resistance that has primarily focused on understanding how bacteria respond to antibiotics in liquid culture [3–5]. While tractable, these conditions lie in stark contrast to the way that bacteria often grow. Both in the environment and in infections, bacteria commonly grow in dense and genetically diverse communities, where competition for space and resources is intense [6–8].
Growth of bacteria in these communities, often known as “biofilms” when on a surface [9], is well known to increase the phenotypic ability of bacteria to resist antibiotics [2, 10]. However, these observations are based on single strains and, to understand the rise and fall of resistance, we need to understand how genetically antibiotic-resistant strains compete with sensitive strains under these conditions. Growth in dense and diverse communities generally means that each cell can much more strongly influence the growth and survival of other cells [11, 12]. This effect is known to promote the evolution of competitive phenotypes, including toxin secretion [13], smothering polymers [14] and adhesion [15]. However, growth in dense communities can also lead to spatiogenetic structure and the evolution of cooperative phenotypes, such as enzymes that digest complex molecules outside the cell [7, 16].
The key insight from these studies is that the details of how cells are arranged and how they affect each other are critical to understanding natural selection in dense colonies. How might colony growth impact selection for antibiotic-resistant strains? If the mechanism of resistance only benefits the cell that carries it, intense competition in colonies is likely to lead to strong selection for resistance. By contrast, if the resistance of one cell protects another, antibiotic resistance can become a cooperative trait. In this case, resistant cells may protect sensitive neighbours from antibiotics, weakening the strength of selection for resistance. Recent work shows that it can be difficult to predict whether a particular mechanism of resistance will have competitive or cooperative effects [17]. Here, we use the term cooperation as it is widely used in social evolution theory, which is a phenotype that provides a benefit to another individual and that evolved, at least in part, because of this benefit [18–20]. Cooperation appears particularly likely for resistance to β-lactams [21, 22], which often arises via the secretion of β-lactamase enzymes that break down antibiotics both in the periplasm and extracellularly [23, 24].
Here, we develop a simple experimental system to understand how competition and cooperation influence the spread of antibiotic-resistant strains in bacterial colonies. Specifically, we directly compete antibiotic-resistant and -sensitive strains of the pathogen P. aeruginosa against each other in dense colonies, that are exposed to variable concentrations of antibiotics, that are broken down by the resistant strain, either intracellularly or extracellularly. The resistant strain carries a clinical, non-conjugative, plasmid (Rms149) [25] that confers resistance to an aminoglycoside (streptomycin), which is adenylated in the cytoplasm, and a β-lactam (carbenicillin), which is thought to be hydrolysed both in the periplasm and extracellularly [23, 24]. As we will show, this allows us to directly compare a purely competitive resistance mechanism, which protects only the cells that carry it, with one that functions cooperatively. This reveals that how cells compete, whether they cooperate, and how they are arranged are all fundamental to the rise, or fall, of resistant strains in bacterial colonies. Crucially, this approach allows us to identity antibiotic dose regimes that inhibit the growth of bacterial colonies without selecting for resistance in vitro.
Materials and methods
In our experiments, we compete susceptible and resistant strains of P. aeruginosa on LB media, either in the form of broth culture or on agar plates. Resistant strains are isogenic with susceptible strains with the exception that they have been transformed with a clinical, multidrug-resistant plasmid, Rms149 [25]. Streptomycin resistance is conferred by the aadA5 gene, which encodes an aminoglycoside adenylyltransferase. The blaA gene provides carbenicillin resistance, in the form of a constitutive class A β-lactamase. Cell divisions are calculated using the following equation:
Relative susceptible fitness is calculated using the following equation [26]:
Further details of the experimental methods and the models used in this paper are provided in the Supplementary Materials and Methods.
Results
Low antibiotic doses select for resistant strains in liquid cultures
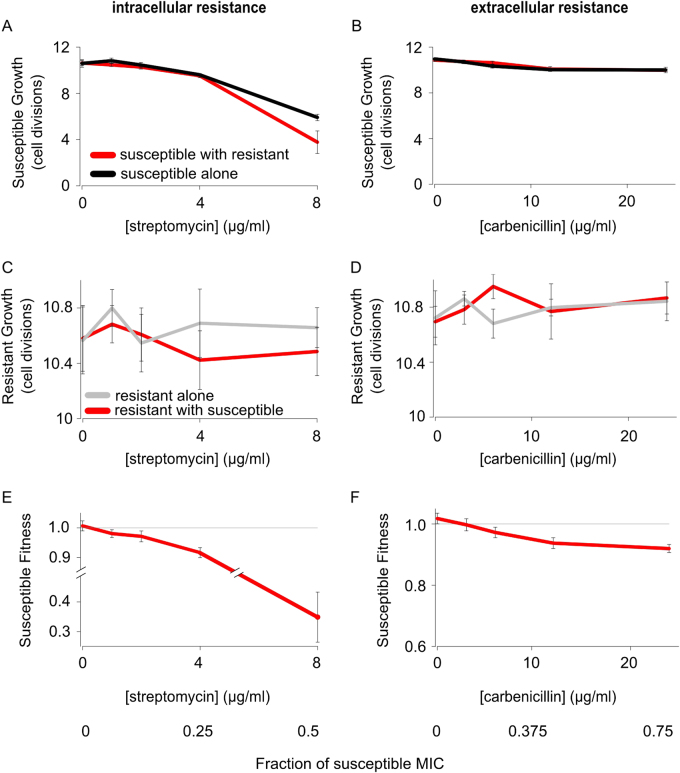
In order to establish our system, we first competed plasmid-bearing and plasmid-free strains against each other in well-mixed broth (Fig. 1 and Supplementary Fig. S1). Plasmid carriage reduced fitness by 3% (s.e. = 0.38%, supplementary experimental procedures) in the absence of antibiotics, demonstrating a cost to resistance (t21 = 9.7, P < 0.001). As expected, supplementing culture medium with either an aminoglycoside (streptomycin) or β-lactam (carbenicillin) caused the fitness of the antibiotic-sensitive strain to rapidly decrease (Fig. 1e, f), because antibiotics suppress the population growth rate of the sensitive strain (Fig. 1a, b), but not the resistant strain (Fig. 1c, d). One way of quantifying this effect is to estimate the minimal concentration of antibiotic required to select for the resistant strain (minimum selective concentration, MSC) as a fraction of the minimal inhibitory concentration (MIC) required to fully suppress the sensitive strain (MIC). We estimate that the MSC was very low for both streptomycin (1.6 μg/ml; 9.7% MIC) and carbenicillin 2.3 μg/ml (7% MIC; Fig. 1). Moreover, we saw no evidence of resistant cells protecting susceptible cells for either antibiotic, such that neither form of resistance is functioning as a cooperative mechanism under these conditions. These findings are in line with previous results [21, 27–30], and demonstrate that the mechanism of resistance has little, if any, impact on selection for resistance in mixed broth cultures.
Fig. 1.
Natural selection for antibiotic resistance in liquid culture. Susceptible and resistant strains were competed in liquid culture supplemented with carbenicillin (extracellular resistance) or streptomycin (intracellular resistance). Both streptomycin (a) and carbenicillin (b) suppress the growth rate of the susceptible strain (red line shows the susceptible growth in the presence of the resistant strain, black line shows the susceptible growth alone). In contrast, the growth rate of the resistant strain is not altered by antibiotic exposure (c, d, red line shows the resistant growth in the presence of the susceptible strain, grey line shows the resistant growth alone). As a result, all antibiotic concentrations select for the resistant strain (e, f). Please note that we use different axis ranges for the susceptible (a, b) and resistant (c, d) cell division data, because the data have very different ranges. The mean and standard error of two experiments, each with n = 6 for co-cultures and n = 3 for monocultures, are shown (Colour figure online). See also Supplementary Fig. S1
Colony growth has complex effects on selection for resistance
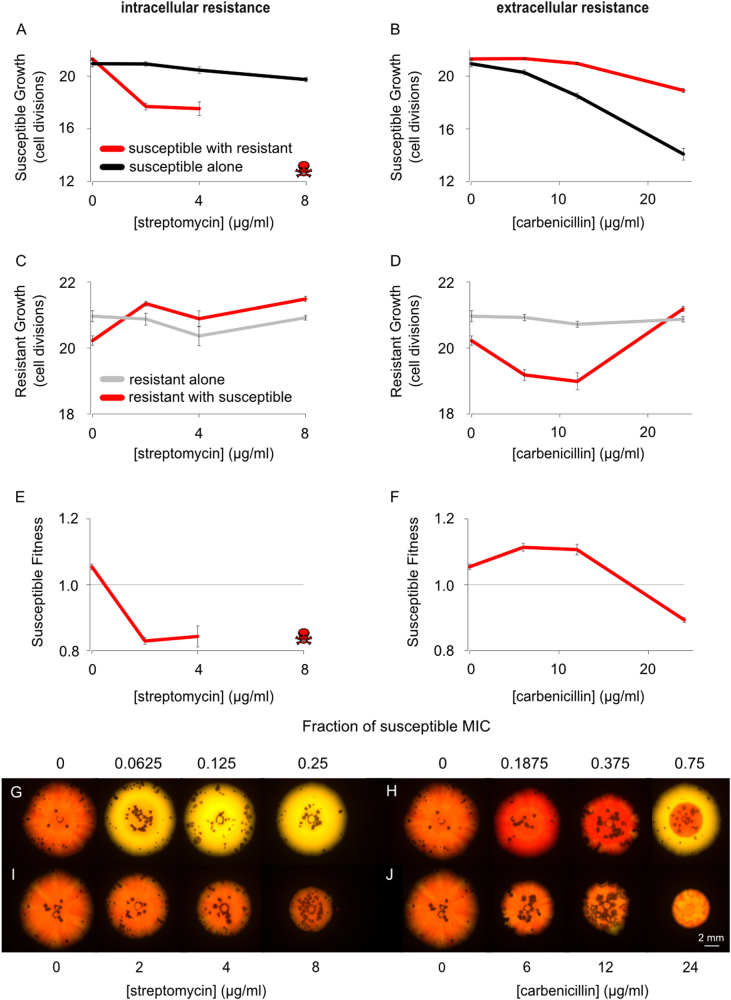
We next asked how life in a dense and genetically diverse community impacts the fate of an antibiotic-resistant strain. To do this, we repeated our competition experiments on agar plates (Fig. 2 and Supplementary Figs. S2 and S3); sometimes known as the “colony biofilm” model [9]. Our first observation was that, without antibiotics, the cost of plasmid carriage is significantly increased to 5.1% (t17.8 = 2.7, P = 0.0157). This increase is consistent with the finding that growth under biofilm conditions increases competition between strains. Cell division in mature colonies and biofilms often only occurs at the very edge [31], and this generates strong competition and natural selection to reach the edge and gain sustained access to nutrients and space for proliferation [14, 32–35]. This effect—competition for the growing edge—is expected to amplify the fitness benefit of rapid cell division because fast-growing strains have the added benefit of preferentially accessing the growing edge. While expected from a large body of previous work, this effect appears to have never been formalised. We, therefore, used an individual-based model of colony growth in order to reconcile our empirical findings with this intuitive effect (Supplementary Fig. S4).
Fig. 2.
Natural selection for resistance in bacterial colonies. Susceptible and resistant strains were competed over 7 days on solid agar with either streptomycin (intracellular resistance) or carbenicillin (extracellular resistance). On streptomycin, resistant strains outcompete susceptible strains at all concentrations of antibiotic (a, red line denotes susceptible growth in the presence of the resistant strain, black line denotes susceptible growth alone), where skulls denote that no colony-forming units of the susceptible strain were observed. However, the resistant strain increased the growth rate of the sensitive strain in the presence of carbenicillin, demonstrating cross-protection (b). Resistant growth increases in the presence of streptomycin when the susceptible strain is present (c, red line denotes resistant growth in the presence of the susceptible strain, grey line denotes resistant growth alone) but is suppressed by the presence of the susceptible strain on carbenicillin (d, red line denotes resistant growth in the presence of the susceptible strain, grey line denotes resistant growth alone). This relative increase in resistant growth, on streptomycin, with the susceptible strain, is expected from a competition effect, specifically the resistant strain outcompeting the susceptible strain for resources more effectively than against another resistant strain. Streptomycin always selects for resistance (e), whereas low doses of carbenicillin select for the sensitive strain (f). Representative images of the colonies in which yellow resistant and red susceptible strains are competed over 6 days, also demonstrate this result (g, h) while control colonies, in which both colours are susceptible, show the effect of these concentrations of antibiotic on non-resistant cells (i, j). The mean and standard error of two experiments, each with n = 6 for co-cultures and n = 3 for monocultures, are shown (Colour figure online). See also Supplementary Figs. S2 and S3
Theoretical considerations suggest that the effect of colony growth on selection for resistance should depend strongly on whether resistance has the potential for cooperative effects. This is because growth in dense colonies should increase the scope for cross-protection, which protects susceptible cells and, thereby, weakens selection for cooperating resistant strains [33]. Moreover, while not guaranteed [17, 36], mechanisms of resistance that occur intracellularly are expected to have weaker effects on other cells than mechanisms that act outside the cell owing to the cell membrane [33].
Consistent with these predictions, the MSC for streptomycin, where resistance is conferred via an intracellular mechanism, was approximately an order of magnitude (0.78 μg/ml; 2.4% MIC) less than the MSC for carbenicillin (20 μg/ml; 62% MIC) in bacterial colonies (Fig. 2). Furthermore, we see no evidence for cross-protection of susceptible cells by resistant cells for streptomycin (Fig. 2a). Rather we see that susceptible cells do worse in direct competition with resistant cells than growing on their own. This is again consistent with streptomycin acting as a non-cooperative mechanism as we saw for the liquid culture experiments.
The carbenicillin data are qualitatively different between the liquid culture and colony experiments. Theory predicts that selection for cooperative resistance will be potentially weak in colonies because the secretion of detoxifying enzymes by plasmid-carrying cells provides cross-protection to nearby sensitive cells [33]. This allows sensitive strains to benefit from reduced antibiotic exposure without paying the fitness cost of resistance [21, 22, 28]. Consistent with this explanation, the growth rate of the susceptible strain is markedly increased by the presence of the resistant strain in colonies at the lower concentrations of carbenicillin (Fig. 2).
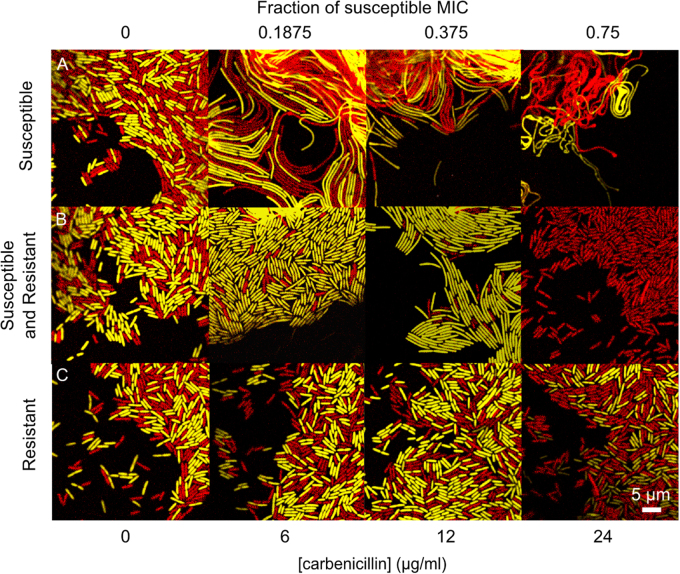
Further evidence of cross-protection comes from comparing the morphology of susceptible cells grown in the presence and absence of resistant cells. Carbenicillin is known to cause cell elongation and filamentation due to induction of the SOS response [37, 38] and inhibition of cell wall synthesis [39]. Cell elongation occurs in susceptible cells both when they grow alone and when they are grown in combination with resistant cells. However, the presence of resistant cells reduces the filamentation of susceptible cells, providing evidence of cross-protection at the single-cell level (Fig. 3a, b). One possible effect of cell elongation is that colony-forming units may underestimate the fitness of susceptible cells if the additional biomass per cell allows more cells to ultimately be produced. However, this would only lead to an underestimation of the extent of cooperation and cross-protection in the experiments.
Fig. 3.
Cross-protection of susceptible cells by resistant cells is supported by the degree of cell filamentation. Confocal images of the colony edge after 2 days show carbenicillin causes susceptible cells to filament, when grown alone on increasing concentrations of antibiotic (a, moving horizontally across the first row, both red and yellow cells are susceptible). The presence of the resistant strain causes susceptible cells to filament less (b, moving across second row, susceptible cells in yellow, resistant cells in red) than when they are grown alone at the same concentration of antibiotic (a, top row). At intermediate concentrations of carbenicillin there is enrichment at the edge for the susceptible strain when in competition with the resistant strain (b, centre panels, susceptible strain in yellow, resistant strain in red). When the resistant cells are grown alone on the antibiotic (c, both colours are resistant) they do not filament at these concentrations of carbenicillin. The MIC (minimum inhibitory concentration) is the lowest concentration of antibiotic that completely inhibits growth of susceptible cells (here measured in a colony (Colour figure online)) (Supplementary Table S1)
Genetic mixing weakens selection for carbenicillin resistance
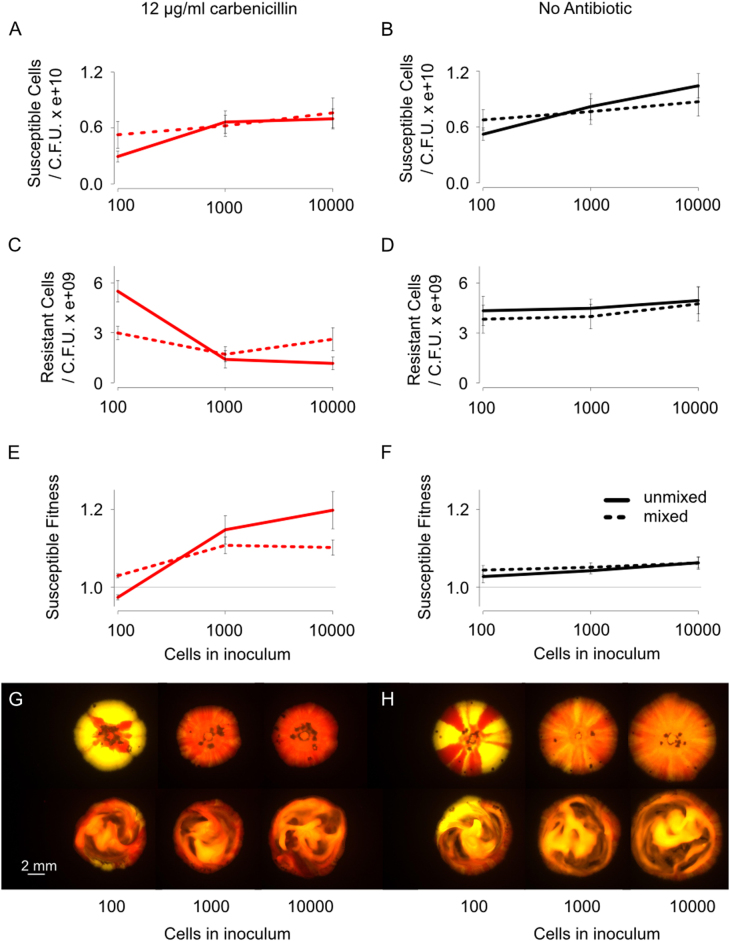
The differences in selection for streptomycin and carbenicillin resistance observed in bacterial colonies suggest that local interactions between neighbouring cells are key to understanding selection for β-lactamase secretion in bacterial colonies. Theoretical work predicts that the spatial organisation of genotypes is fundamental for the evolution of cell–cell interactions in biofilms [32–35, 40, 41]. Specifically, it predicts that the formation of clonal patches within colonies will limit cross-protection, which will favour resistant cells over susceptible cells. We, therefore, tested the importance of spatial structure for the competitiveness of our antibiotic-resistant strain. We inoculated colonies with a varying number of cells, at a concentration of carbenicillin that selected against antibiotic resistance in our earlier experiments (Fig. 2). Inoculating with fewer cells leads to a greater degree of spatial structure in the form of larger clonal patches (Fig. 4 and Supplementary Fig. S5) [42]. In line with our prediction, the fate of the resistant cells was reversed under conditions of low inoculation density and they were able to outcompete the susceptible cells (two-way ANOVA, F1, 15 = 10.9, P = 0.005), (left hand data point, Fig. 4e). By contrast, at higher inoculation densities, we observe that susceptible cells can outcompete resistant cells, replicating our earlier results. Moreover, the effect of density on competition is not seen until after 2 days of growth, once the communities are fully confluent (Supplementary Fig. S5). This suggests it is spatiogenetic structure within the colonies that is key to the observed effects, not simply the fact susceptible cells are physically isolated for longer at a low inoculation density (Fig. 4).
Fig. 4.
Spatiogenetic structure and the evolution of antibiotic resistance. Susceptible (red) and resistant (yellow) strains were co-inoculated onto plates containing either no antibiotic (black line) or 12 μg/ml carbenicillin (red line) (a–d). Final cell counts are shown here, as opposed to cell divisions (Figs. 1 and 2), because the use of different densities itself changes the number of cell divisions that occur (low cell density treatments experienced reduced nutrient competition and increased cell division). The final cell count removes this effect and is an easier metric to interpret. The growth data is shown in the Supplementary Information (Supplementary Fig. S5). From this data, the relative susceptible fitness was calculated (e, f). The extent of separation between susceptible and resistant strains in a colony can be varied with the number of inoculating cells, as can be seen in images taken after 6 days (g, h). Mixing (dotted line, bottom row of g and h) decreases the fitness of the resistant strain in colonies with a low initial density (colonies in LHS row in g go redder upon mixing (bottom row)) and increases fitness in colonies with a high initial density (colonies in middle and RHS row go yellower upon mixing, bottom row). The mean and standard error of two experiments, each with n = 3, are shown (Colour figure online). See also Supplementary Fig. S5
If spatial structure within colonies is capable of causing resistant strains to outcompete susceptible strains, then removing spatial structure should remove the competitive advantage of resistant strains. In order to test this prediction, we mixed one set of colonies daily using an inoculation loop [14]. When there was no antibiotic present, this mixing had no effect on susceptible fitness (three-way ANOVA, F1, 31 = 0.978, P = 0.33). However, as predicted, mixing the low-density treatment reversed the outcome of the competition such that the resistant strain was no longer able to outcompete the susceptible strain (three-way ANOVA, F1, 28 = 5.2, P = 0.03), (Fig. 4e, g). Simply disordering a bacterial colony, therefore, can remove the competitive advantage of an antibiotic-resistant strain. In sum, our data show that spatial structure in bacterial communities can be critical for the spread of antibiotic resistance.
An unexpected benefit to antibiotic susceptibility
Our experiments demonstrate that density and spatial structure can be fundamentally important for the rise of resistance to β-lactam antibiotics. Moreover, our data generally support a simple model where resistant cells experience a growth rate cost from carrying a plasmid, and will cross-protect susceptible cells when they are nearby (high density and low spatiogenetic structure). However, there are a number of intriguing patterns in our data, which all suggest that this simple view is insufficient.
The first observation is that low doses of carbenicillin can actually benefit the sensitive strain. Specifically, the fitness advantage of the susceptible strain doubles from 5.5 to 11 % upon the addition of low doses of carbenicillin (t16 = −2.9, P = 0.01). One explanation for this strong effect could be that carbenicillin promotes the growth of susceptible cells. However, we see no evidence for growth promotion in pure cultures (Figs. 1b and 2b) and, more importantly, the increase in fitness in mixed culture is due to a decrease in the growth rate of resistant cells, not an increase in the susceptible cells (red line in Fig. 2b, d). This effect on resistant cells is also not a direct effect of the antibiotic, as this depression of growth rate is not seen in pure resistant cultures (grey line in Fig. 2d). Instead, the antibiotic is somehow shifting the balance of competition in mixed culture towards the susceptible strain and allowing it to suppress the growth of the resistant strain.
Competition within colonies is intense and a large body of theoretical and empirical work has shown that the effects of natural selection occur primarily at the growing edges [14, 43–45]. In particular, this shows that it is the lateral growing edge—forming the circumference of a colony—that is most critical (upwards growth is much more limited than lateral growth). This led us to hypothesise that susceptible cells were somehow able to obtain better access to the lateral growing edge, allowing them to grow outwards and inhibit the growth of resistant cells. All of our frequency data so far has been from measurements of the whole colony (Figs. 1, 2, 4). Focussing in on the growing edges of colonies does indeed reveal that the edge becomes increasingly dominated by susceptible cells over time (Fig. 3b, middle row). However, this might happen simply because susceptible cells divide faster (above, Supplementary Fig. S4) and does not demonstrate an additional mechanism that allows them to dominate at the edge. To assess the importance of positioning, we need to experimentally test whether susceptible cells do better with antibiotic because they can influence spatial structure.
This test was inadvertently performed in the experiment in Fig. 4, which examines whether increases or decreases in genetic mixing affect cross-protection. As discussed above, mixing up the colonies enabled cross-protection and benefitted susceptible cells in the low-density treatment. However, there was a second and unexpected effect of mixing. In high-density inoculations, where cells start off well-mixed, physically mixing colonies reduces susceptible fitness (Fig. 4e, right hand points). Moreover, this effect is caused by increasing the resistant cell abundance (Fig. 4c). Disrupting spatial structure in colonies where susceptible cells are already winning, therefore, reduces susceptible fitness. This is exactly what is predicted from a model where antibiotic treatment somehow helps susceptible cells to obtain a better position in the colony. This model also predicts that the effect of mixing is not seen in the absence of antibiotics, which is also the case (Fig. 4f).
Cell shape and the increased fitness of susceptible cells
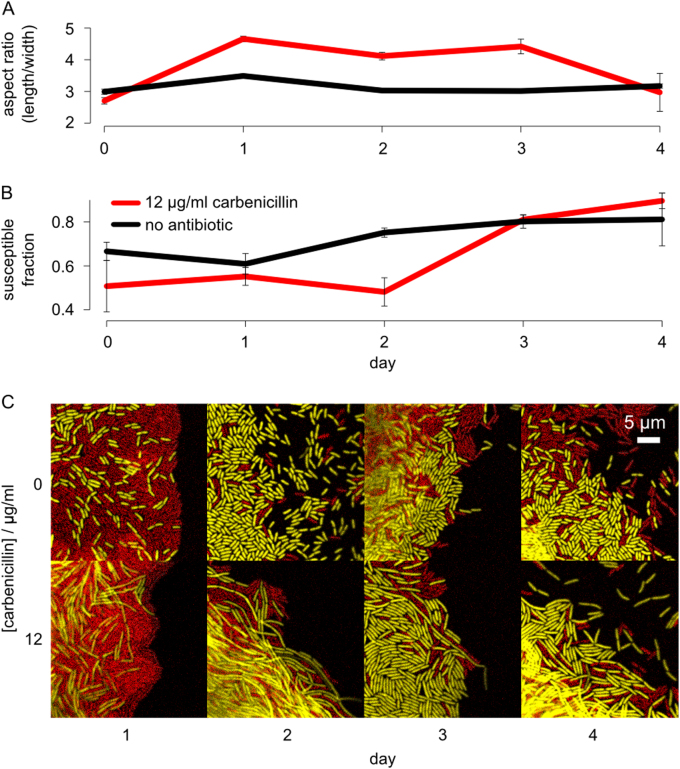
How does antibiotic exposure allow susceptible cells to find more favourable positions in the colony? Inspection of the colony edge gave a clear candidate: susceptible cells in mixed colonies change shape upon the addition of antibiotics (Fig. 3). If shape is important, then a key prediction is that changes in cell shape should proceed any increase in the frequency of susceptible cells. To test this prediction we tracked the aspect ratio of cells during competitions and compared this with the frequency of susceptible and resistant cells at each time point. Consistent with the shape-drives-fitness model, the elongation of susceptible cells occurs prior to their competitive gains against resistant cells (Fig. 5).
Fig. 5.
Susceptible cells increase in aspect ratio before outcompeting resistant cells in carbenicillin-treated colonies. Yellow susceptible and red resistant cells were competed on either 12 μg/ml carbenicillin (red) or no antibiotic (black). The edge of these colonies was then imaged during growth to measure cell length and relative cell numbers (a, b). The bottom images are micrographs of the colony edge under the different conditions showing the shifts in aspect ratio in susceptible cells, in the presence of resistant cells, during carbenicillin treatment (c). The mean and standard error of two experiments, each with n = 3, are shown. (Colour figure online)
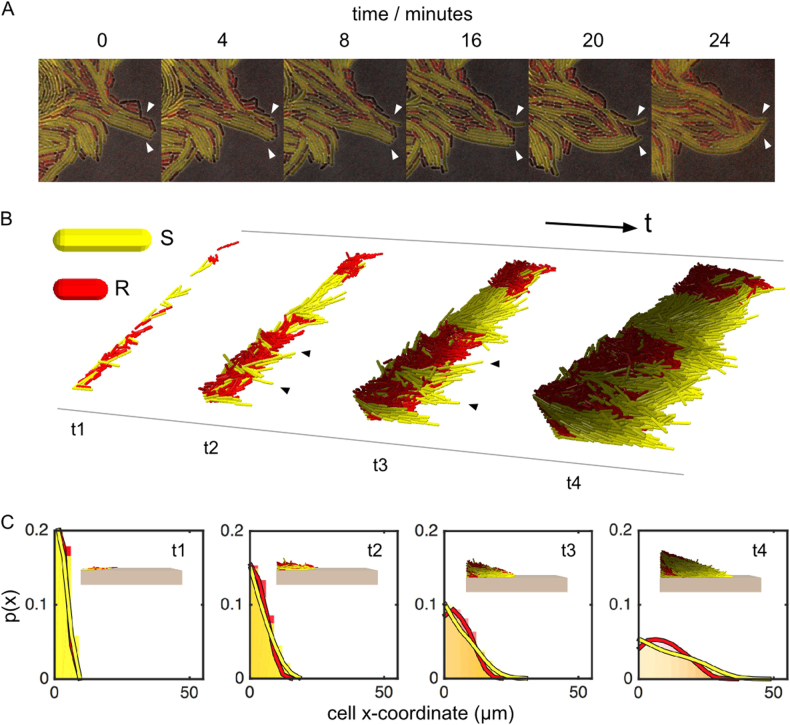
Time-lapse imaging of the colony edge highlighted a candidate mechanism for the advantage of elongated cells. We see sets of elongated cells that align orthogonally to the edge of the colony and push themselves outwards. This enables cells to grow ahead of the resistant strain and cut it off from the growing edge (Fig. 6a). The clearest support for the effect of cell shape would come from manipulating the shape of susceptible cells to ask how shape affects the ability to spatially organise and compete. We do not know of any route to prevent susceptible cells from elongating under antibiotic treatment without strong pleiotropic effects on growth rate and other phenotypes. However, a recent study that combined individual-based modelling with the study of cell shape mutants in Escherichia coli also suggests that cell shape can be important for positioning in bacterial biofilms [46]. Moreover, filamentation of Bacillus subtilis can increase migration away from the colony [47].
Fig. 6.
Cell shape is important for competition at the edge of colonies. Micrographs show longer susceptible cells (yellow) extending from the colony edge and occluding shorter resistant cells (red). This is shown in time-lapse images of growth at the edge of a representative colony after 10 h, growing on 12 μg/ml carbenicillin (a). These images differ in quality from earlier images (e.g., Fig. 5) because we had to use a lower fluorescence intensity to avoid bleaching during time-lapse imaging. An individual-based model shows the same effect (b). In the model, we can alter cell shape alone and follow its effects on cell sorting. Starting from a random configuration of Susceptible (S) and Resistant (R) cells in a 1:1 mixture, at t1, we observe that long cells extend from the colony edge (t2, black arrows). This improves their resource access and, as in our experiments, allows them to occlude the shorter cells from the edge. This, in turn stimulates further growth (t3, black arrows). By t4, S cells have become enriched at the colony edge through positive feedback. Histograms of cell x-coordinates track the growth of the colony edge and quantify the shape-driven enrichment process in the individual-based model, taken at the same 4 time points as in b (c). p(x) shows the probability of finding a given strain at a given point in the colony. Data merged from a sample of 20 simulations; times t1–t4 correspond to 12, 24, 36 and 56 h of growth, respectively. The colour brightness intensity of each cell corresponds to its growth rate, with brighter cells indicating faster growth (Colour figure online). See also Supplementary Fig. S6
We, therefore, extended the simulation models of Smith et al. [46] in order to study the effect of cell shape on positioning at the edge of a colony (Fig. 6 and Supplementary Fig. S6). We focused on the growing edge because this is the critical area of competition in an expanding colony and the region where we see strong enrichment for elongated susceptible cells. As we observe in the time-lapse imaging (Fig. 6a and Supplementary Movie S1), the individual-based model shows how long cells can align themselves orthogonally to the colony edge and push outwards occluding shorter cells from the edge. This gives them a competitive advantage as they can spread outwards, allowing them to align with the direction of growth ahead of resistant strains and hence to dominate the growing edge (Fig. 6b and Supplementary Fig. S6).
To summarise, the combination of competition data, imaging and our individual-based simulations all support a model where carbenicillin drives cell elongation and this provides an unexpected, but strong, fitness advantage to susceptible cells during biofilm growth. In combination with the effects of cross-protection, this means that sub-MIC doses of carbenicillin can inhibit bacterial growth while selecting against an antibiotic-resistant strain.
Discussion
Understanding the spread of antibiotic-resistant strains is a major goal for both science and society. In well-mixed broth cultures, we find that selection for resistance is strong, irrespective of the mechanism of resistance. In contrast, we find that selection for resistance in colonies depends critically on the mechanism of resistance, the spatiogenetic structure of colonies and the regulatory responses of sensitive cells to antibiotic exposure. Although bacterial colonies are a simplification of natural communities, we find they are sufficient to show that structured communities have both complex and surprising effects on selection for antibiotic resistance.
Consistent with theory ([14, 32–35], Supplementary Fig. S4), we see that growth in colonies greatly increases the strength of competition between strains, because rapidly growing bacteria preferentially gain access to the nutrient-rich edge of the colony. All else being equal, this should increase the strength of selection for resistance in colonies. It is difficult to directly compare the results of broth and colony competition experiments with each other, because there are many differences between these two modes of growth other than spatial structure. However, it is clear from our colony competition experiments that selection for carbenicillin resistance was notably weak in colonies. Indeed, of the four conditions we consider (two growth conditions, two antibiotics), we only see evidence for cooperative resistance for colony growth on carbenicillin. In the other three conditions, antibiotic resistance serves only to protect the cells that carry it.
Our experiments, which included disrupting spatiogenetic structure, are consistent with extracellular β-lactamase production protecting the sensitive strain under colony growth. Indeed, this protection can be so strong that the susceptible strain outcompetes and outnumbers the resistant strain in the colony (Figs. 3 and 4), as predicted by evolutionary theory for costly cooperative traits [20, 48]. Furthermore, by focussing on the growing edge, where natural selection is at its most powerful [34, 49], we discovered a second unexpected effect. Our data and theory suggest that exposure to low levels of carbenicillin leads to cell elongation, which allows susceptible bacteria to preferentially access the growing edge of treated colonies (Figs. 5 and 6).
Importantly, these two factors are also associated with greatly weakened natural selection for antibiotic resistance. Specifically, the dose of carbenicillin required to generate selection for resistance is over an order of magnitude higher than for streptomycin that is chemically modified inside the cell (20 μg/ml; 62% MIC versus 0.78 μg/ml; 2.4% MIC); an effect not seen in the liquid experiments (Supplementary Table S2). The broader consequence of this is that natural selection for a multidrug-resistant strain may be reduced by treatment with an antibiotic that targets a cooperative resistance mechanism. More generally, it suggests that drugs with strong extracellular effects, like β-lactamases, may be particularly amenable to treatment strategies intended to slow the rate of resistance evolution. However, this requires the identification of doses that are high enough to limit bacterial growth but low enough to allow susceptible strains to remain competitive relative to resistant strains. We have identified these conditions in our experiments, but additional work is needed to see if they can be harnessed as part of a treatment strategy.
In conclusion, our work cautions against the focus on liquid culture assays for understanding antibiotic resistance evolution. Few pathogenic bacteria live like this and we have shown that competitive outcomes in dense communities can be very different. Encouragingly, this shift in perspective may help to identify exposure regimes that suppress the growth of sensitive strains without favouring resistant strains [50–52].
Electronic supplementary material
Acknowledgements
We thank Christopher Thomas for kindly providing plasmids. IF and WPJS received funding from the Systems Biology Doctoral Training Centre, funded by the EPSRC (grant number EP/G03706X/1). SM was supported by a Marie Curie Intra-European Fellowship and an Ambizione grant from the Swiss National Science Foundation. ASM is supported by a Miguel Servet fellowship from the Instituto de Salud Carlos III (MS15/00012) co-financed by the European Social Fund and The European Development Regional Fund “A way to achieve Europe” (ERDF). KRF was supported by European Research Council Grant 242670 and a grant from the Calleva Research Centre for Evolution and Human Science (Magdalen College, Oxford). RCM was funded by the Royal Society, European Research Council grant 281591, and Wellcome Trust Grant 106918/Z/15/Z.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0090-4) contains supplementary material, which is available to authorised users.
Contributor Information
R. Craig MacLean, Email: craig.maclean@zoo.ox.ac.uk.
Kevin R. Foster, Email: kevin.foster@zoo.ox.ac.uk
References
- 1.Howard DH, Scott RD, Packard R, Jones D. The global impact of drug resistance. Clin Infect Dis. 2003;36:S4–10. doi: 10.1086/344656. [DOI] [PubMed] [Google Scholar]
- 2.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–8. doi: 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 3.Hughes D, Andersson DI. Evolutionary consequences of drug resistance: shared principles across diverse targets and organisms. Nat Rev Genet. 2015;16:459–71. doi: 10.1038/nrg3922. [DOI] [PubMed] [Google Scholar]
- 4.MacLean RC, Hall AR, Perron GG, Buckling A. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat Rev Genet. 2010;11:405–14. doi: 10.1038/nrg2778. [DOI] [PubMed] [Google Scholar]
- 5.zur Wiesch PA, Kouyos R, Engelstädter J, Regoes RR, Bonhoeffer S. Population biological principles of drug-resistance evolution in infectious diseases. Lancet Infect Dis. 2011;11:236–47. doi: 10.1016/S1473-3099(10)70264-4. [DOI] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 7.Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. 2009;33:206–24. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 8.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The calgary biofilm device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37:1771–76. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 12.Costerton JW, Lewandowski Z, Caldwell D, Korber D. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–45. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 13.Koch G, Yepes A, Forstner KU, Wermser C, Stengel ST, Modamio J, et al. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell. 2014;158:1060–71. doi: 10.1016/j.cell.2014.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim W, Racimo F, Schluter J, Levy SB, Foster KR. Importance of positioning for microbial evolution. PNAS. 2014;111:E1639–47. doi: 10.1073/pnas.1323632111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schluter J, Nadell CD, Bassler BL, Foster KR. Adhesion as a weapon in microbial competition. ISME J. 2015;9:139–49. doi: 10.1038/ismej.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 17.Sorg RA, Lin L, Sander Van Doorn G, Sorg M, Olson J, Nizet V, et al. Collective resistance in microbial communities by intracellular antibiotic deactivation. PLoS Biol. 2016;14:1–19. doi: 10.1371/journal.pbio.2000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–9. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West SA, Griffin AS, Gardner A. Social semantics: altruism, cooperation, mutualism, strong reciprocity and group selection. J Evol Biol. 2007;20:415–32. doi: 10.1111/j.1420-9101.2006.01258.x. [DOI] [PubMed] [Google Scholar]
- 20.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 21.Yurtsev Ea, Chao HX, Datta MS, Artemova T, Gore J. Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Mol Syst Biol. 2013;9:683. doi: 10.1038/msb.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dugatkin LA, Perlin M, Lucas JS, Atlas R. Group-beneficial traits, frequency-dependent selection and genotypic diversity: an antibiotic resistance paradigm. Proc R Soc B Biol Sci. 2005;272:79–83. doi: 10.1098/rspb.2004.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iredell J, Brown J, Tagg K. Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ. 2016;352:6420. doi: 10.1136/bmj.h6420. [DOI] [PubMed] [Google Scholar]
- 24.Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-rasmussen J, Høiby N. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J Antimicrob Biol. 2000;45:9–13. doi: 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- 25.Haines AS, Jones K, Cheung M, Thomas CM. The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J Bacteriol. 2005;187:4728–38. doi: 10.1128/JB.187.14.4728-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–41. doi: 10.1086/285289. [DOI] [Google Scholar]
- 27.Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio. 2014;5:19–23. doi: 10.1128/mBio.01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bottery MJ, Wood AJ, Brockhurst MA. Selective conditions for a multidrug resistance plasmid depend on the sociality of antibiotic resistance. Antimicrob Agents Chemother. 2016;60:2524–7. doi: 10.1128/AAC.02441-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogwill T, MacLean RC. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl. 2015;8:284–95. doi: 10.1111/eva.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yurtsev EA, Conwill A, Gore J. Oscillatory dynamics in a bacterial cross-protection mutualism. Proc Natl Acad Sci USA. 2016;113:6236–41 [DOI] [PMC free article] [PubMed]
- 31.Mitri S, Clarke E, Foster KR. Resource limitation drives spatial organization in microbial groups. ISME J. 2015;10:1471–82. doi: 10.1038/ismej.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitri S, Xavier JB, Foster KR. Social evolution in multispecies biofilms. Proc Natl Acad Sci. 2011;108:10839–46. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadell C, Drescher K, Foster KR. Spatial structure, cooperation, and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 34.Nadell CD, Foster KR, Xavier JB. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol. 2010;6:e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Dyken JD, Müller MJI, Mack KML, Desai MM. Spatial population expansion promotes the evolution of cooperation in an experimental Prisoner’s Dilemma. Curr Biol. 2013;23:919–23. doi: 10.1016/j.cub.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicoloff H, Andersson DI. Indirect resistance to several classes of antibiotics in cocultures with resistant bacteria expressing antibiotic-modifying or -degrading enzymes. J Antimicrob Chemother. 2016;71:100–10. doi: 10.1093/jac/dkv312. [DOI] [PubMed] [Google Scholar]
- 37.Blázquez J, Gómez-Gómez JM, Oliver A, Juan C, Kapur V, Martín S. PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol Microbiol. 2006;62:84–99. doi: 10.1111/j.1365-2958.2006.05366.x. [DOI] [PubMed] [Google Scholar]
- 38.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science (80-) 2004;305:1629–31. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 39.Kohanski Ma, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–35. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliveira NM, Niehus R, Foster KR. Evolutionary limits to cooperation in microbial communities. Proc Natl Acad Sci. 2014;111:17941–6. doi: 10.1073/pnas.1412673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci. 2007;104:876–81. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Gestel J, Weissing FJ, Kuipers OP, Kovács AT. Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 2014;10:2069–79. doi: 10.1038/ismej.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korolev KS, Xavier JB, Nelson DR, Foster KR. A quantitative test of population genetics using spatiogenetic patterns in bacterial colonies. Am Nat. 2011;178:538–52. doi: 10.1086/661897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitri S, Clarke E, Foster KR. Resource limitation drives spatial organization in microbial groups. ISME J. 2016;10:1471–82. doi: 10.1038/ismej.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Ditmarsch D, Boyle KE, Sakhtah H, Oyler JE, Nadell CD, Déziel Eacute, et al. Convergent evolution of hyperswarming leads to impaired biofilm formation in pathogenic bacteria. Cell Rep. 2013;4:697–708. doi: 10.1016/j.celrep.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith WPJ, Davit Y, Osborne JM, Kim W, Foster KR, Pitt-Francisa JM. Cell morphology drives spatial patterning in microbial communities. PNAS. 2016;114:E280–86 [DOI] [PMC free article] [PubMed]
- 47.van Gestel J, Vlamakis H, Kolter R. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLOS Biol. 2015;13:e1002141. doi: 10.1371/journal.pbio.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitri S, Foster KR. The genotypic view of social interactions in microbial communities. Annu Rev Genet. 2013;47:247–73. doi: 10.1146/annurev-genet-111212-133307. [DOI] [PubMed] [Google Scholar]
- 49.Hallatschek O, Nelson DR. Life at the front of an expanding population. Evolution. 2010;64:193–206. doi: 10.1111/j.1558-5646.2009.00809.x. [DOI] [PubMed] [Google Scholar]
- 50.Kouyos RD, Metcalf CJE, Birger R, Klein EY, Abel zur Wiesch P, Ankomah P, et al. The path of least resistance: aggressive or moderate treatment? Proc Biol Sci. 2014;281:20140566. doi: 10.1098/rspb.2014.0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Read AF, Day T, Huijben S. The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proc Natl Acad Sci. 2011;108:10871–7. doi: 10.1073/pnas.1100299108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drusano GL. Antimicrobial pharmacodynamics: critical interactions of ‘bug and drug’. Nat Rev Microbiol. 2004;2:289–300. doi: 10.1038/nrmicro862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.