Abstract
The amount of Panton-Valentine leukocidin (PVL) is diverse among Staphylococcus aureus isolates from different geographical regions, and its significance in some infections is disputed. However, data concerning this information in China are limited. Fifty-one lukSF-PV+ methicillin-resistant Staphylococcus aureus (MRSA) isolates gathered from varying infections were used for PVL production using enzyme-linked immunosorbent assay, and the quantity was analyzed in correlation with PVL isoform, genetic background of the isolate, and disease category. All isolates generated PVL with a range of 0.43–360.87 μg/mL, of which 56.9% isolates (29/51) generated 51–200 μg/mL of PVL; 11.8% (6/51) yielded PVL more than 200 μg/mL, and the rest (31.4%, 16/51) produced PVL of ≤50 μg/mL. The amount of PVL was not related to its variant and infection type, although isolates from skin and soft tissue infection had relatively high mean and median. Clonal complex (CC) 22 isolates might be the producer of relatively high concentrations of PVL; however, the difference among CCs was not analyzed due to a small number of CC isolates. The relevance of PVL production with the infection type, toxin isoform, and genetic characteristic of isolates may vary by clone type and also needs to be further evaluated using a large sample size and best concentration on in vivo environment.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) represents a major cause of infections in both the hospital and the community. The virulence of this pathogen partially relies on extracellular molecules. Panton-Valentine leukocidin (PVL, composed of LukS-PV and LukF-PV), a pore-forming toxin causing leukocytolysis and tissue necrosis, is one of these virulence factors that may have a significant influence in some serious Staphylococcus aureus (S. aureus) infections, such as severe skin and soft tissue infection, necrotizing pneumonia, and necrotizing fasciitis1,2. To date, PVL-producing MRSA isolates have emerged throughout the world2–10, and pose a huge threat to the health of patients. In Britain and France, therapeutic regimens may be regulated based on the existence or the absence of S. aureus PVL toxin11. The toxin-suppressing antibiotics, such as clindamycin, linezolid, and rifampin, are suggested for treating patients with serious PVL-expressing isolate infections11.
Previous reports have illustrated sequence variations within the lukSF-PV genes8,12–15. Molecular modeling indicates that a single amino acid replacement at site 176 [histidine (His) to arginine (Arg), namely H isoform changing into R isoform] may increase the leukotoxicity of PVL8. However, no evidence supports this assertion in the laboratory and clinical studies13,16. Genestier et al.17 reported that the role of PVL depends on the amount of toxin generated by S. aureus. Moreover, the only existence of a virulence gene does not imply that the toxin will be transcribed and/or translated and, if it is transcribed and/or translated, the toxin yield can be significantly different among isolates6. Therefore, this study detected the in vitro PVL production in clinical lukSF-PV positive (lukSF-PV+) MRSA isolates and explored whether the quantitative generation of PVL is correlated with specific isoform. In addition, the study also further analyzed the correlation of PVL production with clonal complexes (CCs) of isolates and type of infection.
Results
Clinical features
Fifty-one lukSF-PV+ MRSA isolates were isolated from 31 adult patients (including 14 young and 17 middle-aged and elderly patients), 1 adolescent, and 19 pediatrics (containing 9 children aged <1 year and 5 neonates aged <28 days). Of the 51 isolates, 19 were related to skin and soft tissue infections (SSTI), 16 to pneumonia, 5 to surgical site infections, and 11 to other infections (1 bacteremia, 1 urinary tract infection, 1 acute bacterial tracheitis, 4 tympanitis, and 4 conjunctivitis). According to the case notes, 22 (43.1%) and 29 (56.9%) MRSA were established as hospital-associated (HA) isolate and community-associated (CA) isolate, respectively (Table 1). Of 34 patients aged ≤44 years, 27 (79.4%) had CA infections. However, among 17 patients aged ≥45 years, only 2 (11.8%, 2/17) had CA infections (Table 1, ages of patients not shown).
Table 1.
Characteristics of 51 lukSF-PV+ MRSA isolates
| Infection type, isolate (n) | CA/HA | Molecular characteristic | ||||
|---|---|---|---|---|---|---|
| Isoform of PVL | SCCmec type | PFGE type | MLST type | CC | ||
| Pneumonia (16) | ||||||
| PVL74 | CA | H2 | III | A1 | ST59 | CC59 |
| PVL130 | HA | H1 | III | F2 | ST59 | CC59 |
| PVL161 | CA | H2 | NT | K | ST188 | CC1 |
| PVL211 | HA | H1 | III | E1 | ST22 | CC22 |
| PVL212 | HA | H2 | IVa | G1 | ST59 | CC59 |
| PVL214 | HA | H2 | III | E1 | ST22 | CC22 |
| PVL217 | HA | H2 | III | G3 | ST59 | CC59 |
| PVL219 | HA | H2 | III | D2 | ST338 | CC59 |
| PVL220 | HA | H2 | III | D3 | ST338 | CC59 |
| PVL222 | HA | H2 | II | M | ST149 | CC5 |
| PVL226 | HA | H2 | IVa | G2 | ST59 | CC59 |
| PVL239 | HA | H2 | III | E2 | ST22 | CC22 |
| PVL244 | HA | H2 | NT | W | ST88 | CC88 |
| PVL254 | HA | H2 | III | B1 | ST59 | CC59 |
| PVL255 | HA | H2 | II | P | ST149 | CC5 |
| PVL256 | CA | H1 | III | B1 | ST59 | CC59 |
| Skin and soft tissue infection (19) | ||||||
| PVL8 | CA | H2 | III | B3 | ST59 | CC59 |
| PVL16 | CA | H2 | III | H1 | ST59 | CC59 |
| PVL34 | CA | H2 | IVa | S | ST59 | CC59 |
| PVL51 | CA | H2 | III | B3 | ST59 | CC59 |
| PVL60 | CA | H2 | IVa | H2 | ST59 | CC59 |
| PVL65 | CA | H2 | IVa | T | ST59 | CC59 |
| PVL79 | CA | H2 | IVa | A3 | ST59 | CC59 |
| PVL104 | CA | R2 | NT | X | ST88 | CC88 |
| PVL108 | CA | H2 | III | F1 | ST59 | CC59 |
| PVL145 | CA | H2 | III | F1 | ST59 | CC59 |
| PVL148 | HA | H2 | IVb | O | ST9 | CC9 |
| PVL150 | CA | R2 | V | J | ST25 | CC5 |
| PVL170 | HA | H2 | III | A2 | ST59 | CC59 |
| PVL195 | CA | H2 | III | N | ST59 | CC59 |
| PVL203 | CA | H1 | IVa | Z | ST1 | CC1 |
| PVL205 | HA | H2 | III | E1 | ST22 | CC22 |
| PVL209 | HA | H1 | IVa | C1 | ST59 | CC59 |
| PVL210 | CA | H2 | III | C3 | ST59 | CC59 |
| PVL233 | CA | H2 | III | Q | ST30 | CC30 |
| Surgical site infection (5) | ||||||
| PVL7 | CA | H2 | V | V | ST1 | CC1 |
| PVL82 | HA | H2 | III | A1 | ST59 | CC59 |
| PVL236 | HA | H2 | III | C4 | ST59 | CC59 |
| PVL237 | CA | H2 | III | C2 | ST59 | CC59 |
| PVL242 | CA | H1 | III | D1 | ST338 | CC59 |
| Other infections (11) | ||||||
| PVL40 | CA | R1 | IVa | R | ST59 | CC59 |
| PVL69 | HA | H2 | III | L | ST217 | CC22 |
| PVL186 | CA | H2 | IVa | A4 | ST59 | CC59 |
| PVL202 | CA | H2 | IVa | Y | ST88 | CC88 |
| PVL204 | CA | H2 | III | C3 | ST59 | CC59 |
| PVL206 | CA | R2 | IVa | U | ST25 | CC5 |
| PVL218 | HA | R2 | IVa | I1 | ST59 | CC59 |
| PVL221 | CA | H2 | III | D1 | ST338 | CC59 |
| PVL238 | CA | H2 | IVa | I2 | ST59 | CC59 |
| PVL246 | CA | H2 | IVa | D4 | ST338 | CC59 |
| PVL253 | HA | H2 | III | B2 | ST59 | CC59 |
CA, Community-acquired isolate; HA, hospital-acquired isolate; MLST, multilocus sequence typing; NT, nontypeable; PFGE, pulsed-field gelelectrophoresis; SCCmec, staphyloccoccal cassette chromosome mec element; ST, sequence type.
In vitro production of PVL
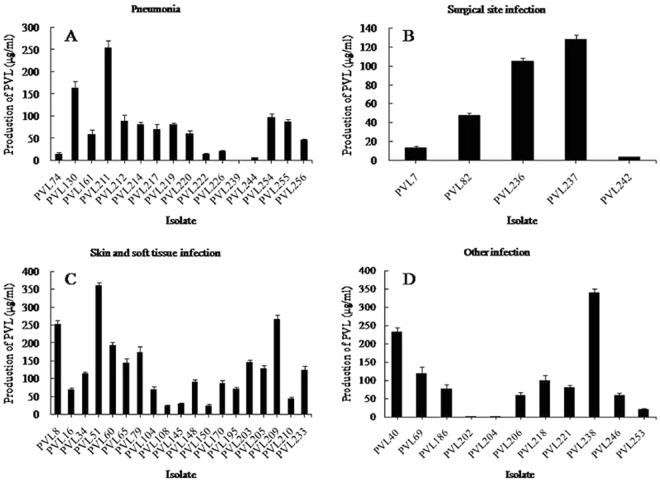
Twenty-nine (56.9%, 29/51) isolates generated PVL between 50 and 200 μg/mL, and 6 isolates had elevated PVL between 201 and 400 μg/mL; the remaining isolates (31.4%, 16/51) produced PVL less than 50 μg/mL. Pneumonia-related isolates yielded a mean [±standard deviation (SD)] of 72.11 ± 64.58 μg/mL and a median of 66.27 μg/mL PVL (Table 2). This group contained one isolate (isolate PVL239) for which extremely low PVL generation (0.94 μg/mL) was measurable (Fig. 1A). SSTI-associated isolates produced an average of 126.58 ± 90.67 μg/mL and a median of 113.63 μg/mL PVL (Table 2), among which one isolate (isolate PVL51) was the highest producer (360.87 μg/mL) (Fig. 1C). For the other infection-related isolates, an average PVL level of 98.34 ± 102.79 μg/mL and a median of 75.79 μg/mL were found (Table 2). This group included one isolate (isolate PVL202), which was the lowest producer (0.43 μg/mL) (Fig. 1D). Surgical site infection–associated isolates generated the lowest PVL quantity (a mean of 59.55 ± 55.11 μg/mL and a median of 47.72 μg/mL) (Table 2 and Fig. 1B). Although the isolates from SSTI had relatively high values, the results from the Kruskal–Wallis test displayed no significant differences in PVL yields among the four different disease groups (P = 0.151).
Table 2.
ELISA data on in vitro PVL production among clinical isolates of lukSF-PV+ MRSA (n = 51).
| Group | Number of isolates (n) | PVL range (μg/mL) | PVL median (μg/mL) | PVL mean ± SD (μg/mL) | P value |
|---|---|---|---|---|---|
| Clonal complex (CC) | |||||
| Isolates belonging to CC59 | 34 | 0.94−360.87 | 80.75 | 107.67 ± 91.92 | NA |
| Isolates belonging to CC22 | 5 | 0.94−254.62 | 118.51 | 116.88 ± 75.98 | NA |
| Isolates belonging to CC88 | 3 | 0.43−68.47 | 6.53 | 25.14 ± 37.64 | NA |
| Isolates belonging to CC1 | 3 | 13.65−145.32 | 59.91 | 72.96 ± 66.80 | NA |
| Isolates belonging to CC5 | 4 | 14.67−88.38 | 40.85 | 46.18 ± 33.72 | NA |
| Isolates belonging to CC9 | 1 | NA | |||
| Isolates belonging to CC30 | 1 | NA | |||
| Type of infection | |||||
| Isolates from SSTI | 19 | 23.31−360.87 | 113.63 | 126.58 ± 90.67 | 0.151 |
| Isolates from pneumonia | 16 | 0.94−254.62 | 66.27 | 72.11 ± 64.58 | |
| Isolates from surgical site infection | 5 | 3.48−128.27 | 47.72 | 59.55 ± 55.11 | |
| Isolates from other infection | 11 | 0.43−339.52 | 75.79 | 98.34 ± 102.79 | |
| CA isolates | 29 | 0.43−360.87 | 68.47 | 105.2 ± 96.6 | 0.754 |
| HA isolates | 22 | 0.94−265.80 | 87.58 | 86.8 ± 68.8 | |
| Isoform of PVL | |||||
| H isoform isolates | 46 | 0.43−360.87 | 80.75 | 96.91 ± 86.41 | 0.975 |
| R isoform isolates | 5 | 23.82−231.74 | 68.47 | 96.09 ± 80.40 | |
CA, Community-associated infection; HA, hospital-associated infection; NA, not applicable for the Kruska–Wallis rank-sum test due to a relatively small number of isolates; PVL, Panton–Valentine leukocidin; SD, standard deviation; SSTI, skin and soft tissue infection.
Figure 1.
PVL yield of 51 clinical lukSF-PV+ MRSA isolates in a given infection classification. Isolates were grouped on the basis of the clinical diagnosis, and PVL generation was examined using ELISA. Error bars signify SDs of the results from three experiments for each isolate.
Among all CCs, the CC22 isolates produced the highest level of PLV (a mean of 116.88 ± 75.98 μg/mL and a median of 118.51 μg/mL), followed by CC59, CC1, CC5, and CC88 isolates (Table 2). Although the PVL levels of isolates with CC88 and CC5 were far lower than those of CC22 and CC59 isolates, the statistical analysis was not performed to analyze the significance of the difference because of the small number of isolates of most CCs (Table 2). In addition, this study also analyzed the correlation of PVL yields of isolates with CA and HA infections, and PVL H and R isoforms. Unfortunately, no statistical differences were found among them (Table 2).
Discussion
Among lukSF-PV+ MRSA, 66.7% (34/51) were isolated from young and pediatric patients (≤44 years old), which included 93.1% (27/29) CA MRSA. This was consistent with the reports that CA MRSA mostly affects young individuals and children, and usually presents PVL18.
Previous reports showed that the lukSF-PV genes were a frequent genetic marker of CA MRSA isolates with SCCmecIV or SCCmecV19,20. Nevertheless, some studies have reported the lukSF-PV+ HA MRSA strains21–25. This study also found that 43.1% (22/51) lukSF-PV+ MRSA were HA MRSA strains (Table 1). The prevalence of lukSF-PV+ MRSA clonal lineages is diverse in different parts of the earth. For example, ST8 (CC8)-SCCmecIV (USA300) clone predominates in USA; ST59 (CC59)-SCCmecVT clone circulates in Asia, ST30 (CC30)-SCCmecIV clone is prevalent in New Zealand, ST93 (CC93)-SCCmecIV clones are found mainly in Australia, ST80 (CC80)-SCCmecIV is the most frequent clone in Europe and the Middle East, ST88 (CC88)-SCCmecIV clone is identified mainly in Africa, and ST22 (CC22)-SCCmecIV and ST772 (CC1)-SCCmecV clones emerge mainly in India18. In the present study, ST59 (CC59)-SCCmecIII was the predominant clone (35.3%, 18/51) of lukSF-PV+ MRSA isolates, followed by ST59 (CC59)-SCCmecIV (IVa/IVb, 25.5%, 13/51). However, for the former, 61.1% (11/18) of isolates were found to be CA MRSA; for the latter, 38.5% (5/13) of isolates were detected as HA MRSA. Historically, SCCmecIII was considered as one of the epidemiological markers of the main HA MRSA clones, and SCCmecIV was often found in CA MRSA isolates24. However, contrary to these views, SCCmecIII and SCCmecIV were found in both HA and CA lukSF-PV+ MRSA isolates in this study. This indicated that the lukSF-PV+ MRSA isolates carrying SCCmecIII or SCCmecIV could disseminate between the hospital and the community in the region studied. The PFGE types of some isolates with SCCmecIII supported the possibility of the transmission of HA strains of SCCmecIII into the community (Table 1). In addition, the reports by Song et al.26 and Yao et al.24 also described this spread of HA and CA MRSA isolates from Taiwan, Hong Kong, Korea, Vietnam, and Wenzhou city of mainland China. Geng et al.27 reported the existence of ST338 (CC59) CA MRSA isolate in Taiwan and the southern region of China. In the present study, five lukSF-PV+ MRSA isolates were also identified to be ST338 type, including three CA isolates and two HA isolates. The investigation results of Song et al.26 showed that ST30 (CC30) was predominant in Asian areas. However, only one HA isolate was ST30 in this study. Additionally, ST22 (CC22) and ST88 (CC88), predominating in India28,29 and Africa3, respectively, were also found in the isolates in this study. However, the ST22 isolates were SCCmecIII type, and not SCCmecIV detected in India. Although ST8 (CC8) and ST80 (CC80) prevailing in the USA and Europe, respectively, have been described in CA MRSA isolates from Japan and Korea (ST8), and Singapore and Malaysia (ST80), both MRSA clones are apparently scarce in Asian regions26 and were not found in the isolates in this study (Table 1).
According to the reports of Dumitrescu et al.12, O’Hara et al.8, and Tong et al.14, the R isoform of PVL was found in CC8 (mainly USA300 strains), CC1, and CC93 isolates, which were mainly from the USA and Australia; while the H isoform was mainly observed in non-USA isolates belonging mainly to CC121, CC30, and CC22. A few isolates belonging to CC1, CC5, CC6, CC25, CC59, and CC88 also carried the H isoform. Table 1 shows that the H isoform was harbored by 90.2% (46/51) of lukSF-PV+ MRSA isolates belonging primarily to CC59 (69.6%, 32/46), followed by CC22 (10.9%, 5/46), CC88 (6.5%, 3/46), CC5 (6.5%, 3/46), CC1 (6.5%, 3/46), CC9 (2.2%, 1/46), and CC30 (2.2%, 1/46); however, the R isoform was found only in five isolates belonging to CC59, CC88, and CC5, which were different from those (CC8, CC1, and CC93) described by the previous studies8,12,14.
PVL can lead to concentration-dependent necrosis and apoptosis of human polymorphonuclear neutrophils and lysis of human monocytes and macrophages12, but whether PVL is pathogenic or a marker of specific infection is controversial. Contradictory data may be associated with the quantity of PVL generated by individual isolates. Up to now, the number of S. aureus isolates producing PVL is mainly determined using polymerase chain reaction (PCR) for lukSF-PV genes or quantitative reverse transcription–PCR for lukSF-PV mRNA levels. However, the findings of these methods may not relate to the actual yield of PVL. Therefore, the present study directly detected the PVL toxin production using enzyme-linked immunosorbent assay (ELISA). This study showed that all the lukSF-PV+ isolates produced PVL in vitro, and the extent of the toxin expression varied among isolates (range from 0.43 to 360.87 μg/mL), even within specific genetic contexts (Table 2).
Undoubtedly, the presence of the genes encoding PVL does not necessarily imply that the toxin is produced in the setting of human infection. While this is true, it is important to note that neither does in vitro production. Additionally, the production of toxin is varied from culture media. Historical documents have reported the inter-isolate variability for ST8, ST80, ST93, and ST121 isolates6,11,30. The same phenomenon was reported for the STs detected in this study. The data showed that two ST59 CA isolates (PVL51 and PVL238) were the strongest producers of PVL (360.87 μg/mL and 339.52 μg/mL, respectively); and three ST59 (PVL8, PVL40, and PVL209) and one ST22 (PVL211) isolates produced 231.74–265.80 μg/mL of PVL (Table 1 and Fig. 1). However, two extremely low producers of PVL also belonged to ST59 and ST22 clones (PVL202 and PVL204, 0.94 μg/mL each) (Table 1 and Fig. 1). The amounts of PVL yielded by other MRSA isolates (including the remaining ST59 and ST22 isolates) varied between 0.43 and 192.96 μg/mL. The cause of the difference in PVL expression is not clear. Also, the correlation of PVL yield with the genetic background of isolates warrants further investigation. The PVL concentration of 0.33 μg/mL evoked the apoptosis of human granulocytes, and the injection of 0.3 μg/mL of toxin into the intracutaneous tissue of rabbits caused local inflammation and necrosis11. According to the in vitro PVL production in this study (≥0.43 μg/mL), all lukSF-PV+ isolates likely had the ability to cause an inflammatory response during infections. Shallcross et al.31 reported that PVL-producing clinical isolates had a link with SSTIs. In the present study, the mean value and median of PVL production of SSTI isolates were relatively high compared with those of isolates from pneumonia, surgical site infections, and other infections. However, the significant difference in the toxin levels was not found among the four groups (Table 2). This result was not consistent with the conclusion that PVL performed a more significant role in S. aureus SSTIs31. In addition, the isolates from invasive infections (such as pneumonia) were even shown to yield low mean and median of PVL (Table 2). The possible explanation was that the clinical outcome was correlated with host factors, or with the existence of other toxins30.
For PVL variants, the H and R isoform isolates generated similar mean concentrations and medians of PVL, and the difference between the two groups was not significant (P = 0.975) (Table 2). This result suggested that point mutations in the open reading frame of lukSF-PV genes might have no effect on the toxin production.
A previous study showed that CA MRSA infection was responsible for rapidly progressive and lethal diseases such as necrotizing pneumonia, severe sepsis, and necrotizing fasciitis. Also, some data sustained the opinion that PVL was accountable at least partially for the enhanced virulence of CA MRSA32. In this study, the CA and HA isolates yielded similar levels of PVL (P = 0.754) (Table 2). This indicated that the lukSF-PV+ HA isolates might lead to the severity of clinical infections similar to that caused by lukSF-PV+ CA isolates.
Being a retrospective study, this study had several limitations. First, the corresponding clinical samples could not be collected to evaluate the actual concentration of PVL in vivo. Because the PVL production depends on many factors, such as environmental factors, host factors, and internal factors of bacteria, it is not yet known which factor impacts mainly on the protein expression level. Second, the representativeness of isolates from only one hospital was insufficient for determining the generation and isoforms of PVL and the association between PVL production and PVL variants or genetic background or infection types. Third, more clinical information could not be collected to evaluate the correlation of the yield and isoform of PVL with the severity of illness.
In summary, the present experimental data did not sustain the direct correlation of PVL production in vitro with clinical types of infection and its isoform. The in vitro production of PVL by lukSF-PV+ MRSA isolates was dramatically changeable, indicating the existence of unknown environmental and host factors in transcriptional and/or translational regulation of gene expression. The regulation mechanisms affecting the differential expression of PVL and the associations among PVL levels and characteristics of isolate and infection require further exploration.
Methods
Clinical isolates
Fifty-one consecutive, nonduplicate lukSF-PV+ MRSA isolates from various clinical specimens of individual patients were obtained from Zhejiang Xiaoshan Hospital (a tertiary hospital), Hangzhou City, China. The isolates were collected between January 2010 and December 2011 and identified using VITEK 32 instrument (bioMérieux, Marcy l′ Etoile, France). Methicillin resistance was determined using the Kirby–Bauer method with cefoxitin disk (30 µg, Oxoid, Basingstoke, UK) and PCR for mecA33,34. All isolates had been previously described by PVL isoform detection and PFGE-SCCmec-MLST typing (including CC) (Table 1)35. The strains ATCC 49775 and MRSA N315 were used as positive and negative controls for the PVL production, respectively.
Quantitative detection of PVL
For quantifying the production of PVL in vitro, a certain amount of lukSF-PV+ MRSA (1 to 3 × 106 CFU/mL) was cultured at 37 °C in 5 mL of casein–casein–yeast extract broth in which the optimal production of PVL could be acquired, with shaking at 200 rpm for 20 h. At this time, all growth isolates had reached a survival plateau of 1 to 4 × 109 CFU/mL6,11. Culture supernatants were acquired by centrifugation at 8000 g for 15 min. The quantification of secreted PVL was performed using ELISA, according to the kit instructions (Shanghai Y-J Biotechnology Co., Ltd., China). Each sample was measured in triplicate. The ELISA detection limit of PVL was 0.3~10 μg/mL. The samples with a concentration of more than 10 μg/mL were diluted for the redetection of PVL. A previous study reported that the intracutaneous injection of 0.3 μg/mL PVL led to local inflammation and necrosis in rabbits, which, like humans, are sensitive to similar PVL concentration11. Therefore, a concentration of 0.3 μg/mL was used in the present study, just the lower detection limit of the ELISA kit, as the cutoff value to determine whether the supernatant was PVL positive.
Definitions
(1) SSTI was described as the existence of a skin abscess or other symptoms of inflammation and a positive MRSA culture36. (2) Pneumonia was described as the existence of fever with imaging changes corresponding to pulmonary infection and MRSA-positive respiratory tract specimens (sputum, endotracheal aspirate, bronchial washing, and bronchoalveolar lavage) or blood cultures36. (3) Surgical site infection was defined as an infected postoperative wound and drainage with a positive MRSA culture36. (4) Primary bacteremia was defined as the existence of a blood culture positive for MRSA without the recognizable origin of infection36. (5) Urinary tract infection was defined as MRSA bacteriuria (≥105 CFU/mL) and pyuria. (6) Acute bacterial tracheitis was diagnosed according to the presentation of upper respiratory tract obstruction, such as stridor, cough, and dysphagia; abundant purulent tracheal excretions; and positive culture for MRSA from the tracheal excretions. (7) Tympanitis was defined as the inflammation of the middle ear with a positive MRSA culture. (8) Conjunctivitis was characterized as a positive culture for MRSA plus two or more of the following symptoms: red, watery, itchy eyes; painful, burning eyes; purulent fluid in eyes; or photophobia. (9) CA MRSA infection was described as MRSA infection emerging in the community or within 48 h after admission to hospital without established risk factors (such as hospitalization history, surgery, dialysis, or living in a long-term care residence within 1 year; existence of a long-term indwelling catheter or other percutaneous medical device; or previous MRSA isolation). (10) HA MRSA infection was characterized as strains isolated from patients in less than 48 h of hospitalization with the aforementioned risk factors or >48 h of admission37.
Clinical information
The clinical data of the patients were collected retrospectively from the case notes. The data included gender, age, outpatient or inpatient, clinical symptoms, sample type, date of strain isolation, clinical diagnosis, and other medical histories related to HA or CA infections.
Statistical analysis
In view of the uneven distribution of PVL production among isolates, the comparisons of two and ≥3 groups were analyzed using the Wilcoxon rank-sum test and the Kruskal–Wallis rank-sum test (SPSS 22.0), respectively. A two-sided P value < 0.05 was considered statistically significant.
Data availability
Most of the experimental data acquired/analyzed during this study have been included in this published version. Information on rest of the data can be obtained from the corresponding author on request.
Ethics approval and consent to participate
The ethics committee of Zhejiang Xiaoshan Hospital provided ethical approval for this study. The approval ID number was XSYY2015012. All study participants provided an informed consent before participation, and patient information was anonymized.
Acknowledgements
This study was supported by grants from the Zhejiang Medical and Health Science and Technology Project (2012KYB171), the Natural Science Foundation, the Science and Technology Commission of Shanghai (12ZR1425000), and the National Natural Science Foundation of China (81371872).
Author Contributions
C.-L.Z. was involved in designing and conducting the study. Y.-Y.G. and X.C. performed data collection, management, and analysis. C.-L.Z. was involved in manuscript preparation, review, and approval.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Miller LG, et al. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med. 2005;352:1445–1453. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 2.Shallcross LJ, et al. Panton-Valentine leukocidin associated staphylococcal disease: a cross-sectional study at a London hospital, England. Clin Microbiol Infect. 2010;16:1644–1648. doi: 10.1111/j.1469-0691.2010.03153.x. [DOI] [PubMed] [Google Scholar]
- 3.Abdulgader SM, Shittu AO, Nicol MP, Kaba M. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol. 2015;6:348. doi: 10.3389/fmicb.2015.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobos-Triguero N, et al. Epidemiology and clinical presentation of Panton-Valentin leukocidin positive methicillin-resistant Staphylococcus aureus. Rev Esp Quimioter. 2010;23:93–99. [PubMed] [Google Scholar]
- 5.Elizur A1, et al. Panton-Valentine Leukocidin-positive methicillin-resistant Staphylococcus aureus lung infection in patients with cystic fibrosis. Chest. 2007;131:1718–1725. doi: 10.1378/chest.06-2756. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton SM, et al. In vitro production of Panton-Valentine leukocidin among strains of methicillin-resistant Staphylococcus aureus causing diverse infections. Clin Infect Dis. 2007;45:1550–1558. doi: 10.1086/523581. [DOI] [PubMed] [Google Scholar]
- 7.Nimmo GR, Coombs GW. Community-associated methicillin-resistant Staphylococcus aureus (MRSA) in Australia. Int J Antimicrob Agents. 2008;31:401–410. doi: 10.1016/j.ijantimicag.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 8.O’Hara FP, et al. A geographic variant of the Staphylococcus aureus Panton-Valentine leukocidin toxin and the origin of community-associated methicillin-resistant S. aureus USA300. J Infect Dis. 2008;197:187–194. doi: 10.1086/524684. [DOI] [PubMed] [Google Scholar]
- 9.Tristan A, et al. Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg Infect Dis. 2007;13:594–600. doi: 10.3201/eid1304.061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu F, et al. Expression of Panton-Valentine leukocidin mRNA among Staphylococcus aureus isolates associates with specific clinical presentations. Plos One. 2013;8:e83368. doi: 10.1371/journal.pone.0083368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badiou C, et al. Rapid detection of Staphylococcus aureus Panton-Valentine leukocidin in clinical specimens by enzyme-linked immunosorbent assay and immunochromatographic tests. J Clin Microbiol. 2010;48:1384–1390. doi: 10.1128/JCM.02274-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumitrescu O, et al. Polymorphism of the Staphylococcus aureus Panton-Valentine leukocidin genes and its possible link with the fitness of community-associated methicillin-resistant S. aureus. J Infect Dis. 2008;198:792–794. doi: 10.1086/590914. [DOI] [PubMed] [Google Scholar]
- 13.Tong SY, et al. Clinical correlates of Panton-Valentine leukocidin (PVL), PVL isoforms, and clonal complex in the Staphylococcus aureus population of Northern Australia. J Infect Dis. 2010;202:760–769. doi: 10.1086/655396. [DOI] [PubMed] [Google Scholar]
- 14.Tong SY, et al. Rapid detection of H and R Panton-Valentine leukocidin isoforms in Staphylococcus aureus by high-resolution melting analysis. Diagn Microbiol Infect Dis. 2010;67:399–401. doi: 10.1016/j.diagmicrobio.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Wolter DJ, Tenover FC, Goering RV. Allelic variation in genes encoding Panton-Valentine leukocidin from community-associated Staphylococcus aureus. Clin Microbiol Infect. 2007;13:827–830. doi: 10.1111/j.1469-0691.2007.01763.x. [DOI] [PubMed] [Google Scholar]
- 16.Besseyre des Horts T, et al. A histidine-to-arginine substitution in Panton-Valentine leukocidin from USA300 community-acquired methicillin-resistant Staphylococcus aureus does not impair its leukotoxicity. Infect Immun. 2009;78:260–264. doi: 10.1128/IAI.00843-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genestier AL, et al. Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. J Clin Invest. 2005;115:3117–3127. doi: 10.1172/JCI22684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shore AC, et al. Panton-Valentine leukocidin-positive Staphylococcus aureus in Ireland from 2002 to 2011: 21 clones, frequent importation of clones, temporal shifts of predominant methicillin-resistant S. aureus clones, and increasing multiresistance. J Clin Microbiol. 2014;52:859–870. doi: 10.1128/JCM.02799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandenesch F, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tristan A, et al. Virulence determinants in community and hospital meticillin-resistant Staphylococcus aureus. J Hosp Infect. 2007;65(Suppl. 2):105–109. doi: 10.1016/S0195-6701(07)60025-5. [DOI] [PubMed] [Google Scholar]
- 21.Goering RV, et al. Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trials. J Clin Microbiol. 2008;46:2842–2847. doi: 10.1128/JCM.00521-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lina G, et al. Involvement of Panton-Valentine leukocidin producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 23.McClure JA, et al. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from resistant staphylococci. J Clin Microbiol. 2006;44:1141–1144. doi: 10.1128/JCM.44.3.1141-1144.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao D, et al. Molecular characterization of Staphylococcus aureus isolates causing skin and soft tissue infections (SSTIs) BMC Infect Dis. 2010;10:133. doi: 10.1186/1471-2334-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang YH, et al. Clonal spread of SCCmec type IV methicillin-resistant Staphylococcus aureus between community and hospital. Clin Microbiol Infect. 2007;13:717–724. doi: 10.1111/j.1469-0691.2007.01718.x. [DOI] [PubMed] [Google Scholar]
- 26.Song JH, et al. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother. 2011;66:1061–1069. doi: 10.1093/jac/dkr024. [DOI] [PubMed] [Google Scholar]
- 27.Geng W, et al. Molecular characteristics of community-acquired, methicillin-resistant Staphylococcus aureus isolated from Chinese children. FEMS Immunol Med Microbiol. 2010;58:356–362. doi: 10.1111/j.1574-695X.2009.00648.x. [DOI] [PubMed] [Google Scholar]
- 28.Brown ML, et al. Prevalence and sequence variation of panton-valentine leukocidin in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains in the United States. J Clin Microbiol. 2012;50:86–90. doi: 10.1128/JCM.05564-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Souza N, Rodrigues C, Mehta A. Molecular characterization of methicillin-resistant Staphylococcus aureus with emergence of epidemic clones of sequence type (ST) 22 and ST 772 in Mumbai, India. J Clin Microbiol. 2010;48:1806–1811. doi: 10.1128/JCM.01867-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monecke S, Slickers P, Ellington MJ, Kearns AM, Ehricht R. High diversity of Panton-Valentine leukocidin-positive, methicillin-susceptible isolates of Staphylococcus aureus and implications for the evolution of community-associated methicillin-resistant S. aureus. Clin Microbiol Infect. 2007;13:1157–1164. doi: 10.1111/j.1469-0691.2007.01833.x. [DOI] [PubMed] [Google Scholar]
- 31.Shallcross LJ, Fragaszy E, Johnson AM, Hayward AC. The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: a systematic review and meta-analysis. Lancet Infect Dis. 2013;13:43–54. doi: 10.1016/S1473-3099(12)70238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle-Vavra S, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab Invest. 2007;87:3–9. doi: 10.1038/labinvest.3700501. [DOI] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; Twenty-Fourth Informational Supplement. CLSI document M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute (2014).
- 34.Kampf G, Adena S, Rüden H, Weist K. Inducibility and potential role of mecA-gene-positive oxacillin-susceptible Staphylococcus aureus from colonized healthcare workers as a source for nosocomial infections. J Hosp Infect. 2003;54:124–129. doi: 10.1016/S0195-6701(03)00119-1. [DOI] [PubMed] [Google Scholar]
- 35.Zhao H, et al. Typing of Panton-Valentine Leukocidin-Encoding Phages and lukSF-PV Gene Sequence Variation in Staphylococcus aureus from China. Front Microbiol. 2016;7:1200. doi: 10.3389/fmicb.2016.01200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muttaiyah S, et al. Incidence, risk factors, and outcomes of Panton-Valentine leukocidin-positive methicillin-susceptible Staphylococcus aureus infections in Auckland, New Zealand. J Clin Microbiol. 2010;48:3470–3474. doi: 10.1128/JCM.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fridkin SK, et al. Active Bacterial Core Surveillance Program of the Emerging Infections Program Network. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352:1436–1444. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Most of the experimental data acquired/analyzed during this study have been included in this published version. Information on rest of the data can be obtained from the corresponding author on request.