Abstract
As a childhood-onset psychiatric disorder, attention deficit hyperactivity disorder (ADHD) is complicated by phenotypic and genetic heterogeneity. Lifelong executive function deficits in ADHD are described in many literatures and have been proposed as endophenotypes of ADHD. However, its genetic basis is still elusive. In this study, we performed a genome-wide association study of executive function, rated with Behavioral Rating Inventory of Executive Function (BRIEF), in ADHD children. We identified one significant variant (rs852004, P = 2.51e-08) for the overall score of BRIEF. The association analyses for each component of executive function found this locus was more associated with inhibit and monitor components. Further principle component analysis and confirmatory factor analysis provided an ADHD-specific executive function pattern including inhibit and monitor factors. SNP rs852004 was mainly associated with the Behavioral Regulation factor. Meanwhile, we found the significant locus was associated with ADHD symptom. The Behavioral Regulation factor mediated its effect on ADHD symptom. Functional magnetic resonance imaging (fMRI) analyses further showed evidence that this variant affected the activity of inhibition control related brain regions. It provided new insights for the genetic basis of executive function in ADHD.
Introduction
Attention deficit hyperactivity disorder (ADHD) is a common psychiatric disorder characterized by age-inappropriate deficiency in sustained attention and/or hyperactive, impulsive behaviors1. The genetic epidemiological studies revealed gene variants constituted the primary etiology of ADHD, with a heritability estimated to be 0.762. Candidate genes were involved in the biosynthesis, release, transmission and metabolism of neurotransmitters3, while genome-wide association studies4–7 suggested genes contributing to the brain development. The complex genetic architecture of ADHD is still far from fully illustrated2,8.
One of the reasons for the complexity of ADHD is that it is a heterogeneous disorder. The same clinical presentation of inattention, hyperactivity and impulsivity may have different etiological contribution. Identification of the genetic contributions to ADHD is likely complicated by phenotypic and genetic heterogeneity, low penetrance, and limited statistical power. One way to enhance power for genetic discovery is to reduce heterogeneity by use of endophenotypes. Presently, the most common endophenotypes under consideration are neuropsychological markers of executive function9,10. Executive function (EF) is a high order cognitive function that provides people the capacity to change and adjust behaviors according to the shifting demands of the complex environment11. Executive dysfunction has been found in many psychiatric disorders, such as bipolar disorder12, depression13, schizophrenia14, autism15 and ADHD16. Candidate genes involved in the neurotransmitter system, including dopaminergic, noradrenergic, serotonergic and cholinergic, have been reported to be associated with some component of executive functions17. Yang et al. identified one significant locus for inhibition in ADHD by using genome-wide association study18.
In this study, we performed a genome-wide association analysis on the global score of executive function using the Behavior Rating Inventory of Executive Function (BRIEF) and further analyzed the association of the significant locus with each component of executive function and ADHD symptoms. Furthermore, we analyzed the function of associated locus via neuroimaging study in human and its underlying molecular mechanism contributing to brain function and behavior. The findings may provide new insights into the pathological mechanisms of ADHD.
Results
Correlation between executive function and ADHD symptoms
We collected three dimensional symptoms for the patients, namely inattention (CDISatt), hyperactivity-impulsivity (CDIShi) and overall assessment (CDISall). We calculated the correlations between the BRIEF scales and three symptom traits (as shown in Table 1). All BRIEF scales were significantly correlated with inattention symptom, but only eight of them were significantly correlated with hyperactivity-impulsivity symptom (Total score, Behavior Regulation Index, Metacognition Index, Inhibit, Emotional Control, Working Memory, Organization of Materials, Monitor), and nine with overall assessment (Total score, Behavior Regulation Index, Metacognition Index, Inhibit, Emotional Control, Working Memory, Plan/Organize, Organization of Materials, Monitor).
Table 1.
Correlations between the BRIEF scales and the three symptom traits from the ADHD Rating scale-IV-patient report.
| CDISatt | CDIShi | CDISall | ||||
|---|---|---|---|---|---|---|
| Cor. | Sig. | Cor. | Sig. | Cor. | Sig. | |
| Total | 0.3748 | <2.2e-16 | 0.1825 | 2.05E-05 | 0.2406 | 1.60E-08 |
| BRI | 0.2206 | 2.37E-07 | 0.2033 | 2.00E-06 | 0.2283 | 8.63E-08 |
| MI | 0.4275 | <2.2e-16 | 0.1317 | 2.21E-03 | 0.2053 | 1.58E-06 |
| IB | 0.2633 | 5.55E-10 | 0.2916 | 5.25E-12 | 0.3177 | 4.43E-14 |
| SFT | 0.1175 | 6.38E-03 | 0.0292 | 5.00E-01 | 0.0496 | 2.51E-01 |
| ECTRL | 0.1348 | 1.72E-03 | 0.1225 | 4.45E-03 | 0.1373 | 1.41E-03 |
| INIT | 0.2619 | 6.87E-10 | 0.0282 | 5.14E-01 | 0.0764 | 7.67E-02 |
| WM | 0.4393 | <2.2e-16 | 0.1186 | 5.87E-03 | 0.1956 | 4.87E-06 |
| PO | 0.3291 | 4.67E-15 | 0.0730 | 9.05E-02 | 0.1322 | 2.12E-03 |
| OM | 0.3644 | <2.2e-16 | 0.1264 | 3.32E-03 | 0.1893 | 9.85E-06 |
| MONI | 0.2790 | 4.45E-11 | 0.1891 | 1.01E-05 | 0.2283 | 8.58E-08 |
R function cor.test is used for this analysis. Cor. is correlation; Sig. is significance. BRI, Behavior Regulation Index; MI, Metacognition Index; IB, Inhibit; SFT, Shift; ECTRL, Emotional Control; INIT, Initiate; WM, Working Memory; PO, Plan/Organize; OM, Organization of Materials; MONI, Monitor.
SNPs associated with executive function components in BRIEF
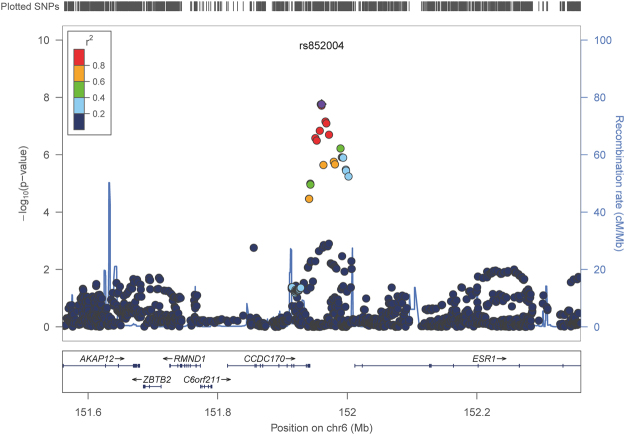
Firstly, we analyzed the association of SNPs with the global score of BRIEF. We identified one significant SNP (rs852004, P = 2.51e-08, A1 = A, frequency = 0.08, BETA (95% CI) = −11.84 (−15.94, −7.734)) located in 6q25.1. The A allele carriers had smaller BRIEF score, which means better executive function. Regional plot for this locus was shown in Fig. 1. This locus was within the upstream of ESR1, RMND1, ZBTB2 and downstream of CCDC170 and C6orf211. Next, we analyzed the association of this significant SNP with each subscale of BRIEF. As shown in Table 2, this SNP is the first top associated SNP with the BRI (Behavioral Regulation Index for Inhibit, Shift and Emotional Control) and MI (Metacognition Index for Initiate, Working Memory, Plan/Organize, Organization of Materials, Monitor). The association with BRI was mainly from Inhibit (top one significant SNP), while the association with MI was mainly from Monitor (top one significant SNP).
Figure 1.
Regional plot for the significant locus with total score in BRIEF. LocusZoom (http://locuszoom.org/) was used to generate the plot using the association analysis result after imputation.
Table 2.
Phenotype information and the association result of the significant SNP rs852004 in each scale of BRIEF.
| Name | Phenotype | Mean (SD) | P-value | Corrected P-valuea | Effect Size (95% CI) | Rank |
|---|---|---|---|---|---|---|
| Total | Total score for all 8 scales of BRIEF | 151.98 (19.56) | 2.51E-08 | 2.51E-08 | −11.84 (−15.94, −7.734) | 1 |
| IB | Inhibit | 20.04 (4.59) | 9.52E-08 | 1.14E-06 | −2.642 (−3.599, −1.685) | 1* |
| SFT | Shift | 12.89 (2.75) | 3.58E-03 | 4.30E-02 | −0.8729 (−1.458, −0.2881) | 2326 |
| ECTRL | Emotional Control | 17.27 (4.48) | 6.92E-04 | 8.30E-03 | −1.661 (−2.615, −0.7071) | 418 |
| INIT | Initiate | 15.69 (2.96) | 5.52E-04 | 6.62E-03 | −1.118 (−1.749, −0.4876) | 333 |
| WM | Working Memory | 23.06 (3.17) | 3.38E-04 | 4.06E-03 | −1.244 (−1.92, −0.568) | 189 |
| PO | Plan/Organize | 27.99 (3.98) | 3.39E-06 | 4.07E-05 | −2.009 (−2.848, −1.17) | 2 |
| OM | Organization of Materials | 14.58 (2.61) | 5.38E-03 | 6.46E-02 | −0.7922 (−1.348, −0.2366) | 3327 |
| MONI | Task-Monitor | 20.46 (2.75) | 5.05E-07 | 6.06E-06 | −1.496 (−2.072, −0.9195) | 1* |
| BRI | Behavioral Regulation Index for IB, SFT and ECTRL | 50.2 (9.68) | 9.11E-07 | 1.09E-05 | −5.176 (−7.218, −3.134) | 1 |
| MI | Metacognition Index for INIT, WM, PO, OM and MONI | 101.78 (12.16) | 4.75E-07 | 5.70E-06 | −6.66 (−9.22, −4.099) | 1 |
| PC1 | Principle component containing IB, INIT, WM, PO, OM and MONI | N.A. | 2.013E-07 | 2.42E-06 | −0.5655 (−0.7759, −0.355) | 1 |
| PC2 | Principle component containing SFT and ECTRL | N.A. | 6.194e-05 | 7.43E-04 | −0.4364 (−0.6483, −0.2246) | 32 |
SD: standard deviation. The association analysis for PC1 and PC2 were performed for the normalized value, so the mean and SD were not available. Rank is the rank of SNP rs852004 in the genome-wide association result for each BRIEF scale. The rank marked with * means the rank is for rs6908732, which is in high LD with rs852004 (r2 = 0.75). A1 = A, allele frequency = 0.08.
aSince we firstly performed the GWAS for the total score of BRIEF, the P-value for Total didn’t need multiple correction. Then, we checked the association of the significant SNP in 8 scales, two indexes and two PCs. The corrected P-value for them were obtained by multiplying 12.
Association of the significant SNP with principle component of BRIEF
The component plot in the rotated space and rotated component matrix from the PCA for the eight subscales of BRIEF was shown in Supplementary Fig. S1. Two components were detected (eigen value >1 in scree plot). The first component is related with Inhibit (IB), Initiate (INIT), Working Memory (WM), Plan/Organize (PO), Organization of Materials (OM) and Monitor (MONI); the second component is related with Shift (SFT) and Emotional Control (ECTRL). Furthermore, we extracted the two components values and conducted association analysis for the two components. As shown in Table 2, rs852004 was the top one SNP in the association result of component 1, while, the SNP ranked 32 in the association result of component 2.
Since the components from PCA is slightly different with the definition of the two indexes in BRIEF (BRI and MI), we further performed confirmatory factor analysis (CFA) for the two candidate models. The Model A is based on the definition of BRIEF indexes: one group constituted of IB, SFT and ECTRL, the other group constituted of INIT, WM, PO, OM and MONI (see Supplementary Fig. S2a). The Model B is based on the PCA result: one group constituted of IB, INIT, WM, PO, OM and MONI, the other group constituted of SFT and ECTRL (see Supplementary Fig. S2b). The result showed the Model B is more suitable for our samples. Next, we further checked a 3-factor model as shown in Fig. 2 (Model C), in which, we grouped IB and MONI into one subgroup. The CFA result showed the 3-factor model was more suitable for our data than the two 2-factor models.
Figure 2.
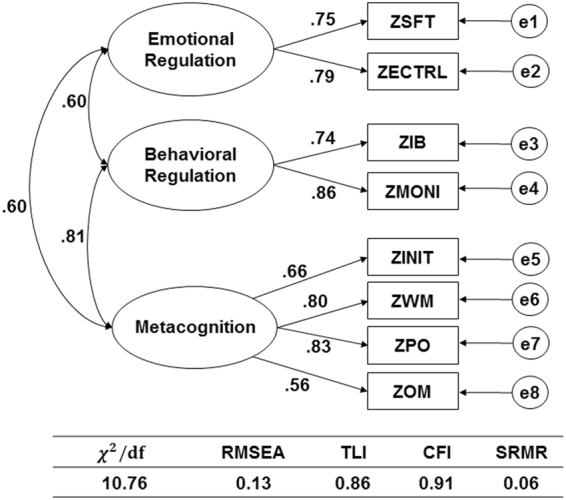
The 3-factor model for the confirmatory factor analysis of the BRIEF data. The fit parameters for the model was shown below the model.
Association of the significant SNP with ADHD symptom
SNP rs852004 was not significant in our Chinese ADHD case-control GWAS (P = 0.1619)4. Association analysis for the significant SNP rs852004 with ADHD symptoms was conducted to further validate the contribution of the significant SNP rs852004. Among the 550 samples, 533 samples have both symptoms data and genotype data for SNP rs852004. The analysis showed it was significantly associated with the total assessment (P = 0.0163, A1 = A, BETA = −0.5733), which denoted the ADHD symptom of A carriers was better.
Furthermore, we examined the role of each BRIEF scale as a mediator to mediate the association between rs852004 with the ADHD symptom. We used the model 4 in PROCESS19 to bootstrap the sampling distribution of the indirect effect (where the indirect effect is the reduction in the strength of the SNP/symptom association that is due to the executive function). The indirect effect of rs852004 on ADHD total symptom score (CDISall) through each BRIEF scale was shown in Supplementary Table S1. The result showed all BRIEF scales have significant indirect effect on ADHD symptom. The mediation effect was different to zero even at the lower bound of the confidence interval. Furthermore, the effect of Inhibit (IB), Monitor (MONI), Working Memory (WM) and Plan/Organize (PO) were bigger among the eight scales. These data showed that SNP rs852004 accounts for significant variation in ADHD symptom, in part through the effects of the SNP on the intermediate phenotype of executive function.
Function validation of associated SNP rs852004 by resting-state fMRI
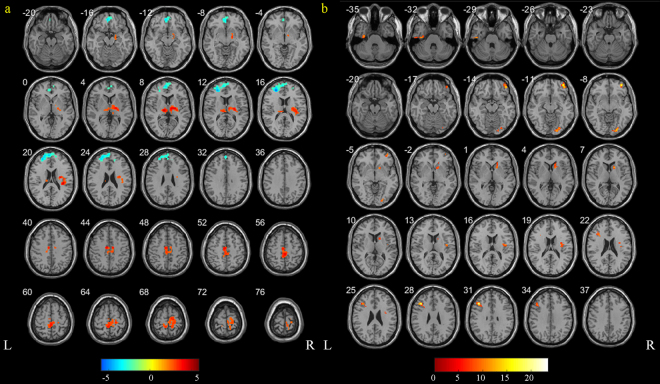
Among 50 ADHD patients, 42 are G homozygotes for rs852004; among 66 control children, 57 are G homozygotes for rs852004. Compared with healthy controls, reduced Regional Homogeneity (ReHo) was uncovered in ADHD in dorsolateral prefrontal cortex, medial prefrontal cortex and increased ReHo in thalamus, anterior cingulate cortex, and anterior insular cortex (Fig. 3a, T values, P < 0.01, cluster size above 202 voxels). The interaction of diagnosis and genotype was found in six clusters, including right orbitofrontal cortex, dorsal striatum, insula, lingual cortex, left inferior frontal cortex, inferior temporal cortex (Fig. 3b, F values, P < 0.01, cluster size above 31 voxels). In term of the simple effect of rs852004, compared to G homozygote, no increased or reduced ReHo was discovered after multiple comparison in either ADHD or control group.
Figure 3.
Regions exhibiting differences in ReHo. (a) regions showed the differences between ADHD individuals and controls (T values, P < 0.01). (b) regions showed the interaction of rs852004 and ADHD (F values, P < 0.01).
Regulatory feature and expression quantitative trait loci (eQTL) analysis of associated SNP rs852004
The significant locus we identified was within the upstream of ESR1, RMND1, ZBTB2 and downstream of CCDC170 and C6orf211 (as shown in Fig. 1). It may affect the nearby genes by regulatory mechanism. We obtained nine SNPs with high LD with rs852004 and checked the related regulatory features of the SNPs by searching rVarBase and RoadMap data. In the LD block of the significant locus, there is only some weak enhancer, DNase hyperactivity site and heterochromatin signal in RoadMap (upper panel of Supplementary Fig. S3, only neuronal cells and brain tissues were shown). rVarBase showed these SNPs were located in chromatin interactive region, which targeted genes RMND1, C6orf211, CCDC170 and ESR1. eQTL data searching found rs852004 and rs6908732 could regulate the expression of ZBTB2 in the cerebellum of bipolar disorder20, and rs852004 could regulate the expression of RMND1 in human prefrontal cortex21. We further searched the expression profiles of these genes in human brain tissues in BRAINEAC database and performed eQTL analyses to elucidate whether SNP rs852004 influences the genes expression in the brain. CCDC170 was not found in this database, and other four genes (RMND1, C6orf211, ESR1, ZBTB2) are expressed in various brain regions, with the highest transcript level in cerebellar cortex (RMND1), thalamus (C6orf211 and ZBTB2), and inferior olivary nucleus (ESR1). We found significant association between rs852004 with these genes separately in thalamus (RMND1, P = 0.0081), cerebellar cortex (C6orf211, P = 0.024; ESR1, P = 0.0021), frontal cortex (ESR1, P = 0.049). All the eQTL data were summarized in Table 3.
Table 3.
The eQTL data summary of significant SNP rs852004.
| Source | Tissue | SNP | Gene | P-value |
|---|---|---|---|---|
| SCAN | cerebellum | rs852004 | ZBTB2 | N.A. |
| SCAN | cerebellum | rs6908732 | ZBTB2 | N.A. |
| Liu C et al.21 | prefrontal cortex | rs852004 | RMND1 | 4.42E-03 |
| BRAINEAC | FCTX | rs852004 | ESR1 | 4.90E-02 |
| BRAINEAC | THAL | rs852004 | RMND1 | 8.10E-03 |
| BRAINEAC | HIPP | rs852004 | RMND1 | 4.70E-02 |
| BRAINEAC | CRBL | rs852004 | C6orf211 | 2.40E-02 |
| BRAINEAC | CRBL | rs852004 | ESR1 | 2.10E-03 |
FCTX, frontal cortex; THAL, thalamus; HIPP, hippocampus; CRBL, cerebellar cortex. N.A. is not available.
Discussion
Dysregulation of executive function is a key deficit of ADHD. In this study, a genome-wide association study was conducted to explore genetic loci associated with impaired executive function evaluated by BRIEF in ADHD. A significant SNP rs852004 was found to be associated with BRIEF behavioral evaluation of executive function. We further checked the association of the significant SNP with each subscale of BRIEF, and found rs852004 was more associated with Inhibit and Monitor. According to the primary description of BRIEF22, Inhibit and Monitor belong to different indexes. Our principle component analysis found different pattern for the eight scales of BRIEF in ADHD patients. In this population, inhibit was more correlated with the metacognitive index, which was further validated by our confirmatory factor analysis result.
By using CFA, we built a three-factor model for BRIEF (Fig. 2). Normally, the model with TLI and CFI > 0.8, χ2/df < 5, RMSEA and SRMR < 0.1 were considered to be acceptable. In Model C, TLI, CFI and SRMR satisfied the criteria, but χ2/df and RMSEA is larger than the threshold. χ2/df is proportioned with the sample size. We tried to use half of the sample size to fit the model, and the χ2/df became almost half of the current value. In addition, both χ2/df and RMSEA were related with the correlation between different groups. Since Inhibit had significant correlation with Working Memory, Initiate and Emotional Control scales, it is one of the reasons for the larger RMSEA23. Gioia et al. had described a 3-factor model for BRIEF by parsing Monitor into Self-Monitor and Task-Monitor23. The 3-factor model included Behavioral Regulation factor (Inhibit, Self-Monitor), Emotional Regulation factor (Emotional Control, Shift), and Metacognition factor (Initiate, Working Memory, Plan/Organize, Organization of Materials, Task-Monitor). The Model C we established based on the association result of rs852004 was similar with their 3-factor model although the Monitor was not parsed in our model. Based on this model, SNP rs852004 was mainly associated with Behavioral Regulation factor. Remarkably, in our model, Behavioral Regulation factor is more correlated with Metacognition factor, but not Emotional Regulation factor as Gioia et al.’s 3-factor model23. One possible explanation for this is disorder-specific executive function profile. This model is more consistent with Barkley’s view of executive function in ADHD24: inhibitory control having a unique and separable role in executive function; inhibition is more primary and plays an underlying role that enables other functions including working memory, emotional regulation and goal directed analysis and synthesis in problem-solving. Furthermore, in ADHD, the underlying “enable” role of inhibition may have more apparent effect on metacognition factor, including working memory, monitor and plan/organize. The indirect mediation effect of rs852004 on ADHD symptom through each BRIEF scale (see Supplementary Table S1) also supported this, which showed inhibit, monitor, working memory and plan/organize had bigger effect on ADHD overall symptom.
Furthermore, our fMRI data on ADHD and controls suggested specific inhibitory activation in dorsal lateral and medial prefrontal cortex, right insular, putman and supplementary motor area which was overlapped with previous literature reports25. The fMRI data combined with genetic data showed that the associated genetic locus might affect the brain function. The interaction effect of gene and diagnosis was uncovered in brain regions of dorsolateral prefrontal striatum circuit, which was known as the basis of inhibition and in accordance with previous meta-analysis26. It suggested that the associated SNP might have modulating effect on the function of brain region responsible for executive inhibition.
SNP rs852004 and its LD-proxy rs6908732 were suggested to be associated with schizophrenia27, major depressive disorder28 and bipolar disorder29, all of these disorders had executive function deficits30–32. Regulatory feature analyses found this locus was within the chromatin interactive region, which targeted genes RMND1, C6orf211, CCDC170 and ESR1. SNP in ESR1 also predicted intracranial volume33. Meanwhile, eQTL data showed SNP rs852004 regulated the expression of ZBTB2 in cerebellum of bipolar disorder patients and human prefrontal cortex. ZBTB2 is a regulator of p53 pathway34. The homolog gene of ZBTB2 – ZBTB20 has been reported to modulate the sequential generation of neuronal layers in developing cortex35. It is also reported that hyper methylation in the ZBTB20 gene is associated with major depressive disorder36. But the report about the association of ZBTB2 with psychiatric disorder was not much. In consist with our fMRI analysis, our eQTL analyses showed that the SNP rs852004 influenced ZBTB2, RMND1, C6orf211 and ESR1 expression in many brain regions especially frontal cortex (ESR1) and thalamus (RMND1) which constitutes dorsolateral prefrontal striatum circuit, which is the basis of inhibition, including response inhibition and interference inhibition37. We hypothesized that SNP rs852004 influenced these genes expression in consequence to affect the executive function, especially inhibition, and ADHD symptoms.
The detailed molecular mechanism of the significant locus on inhibition related brain function and behavior need further validation study. In consideration of the complexity of cognition and behavior, it’s likely that more genes are still needed to be discovered in larger samples, and more aspects of executive function should be discussed.
Methods
Genome wide association study samples
Totally, 550 subjects finished the Behavior Rating Inventory of Executive Function (BRIEF) (470 boys, 80 girls) aged between 6 and 16 years (average 9.77 ± 2.44 years). All participants in this study were recruited from the Child and Adolescent Psychiatric Outpatient Department of Peking University Sixth Hospital. All cases met DSM-IV ADHD diagnostic criteria. A clinical diagnosis was first made by a senior child and adolescent psychiatrist based on the ADHD Rating Scale-IV (ADHD-RS-IV) completed by parents (and teacher when available), and then confirmed by semi-structured interview using the Chinese version of the Clinical Diagnostic Interview Scale38. Those comorbidities with major neurological or psychiatric disorders including epilepsy, schizophrenia, pervasive development disorder, and mental retardation (IQ < 70) were excluded.
Resting-state fMRI study samples
A total of 50 ADHD patients (47 boys, 3 girls, 42 G homozygotes for rs852004) and 66 control children (37 boys, 29 girls, 57 G homozygotes for rs852004) aged between 8 and 16 years were enrolled from the Child and Adolescent Psychiatric Outpatient Department of Peking University Sixth Hospital in this section. All the participants were right-handed. The ADHD samples are part of the above ADHD samples for BRIEF. The controls were also interviewed to ensure that they were free of any Axis I psychiatric disorders. This study was approved by Institutional Review Board of Peking University Sixth Hospital and written informed consent was signed by parents.
Executive function test and three dimensional symptoms in ADHD patients
We assessed the executive function using BRIEF, an 86-item questionnaire designed for parents of children aged 5–18 years to assess executive function behaviors. In the questionnaire, the parent responds whether their child exhibits problems with specific behaviors: Never, Sometimes or Often, scored as 1, 2, or 3, respectively. The questionnaire includes two domains: Metacognition Index (MI) comprising of five subscales, i.e. Initiate (INIT), Working Memory (WM), Plan/Organize (PO), Organization of Materials (OM) and Monitor (MONI); while Behavior Regulation Index (BRI) comprising of three subscales, i.e. Inhibit (IB), Emotional Control (ECTRL) and Shift (SFT)23,39. The score of each subscale equals to the sum of the scores of all items belonged to that subscale; the scores of two indexes equals to the sum of the scores of all subscales belonged to the index. The score of the Global Executive Composite equals to the sum of the scores of the two indexes. All these traits and their name abbreviation used in this study were shown in Table 2. Besides the BRIEF, we have collected three dimensional symptoms, namely inattention (CDISatt), hyperactivity-impulsivity (CDIShi) and overall score (CDISall), for the patients according to the Clinical Diagnostic Interview Scale40. CDISatt denotes the inattention symptom score, CDIShi denotes the hyperactivity-impulsivity symptom score, and CDISall denotes the sum of CDISatt score and CDIShi score.
Genotyping and quality control
Genomic DNA was extracted from peripheral blood using Omega DNA extraction Kit (Omega Bio-tek Inc., Doraville, GA). Genotypes were obtained using the Affymetrix6.0 array at CapitalBio Ltd. (Beijing) using the standard Affymetrix protocol. The Affymetrix6.0 array included 906,600 SNP probes. After mapping them to SNPs with #rs, 653,428 SNPs were left. For quality control, the individuals (1) with per-individual autosomal heterozygosity >5 s.d. away from the mean, (2) having no age or IQ information, (3) per-individual call rate <95% and (4) with relatives having genome identity PI_HAT ≥ 0.185 were removed. Then, the remaining samples were assessed for population stratification using Principal Component Analysis (PCA) implemented in EIGENSOFT4.241,42. Tracy-Widom test was employed to detect significant eigenvectors (P < 0.05). Only the first eigenvector (eigenvector 1) was significant, which was used as a covariate in the subsequent statistical analysis. When controlling for the quality of SNPs, we removed SNPs if (1) per-SNP call rate <98%, (2) Hardy-Weinberg equilibrium test P < 0.001, (3) MAF < 1%. Totally, 644,166 autosomal SNPs were analyzed in 547 ADHD patients after quality control.
Resting-state fMRI data collection
Resting-state fMRI data acquisition, preprocessing and quality control detailed is described in the supplementary material.
Statistical Analyses
Genome wide association test and imputation
Association analysis for each quantitative trait was conducted using an additive model in linear regression in PLINK43 with age, IQ, sex and eigenvector 1 of PCA as covariates. Two-sided P < 5 × 10−8 was considered as genome-wide significance. We used MACH-admix 1.044 to impute non-genotyped SNPs using the ASN data (286 individuals) from the 1000 Genomes Project Integrated Phase 1 Release45 as the reference panel. Imputed SNPs with squared correlation between imputed and true genotypes (rsq) <0.6 or SNPs with MAF < 0.01 were removed. Association analysis after imputation was done using mach2qtl46.
Principle component analysis and confirmatory factor analysis
All scores of the eight scales of BRIEF were normalized prior the principle component analysis (PCA) and confirmatory factor analysis (CFA). The PCA was conducted in SPSS. CFA for the several candidate models of BRIEF was conducted using AMOS. The maximum likelihood method was used. Fit of all models was evaluated using several indexes, including the χ2/df value, the root mean square error of approximation (RMSEA), the Comparative Fit Index (CFI), Tucker-Lewis Index (TLI) and standardized root mean-square residual (SRMR). Smaller χ2/df, RMSEA and SRMR values and larger CFI and TLI values indicate a better fit.
Resting-state fMRI data analysis
Individual ReHo map was generated in DPABI by calculating the Kendall coefficient of concordance (KCC) of the time series of a given voxel with those of its neighbors (26 voxels) in a voxel-wise way47 and the inclusive threshold for each voxel was set to P < 0.01. The following statistic analysis was conducted in SPM (http://www.fil.ion.ucl.ac.uk/spm/software/spm12/). A T test was applied to identify the differences between ADHD and control, and then a full factor model was built to test the main effect of the interaction effect of genotype and diagnosis, with mean framewise displacement (FD)48, gender, age, cohort used as covariates. Post hoc t tests were performed to further investigate the effect of genotype in different diagnostic groups (ADHD or control). The cluster-level analysis threshold was set to P < 0.01 determined by Monte Carlo simulation correction which was also utilized for multiple comparison correction, and recalculated the kernel of smoothness.
Expression quantitative trait loci (eQTL)
To explore the affected gene expression, we got the SNPs in LD with the significant SNP (r2 > = 0.75) using the 1000 Genomes Project ASN population data. The regulatory features related with these SNPs were searched in rVarBase49, HaploReg50 and RoadMap WashU EpiGenome Browser51. The eQTL data were searched in GTEx Portal52, SCAN53, seeQTL54, SMRI human prefrontal cortex eQTL data21, and BRAINEAC (http://caprica.genetics.kcl.ac.uk/BRAINEAC/).
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Ethic approval
The study was approved by the Institutional Review Board of Peking University Sixth Hospital and carried out in accordance with relevant guidelines and regulations. Written informed consent was obtained from parents of the ADHD probands and controls.
Electronic supplementary material
Acknowledgements
We thank all patients and controls for participating in this study. This work was supported by grants from the National Natural Science Foundation of China (81671358 to Li Yang), the Major State Basic Research Development Program of China (973 Program, 2014CB846100 to Yufeng Wang), the National Natural Science Foundation of China (31401139 to Suhua Chang), Sanming Project of Medicine in Shenzhen “The ADHD research group from Peking University Sixth hospital”(SZSM201612036), and National Key R&D Program of China (2016YFC1306103 to Qingjiu Cao).
Author Contributions
S.C., L.Y. and Y.W. participated in the design of this study, they carried out the genome-wide association study together. With the instruction of Y.Q. and S.C., X.S. and Y.L. carried out principle component analysis and confirmatory factor analysis. X.S., Z.W. participated in the fMRI data analysis, and Q.C. revised the analysis. X.S., Y.L., Z.W., B.Y. drafted the manuscript, and S.C., L.Y., Y.W., Q.C., and Y.Q. performed manuscript review. All authors contributed to the data acquisition.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-26042-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Suhua Chang, Email: changsh@psych.ac.cn.
Li Yang, Email: yangli_pkuimh@bjmu.edu.cn.
References
- 1.Banaschewski T, Becker K, Scherag S, Franke B, Coghill D. Molecular genetics of attention-deficit/hyperactivity disorder: an overview. European child & adolescent psychiatry. 2010;19:237–257. doi: 10.1007/s00787-010-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faraone SV, et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biological psychiatry. 2005;57:1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 3.Gizer IR, Ficks C, Waldman ID. Candidate gene studies of ADHD: a meta-analytic review. Hum Genet. 2009;126:51–90. doi: 10.1007/s00439-009-0694-x. [DOI] [PubMed] [Google Scholar]
- 4.Yang L, et al. Polygenic transmission and complex neuro developmental network for attention deficit hyperactivity disorder: genome-wide association study of both common and rare variants. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2013;162B:419–430. doi: 10.1002/ajmg.b.32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neale BM, et al. Meta-Analysis of Genome-Wide Association Studies of Attention-Deficit/Hyperactivity Disorder. Journal of the American Academy of Child & Adolescent Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinney A, et al. Genome-wide association study in German patients with attention deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:888–897. doi: 10.1002/ajmg.b.31246. [DOI] [PubMed] [Google Scholar]
- 7.Stergiakouli E, et al. Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am J Psychiatry. 2012;169:186–194. doi: 10.1176/appi.ajp.2011.11040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smoller JW, et al. Association between the 5HT1B receptor gene (HTR1B) and the inattentive subtype of ADHD. Biol Psychiatry. 2006;59:460–467. doi: 10.1016/j.biopsych.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Gau SS, Shang CY. Executive functions as endophenotypes in ADHD: evidence from the Cambridge Neuropsychological Test Battery (CANTAB) Journal of child psychology and psychiatry, and allied disciplines. 2010;51:838–849. doi: 10.1111/j.1469-7610.2010.02215.x. [DOI] [PubMed] [Google Scholar]
- 10.McAuley T, Crosbie J, Charach A, Schachar R. The persistence of cognitive deficits in remitted and unremitted ADHD: a case for the state-independence of response inhibition. Journal of child psychology and psychiatry, and allied disciplines. 2014;55:292–300. doi: 10.1111/jcpp.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funahashi S, Andreau JM. Prefrontal cortex and neural mechanisms of executive function. Journal of physiology, Paris. 2013;107:471–482. doi: 10.1016/j.jphysparis.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Baune BT, Malhi GS. A review on the impact of cognitive dysfunction on social, occupational, and general functional outcomes in bipolar disorder. Bipolar Disord. 2015;17(2):41–55. doi: 10.1111/bdi.12341. [DOI] [PubMed] [Google Scholar]
- 13.Bortolato B, Carvalho AF, McIntyre RS. Cognitive dysfunction in major depressive disorder: a state-of-the-art clinical review. CNS & neurological disorders drug targets. 2014;13:1804–1818. doi: 10.2174/1871527313666141130203823. [DOI] [PubMed] [Google Scholar]
- 14.Kitchen H, Rofail D, Heron L, Sacco P. Cognitive impairment associated with schizophrenia: a review of the humanistic burden. Advances in therapy. 2012;29:148–162. doi: 10.1007/s12325-012-0001-4. [DOI] [PubMed] [Google Scholar]
- 15.Wallace GL, et al. Real-World Executive Functions in Adults with Autism Spectrum Disorder: Profiles of Impairment and Associations with Adaptive Functioning and Co-morbid Anxiety and Depression. Journal of autism and developmental disorders. 2016;46:1071–1083. doi: 10.1007/s10803-015-2655-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coghill DR, Seth S, Matthews K. A comprehensive assessment of memory, delay aversion, timing, inhibition, decision making and variability in attention deficit hyperactivity disorder: advancing beyond the three-pathway models. Psychol Med. 2014;44:1989–2001. doi: 10.1017/S0033291713002547. [DOI] [PubMed] [Google Scholar]
- 17.Logue SF, Gould TJ. The neural and genetic basis of executive function: Attention, cognitive flexibility, and response inhibition. Pharmacol Biochem Be. 2014;123:45–54. doi: 10.1016/j.pbb.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Yang et al. A new locus regulating MICALL2 expression was identified for association with executive inhibition in children with attention deficit hyperactivity disorder. Molecular Psychiatry in press (2017). [DOI] [PubMed]
- 19.Hayes, A. F. Introduction to Mediation, Moderation, and Conditional Process Analysis A Regression-Based Approach. 507 (Guilford Press, 2013).
- 20.Gamazon ER, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry. 2013;18:340–346. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C, et al. Whole-genome association mapping of gene expression in the human prefrontal cortex. Mol Psychiatry. 2010;15:779–784. doi: 10.1038/mp.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baron IS. Behavior rating inventory of executive function. Child neuropsychology: a journal on normal and abnormal development in childhood and adolescence. 2000;6:235–238. doi: 10.1076/chin.6.3.235.3152. [DOI] [PubMed] [Google Scholar]
- 23.Gioia GA, Isquith PK, Retzlaff PD, Espy KA. Confirmatory factor analysis of the Behavior Rating Inventory of Executive Function (BRIEF) in a clinical sample. Child neuropsychology: a journal on normal and abnormal development in childhood and adolescence. 2002;8:249–257. doi: 10.1076/chin.8.4.249.13513. [DOI] [PubMed] [Google Scholar]
- 24.Barkley, R. A. ADHD and the nature of self-control. (Guilford Press, 1997).
- 25.Norman LJ, et al. Structural and Functional Brain Abnormalities in Attention-Deficit/Hyperactivity Disorder and Obsessive-Compulsive Disorder: A Comparative Meta-analysis. Jama Psychiat. 2016;73:815–825. doi: 10.1001/jamapsychiatry.2016.0700. [DOI] [PubMed] [Google Scholar]
- 26.Hart H, Radua J, Nakao T, Mataix-Cols D, Rubia K. Meta-analysis of functional magnetic resonance imaging studies of inhibition and attention in attention-deficit/hyperactivity disorder: exploring task-specific, stimulant medication, and age effects. JAMA psychiatry. 2013;70:185–198. doi: 10.1001/jamapsychiatry.2013.277. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda M, et al. Genome-wide association study of schizophrenia in a Japanese population. Biol Psychiatry. 2011;69:472–478. doi: 10.1016/j.biopsych.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 28.McMahon FJ, et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat Genet. 2010;42:128–131. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen DT, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry. 2013;18:195–205. doi: 10.1038/mp.2011.157. [DOI] [PubMed] [Google Scholar]
- 30.Heinrichs RW, Zakzanis KK. Neurocognitive deficit in schizophrenia: A quantitative review of the evidence. Neuropsychology. 1998;12:426–445. doi: 10.1037/0894-4105.12.3.426. [DOI] [PubMed] [Google Scholar]
- 31.Hasler G, Drevets WC, Manji HK, Charney DS. Discovering endophenotypes for major depression. Neuropsychopharmacol. 2004;29:1765–1781. doi: 10.1038/sj.npp.1300506. [DOI] [PubMed] [Google Scholar]
- 32.Bora E, Yucel M, Pantelis C. Cognitive endophenotypes of bipolar disorder: A meta-analysis of neuropsychological deficits in euthymic patients and their first-degree relatives. J Affect Disorders. 2009;113:1–20. doi: 10.1016/j.jad.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Knickmeyer RC, et al. Common variants in psychiatric risk genes predict brain structure at birth. Cereb Cortex. 2014;24:1230–1246. doi: 10.1093/cercor/bhs401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeon BN, et al. ZBTB2, a novel master regulator of the p53 pathway. J Biol Chem. 2009;284:17935–17946. doi: 10.1074/jbc.M809559200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonchev AB, Tuoc TC, Rosenthal EH, Studer M, Stoykova A. Zbtb20 modulates the sequential generation of neuronal layers in developing cortex. Molecular brain. 2016;9:65. doi: 10.1186/s13041-016-0242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies MN, et al. Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biol. 2014;15:R56. doi: 10.1186/gb-2014-15-4-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallo EF, Posner J. Moving towards causality in attention-deficit hyperactivity disorder: overview of neural and genetic mechanisms. The lancet. Psychiatry. 2016;3:555–567. doi: 10.1016/S2215-0366(16)00096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, Wang YF, Qian QJ, Biederman J, Faraone SV. DSM-IV subtypes of ADHD in a Chinese outpatient sample. J Am Acad Child Adolesc Psychiatry. 2004;43:248–250. doi: 10.1097/00004583-200403000-00004. [DOI] [PubMed] [Google Scholar]
- 39.Mahone EM, Hoffman J. Behavior ratings of executive function among preschoolers with ADHD. The Clinical neuropsychologist. 2007;21:569–586. doi: 10.1080/13854040600762724. [DOI] [PubMed] [Google Scholar]
- 40.Barkley, R. Attention-deficit hyperactivity disorder (third edition): A handbook for diagnosis and treatment., (The Guilford Press, 2006).
- 41.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 43.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu EY, Li M, Wang W, Li Y. MaCH-admix: genotype imputation for admixed populations. Genet Epidemiol. 2013;37:25–37. doi: 10.1002/gepi.21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Genomes Project C, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annual review of genomics and human genetics. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zang Y, Jiang T, Lu Y, He Y, Tian L. Regional homogeneity approach to fMRI data analysis. Neuroimage. 2004;22:394–400. doi: 10.1016/j.neuroimage.2003.12.030. [DOI] [PubMed] [Google Scholar]
- 48.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1006/nimg.2002.1132. [DOI] [PubMed] [Google Scholar]
- 49.Guo L, Du Y, Qu S, Wang J. rVarBase: an updated database for regulatory features of human variants. Nucleic acids research. 2016;44:D888–893. doi: 10.1093/nar/gkv1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roadmap Epigenomics C. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gamazon ER, et al. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26:259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia K, et al. seeQTL: a searchable database for human eQTLs. Bioinformatics. 2012;28:451–452. doi: 10.1093/bioinformatics/btr678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.