Abstract
Advances in molecular biology have improved crops through transferring genes from one organism to new hosts, and these efforts have raised concerns about potential unexpected outcomes. Here, we provide evidence that a gene with a specific function in one organism can yield completely different effects in a new host. CaRZFP1 is a C3HC4-type RING zinc finger protein gene previously isolated from a cDNA library for heat-stressed hot pepper. In our previous work investigating in vivo CaRZFP1 function, we transferred CaRZFP1 into tobacco; transgenic tobacco exhibited enhanced growth and tolerance to abiotic stresses. As further analysis of CaRZFP1 ectopic expression in a heterologous host plant, here we mobilized and constitutively overexpressed CaRZFP1 in lettuce. In contrast to tobacco, transgenic lettuce exhibited poorer growth and delayed flowering compared with vector-only controls. To identify genes that might be involved in this phenotypic effect, transcriptome analyses on transgenic plants of both species were performed, uncovering dozens of genes that reflect the different outcomes between tobacco and lettuce. These included protein kinase, transcriptional factor, transporter protein, hormone and metabolism-related genes, and some unannotated genes. The opposite effects of CaRZFP1 ectopic expression in lettuce and tobacco address concerns of unexpectedly different outcomes in different host species.
Electronic supplementary material
The online version of this article (10.1007/s11032-018-0812-3) contains supplementary material, which is available to authorized users.
Keywords: Ectopic expression, RING zinc finger, Lettuce, Tobacco, Hot pepper, Gene expression pattern
Introduction
Advances in molecular biology have improved crops through transferring genes from one organism to new hosts. Studies showed that GM crops have increased crop yields by 21% and reduced pesticide usage by 37%. The agronomic and economic benefits of GM crops are apparently large and significant (Klumper and Qaim 2014). Until now, US-based companies have been only developers of GM crops which cultivated on a large scale as authorized. Other countries are now producing GM crops with little or no information systematically provided. The US regulatory authorities are beginning to accept the possible need of European style GM crop regulations to mitigate or avoid future risks to the environment and human health. A group of scientists did an extensive review of research on the safety of GM crops over the past years. They found that the scientific research conducted so far has not documented any significant adverse effects directly associated with the use of GM crops on human health and environment (Nicolia et al. 2014).
The zinc finger domain in zinc finger proteins (ZFPs) consists of 20–100 amino acid residues and is found ubiquitously from prokaryotes to eukaryotes (Krishna et al. 2003). Cysteine and histidine residues in the domain stabilize the structure through binding to one or more zinc ions (Laity et al. 2001). Comprising a large and diverse gene family, ZFPs have considerable variation in structure and recognition sequences that interact with DNA, chromatin, RNA, and proteins. Thus, ZFPs are linked to major, diverse biological functions, including transcriptional activation, DNA recombination, translational processes, signal transduction, programmed cell death, membrane association, as well as protein folding and assembly (Gamsjaeger et al. 2007; Zhang et al. 2013). ZFPs participate in various fundamental aspects of plant growth and development, such as phytohormone response, signal transduction, and responses to abiotic or biotic stimuli (Chai et al. 2015; Larrieu and Vernoux 2009; Liu et al. 2016). Originally, zinc fingers were classified based on the number and order of cysteine and histidine residues, resulting in Cys2His2, Cys4, Cys6, and other similarly named types. Recent ZFP groupings instead use unique characteristics of the folded domain from the protein backbone. The most common zinc finger “fold groups” are Cys2His2-like (classic zinc finger), treble clef, and zinc ribbon. Most of the Cys2His2-like ZFPs are also commonly called “Really Interesting New Gene (RING)” finger proteins. They have been further grouped into several classes (C2H2, C2C2, C2C2C2C2, C2HCC2C2, C2HC, C2HC5, C3HC4, C3H2C3, C3H, C3HDC3, C3HGC3, C3H2SC2, C4HC3, C4, C4C4, C5HC2, C6, and C8) according to the number of conserved cysteine and histidine residues, the spacing between them, and their specific molecular functions (Laity et al. 2001; Krishna et al. 2003; Gamsjaeger et al. 2007).
Previously, we isolated a C3HC4-type RING zinc finger protein gene, CaRZFP1, from a cDNA library of heat-stressed hot pepper (Capsicum annuum). Various environmental stresses (e.g., heat, cold, dehydration, and high salinity) induce CaRZFP1 transcription in hot pepper. When the CaRZFP1 open reading frame (ORF) was transformed into tobacco (Nicotiana tabacum) and ectopically expressed, growth and tolerance to abiotic stresses were enhanced in the transgenic tobacco plants (Zeba et al. 2009). In this study, we mobilized and expressed CaRZFP1 in lettuce (Lactuca sativa) to further analyze the effect of CaRZFP1 ectopic expression in a heterologous host plant. Contrasting with tobacco, transgenic lettuce exhibited poorer growth than vector-only controls, specifically delayed flowering, weakened leaf growth, shorter plant height, and stunted root growth. This report examines possible mechanisms underlying the different effects caused by the same RING zinc finger protein gene expressed in transgenic lettuce versus transgenic tobacco.
Materials and methods
Plant material and growth condition
Seeds of hot pepper (C. annuum L. cv. Bu Gang), tobacco (N. tabacum L. cv. Wisconsin 38), and lettuce (L. sativa L. cv. Chung Chima) were sown on soil in plastic pots and reared in a growth chamber under controlled conditions of 25 °C, 60% relative humidity, and a 16-h photoperiod from white fluorescent lamps (200 μmol photons m−2 s−1). Vector-only or CaRZFP1-transgenic tobacco and lettuce plants were further grown in a greenhouse insulated with a dual door at 25 ± 2 °C under natural lighting with additional fluorescent lighting to maintain a 16-h photoperiod. Transgenic tobacco and lettuce plants carrying recombinant expression construct of CaRZFP1 and the vector-only were self-fertilized, and T1 generation seeds were harvested. T1 generation seeds with the transgene expressed were selected on a kanamycin-containing medium, raised for flowering, and self-fertilized again to obtain T2 generation seeds. This selection, self-fertilization, and rearing in a greenhouse was repeated to get further generations, up to T5 generation. For the growth assay, transgenic lines were transferred to soil in plastic pots and reared in a growth chamber as described above. Four- to 5-week-old plants were used for nucleic acid extraction.
Generation of transgenic lettuce plants overexpressing CaRFZP1
The open reading frame of CaRZFP1 was amplified by PCR with a primer pair covering both termini. The 5′ primer was 5′-ATATGGATCCATGCAGAAGTCAACTGCTACG-3′ and the 3′ primer was 5′-ATATGGATCCCTAACCAAACAAATATAGGAATAC-3′ with the underlined BamHI restriction site. PCR was carried out with the initial reaction of 94 °C for 5 min; followed by 30 cycles of 94 °C for 1 min, 55 °C for 30 s, and 72 °C for 1 min; with a final reaction of 10 min at 72 °C. The amplified PCR product was then digested with BamHI and ligated into the pBKS1-1 plant expression vector (Suh et al. 1994) at the BamHI site to locate the open reading frame under the control of the CaMV35S promoter. Nucleotide sequence of the cloned coding region in pBKS1-1 was confirmed by an automated DNA sequencer (3730xI DNA Analyzer, Applied Biosystems). The CaRZFP-pBKS1-1 plasmid was electroporated into the Agrobacterium tumefaciens strain LBA4404 and used for transformation of lettuce. In brief, sterilized hypocotyls of lettuce were cut to 1–2 cm length and infected with the Agrobacterium cells carrying the expression construct. After co-cultivation for 24 h, explants were washed with sterilized MS medium and placed in a shoot induction medium containing 200 mg/ml kanamycin and 100 mg/ml cefotaxime. Kanamycin-resistant shoots were selected and transferred to a root induction medium containing 200 mg/ml kanamycin (Horsch et al. 1985). The putative CaRZFP1-transgenic plants were then transferred to soil and reared in a growth chamber, then in a greenhouse as described above.
RNA and DNA blot analyses
Total RNA was extracted from plant tissues frozen in liquid nitrogen. Briefly, frozen tissue was ground to powder, homogenized in 3 ml of extraction buffer (100 mM LiCl, 100 mM Tris-Cl pH 8.0, 10 mM EDTA, and 1% SDS), a mixture of 3 ml of chloroform-isoamyl alcohol (24:1) was added, followed by vortexing and centrifugation at 10,000×g for 25 min at 4 °C. The supernatant was transferred to a 1.5-ml microcentrifuge tube, extraction was repeated with 1.5 ml chloroform-isoamyl alcohol (24:1) mixture, and precipitated in an equal volume of 4 M LiCl at − 70 °C for 2 h. After centrifugation, the pellet was washed with cold 70% ethanol and dissolved in DEPC-treated distilled water. Twenty micrograms of total RNA, for each sample, was loaded onto a 1.2% agarose gel with formaldehyde. To check the integrity of the sample, RNA was visualized by staining with ethidium bromide and UV illumination after electrophoresis, and RNA was transferred onto nylon membranes (Hybond-N+, GE Healthcare Bio-Sciences), followed by cross-linking with UV illumination. To generate CaRZFP1-specific probe, the coding sequence of CaRZFP1 was amplified by PCR and labeled with α32P-dCTP. Pre-hybridization for 2 h and hybridization for 16 to 22 h were done in a solution of 1 M dibasic sodium phosphate (pH 7.2), 14% SDS, and 20 μl of 1 mM EDTA (pH 8.0) at 65 °C. For RNA blot analysis using oligonucleotides, 60 mer oligonucleotides were end labeled with γ32P-dATP and polynucleotide kinase. The hybridization procedure was the same as the PCR-generated probe, except the hybridization and washing temperature which was at 58 °C. After hybridization, the membrane was washed twice with 1X SSPE and 0.1% SDS for 15 min, once at room temperature, then at 65 °C, and the membrane was washed several times in 0.5X SSPE and 0.1% SDS at 65 °C (Sambrook and Russell 2001). The blots were exposed to a phosphoimager screen and an image was developed in a phosphoimager (Typhoon 8600, Molecular Dynamics). For RT-PCR analysis, total RNA preparation was predigested with DNase I (Takara Bio.) at 37 °C for 30 min, and the first cDNA strand was generated by reverse transcribing RNA using AMV reverse transcriptase (Promega) at 42 °C for 1 h. cDNA was quantified using a spectrophotometer (ND-1000, NanoDrop Technologies) and subjected to PCR using a primer pair of 5′-ATGCAGAAGTCAACTGCTACG-3′ and 5′-CTAACCAAACAAATATAGGAATAC-3′ covering the open reading frame of CaRZFP1 in a PCR mixer (Promega) on a DNA thermal cycler (MJ Mini thermal cycler, Bio-Rad) with a following profile: initial step at 94 °C for 5 min followed by 30 cycles of 94 °C for 1 min, 55 °C for 30 s, and 72 °C for 1 min and a final step at 72 °C for 10 min. Amplified RT-PCR products were confirmed by DNA nucleotide sequencing. All chemicals used were from Sigma-Aldrich, Becton, Dickinson and Co. and Duchefa Biochemie, otherwise mentioned.
Phenotypic assay
Seeds of CaRZFP1-transgenic and vector-only lettuce plants were sown in plastic pots and reared in a growth chamber or a greenhouse under the controlled conditions as described above for the phenotypic examination during the vegetative growth. The plants were moved to a greenhouse under the controlled conditions as described above to follow the full life cycle. Evaluated parameters were leaf length, leaf width, leaf fresh weight, root mass, root fresh weight, plant height, flowering time, flower size, and seed morphology.
Histological and in situ hybridization analyses
To examine the effect of ectopic expression of CaRZFP1 at the cellular level, the CaRZFP1-transgenic and the vector-only lettuce and tobacco (Zeba et al. 2009) plants were anatomically assayed. Leaves, stems, and roots of the plants were taken and placed them immediately in 4% paraformaldehyde and vacuum infiltrated until the tissues sank. The samples were washed twice with 1X PBS (130 mM NaCl, 7 mM Na2HPO4, and 3 mM NaH2PO4, pH 7.0) for 30 min each and dehydrated using increasing concentrations of ethanol series (10, 30, 40, 50, 60, 70, and 85%), 60 min for each step. The samples were then incubated in 95% ethanol plus 0.1% eosin-Y overnight. Next day, samples were incubated in 100% ethanol, histoclear/ethanol series of 25, 50, and 75%, and twice in 100% histoclear, 60 min for each step. Histoclear was replaced with paraplast and kept the samples at 60 °C overnight. Paraplast was changed three times in a day with fresh molten wax and cool to room temperature to solidify. Samples were cross-sectioned in 6- to 8-μm thickness using a microtome (Microm HM340E, Microm International GmbH). The cross sections were floated on DEPC-treated water and dried overnight at 40 °C to fix the sections onto Superfrost Plus Microscope Slides (Thermo Fisher Scientific). The cross sections were deparaffinized by dipping the slides in xylene for 10 min and rehydrated with a graded ethanol series (100, 95, 90, 80, 60, and 30%). After rinsing with distilled water, the sections were stained with 1% safranin and observed under light microscopes (Leica DC500, Leica Microsystems; Olympus BX51, Olympus Corporation). In situ hybridization to visualize CaRZFP1 transcript was carried out as described by Brewer et al. 2006. Briefly, the cross sections were fixed onto Superfrost Plus Microscope Slides and deparaffinized. The sections were incubated in 1X PBS for 5 min, permeabilized by proteinase K treatment for 30 min, fixed in 4% paraformaldehyde in phosphate buffer (pH 7.0) for 10 min, acetylated twice with 0.5% acetic anhydride in 0.1 M triethanolamine for 10 min each, and the slides were washed in 1X PBS for 5 min. Then, the sections were hybridized with in vitro transcribed digoxigenin-labeled CaRZFP1 sense or antisense riboprobes which were synthesized from linearized pBluescript plasmids containing CaRZFP1 as described in the (SP6/T7) DIG RNA labelling Kit (Roche Applied Science). Briefly, the root sections were hybridized with equal concentrations of either sense or antisense RNA probes in a hybridization buffer [50X Denhardt’s, 50% dextran sulfate, 100 mg/ml tRNA, 50% formamide, 10X in situ hybridization salts (3 M NaCl, 100 mM Tris-HCl pH 8.0, 100 mM Na-phosphate pH 6.8, and 50 mM EDTA pH 8.0)], covered with parafilm, and incubated at 53 °C overnight in a humidity chamber. After hybridization, the slides were put into 2X SSC for 60 min to allow parafilm to float off. Then, the slides were washed twice in 0.2X SSC at 55 °C for 1 h each and equilibrated twice in 1X NTE (2.5 M NaCl, 50 mM Tris-Cl pH 8.0, and 5 mM EDTA pH 8.0) at 37 °C with gentle agitation for 5 min each. The slides were then put into 1X NTE buffer with 20 μg/ml RNAse A for 30 min at 37 °C with gentle agitation, rinsed twice in 1X NTE for 2 min each, washed in 0.2X SSC at 55 °C for 1 h, equilibrated in 1X PBS for 5 min at room temperature, incubated in blocking solution [1% blocking reagent (Roche Diagnostics Gmbh), 100 mM Tris-Cl pH 7.5, and 150 mM NaCl] for 45 min at room temperature, and washed with a washing buffer (1% BSA, 100 mM Tris-Cl pH 7.5, 150 mM NaCl, and 0.3% Triton X-100) for 45 min. The anti-digoxigenin antibody (Roche Applied Science) was diluted to 1:1250 ratio in the washing buffer, and 200 μl of anti-digoxigenin antibody solution was directly applied onto the slide carrying the parafilm strips. The slides were incubated in the dark for 2–3 h at room temperature or overnight at 4 °C, washed four times in washing buffer for 15 min each on a shaking platform, and the slides were transferred into 1X PBS, incubated for 2 min, and equilibrated in TN buffer (100 mM Tris-HCl pH 9.5 and 100 mM NaCl) twice for 2 min each. Staining solution was prepared immediately before use by adding 20 μl nitro-blue tetrazolium/5-bromo-4-chloro-3′-indolyphosphate per 1 ml TN buffer. The slides were covered with the staining solution and kept in a plastic box in the dark at room temperature for 2–3 days. Development of staining was monitored under a light microscope, and once the color reaction was complete, the slides were placed in TE buffer for 5–10 min to stop the staining reaction. The slides were washed in 1X PBS for 5 min, and images were captured with a light microscope (Olympus BX51).
Transcriptome analysis
Four T4 generation CaRZFP1-transgenic lettuce lines (no. 6, no. 12, no. 14, and no. 16) and three vector-only control lettuce lines (V10, V30, and V38) were selected for transcriptome analysis using Arabidopsis 44K oligo microarray (Agilent Technology) to maintain the methods used for the transcriptome profiling of CaRZFP1-transgenic tobacco (Zeba et al. 2009). Total RNA was extracted from 4-week-old plants, and genomic DNA was removed by DNase I (Takara Bio Inc.) digestion. Synthesis of cRNA probes and hybridization were carried out using Agilent’s low RNA Input linear amplification kit (Agilent Technologies) according to the manufacturer’s instructions. Briefly, total RNA 1 μg was mixed with T7 promoter primer mix and incubated at 65 °C for 10 min, cDNA master mix (5X first strand buffer, 0.1 M DTT, 10 mM dNTP mix, RNase-Out, and MMLV-RT) was added, incubated at 40 °C for 2 h, and reverse transcription and dsDNA synthesis were terminated by incubating at 65 °C for 15 min. Transcription of the dsDNA was done by adding transcription master mix (4x transcription buffer, 0.1 M DTT, NTP mix, 50% PEG, RNase-Out, inorganic pyrophosphatase, T7-RNA polymerase, and cyanine 3-CTP) and incubating at 40 °C for 2 h. The labeled cRNA was purified on cRNA cleanup module and hybridized to the microarrays at 65 °C for 17 h. After hybridization, microarrays were washed for 1 min at room temperature with GE Wash Buffer 1, again for 1 min at 37 °C with GE Wash buffer 2, and dried immediately by centrifugation at 400×g for 2 min at room temperature. The hybridization images were scanned using a DNA microarray scanner and quantified with the feature extraction software 9.3.2.1 (Agilent Technologies). Data normalization and calculation of fold change were performed using GeneSpringGX 7.3 (Agilent Technologies). Greater than twofold changes with p < 0.05 were set as the threshold for statistical significance (McCarthy and Smyth 2009).
Data availability statement
All the data in this publication will be available upon request.
Results
CaRZFP1 overexpressing transgenic lettuce plants showed hampered growth and development
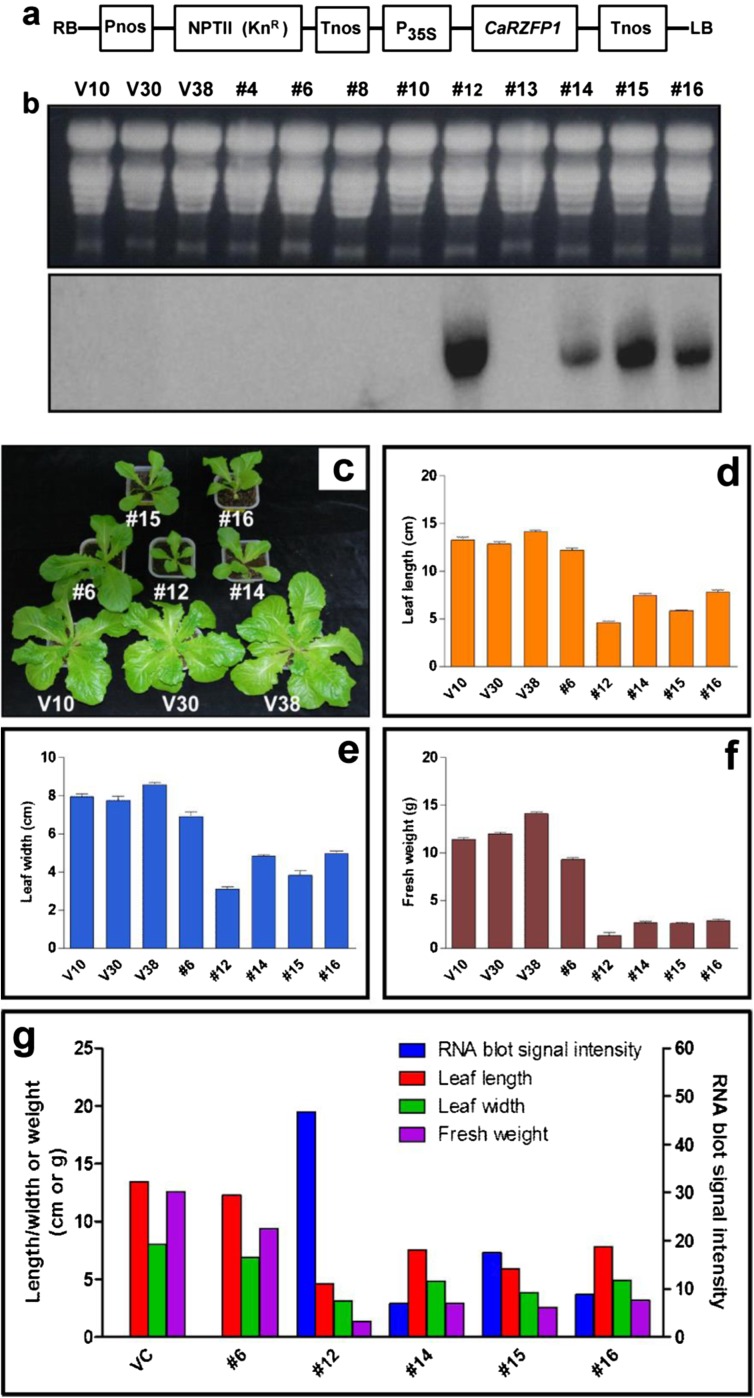
To investigate the effect of CaRZFP1-ectopic expression, transgenic lettuce was generated that overexpressed CaRZFP1 under the control of cauliflower mosaic virus 35S promoter (Fig. 1a). This construct and the vector without CaRZFP1 were mobilized into the lettuce genome. The putative CaRZFP1-transgenic lettuce plants were selected on kanamycin-containing medium and planted in soil. Next, T1 generation lines were screened with RNA blot analysis to evaluate CaRZFP1 transcript levels and confirm transgenicity (Fig. 1b). In the representative examples shown, T1 transgenic line no. 12, no. 14, no. 15, and no. 16 exhibited different but significant levels of ectopically expressed CaRZFP1 transcript under normal growth conditions. Line no. 6 expressed very low CaRZFP1 levels undetectable via RNA blot hybridization; transcripts were thus confirmed with RT-PCR and DNA blot analyses (Fig. S1). Confirmed T1 lines were reared in a greenhouse and repeatedly self-fertilized to obtain next-generation seeds (T2 to T5). Transgenicity of all putative transgenic plants from different generations was confirmed with RNA blot analysis. We noticed that most CaRZFP1-transgenic lettuce plants with detectable CaRZFP1 transcript levels exhibited growth impairment, more strongly in some lines than in others. Starting from T2, we categorized transgenic lettuce lines according to CaRZFP1 transcript expression levels (low, medium, and high). Five independent T2 lines were selected based on CaRZFP1 transcript levels and denoted with numerals 6, 12, 14, 15, and 16. Additionally, three vector-only lines (V10, V30, and V38) with morphology typical of nontransgenic lettuce were also selected for further analyses (Fig. 1b).
Fig. 1.
CaRZFP1 overexpressing transgenic lettuce plants showed hampered growth and development. a Diagrammatic representation of pBKS1-1-CaRZFP1 construct used for lettuce transformation. For the expression vector pBKS1-1, only the region inside of the border sequences, RB and LB, that was actually transferred into the lettuce genome is shown. b RNA blot hybridization results for vector-only and putative T1 transgenic plant lines. Total RNA was separated by electrophoresis on a 1.2% formaldehyde agarose gel and blotted to a Hybond-N nylon membrane. Separated RNA was stained with ethidium bromide for visualization with UV illumination. The blots were hybridized to 32P-labeled CaRZFP1 probe. c Typical examples of transgenic lettuce plant lines no. 6, no. 12, no. 14, no. 15, and no. 16 and lettuce plants carrying only the vector after 4 weeks since seed imbibition. d Comparison of leaf length of the plants in c. e Comparison of leaf width of the plants in c. f Comparison of fresh weight of the plants in c. g The data in d, e and f were aligned with CaRZFP1 transcript level analyzed by RNA blot hybridization. V10, V30 and V38, lettuce plants carrying only the expression vector. Error bars show standard deviation. VC, average of V10, V30 and V38. h RNA blot hybridization results of vector-only or putative T3 CaRZFP1-transgenic lettuce lines that are shown in i. i Typical examples of T3 transgenic lettuce plant lines no. 6, no. 12, no. 14, no. 15 and no. 16 and lettuce plants carrying only the vector after 12 weeks since seed imbibition. j Typical roots of T3 transgenic lettuce plant lines no. 6, no. 12, no. 14, no. 15 and no. 16 and lettuce plants carrying only the vector after 4 weeks since seed imbibition. k Comparison of root mass of the plants in j. l RNA blot hybridization results for the plants in j. m RT-PCR results for the same samples in l. n RT-PCR results in m was further confirmed by DNA blot analysis with 32P-labeled CaRZFP1 probe. M, size marker. Error bars show standard deviation
A strong negative correlation existed between CaRZFP1 transcript level and growth robustness in the vegetative stage, i.e., stronger CaRZFP1 expression, was tightly linked to severe growth retardation. Growth and development retardation was noticeable soon after germination and continued thereafter; transgenic lettuce exhibited poorer overall growth compared with vector-only plants, including diminishing leaf growth, shorter plant height, and lower fresh weight (Fig. 1c–f). Line no. 12 exhibited the highest CaRZFP1 expression and the most impeded growth among all transgenic lettuce lines. Line no. 14, no. 15, and no. 16 were more moderate in terms of both CaRZFP1 expression and hampered growth. Line no. 6’s very low, near-undetectable CaRZFP1 expression corresponded to a negligible effect on growth and development; its phenotype was similar to vector-only controls. Ectopic CaRZFP1 expression thus caused pleiotropic developmental changes. After 28 days of germination, the average leaf lengths of CaRZFP1-transgenic lettuce line no. 6, no. 12, no. 14, no. 15, and no. 16 were 13.8 ± 0.47, 9.3 ± 0.19, 11.0 ± 0.62, 10.4 ± 0.74, and 11.5 ± 0.89 cm, while leaves from vector-only V10, V30, and V38 averaged 15.4 ± 0.53, 14.4 ± 0.30, and 14.5 ± 0.32 cm (Fig. 1d). Average leaf widths of no. 6, no. 12, no. 14, no. 15, and no. 16 were 6.9 ± 0.41, 4.0 ± 0.22, 5.6 ± 0.78, 5.1 ± 0.24, and 5.0 ± 0.74 cm, whereas V10, V30, and V38 leaf widths averaged 9.0 ± 0.25, 8.4 ± 0.31, and 8.6 ± 0.29 cm (Fig. 1e). Average fresh weights of no. 6, no. 12, no. 14, no. 15, and no. 16 were 4.4 ± 0.23, 1.3 ± 0.34, 2.9 ± 0.15, 2.7 ± 0.21, and 3.4 ± 0.24 g, while those of V10, V30, and V38 were 6.6 ± 0.43, 5.6 ± 0.34, and 5.7 ± 0.20 g (Fig. 1f). The correlation between CaRZFP1 transcript level and a negative phenotypic effect was repeatedly maintained through the later generations of CaRZFP1-transgenic lettuce (Fig. 1g–i). Overall, growth of CaRZFP1-transgenic lettuce plants was dramatically impeded by CaRZFP1 expression in a dose-dependent fashion.
This negative correlation extended to the root growth. Transgenic lettuce possessed shorter, less-developed roots, and this phenotype was closely correlated to CaRZFP1 transcript levels. Compared with vector-only plants, primary root growth as well as total root-system length and lateral/adventitious root formation were strongly decreased in line no. 12 and mildly impaired in line no. 14, no. 15, and no. 16 but only slightly affected in line no. 6 (Fig. 1j). Average root fresh weights in line no. 6, no. 12, no. 14 no. 15, and no. 16 were 1.05 ± 0.15, 0.17 ± 0.01, 0.55 ± 0.09, 0.39 ± 0.04, and 0.52 ± 0.08 g, while those in V10, V30, and V38 were 1.71 ± 0.20, 1.64 ± 0.28, and 1.40 ± 0.28 g (Fig. 1k). Thus, CaRZFP1 expression was strongly correlated with root underdevelopment (Fig. 1k and l). Again, CaRZFP1 expression in line no. 6 was very low in the root and required confirmation with RT-PCR and DNA blot analysis (Fig. 1m, n).
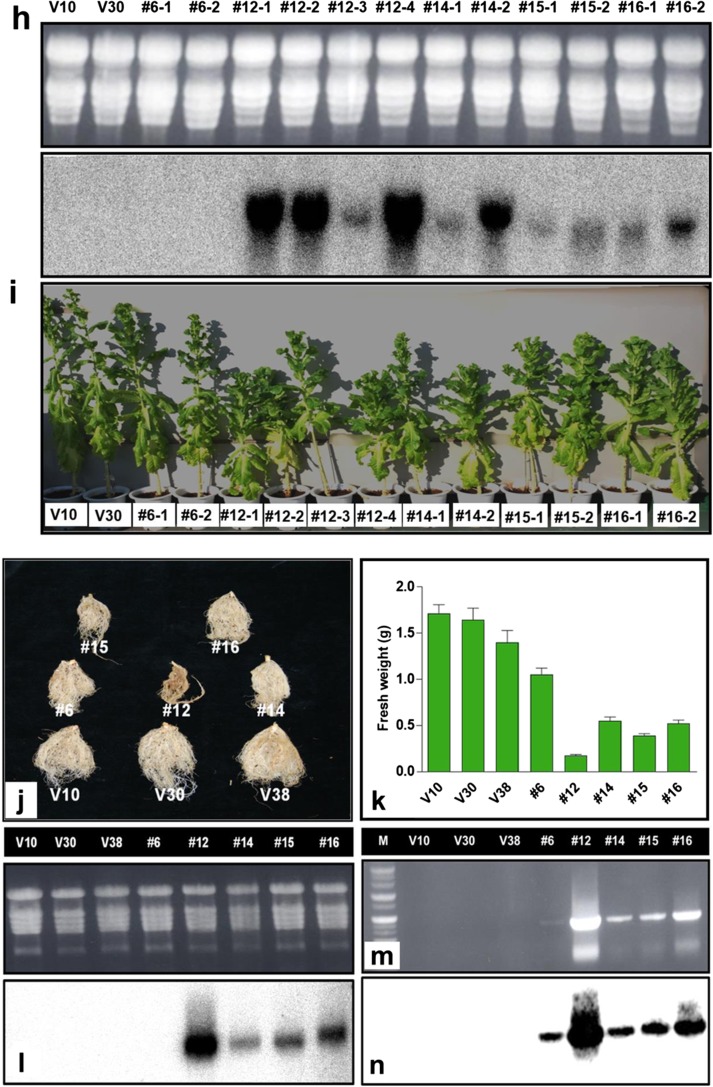
The weak growth of CaRZFP1-transgenic lettuce plants continued late in development, in contrast to vector-only plants, which appeared normal from the vegetative phase to flowering. At full growth, CaRZFP1-transgenic lettuce was shorter than vector-only plants (Fig. 2a). Additionally, vector-only plants began to flower at 115 days after sowing (DAS), whereas CaRZFP1-transgenic lettuce began to flower 123 to 130 DAS (Fig. 2b). Inflorescence size was smaller in transgenic lettuce than in vector-only plants (Fig. 2c). Flower size did not differ significantly between the transgenic and control lines, but the former had significantly fewer flowers per inflorescence, a characteristic that was again correlated with CaRZFP1 transcript levels. Transgenicity at full growth was again confirmed with RNA blot hybridization for most lines and with RT-PCR and DNA blot analysis for line no. 6 (Fig. 2d–f). Seed development in each flower appeared normal in transgenic lettuce (Fig. 2g–i), although the total seed number per inflorescence was negatively correlated with CaRZFP1 expression levels. Seed size, morphology, and weight in the transgenic lettuce lines did not differ significantly from those in the vector-only plants (Fig. 2g–j). We next selected four CaRZFP1-transgenic lettuce lines (no. 6, no. 12, no. 14, and no. 16) for further investigation.
Fig. 2.
Hampered growth and development CaRZFP1 overexpressing transgenic lettuce plants extended to the reproductive stage. a Typical examples of T3 transgenic lettuce plant lines no. 6, no. 12, no. 14, no. 15 and no. 16 and lettuce plants carrying only the vector at the full growth. b Typical examples of T3 transgenic lettuce plant lines and vector-control lines at the flowering stage. c Fully developed inflorescences. d RNA blot hybridization results for the plants in b. e RT-PCR results for the same samples in d. f RT-PCR results in e was further confirmed by DNA blot analysis with 32P-labeled CaRZFP1 probe. M, size marker. g Typical examples of mature T2 generation seeds. h Typical examples of mature T3 generation seeds. i Typical examples of mature T4 generation seeds. j Average weight of 100 mature seeds and number of seeds per inflorescence of CaRZFP1-transgenic and vector-only lettuce lines from T2, T3 and T4 generation. Error bars show standard deviation. Student t test analyses showed that there was statistically significant at the <0.05 and < 0.001 level (*P < 0.05 and **P < 0.001) difference between the average weight of 100 mature seeds and number of seeds per inflorescence of CaRZFP1-transgenic and vector-only lettuce lines
CaRZFP1-transgenic lettuce plants were differentially damaged in root internal structures
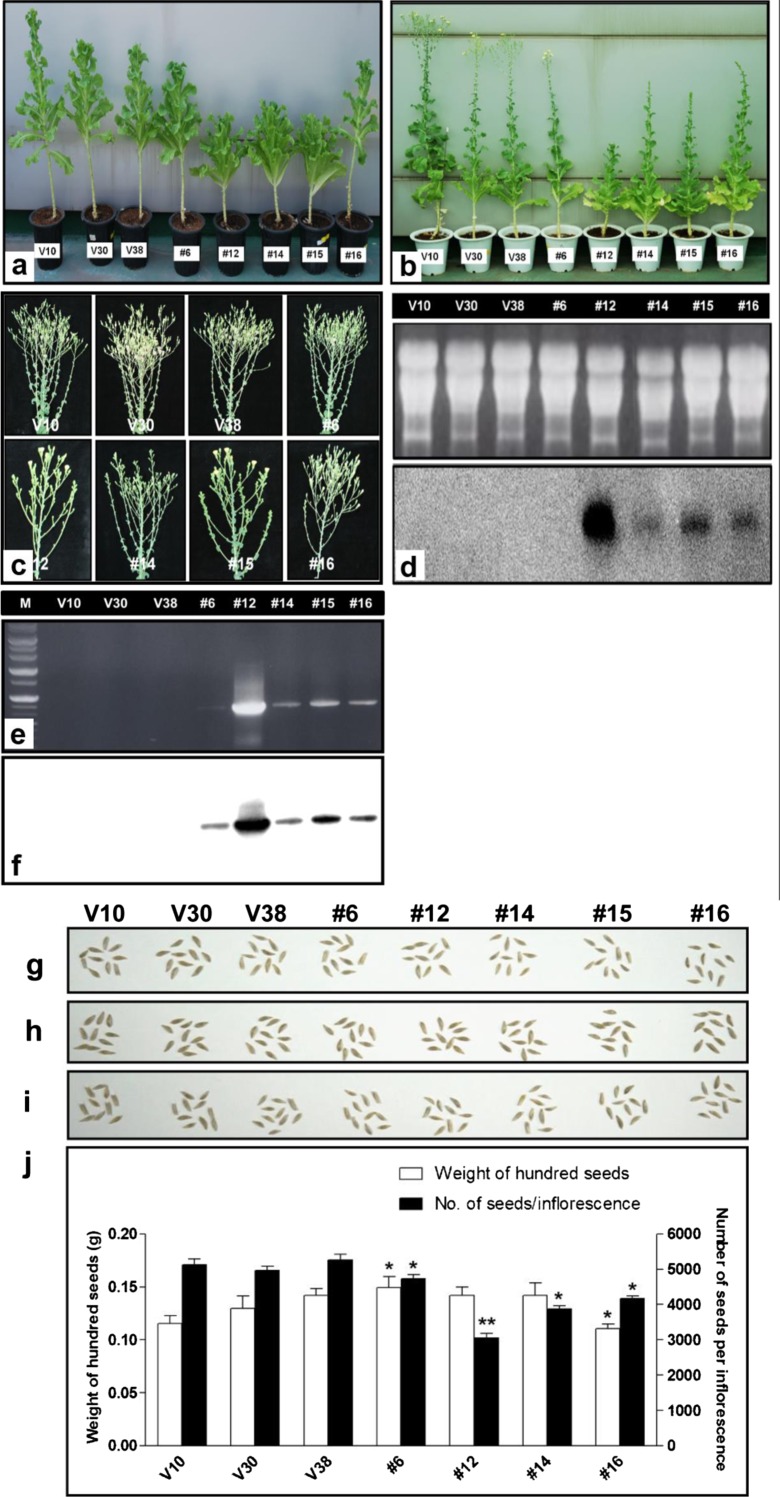
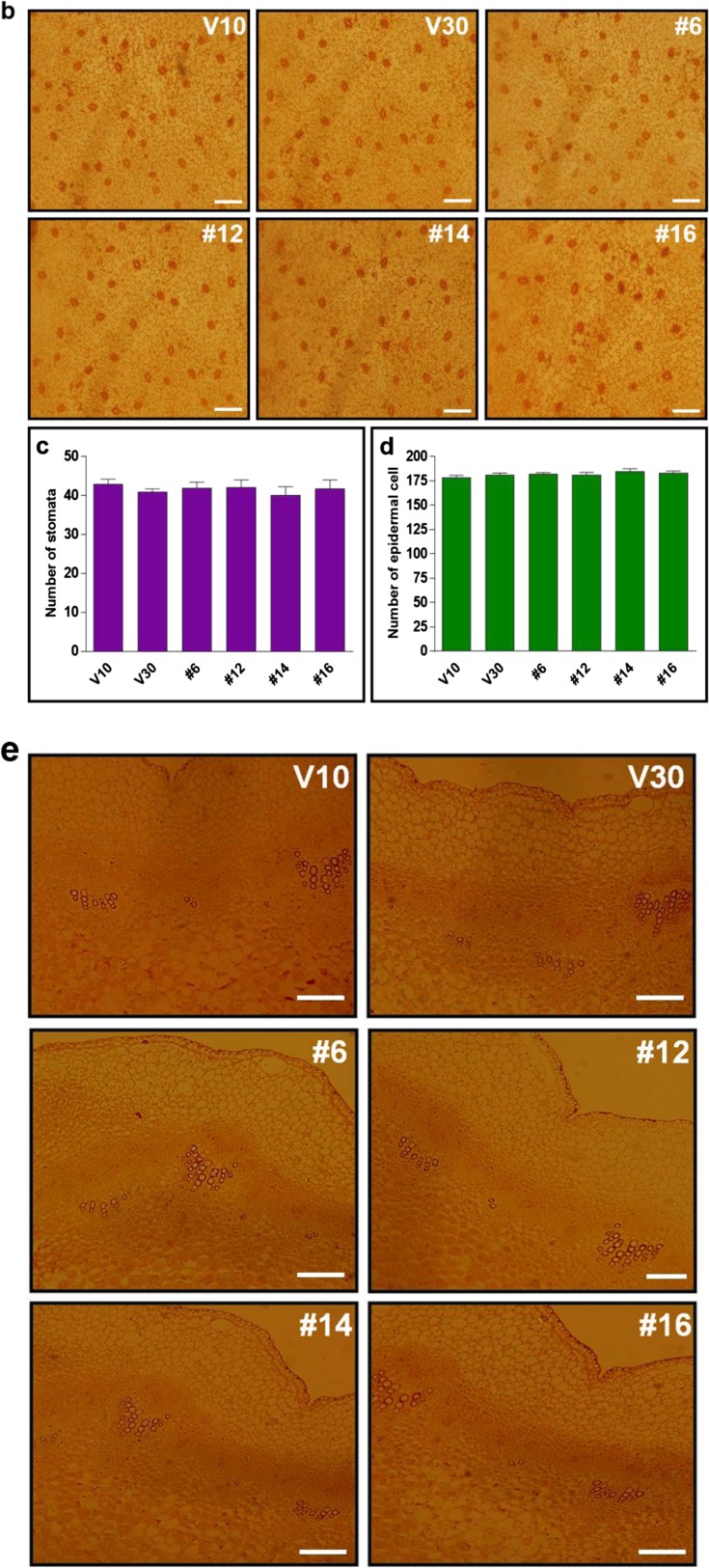
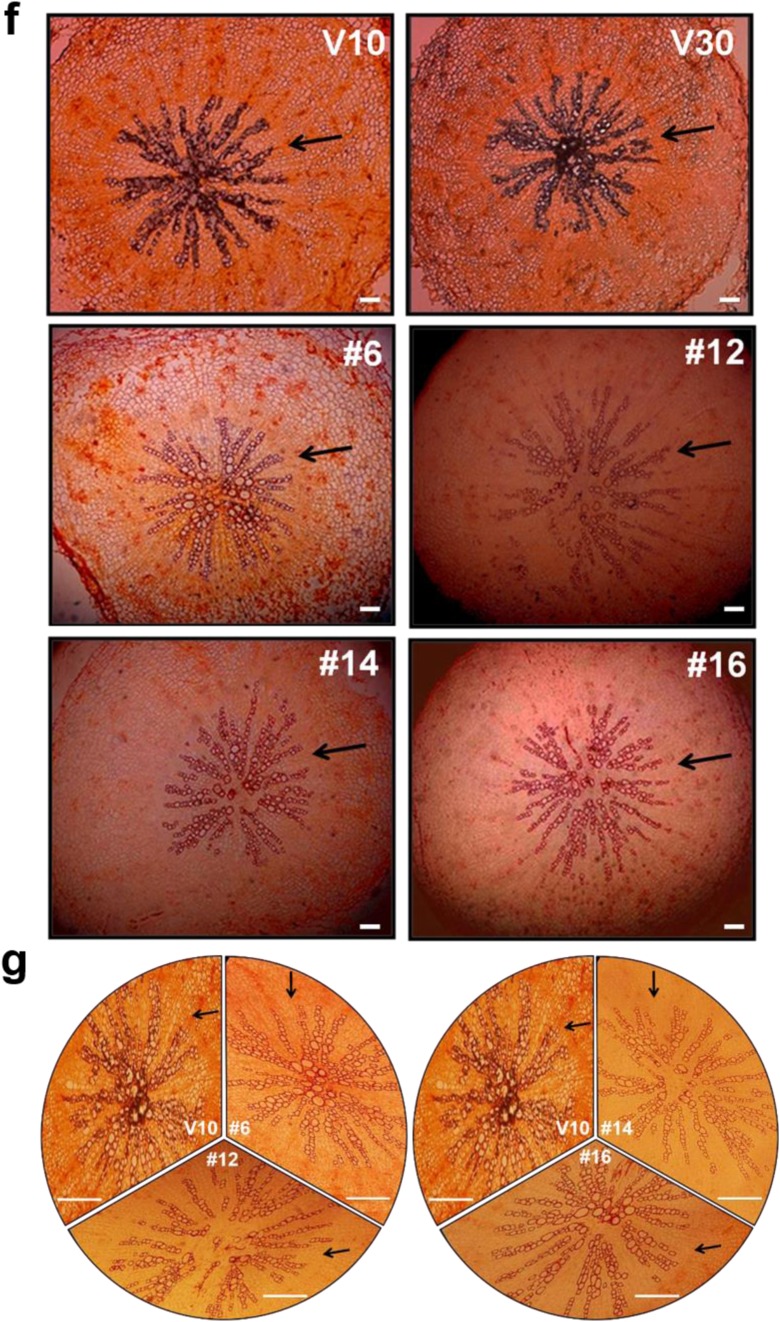
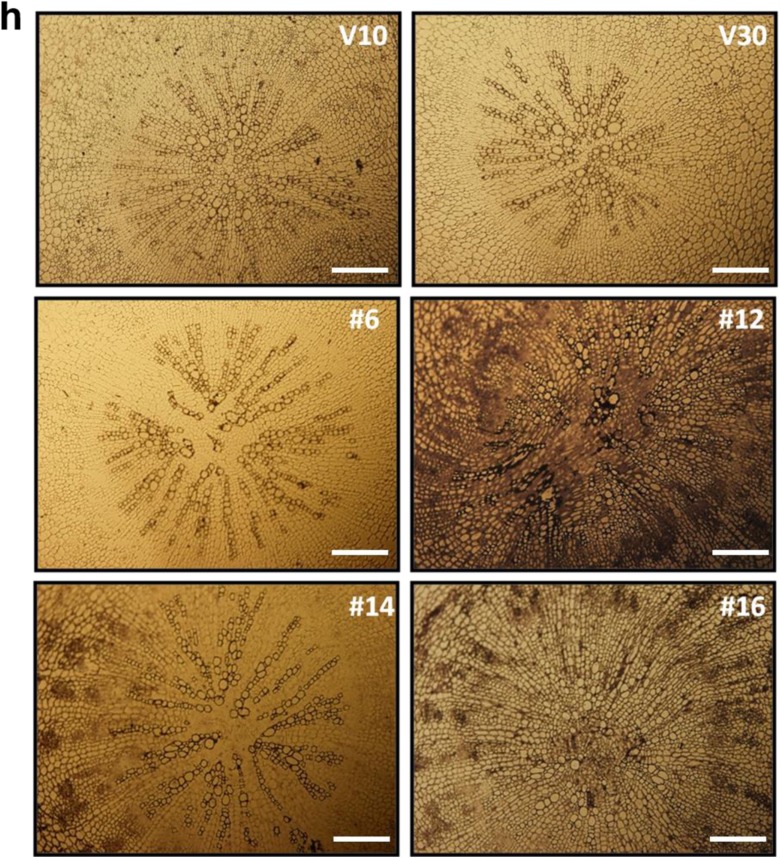
To examine the retarded growth of CaRZFP1-transgenic lettuce and the robust growth of CaRZFP1-transgenic tobacco (Zeba et al. 2009) on the cellular level, we analyzed leaf, stem, and root sections of both plants and compared them with vector-only controls. Leaves of transgenic lettuce and vector-only lettuce were not distinguishable in terms of morphology, cell size, or tissue organization. Cross sections of transgenic and control lines revealed basically identical mesophyll cell size and number, vascular bundle structure, chloroplast distribution in the mesophyll cells, as well as epidermal cell size and morphology (Fig. 3a). Top views of epidermal cells and stomata of transgenic and vector-only lettuce did not reveal any differences in epidermal cell size, stomatal size, or stomatal density (Fig. 3b–d). Likewise, stem morphology, cell size, and tissue organization in transgenic versus vector-only lettuce were indistinguishable (Fig. 3e). Significant defects in endodermis and vascular bundle development were observed in CaRZFP1-transgenic lettuce roots. The internal structure of vector-only lettuce roots was typical and well defined; visible structures included the surrounding epidermal layer, parenchyma cells inside the epidermis, endodermal layer, and pericycle surrounding the internal vascular bundles, as well as radially lined vessel elements and phloem between the xylem elements (Fig. 3f, g). The internal root structures of transgenic lettuce plants significantly deviated from typical. In line with the strongest CaRZFP1 expression (no. 12), the endodermis and pericycle were barely defined and vessel elements were not compactly structured and sometimes even disconnected, while the xylem element did not appear to be formed at the root center (Fig. 3f, g). These root development defects were also present in other transgenic lettuce lines to a lesser extent and were strongly correlated with CaRZFP1 expression levels; stronger CaRZFP1 expression resulted in greater impairment in endodermis, pericycle, and vascular bundle structure. However, CaRZFP1 expression did not seem to affect root cortex cells; parenchymal cell size and overall shape were normal in CaRZFP1-transgenic lettuce (Fig. 3f, g). To identify spatial CaRZFP1 expression patterns in CaRZFP1-transgenic and vector-only lettuce roots, we performed in situ hybridization assays. The antisense probe of CaRZFP1 detected the transcript strongly from the roots of line no. 12, at medium levels from line no. 14 and no. 16 and at low levels from line no. 6; however, CaRZFP1 mRNA was expressed equally within all root cell types in every transgenic plant. In the vector-only plants, no significant signal could be detected (Fig. 3h). Hybridization of the root sections with the CaRZFP1 sense probe found no significant signal over background, failing to distinguish between the transgenic and vector-only roots (data not shown). Previously, we engineered transgenic lines of tobacco overexpressing CaRZFP1 under the control of CaMV35S promoter. These CaRZFP1-transgenic tobacco plants exhibited robust growth and abiotic stress tolerance (Zeba et al. 2009). When these CaRZFP1-transgenic tobacco plants were examined for the development of roots at the cellular level, the overall root cell structure, tissue patterns, or morphology, including the endodermis and vascular bundle, did not differ from vector-only plants (Fig. S2a–c).
Fig. 3.
Cytological comparisons of CaRZFP1-transgenic and vector-only lettuce plants. a Leaf cross sections of CaRZFP1-transgenic lettuce and vector-only lettuce. Scale bar is 10 μm. b Leaf epidermal layer with stomata. Scale bar is 25 μm. c Number of stomata in CaRZFP1-transgenic lettuce lines and vector-only lettuce lines. d Number of epidermal cells in CaRZFP1-transgenic lettuce lines and vector-only lines. Error bars show standard deviation. e Stem cross sections of CaRZFP1-transgenic lettuce lines and vector-only lines. Scale bar is 25 μm. f Root cross sections of CaRZFP1-transgenic lettuce lines and vector-only lines. Arrows point endodermis layer. Scale bar is 25 μm. g Magnified view of f. Scale bar is 25 μm. h In situ localization of CaRZFP1 transcript in CaRZFP1-transgenic lettuce plant roots. Cross sections of roots were hybridized with digoxigenin-labeled CaRZFP1 antisense RNA probes. Scale bar is 25 μm
Different genes were induced in CaRZFP1-transgenic lettuce and CaRZFP1-transgenic tobacco plants
To elaborate on the potential underlying genes, whole transcriptome profiling was performed on transgenic lettuce that differed in CaRZFP1 transcript levels (low, moderate, and high; line no. 6, no. 12, no. 14, and no. 16) and the vector-only lettuce. Agilent Arabidopsis GE 4 X 44K microarray was used for the transcriptome profiling of the lettuce lines because a lettuce microarray was unavailable and our previous work on tobacco also used the Arabidopsis microarray (Zeba et al. 2009). Differentially expressed (either up- or down-regulated) transcripts between line no. 12 (with the strongest CaRZFP1 expression) and control lines were determined via a twofold change threshold (P < 0.05). Among 87 genes identified as significantly differentially expressed, 73 were up-regulated (Tables 1a and S1) and 14 were down-regulated (Tables 1b and S2). Differentially expressed genes in transgenic line no. 12 were again screened for the correlation between CaRZFP1 expression and growth impairment in line no. 14, no. 16 (medium CaRZFP1 expression), and no. 6 (low CaRZFP1 expression). This analysis verified the presence of correlative change in fold values from the highest in line no. 12 to the lowest in vector-only lines. We separated the up- and down-regulated genes into two groups: “Group 1” included genes with correlative changes in expression level among all four CaRZFP1-transgenic lines, while “Group 2” included only genes with significant expression level changes mainly in line no. 12. Group 1 contained 24 up-regulated putatively annotated genes, seven up-regulated unannotated genes, seven down-regulated putatively annotated genes, and one down-regulated unannotated gene (Tables 1a, b, and S1 and S2). Group 2 contained 39 up-regulated putatively annotated genes, three up-regulated unannotated genes, four down-regulated putatively annotated genes, and two down-regulated unannotated genes (Tables 1a, b, and S1 and S2).
Table 1.
Transcriptome profiling of CaRZFP1 overexpressing lettuce and tobacco plants by microarray analysis
| Gene symbol | Gene description | CaRZFP1 -transgenic lettuce lines/vector controls (log 2 fold change) | |||
|---|---|---|---|---|---|
| No. 6 | No. 14 | No. 16 | No. 12 | ||
| a. Genes up-regulated in CaRZFP1-overexpressing T4 generation lettuce plants | |||||
| At5g53170 | Cell division protease ftsH-11 (FTSH11) | 0.28 | 0.38 | 1.36 | 2.36 |
| At5g52910 | Timeless family protein (ATIM) | 0 | 0.52 | 0.42 | 3.35 |
| At1g08260 | DNA polymerase epsilon subunit 1 (TIL1) | 0 | 0 | 0 | 3.35 |
| At1g80490 | Topless-related protein 1 (TPR1) | 0.50 | 0.75 | 0.37 | 3.31 |
| At1g55110 | Indeterminate-domain 7 protein (IDD7) | 0.03 | 1.33 | 0.76 | 3.09 |
| At4g31920 | Arabidopsis response regulator 10 (ARR10) | 0.00 | 0.30 | 0.11 | 3.31 |
| At5g08130 | BES1-interacting MYC-like 1 (BIM1) | 0 | 0.20 | 0.18 | 3.08 |
| At1g80580 | Ethylene-responsive factor (ERF) | 0 | 0 | 0 | 4.32 |
| At1g08465 | Putative axial regulator YABBY 2 (YAB2) | 0 | 0 | 0 | 5.97 |
| At1g20910 | ARID/BRIGHT DNA-binding domain-containing protein | 0 | 0 | 0 | 4.38 |
| At3g44370 | Membrane insertion protein, OxaA/YidC with tetratricopeptide repeat domain-containing protein | 0.21 | 1.23 | 1.01 | 2.68 |
| At1g21310 | Extensin 3 (EXT3) | 0.73 | 1.44 | 1.02 | 3.50 |
| At4g33610 | Glycine-rich protein | 0 | 0 | 0 | 3.08 |
| At3g28550 | Proline-rich extensin-like family protein | 0 | 0 | 0 | 3.04 |
| At3g24860 | Hydroxyproline-rich glycoprotein family protein | 0 | 0 | 0 | 2.95 |
| At4g30170 | Peroxidase 45 | 0.12 | 1.34 | 0.83 | 2.65 |
| At5g48300 | Glucose-1-phosphate adenylyltransferase small subunit (ADG1) | 0.02 | 0.26 | 0.41 | 2.79 |
| At5g08530 | NADH dehydrogenase (ubiquinone) flavoprotein 1(CI51) | 0.02 | 1.22 | 1.53 | 3.02 |
| At3g03890 | FMN binding protein | 1.39 | 1.59 | 2.08 | 3.12 |
| At4g10120 | Sucrose-phosphate synthase | 0 | 0 | 0 | 2.44 |
| At5g66230 | Chalcone-flavanone isomerase family protein | 0 | 0 | 0 | 4.34 |
| At1g17420 | Lipoxygenase 3 (LOX3) | 0 | 0 | 0 | 4.60 |
| At5g08100 | Isoaspartyl peptidase/l-asparaginase 1 subunit beta | 0 | 0 | 0 | 4.63 |
| At5g41080 | Glycerophosphoryl diester phosphodiesterase family protein | 0 | 0 | 0 | 4.10 |
| At1g54620 | Pectin methylesterase inhibitor superfamily protein | 0 | 0 | 0 | 4.30 |
| At1g06700 | Protein kinase domain-containing protein | 0.50 | 0.48 | 0.06 | 4.08 |
| At3g48190 | Serine/threonine-protein kinase (ATM) | 0.00 | 1.50 | 0.86 | 2.74 |
| At4g04960 | L-type lectin receptor kinase VII.1 (LECRK-VII.1) | 0 | 0 | 0 | 4.31 |
| At3g20860 | NIMA-related kinase 5 (NEK5) | 0 | 0 | 0 | 2.44 |
| At3g24540 | Proline-rich extensin-like receptor kinase (PERK) | 0 | 0 | 0 | 3.46 |
| At5g11360 | Interleukin-1 receptor-associated kinase 4 protein | 0 | 0 | 0 | 3.33 |
| At5g12235 | CLAVATA3/ESR-related 22 protein (CLE22) | 0 | 0 | 0 | 3.99 |
| At3g55840 | Hs1pro-1 protein | 0.54 | 1.16 | 0.87 | 4.62 |
| At3g61185 | Defensin-like (DEFL) family protein | 0 | 0 | 0 | 5.43 |
| At4g10850 | Nodulin MtN3-like protein | 0.32 | 0.40 | 0.39 | 3.39 |
| At3g07100 | Sec24-like transport protein (ERMO2) | 0.69 | 1.27 | 2.14 | 3.49 |
| At5g27100 | Glutamate receptor 2.1 (GLR2.1) | 0.06 | 0.24 | 1.97 | 2.52 |
| At5g01990 | Auxin efflux carrier family protein | 0.87 | 1.44 | 1.48 | 2.32 |
| At3g26520 | Tonoplast intrinsic protein 2 (TIP2) | 0 | 0 | 0 | 2.86 |
| At2g39890 | Proline transporter 1 (PROT1) | 0 | 0 | 0 | 3.27 |
| At5g51710 | K(+) efflux antiporter 5 (KEA5) | 0 | 0 | 0 | 2.47 |
| At1g79520 | Cation efflux family protein | 0 | 0 | 0 | 2.47 |
| At3g55740 | Proline transporter 2 (PROT2) | 0 | 0 | 0 | 2.55 |
| At1g80300 | Nucleotide transporter 1 (NTT1) | 0 | 0 | 0 | 3.64 |
| At1g23910 | Polyketide cyclase/dehydrase and lipid transport superfamily protein | 0 | 0 | 0 | 4.77 |
| At4g19680 | Fe(2+) transport protein 2 (IRT2) | 0 | 0 | 0 | 4.06 |
| At5g59030 | Copper transporter 1 (COPT1) | 0 | 0 | 0 | 4.65 |
| At4g08290 | Nodulin MtN21/EamA-like transporter family protein | 0 | 0 | 0 | 4.88 |
| At1g31920 | Pentatricopeptide repeat-containing protein | 0.14 | 1.39 | 1.29 | 3.76 |
| At2g17600 | Cysteine/histidine-rich C1 domain-containing protein | 0.46 | 0.62 | 0.48 | 2.77 |
| At5g45428 | Conserved peptide upstream open reading frame 24 (CPuORF24) | 0.53 | 0.54 | 0.63 | 3.23 |
| At5g42990 | Putative ubiquitin-conjugating enzyme E2 18 (UBC18) | 0.27 | 0.77 | 1.01 | 2.17 |
| At1g71410 | Armadillo/beta-catenin-like repeats-containing protein | 0.75 | 1.88 | 2.04 | 3.09 |
| At4g01640 | F-box associated ubiquitination effector family protein | 0.04 | 0.66 | 0.80 | 6.21 |
| At2g35140 | Development and cell death domain protein (DCD) | 0 | 0.31 | 0.88 | 3.53 |
| At1g31090 | F-box domain-containing protein | 0 | 0 | 1.26 | 2.97 |
| At4g03510 | E3 ubiquitin-protein ligase (RMA1) | 0 | 0 | 2.05 | 3.47 |
| At3g62940 | Cysteine proteinases family protein | 0 | 0 | 0.17 | 4.72 |
| At3g26805 | Aspartic protease family protein | 0 | 0 | 0 | 4.47 |
| At4g30020 | PA-domain-containing subtilase family protein | 0 | 0 | 0 | 4.99 |
| At1g06630 | F-box domain-containing protein | 0 | 0 | 0 | 3.43 |
| At3g23880 | F-box/kelch-repeat protein | 0 | 0 | 0 | 3.69 |
| At3g11000 | Development and cell death domain protein (DCD) | 0 | 0 | 0 | 5.13 |
| At2g18970 | Uncharacterized gene | 0.83 | 1.92 | 0.95 | 3.40 |
| At5g04550 | Uncharacterized gene | 0.11 | 1.43 | 1.67 | 3.29 |
| At1g55160 | Uncharacterized gene | 0.63 | 1.13 | 1.86 | 3.54 |
| At2g11630 | Uncharacterized gene | 0.02 | 0.23 | 0.59 | 6.04 |
| At2g24755 | Uncharacterized gene | 0 | 0.32 | 0.51 | 4.97 |
| At5g26775 | Uncharacterized gene | 0 | 1.02 | 1.71 | 6.71 |
| At3g11300 | Uncharacterized gene | 0 | 0 | 1.09 | 4.25 |
| At1g43195 | Uncharacterized gene | 0 | 0 | 0 | 4.21 |
| At2g22122 | Uncharacterized gene | 0 | 0 | 0 | 4.47 |
| At1g63610 | Uncharacterized gene | 0 | 0 | 0 | 4.38 |
| b. Genes down-regulated in CaRZFP1-overexpressing T4 generation lettuce plants | |||||
| At2g47850 | Zinc finger CCCH domain-containing protein 32 | − 0.90 | − 0.54 | − 0.43 | − 3.62 |
| At4g10300 | RmlC-like cupins super family protein | − 0.85 | − 0.92 | − 1.69 | − 2.02 |
| At3g55850 | Amidohydrolase family protein (LAF3) | − 0.12 | − 0.19 | − 0.51 | − 2.63 |
| At4g15550 | Indole-3-acetate beta-D-glucosyltransferase (IAGLU) | − 0.05 | − 0.15 | − 0.11 | − 3.41 |
| At5g37600 | Glutamine synthetase cytosolic isozyme 1–1 (GSR 1) | − 1.68 | − 1.31 | − 1.85 | − 3.22 |
| At3g44200 | Serine/threonine-protein kinase Nek5 (NEK6) | − 1.44 | − 1.46 | − 1.92 | − 2.06 |
| At1g60800 | NSP-interacting kinase 3 (NIK3) | −− 1.30 | −− 1.32 | − 1.63 | − 2.96 |
| At2g47990 | Transducin family protein/WD-40 repeat family protein | − 0.59 | − 0.92 | − 1.65 | − 2.00 |
| At4g08950 | Phosphate-responsive 1 family protein (EXO) | − 1.29 | − 1.35 | − 1.09 | − 4.47 |
| At3g02490 | Pentatricopeptide repeat (PPR) superfamily protein | − 1.40 | − 0.75 | − 0.81 | − 3.11 |
| AtCG00290 | tRNA-Ser | − 0.17 | − 0.74 | − .074 | − 2.64 |
| At5g37650 | Uncharacterized gene | − 0.71 | − 1.07 | − 1.38 | − 3.17 |
| At3g52742 | Uncharacterized gene | − 0.41 | − 1.10 | − 0.12 | − 2.14 |
| At3g15518 | Uncharacterized gene | − 2.26 | − 2.12 | − 1.73 | − 3.95 |
| Gene symbols | Gene description | CaRZFP1 -transgenic tobacco/vector controls (log 2 fold change) | |||
| c. Up-regulated genes in CaRZFP1-overexpressing T2 generation tobacco plants (Zeba et al. 2009) | |||||
| At5g55280 | Cell division protein ftsZ | 2.21 | |||
| At3g12400 | Tumor susceptibility gene 101 (tsg101) family protein | 3.21 | |||
| At1g70490 | ADP-ribosylation factor | 6.88 | |||
| At1g13740 | ABI five binding protein 2 | 5.80 | |||
| At5g39650 | DUO1-activated unknown 2 (DUO2) | 2.03 | |||
| At5g10430 | Arabinogalactan-protein (agp4) | 4.42 | |||
| At5g64310 | Arabinogalactan-protein (agp1) | 2.52 | |||
| At4g37450 | Arabinogalactan-protein (agp18) | 3.50 | |||
| At2g46330 | Arabinogalactan-protein (agp16) | 2.87 | |||
| At5g49080 | Proline-rich extensin-like family protein | 2.18 | |||
| At3g49300 | Proline-rich family protein | 1.78 | |||
| At3g49305 | Hypothetical protein contains proline-rich extensin domains | 2.17 | |||
| At4g38770 | Proline-rich family protein (prp4) | 6.05 | |||
| At4g08380 | Proline-rich extensin-like family protein | 2.48 | |||
| At5g19800 | Hydroxyproline-rich glycoprotein family protein | 2.72 | |||
| At2g05380 | Glycine-rich protein (grp3) | 3.71 | |||
| At1g11580 | Pectin methylesterase | 2.32 | |||
| At5g38410 | Ribulose bisphosphate carboxylase small chain 3b | 3.49 | |||
| At3g23820 | NAD-dependent epimerase | 2.21 | |||
| At3g25140 | Glycosyl transferase family 8 protein | 2.86 | |||
| At4g15233 | ABC transporter family protein | 7.31 | |||
| At1g11860 | Aminomethyltransferase | 2.72 | |||
| At5g58050 | Glycerophosphoryl diester phosphodiesterase family protein | 2.43 | |||
| At4g13090 | Xyloglucan:xyloglucosyl transferase/xyloglucan endotransglycosylase | 2.27 | |||
| At3g63200 | Patatin-like protein 9 | 4.88 | |||
| At1g80380 | Phosphoribulokinase/uridine kinase-related | 2.51 | |||
| At4g36360 | Beta-galactosidase | 4.55 | |||
| At2g46820 | Curvature thylakoid 1B (CURT1B) | 3.07 | |||
| At1g74670 | Gibberellin-responsive protein | 4.46 | |||
| At2g45770 | Signal recognition particle receptor protein/chloroplast (ftsY) similar to cell division protein | 3.06 | |||
| At1g55480 | Plant protein family containing a PDZ, a K-box, and a TPR motif (ZKT) | 2.61 | |||
| At5g48760 | 60s ribosomal protein L13A (RPL13aD) | 1.89 | |||
| At3g59760 | Cysteine synthase c/O-acetylserine (thiol)-lyase isoform c | 2.22 | |||
| At2g17360 | 40s ribosomal protein s4 (rps4a) | 2.32 | |||
| At1g02780 | 60s ribosomal protein | 3.31 | |||
| At5g13650 | Elongation factor family protein | 2.38 | |||
| At4g33510 | 3-Deoxy-D-arabino-heptulosonate-7-phosphate 2 (dahp2) | 2.58 | |||
| At5g64680 | Uncharacterized gene | 2.23 | |||
| At5g36070 | Uncharacterized gene | 5.56 | |||
| At3g43684 | Uncharacterized gene | 2.58 | |||
| At2g36885 | Uncharacterized gene | 4.09 | |||
| At2g05752 | Uncharacterized gene | 4.38 | |||
| At5g02240 | Uncharacterized gene | 3.00 | |||
| At1g67700 | Uncharacterized gene | 1.99 | |||
| At4g18070 | Uncharacterized gene | 2.28 | |||
| d. Down-regulated genes in CaRZFP1-overexpressing T2 generation tobacco plants (Zeba et al. 2009) | |||||
| At1g61780 | Postsynaptic protein-related | − 1.94 | |||
| At5g60870 | Regulator of chromosome condensation (rcc1) family protein | − 2.43 | |||
| At3g12090 | Tetraspanin gene family (TET6) | − 2.43 | |||
| At5g66070 | Zinc finger (C3HC4-type ring finger) family protein | − 2.03 | |||
| At1g54060 | Trihelix DNA-binding protein family | − 1.80 | |||
| At4g02720 | NF-kB activating protein (NKAP) proteins | − 2.22 | |||
| At5g02820 | Brassinosteroid insensitive 5 (BIN5) | − 2.55 | |||
| At3g13520 | Arabinogalactan-protein (agp12) | − 2.36 | |||
| At2g05540 | Glycine-rich protein | − 1.69 | |||
| At1g29050 | Trichome birefringence-like 38 | − 2.25 | |||
| At2g18700 | Trehalose phosphatase/synthase 11 (TPS11) | − 2.40 | |||
| At2g18600 | Rub1-conjugating enzyme | − 2.23 | |||
| At3g47950 | ATPase | − 2.03 | |||
| At3g19700 | Leucine-rich repeat transmembrane protein kinase | − 2.17 | |||
| At5g63870 | Serine/threonine-protein phosphatase (pp7) | − 2.16 | |||
| At5g56460 | Protein kinase | − 2.86 | |||
| At5g60460 | Sec61-beta subunit family protein | − 2.78 | |||
| At2g37470 | Histone h2b | − 3.19 | |||
| At4g27960 | Ubiquitin-conjugating enzyme 9 (ubc9) | − 2.35 | |||
| At3g01400 | Armadillo/beta-catenin repeat family protein | − 2.89 | |||
| At5g66040 | Protein with thiosulfate sulfurtransferase/rhodanese activity (STR16) | − 1.91 | |||
| At3g09440 | Heat shock protein 70 (hsp70) | − 1.83 | |||
| At2g46550 | Uncharacterized gene | − 2.66 | |||
| At1g63930 | Uncharacterized gene | − 2.72 | |||
Group 1 genes are in bold
To validate the transcriptome analysis results, total RNA from the same transgenic lettuce lines used for transcriptome experiments was subjected to RNA blot analyses with oligonucleotides corresponding to those on the microarray. Nine genes that were either significantly up- or down-regulated in transgenic lettuce lines were randomly selected for analysis. The genes encoded an FMN binding protein, Hs1pro-1 protein, EXT3, F-box, auxin efflux carrier protein, ERMO2, ATM, NIK3, and NEK6. Although some variation existed between the microarray and RNA blot results, trends in the differentially expressed genes were generally consistent across the two different approaches (Fig. S3).
Discussion
Genes significantly up-regulated in CaRZFP1-transgenic lettuce compared with vector-only plants could be grouped into two, Group 1 and Group 2. Among Group 1 genes, cell division protease ftsH-11 (FTSH11) has been implicated in housekeeping proteolysis of membrane proteins (Wagner et al. 2016). Timeless family protein (ATIM) regulates circadian rhythm mechanism in the files (Rosato et al. 2006). Indeterminate-domain (IDD) protein family members have been implicated in auxin production, gravitropism and lateral organ differentiation, heat stress responses, regulation of sugar transporter regulation, as well as promotion of seed germination (Cui et al. 2013). In maize and rice, the ID1 gene, a member of IDD family, acts as a master switch for transitioning from the vegetative to the reproductive phase. Plants with loss-of-function id1 remain in a prolonged state of vegetative growth and form aberrant flowers (Park et al. 2008). Flowering time and inflorescence development were significantly altered in CaRZFP1-transgenic lettuce. Arabidopsis response regulator 10 (ARR10) has been implicated in the cytokinin signaling (To et al. 2007). BES1-interacting MYC-like 1 (BIM1) played an essential role in brassinosteroid signaling (Xing et al. 2013). The ectopic expression of CaRZFP1 may regulate phytohormone signaling and subsequently affect the growth and development of CaRZFP1-transgenic lettuce plants. OxaA/YidC is essential for protein insertion into bacterial and mitochondrial inner membranes, as well as thylakoid membranes of chloroplasts (Hennon et al. 2015). Extensin 3 (EXT3) expression was also up-regulated over threefold in CaRZFP1-transgenic lettuce; these proteins are critical structural components of the cell wall during plant growth and development (Lamport et al. 2011). Peroxidases are among the largest protein families; they function in the cross-linking of cell wall proteins, lignin biosynthesis, suberization, auxin catabolism, oxidative stress, and defense responses (Marjamaa et al. 2009). Glucose-1-phosphate adenylyltransferase small subunit (ADG1) is a key regulatory enzyme in the plant starch and bacterial glycogen biosynthesis pathway (Bahaji et al. 2011). NADH:ubiquinone oxidoreductase in the Complex I of the electron transport chain is a central component in cellular respiration, the main process providing energy in most heterotrophic eukaryotes and in autotrophic organisms during their heterotrophic phase (Kuhn et al. 2015). FMN binding proteins are critical in the electron transport process. Overexpression of FMN binding protein (AtHal3) altered growth rates while improving salt and drought tolerance in Arabidopsis (Espinosa-Ruiz et al. 1999). The ectopic expression of CaRZFP1 may deregulate genes involved in vital metabolic processes. Transcriptome analysis also showed that many genes with known function in plant development were altered in the CaRZFP1-transgenic lettuce. A serine/threonine-protein kinase (ATAXIA-TELANGIECTASIA MUTATED, ATM) that is important in DNA damage response (Liu et al. 2015) was also elevated in CaRZFP1-transgenic lettuce plants. The up-regulated Hs1pro-1 gene has been implicated in defense response in plants (Yuan et al. 2008). Nodulin MtN3-like protein is member of Sugars Will Eventually be Exported Transporters (SWEETs) family that is essential for the maintenance of animal blood glucose levels, plant nectar production, plant seed, and pollen development (Chandran 2015). Another significantly up-regulated gene in transgenic lettuce encoded sec24-like transport protein (ERMO2); SEC24A encodes a coat protein complex II vesicle coat subunit involved in endoplasmic reticulum-to-Golgi trafficking during the early secretory pathway. In plants, secretory pathway defects often lead to cell division defects (Qu et al. 2014). Glutamate receptor 2.1 (GLR2.1) is a member of ligand-gated ion channel family, and it functions in coordination of mitotic activity during root development, sensing carbon-to-nitrogen status, cellular calcium ion homeostasis, response to light, regulation of plant hormone biosynthesis, and signaling pathways (Weiland et al. 2016). Because an optimum auxin concentration is required for shoot and root growth, lateral root development, and differentiation of vascular strands (Aremu et al. 2016), excessive expression of auxin efflux carrier family genes could be harmful for plant growth and development. Pentatricopeptide repeat proteins function in multiple processes as modular RNA-binding proteins that mediate gene expression through altering RNA sequence, turnover, and processing. They exhibit profound effects on organelle biogenesis and function; consequently, they are influential in major plant processes, including photosynthesis, respiration, development, and environmental responses (Manna 2015). Thus, the ectopic expression of CaRZFP1 may trigger secondary effects that are critical to plant growth and development. Cysteine/histidine-rich C1 domain-containing protein also has been implicated for its important role in regulating plant growth and development (Hwang et al. 2014). Conserved peptide upstream open reading frame 24 (CPuORF24) regulates polyamine and sucrose concentrations in response to starvation (Hayden and Jorgensen 2007). Ubiquitin-conjugating enzyme E2 18 (UBC18), armadillo/beta-catenin-like repeats-containing protein (ARM), F-box associated ubiquitination effector family protein, and development and cell death domain protein (DCD), all have been implicated in ubiquitin-mediated protein degradation. Ubiquitin-mediated protein degradation plays a key regulatory role during plant growth and development, as well as being implicated in plant hormone signaling (Mudgil et al. 2004). Thus, CaRZFP1 might also function at the post-translational level in various developmental processes through ubiquitin-dependent protein degradation.
Some genes were significantly down-regulated (< 2-fold) in CaRZFP1-transgenic lettuce compared with vector-only plants. RmlC-like cupin super family proteins have both enzymatic and non-enzymatic functions, the former including decarboxylases, isomerases, epimerases, oxidoreductases, disomerases, dioxygenases, and hydrolases, the latter including auxin binding, seed storage, and nuclear transcription factors (Uberto and Moomaw 2013). Amidohydrolase family protein (LAF3) is involved in phytochrome A signal transduction (Hare et al. 2003). Indole-3-acetate beta-D-glucosyltransferase (IAGLU) plays a crucial role in auxin conjugation pathway and auxin metabolism (Jackson et al. 2002). The NIMA-related Kinase 6 (NEK6) in Arabidopsis organizes microtubules, thus regulating cellular expansion, directional growth of roots and hypocotyls, petiole elongation, cell file formation, and morphogenesis (Takatani et al. 2015). NSP-interacting kinase 3 (NIK3) is involved in plant defense response and developmental processes (Zorzatto et al. 2015). WD40 domains are present in eukaryotic proteins linked to scaffolding, cooperative assembly, chaperoning other proteins, and regulation of multicellular processes. WD40 repeat protein NEDD1 regulates microtubule development during mitotic cell division in Arabidopsis (Gachomo et al. 2014). The transcript level of tRNA-Ser was also significantly repressed in CaRZFP1-transgenic lettuce plants.
These 24 up-regulated genes and seven down-regulated Group 1 genes are all necessary for plant growth and development. Thus, the negative phenotypic results related to CaRZFP1 expression in lettuce are probably caused by imbalance among growth and development regulators. Ectopically expressed CaRZFP1 induced and suppressed various genes in lettuce, leading to composite negative effects on growth and development, especially in the root and floral meristem division in the inflorescence. Although root endodermal development was significantly hampered in the CaRZFP1-transgenic lettuce plants, none of the annotated genes exhibited a direct relationship in their regulation with suberin biosynthetic processes, except one peroxidase that was expressed, albeit not significantly. We note that among Group 1, seven uncharacterized genes were correlatively up-regulated and one, correlatively down-regulated; it is impossible to speculate on their functions at present (Tables 1a, b, S1 and S2). In Group 2, 43 genes were strongly overexpressed and six genes strongly down-regulated in the line no. 12 (Tables 1a, b, S1 and S2). Because the regulation of these genes was not correlated with CaRZFP1 expression in the line no. 14, no. 16, and no. 6, their overexpression and suppression in the line no. 12 were likely the result of composite effects from the biased expression of Group 1 genes, rather than due directly to CaRZFP1 expression.
Ectopic expression of CaRZFP1 in tobacco enhanced plant growth (larger leaves, longer hypocotyls, longer primary roots, and increased lateral roots), leading to heavier fresh weight (Zeba et al. 2009). Transcriptome analysis revealed that growth-related genes were widely altered, i.e., 37 up-regulated and 22 down-regulated annotated genes, in CaRZFP1-overexpressing transgenic tobacco (Tables 1c, d, S3 and S4). Up-regulated ADP-ribosylation factors are important in regulating intracellular membrane trafficking, a process linked to root development and the polar localization of PIN-FORMED (PIN) family auxin efflux facilitators (Yuan et al. 2015). Dramatic elevation (up to sixfold) was observed in the expression of growth-related cell wall proteins, four of which were arabinogalactan proteins (AGPs) (Showalter and Basu 2016). These results suggest that CaRZFP1 activates and positively regulates cell wall protein expression to modify cell wall plasticity, effectively promoting the growth of transgenic tobacco plants. Proline-rich proteins are important in Arabidopsis root-hair formation (Boron et al. 2014), and transcriptome analysis revealed that four proline-rich protein genes were up-regulated in CaRZFP1-transgenic tobacco, and CaRZFP1-transgenic tobacco plants exhibited more developed root hairs (Zeba et al. 2009). Plant ABC transporter family proteins experienced an over sevenfold up-regulation; these proteins are implicated in chlorophyll biosynthesis, Fe-S cluster formation, stomatal movement, and ion fluxes (Hwang et al. 2016). Gibberellin-responsive proteins were also highly induced in transgenic tobacco; these proteins have been implicated in hypocotyl and stem elongation (Achard et al. 2007). Ectopic expression of CaRZFP1 in tobacco also down-regulated two dozens of genes but to a lesser degree than the up-regulation experienced by the genes described above (Tables 1d, and S4). Finally, eight up- and two down-regulated genes were unannotated in CaRZFP1-transgenic tobacco (Tables 1c, d, S3 and S4), and their possible effects on tobacco growth and development are not feasible to be suggested.
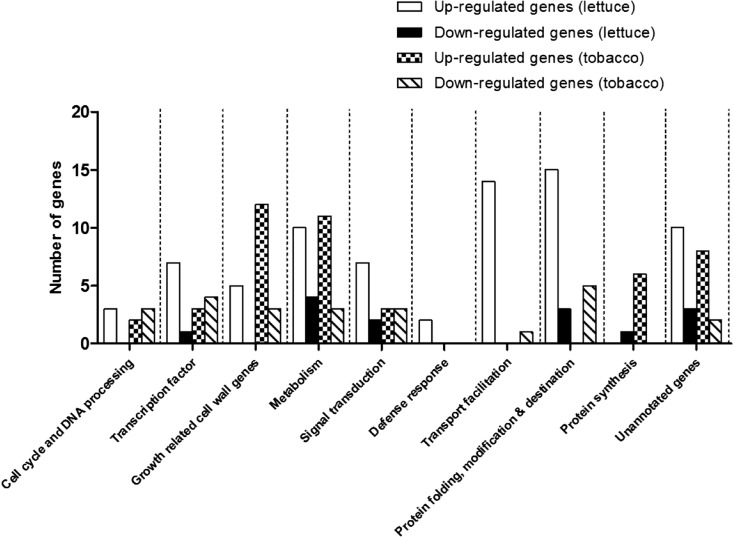
The expression profiles, either significantly up-regulated or significantly down-regulated, of lettuce and tobacco plants overexpressing CaRZFP1 were dissimilar. Transgenic lettuce and tobacco did not share any genes with significantly altered expression (Table 1). And, overall expression profiles, either significantly up-regulated or significantly down-regulated, were largely different between transgenic lettuce and tobacco. In transgenic tobacco, more genes involved in protein synthesis and growth-related cell wall proteins experienced altered expression than in lettuce. However, transgenic lettuce contained more highly altered genes that were involved in transcription factors, transport facilitation, as well as protein folding, modification, and destination (Fig. 4 and Tables S1–S6).
Fig. 4.
Differentially expressed genes in CaRZFP1-transgenic lettuce and CaRZFP1-transgenic tobacco plants were categorized into several groups according to their putative functions. The x-axis shows putative functional categories, and the y-axis shows the number of genes altered in expression
The development of multicellular organisms depends on cellular growth and morphogenesis, involving the expression of numerous genes and intricate gene regulatory networks. RING zinc finger proteins have been closely and repeatedly implicated in the development of multiple organisms (Laity et al. 2001; Krishna et al. 2003; Gamsjaeger et al. 2007; Zhang et al. 2013), but cases reported in plants are comparatively few (Chai et al. 2015; Larrieu and Vernoux 2009; Liu et al. 2016). Enhanced growth and tolerance to abiotic stresses were observed in CaRZFP1-transgenic tobacco (Zeba et al. 2009), but CaRZFP1-transgenic lettuce displayed impaired growth (weakened leaf growth, stunted root growth, shorter plant height, and delayed flowering) compared with vector-only plants. This poor growth was strongly correlated with CaRZFP1 expression levels. When the poor growth phenotype was investigated at the tissue and cellular level, CaRZFP1 expression led to specific effects in lettuce. Although overall height of transgenic lettuce decreased parallel to CaRZFP1 expression, the development of shoot internal structure was not significantly hampered. Shoot and leaf cross sections also exhibited normal development overall, as did vascular bundles, mesophyll cells, and epidermal cells. However, inflorescence development was dramatically affected by CaRZFP1 expression, with overall size decreasing with increasing CaRZFP1 expression. Yet transgenic flowers did not differ from control plants, although flower number per inflorescence was drastically reduced. Likewise, while transgenic seeds exhibited normal morphology, their total number per inflorescence decreased drastically with increasing CaRZFP1 expression. Transgenic lettuce roots were strongly affected by CaRZFP1 expression, increasing in disorganization of vascular bundles as CaRZFP1 expression levels rose. Moreover, the xylem ray became irregular and disconnected, while the endodermis and pericycle were ill-defined and eventually unidentifiable. However, cells in root ground tissue were unaffected. Further, transgenic root length was drastically shortened, but root diameter was not significantly changed. Overall root size, mass, and branching in transgenic lettuce were reduced considerably with rising CaRZFP1 expression levels.
In-depth studies are necessary to clearly understand the basis of the described differences in CaRZFP1-transgenic lettuce and tobacco. However, several possible explanations can be proposed. Because tobacco is closely related to hot pepper (both Solanaceae), hot pepper-derived CaRZFP1 was likely expressed in a similar genetic environment when mobilized to tobacco. In contrast, CaRZFP1 in lettuce was probably expressed in a very different genetic environment, likely even interacting with proteins that are not normal counterparts or binding at a DNA domain that is extremely dissimilar from any in hot pepper or tobacco. These new interactions probably caused the expression of unexpected downstream genes. Because signal transduction pathways are complex and extensively interconnected (Larrieu and Vernoux 2009), unpredictable outcomes may result once novel interactions trigger a different network of signal transductions or gene sets.
This study provides important insight into the function of C3HC4-type RING finger proteins in lettuce. We provided candidate genes downstream of the ectopically expressed CaRZFP1 in lettuce that specifically affected root development, especially of the endodermis, pericycle, and vascular structures. We also provided strong evidence that the same gene can yield completely different outcomes depending on its host species. These results are significant because unexpected outcomes of gene mobilization to improve crops are a major safety concern. In addition to the extreme cares to control the unwanted and uncontrolled spread of transgenes between closely related wild species and the newly introduced transgenic crops through pollen diffusion, the results in this report asks for another level of concerns due to the chance of occurring unexpected phenotype appearance from the gene transfer. Likewise, the concern can be extended to the general molecular approach to decipher the function of a genetic component. Scientific researches heavily rely on a model system, for an example Arabidopsis, to decipher the function of a gene/protein, pathways, and networks in plants. If different plant species carry quite different scaffolding network for a protein or gene, the outcome can be quite different. Since plasticity of development is one important nature in plants, we tempt to see much heavier emphasis on each plant species as a host for the common and for the specificity.
Electronic supplementary material
(DOCX 42 kb)
(DOCX 28 kb)
(DOCX 22 kb)
(DOCX 19 kb)
(DOCX 22 kb)
(DOCX 21 kb)
(DOC 3.88 MB)
Acknowledgements
This work was supported by grants allocated to C.B. Hong from the Basic Science Research Program (Grant no. 0409-20130046), supported through the National Research Foundation of Korea, which is funded by the Ministry of Education, Science and Technology, and from the Technology Development Program for Agriculture and Forestry, Ministry for Agriculture, Forestry and Fisheries, Korea. Transformation of lettuce and rearing of the transgenic lettuce and tobacco plants were supported by Planta Company, Korea.
Author contribution
Choo Bong Hong designed and supervised research and wrote the manuscript. Mahipal Singh Kesawat carried out all the molecular analyses and phenotypic assays for the lettuce plants. Dong Kyun Kim did transformation experiments including construction of the expression vectors to build target plants. Naheed Zeba carried out most of the molecular analyses and phenotypic assays for the tobacco plants. Mi Chung Suh and Xinli Xia reared and bred the plants and also helped Naheed Zeba for the analysis of the transgenic tobacco plants.
Compliance with ethical standards
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s11032-018-0812-3) contains supplementary material, which is available to authorized users.
References
- Aremu AO, Plačková L, Pěnčík A, Novák O, Doležal K, van Staden J. Auxin-cytokinin interaction and variations in their metabolic products in the regulation of organogenesis in two Eucomis species. New Biotechnol. 2016;33:883–890. doi: 10.1016/j.nbt.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 2007;143:1163–1172. doi: 10.1104/pp.106.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahaji A, Li J, Ovecka M, Ezquer I, Muñoz FJ, Baroja-Fernández E, Romero JM, Almagro G, Montero M, Hidalgo M, Sesma MT, Pozueta-Romero J. Arabidopsis thaliana mutants lacking ADP-glucose pyrophosphorylase accumulate starch and ADP-glucose: further evidences for the occurrence of important sources, other than ADP-glucose pyrophosphorylase, of ADP-glucose linked to leaf starch biosynthesis. Plant Cell Physiol. 2011;52:1162–1176. doi: 10.1093/pcp/pcr067. [DOI] [PubMed] [Google Scholar]
- Boron AK, van Orden J, Nektarios Markakis M, Mouille G, Adriaensen D, Verbelen JP, Höfte H, Vissenberg K. Proline-rich protein-like PRPL1 controls elongation of root hairs in Arabidopsis thaliana. J Exp Bot. 2014;65:5485–5495. doi: 10.1093/jxb/eru308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Heisler MG, Hejatko J, Friml J, Benkova E. In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nat Protoc. 2006;1:1462–1467. doi: 10.1038/nprot.2006.226. [DOI] [PubMed] [Google Scholar]
- Chai G, Kong Y, Zhu M, Yu L, Qi G, Tang X, Wang Z, Cao Y, Yu C, Zhou G. Arabidopsis C3H14 and C3H15 have overlapping roles in the regulation of secondary wall thickening and anther development. J Exp Bot. 2015;66:2595–2609. doi: 10.1093/jxb/erv060. [DOI] [PubMed] [Google Scholar]
- Chandran D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life. 2015;67:461–471. doi: 10.1002/iub.1394. [DOI] [PubMed] [Google Scholar]
- Cui D, Zhao J, Jing Y, Fan M, Liu J, Wang Z, Xin W, Hu Y. The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet. 2013;9:e1003759. doi: 10.1371/journal.pgen.1003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Ruiz A, Belles JM, Serrano R, Culianez-Macia FA. Arabidopsis thaliana AtHAL3: a flavoprotein related to salt and osmotic tolerance and plant growth. Plant J. 1999;20:529–539. doi: 10.1046/j.1365-313X.1999.00626.x. [DOI] [PubMed] [Google Scholar]
- Gachomo EW, Jimenez-Lopez JC, Baptiste LJ, Kotchoni SO. GIGANTEUS (GTS1), a member of Transducing/WD40 protein superfamily, controls seed germination, growth and biomass accumulation through ribosome-biogenesis protein interactions in Arabidopsis thaliana. BMC Plant Biol. 2014;14:37. doi: 10.1186/1471-2229-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci. 2007;32:63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Hare PD, Moller SG, Huang LF, Chua NH. LAF3, a novel factor required for normal phytochrome A signaling. Plant Physiol. 2003;133:1592–1604. doi: 10.1104/pp.103.028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden C, Jorgensen R. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol. 2007;5:32. doi: 10.1186/1741-7007-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennon SW, Soman R, Zhu L, Dalbey RE. YidC/Alb3/Oxa1 family of invertases. J Biol Chem. 2015;290:14866–14874. doi: 10.1074/jbc.R115.638171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R, et al. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Hwang IS, Choi DS, Kim NH, Kim DS, Hwang BK. The pepper cysteine/histidine-rich DC1 domain protein CaDC1 binds both RNA and DNA and is required for plant cell death and defense response. New Phytol. 2014;201:518–530. doi: 10.1111/nph.12521. [DOI] [PubMed] [Google Scholar]
- Hwang JU, Song WY, Hong D, Ko D, Yamaoka Y, Jang S, Yim S, Lee E, Khare D, Kim K, Palmgren M, Yoon HS, Martinoia E, Lee Y. Plant ABC transporters enable many unique aspects of a terrestrial plant's lifestyle. Mol Plant. 2016;9:338–355. doi: 10.1016/j.molp.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J, Sandberg G, Bowles DJ. Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterization of transgenic lines. Plant J. 2002;32:573–583. doi: 10.1046/j.1365-313X.2002.01445.x. [DOI] [PubMed] [Google Scholar]
- Klumper W, Qaim M. A meta-analysis of the impacts of genetically modified crops. PLoS One. 2014;9:e111629. doi: 10.1371/journal.pone.0111629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn K, et al. Complete mitochondrial complex I deficiency induces an up-regulation of respiratory fluxes that is abolished by traces of functional complex I. Plant Physiol. 2015;168:1537–1549. doi: 10.1104/pp.15.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/S0959-440X(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Lamport DT, Kieliszewski MJ, Chen Y, Cannon MC. Role of the extension superfamily in primary cell wall architecture. Plant Physiol. 2011;156:11–19. doi: 10.1104/pp.110.169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieu A, Vernoux T. Comparison of plant hormone signalling systems. Essays Biochem. 2009;58:165–181. doi: 10.1042/bse0580165. [DOI] [PubMed] [Google Scholar]
- Liu CH, Finke A, Díaz M, Rozhon W, Poppenberger B, Baubec T, Pecinka A. Repair of DNA damage induced by the cytidine analog zebularine requires ATR and ATM in Arabidopsis. Plant Cell. 2015;27:1788–1800. doi: 10.1105/tpc.114.135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang C, Wei C, Liu X, Wang M, Yu F, Xie Q, Tu J. The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice. Plant Physiol. 2016;170:429–443. doi: 10.1104/pp.15.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S. An overview of pentatricopeptide repeat proteins and their applications. Biochimie. 2015;113:93–99. doi: 10.1016/j.biochi.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Marjamaa K, Kukkola EM, Fagerstedt KV. The role of xylem class III peroxidases in lignification. J Exp Bot. 2009;60:367–376. doi: 10.1093/jxb/ern278. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Smyth GK. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics. 2009;25:765–771. doi: 10.1093/bioinformatics/btp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 2004;134:59–66. doi: 10.1104/pp.103.029553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolia A, Manzo A, Veronesi F, Rosellini D. An overview of the last 10 years of genetically engineered crop safety research. Crit Rev Biotechnol. 2014;34:1–12. doi: 10.3109/07388551.2013.823595. [DOI] [PubMed] [Google Scholar]
- Park SJ, Kim SL, Lee S, Je BI, Piao HL, Park SH, Kim CM, Ryu CH, Park SH, Xuan YH, Colasanti J, An G, Han CD. Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod. Plant J. 2008;56:1018–1029. doi: 10.1111/j.1365-313X.2008.03667.x. [DOI] [PubMed] [Google Scholar]
- Qu X, Chatty PR, Roeder AH. Endomembrane trafficking protein SEC24A regulates cell size patterning in Arabidopsis. Plant Physiol. 2014;166:1877–1890. doi: 10.1104/pp.114.246033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato E, Tauber E, Kyriacou CP. Molecular genetics of the fruit-fly circadian clock. Eur J Hum Genet. 2006;14:729–738. doi: 10.1038/sj.ejhg.5201547. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. (3rd ed. Sambrook J, Russell DW) 7.35–7.41 (Cold Spring Harbor Laboratory Press)
- Showalter AM, Basu D. Glycosylation of arabinogalactan-proteins essential for development in Arabidopsis. Commun Integr Biol. 2016;9:e1177687. doi: 10.1080/19420889.2016.1177687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MC, Hong CB, Kim SS, Sim WS. Transgenic tobacco plants with Bacillus thuringiensis δ-endotoxin gene resistant to Korean-born tobacco budworms. Mol Cells. 1994;4:211–219. [Google Scholar]
- Takatani S, Otani K, Kanazawa M, Takahashi T, Motose H. Structure, function, and evolution of plant NIMA-related kinases: implication for phosphorylation-dependent microtubule regulation. J Plant Res. 2015;128:875–891. doi: 10.1007/s10265-015-0751-6. [DOI] [PubMed] [Google Scholar]
- To JP, et al. Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell. 2007;19:3901–3914. doi: 10.1105/tpc.107.052662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberto R, Moomaw EW. Protein similarity networks reveal relationships among sequence, structure, and function within the cupin superfamily. PLoS One. 2013;8:e74477. doi: 10.1371/journal.pone.0074477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R, von Sydow L, Aigner H, Netotea S, Brugière S, Sjögren L, Ferro M, Clarke A, Funk C. Deletion of FtsH11 protease has impact on chloroplast structure and function in Arabidopsis thaliana when grown under continuous light. Plant Cell Environ. 2016;39:2530–2544. doi: 10.1111/pce.12808. [DOI] [PubMed] [Google Scholar]
- Weiland M, Mancuso S, Baluska F. Signalling via glutamate and GLRs in Arabidopsis thaliana. Funct Plant Biol. 2016;43:1–25. doi: 10.1071/FP15109. [DOI] [PubMed] [Google Scholar]
- Xing SP, Quodt V, Chandler J, Höhmann S, Berndtgen R, Huijser P. SPL8 acts together with the brassinosteroid-signaling component BIM1 in controlling Arabidopsis thaliana male fertility. Plants. 2013;2:416–428. doi: 10.3390/plants2030416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C, Zhou G, Li Y, Wang K, Wang Z, Li X, Chang R, Qiu L. Cloning and sequence diversity analysis of GmHs1 pro-1 in Chinese domesticated and wild soybeans. Mol Breed. 2008;22:593–602. doi: 10.1007/s11032-008-9201-7. [DOI] [Google Scholar]
- Yuan J, et al. Ectopic expression a maize ADP-ribosylation factor gene in Arabidopsis, increase plant size and growth rate. J Plant Biochem Biotechnol. 2015;24:161–166. doi: 10.1007/s13562-013-0248-0. [DOI] [Google Scholar]
- Zeba N, Isbat M, Kwon NJ, Lee MO, Kim SR, Hong CB. Heat-inducible C3HC4 type RING zinc finger protein gene from Capsicum annuum enhances growth of transgenic tobacco. Planta. 2009;229:861–871. doi: 10.1007/s00425-008-0884-0. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang H, Zhao Y, Jiang H, Zhu S, Cheng B, Xiang Y. Genome-wide analysis of the CCCH zinc finger gene family in Medicago truncatula. Plant Cell Rep. 2013;32:1543–1555. doi: 10.1007/s00299-013-1466-6. [DOI] [PubMed] [Google Scholar]
- Zorzatto C, Machado JPB, Lopes KVG, Nascimento KJT, Pereira WA, Brustolini OJB, Reis PAB, Calil IP, Deguchi M, Sachetto-Martins G, Gouveia BC, Loriato VAP, Silva MAC, Silva FF, Santos AA, Chory J, Fontes EPB. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature. 2015;520:679–682. doi: 10.1038/nature14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 42 kb)
(DOCX 28 kb)
(DOCX 22 kb)
(DOCX 19 kb)
(DOCX 22 kb)
(DOCX 21 kb)
(DOC 3.88 MB)
Data Availability Statement
All the data in this publication will be available upon request.