Abstract
The importance of biodiversity effects on ecosystem functioning across trophic levels, especially via predatory–prey interactions, is receiving increased recognition. However, this topic has rarely been explored for marine microbes, even though microbial biodiversity contributes significantly to marine ecosystem function and energy flows. Here we examined diversity and biomass of bacteria (prey) and nanoflagellates (predators), as well as their effects on trophic transfer efficiency in the East China Sea. Specifically, we investigated: (i) predator diversity effects on prey biomass and trophic transfer efficiency (using the biomass ratio of predator/prey as a proxy), (ii) prey diversity effects on predator biomass and trophic transfer efficiency, and (iii) the relationship between predator and prey diversity. We found higher prey diversity enhanced both diversity and biomass of predators, as well as trophic transfer efficiency, which may arise from more balanced diet and/or enhanced niche complementarity owing to higher prey diversity. By contrast, no clear effect was detected for predator diversity on prey biomass and transfer efficiency. Notably, we found prey diversity effects on predator–prey interactions; whereas, we found no significant diversity effect on biomass within the same trophic level. Our findings highlight the importance of considering multi-trophic biodiversity effects on ecosystem functioning in natural ecosystems.
Introduction
Anthropogenic disturbance on natural environments demands a greater understanding of the consequences of biodiversity change on ecosystem functioning [1, 2]. The majority of studies on this topic, however, have focused on single-trophic levels [3]. Over the last decade, increasing evidence of significant biodiversity effects on ecosystem functioning through trophic interactions (e.g., predator–prey interactions and flux of energy and matter) has highlighted the need to go beyond single-trophic-level diversity effects (e.g., [4–8]). However, a point of concern is that multi-trophic biodiversity effects are complex (e.g., the direction and strength of effect) and the issue remains controversial (e.g., [9–12]). Disagreements over these biodiversity effects between previous studies are partly attributed to the differences in trophic structure of the studied predator–prey interactions [3, 13–15]. Examples of these differences include: the degree of intraguild predation in the food web; the proportion of generalists present at the predator level [16, 17]; and the amount of edible species at the prey level [18].
Multiple hypotheses have been established in terms of trophic interactions mediated by diversity effects of predators and prey, respectively (Fig. 1). These hypotheses predict opposing effects of diversity on trophic transfer as follows: from the aspect of predator diversity, Hypothesis I-1 predicts that higher predator diversity enhances the total amount of consumption and thus promotes trophic transfer. The assumption underlying this hypothesis is that higher predator diversity strengthens the top-down control on prey through diet niche partitioning and thus increases the total consumption by predators [19–21]. In contrast, Hypothesis I-2 predicts that predator diversity decreases consumption and trophic transfer. This prediction considers interference, such as intraguild predation among predators, as a potential consequence of elevated predator diversity [22, 23]. There are also two contrasting hypotheses about how prey diversity affects predators through bottom-up controls. That is, Hypothesis II-1 predicts that higher prey diversity increases predator consumption and trophic transfer between predators and prey via wider diet breadth [19–21] or more diverse and balanced resources for predators (the balance diet hypothesis [24]). By contrast, Hypothesis II-2 predicts that a greater prey diversity hinders predator consumption and thus decreases the trophic transfer between trophic levels. This hypothesis involves the scenario that higher prey diversity may also be associated with greater inedible prey abundance, positive interaction (such as facilitation), higher prey recovery rate (through enhanced resource utilization), and antagonist activity in the prey community, all of which can strengthen prey resistance to predation [11, 25–27].
Fig. 1.
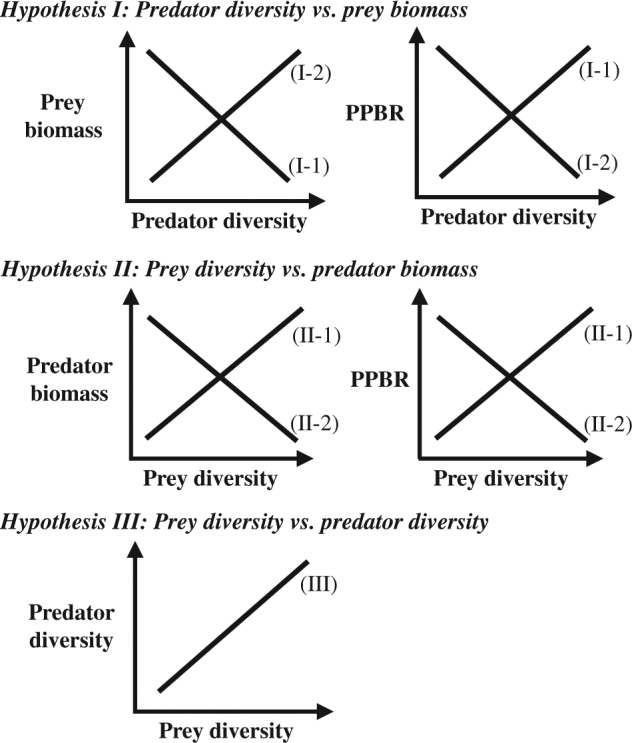
Conceptual diagram illustrating the studied hypotheses associated with diversity effects on trophic interactions. Hypothesis I: predator diversity (I-1) increases predator consumption and consequently prey biomass, which in turn elevates predator/prey biomass ratio (PPBR, a proxy for trophic transfer efficiency), or (I-2) weakens grazing pressure, and as a consequence releasing prey biomass from depression and reducing PPBR. Hypothesis II: prey diversity (II-1) promotes predator biomass and thus increases PPBR or (II-2) reduces predator production and biomass by hindering predation through diluting the proportion of edible prey and thus decreases PPBR. Hypothesis III: (III) prey diversity promotes predator diversity
In addition, focusing on the interplay between predator and prey diversity, Hypothesis III predicts that predator diversity is promoted by prey diversity: greater diversity of prey could increase the opportunities for niche specialization (e.g., [13]). Furthermore, based on an extension of the balanced diet hypothesis, predator coexistence may be maintained via balanced food resources. In fact, positive predator–prey diversity relationships have been commonly observed in terrestrial ecosystems [28–30]. However, in other ecosystems, such as marine and freshwater systems, the nature of predator-prey (e.g., zooplankton–phytoplankton) diversity relationships are thus far unclear (weak, neutral, or even negative; [31–33]).
In spite of the recognized importance of multi-trophic diversity effects, very few studies have been conducted to test these aforementioned, contrasting hypotheses in open ocean systems [34, 35]. To fill this knowledge gap, we investigated nanoflagellates (predator) and bacteria (prey) communities in the East China Sea (ECS). The ECS features a wide gradient of microbial diversity owing to the substantial environmental variations from coastal to oceanic regions [36, 37]. Regarding trophic interaction, the grazing effect of nanoflagellates on bacterial biomass has been studied in the ECS [38], and has potentially profound impacts on biogeochemical cycling and energy transfer through the microbial loop [39–41]. Clear biodiversity effects on the trophic interaction between protist and bacterial communities had been examined in laboratory experiments [42, 43]. Yet, to our knowledge, the diversity relationship between nanoflagellates and bacteria, and the bi-trophic diversity effects on the functioning of marine ecosystems has not been explored in marine pelagic systems.
Here we tested the aforementioned hypotheses (Fig. 1) by exploring the relationship between nanoflagellate (predator) and bacterial (prey) biodiversity, and biodiversity effects on ecosystem functioning. We obtained predator and prey diversity estimates by performing MiSeq paired-end sequencing and used predator and prey biomass as the response variables to the biodiversity change: the predator biomass as a consequence of predator production, the prey biomass as a result of predator’s consumption, and the predator-prey biomass ratio (PPBR) as a proxy for trophic transfer efficiency [33, 44, 45]. This study is expected to provide new insight into the importance of biodiversity effects at multiple trophic levels in marine ecosystems.
Materials and methods
Sample collection and environmental variables
A total of 70 surface layer samples (3–5-m depth) were collected in April, May, July, and August of 2014 and May, June, and July of 2015 (Supplementary Fig. S1), using Go-Flo bottles mounted on a CTD-equipped rosette (Sea-Bird Electronics, Bellevue, WA, USA). The water samples were pre-filtered through a 20-μm pore size mesh to exclude large plankton. Approximately 20 liters of pre-filtered seawater was sequentially filtered through two size fractions (1.2-μm and 0.2-μm) of polycarbonate membranes (Millipore, USA) for a coarse separation of predator (protists, including nanoflagellates) and prey (bacteria). To estimate bacterial abundance, 2 ml of the pre-filtered seawater was fixed using paraformaldehyde solution with a final concentration 0.2% [46]. For the nanoflagellate abundance counts, 50 ml of the pre-filtered seawater was fixed using glutaraldehyde with a final concentration 1% [47]. All samples were stored in liquid nitrogen onboard.
Temperature and salinity were recorded by the CTD profiler. Nitrite and nitrate concentrations were measured by the pink dye method, and phosphate concentrations were measured by the molybdenum blue, according to standard methods [48].
DNA extraction, sequencing, and sequences processing
Total DNA was extracted separately from the 1.2-μm and 0.2-μm pore size membranes using the Meta-G-Nome™ DNA Isolation Kit (Epicentre Biotechnologies, Madison, WI, USA) according to the manufacturer’s instructions. DNA extracts from 1.2-μm and 0.2-μm pore size membranes were separately used as templates of polymerase chain reaction (PCR) to amplify the V4 region [49] of 18S rDNA (for protists including nanoflagellates) and the V5–V6 region [50] of 16S rDNA (for bacteria), respectively. PCR was performed in two-steps to avoid barcode bias [51]. Specifically, we amplified the target region in the first PCR, and generated amplicons with unique dual-index for each sample in the second PCR. Sequencing was carried out using the Illumina MiSeq platform (Illumina, CA, USA), producing 2 × 300 bp paired-end reads. More experimental details were provided in Supplementary Information. The sequence data have been deposited in the NCBI Sequence Read Archive (SRA) under the accession numbers: PRJNA378895, PRJNA378896, PRJNA387529, and PRJNA387530.
Sequence processing
To minimize sequencing errors, low-quality sequences (<Q30) were first trimmed out with Trimmomatic 0.35 [52]. The paired-reads were merged (with minimum 100-bp overlap) and filtered, using PANDAseq [53]. The following filtering parameters were used: (i) non-overlapping paired-reads; (ii) sequences <120 nucleotides; (iii) with incomplete or incorrect primer sequences; and (iv) with more than one undetermined nucleotide. Qualified reads were processed using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline [54]. For chimera checking, VSEARCH [55] and UCHIME [56] were used in de novo mode. Operational taxonomic units (OTUs) were identified at the 97% sequence similarity level using sumaclust [57]. Representative sequences for OTUs were aligned using PyNAST [58] and then phylogenetic trees were constructed using FastTree [59]. Taxonomy assignments for the 18S and 16S rDNA sequences were generated using uclust [60] based on Silva 123 database [61]. The singletons at regional level and unassigned OTUs at Domain level were excluded.
Estimating predator and prey diversity
To obtain predator diversity estimates, we extracted the OTUs from the 18S rDNA pool that was affiliated to the common bacterivorous nanoflagellates according to the literature (Supplementary Table S1), which contained mostly the heterotrophic nanoflagellates (HNF). Although a number of pigmented nanoflagellates are bacterivorous (known as mixotrophic nanoflagellates), we did not include them in the predator assemblages, given controversy surrounding the extent to which these groups contribute to the top-down control on bacterial communities [62–65].
For prey diversity estimates, we considered two functional groups: autotrophic (cyanobacteria) and heterotrophic bacteria communities (cyanobacteria excluded). This distinction was motivated by observations that these two functional groups show different responses to environmental factors (Supplementary Table S2). Moreover, it has been suggested that the HNF have a different diet preference on cyanobacteria and heterotrophic bacteria (e.g., ref. [66]). We note, for autotrophic bacteria, one sample was excluded due to low read number (<100 reads), and thus only 69 out of 70 samples were included in the statistical analysis.
Diversity indices and standardization of diversity estimates
To achieve a more comprehensive investigation on diversity effects, we compared various indices with different emphases on relative abundance: OTU richness, the exponential of Shannon’s entropy index (hereafter, Shannon diversity), the inverse of Simpson’s concentration index (hereafter, Simpson diversity), as well as phylogenetic diversity [67] that incorporates evolutionary influence. To have a fair among-site comparison, we employed the coverage-based approach for diversity estimation [68, 69]. Different from the traditional sample size-based approach, the coverage-based approach yields less biased comparisons of diversity among communities resulted from unequal sampling coverage [68]. The fixed-coverage of heterotrophic (97.9%) and autotrophic (99.3%) bacterial communities was determined by the minimum coverage among samples. For the HNF communities, the fixed-coverage (98%) was decided by the coverage of the double of the minimum reference reads among all samples [68]. To achieve this coverage, exploration prediction was conducted for some samples (Supplementary Table S3). Results from the sample size-based approach were provided (Supplementary Table S2 and S4) and showed consistent diversity patterns as those of the coverage-based approach.
Quantification of predator and prey biomass and trophic transfer efficiency
The numbers of the HNF used as the predator abundance were counted under an epifluorescence microscope (Olympus, Tokyo, Japan). Glutaraldehyde-fixed water samples of 50 ml were filtered onto black nucleopore filters (0.8-μm pore size) and stained with 4′6-diamidino-2-phenylindole (DAPI) at a final concentration of 1 μg ml–1 [70]. The HNF were recognized and counted by blue cell nuclear fluorescence under UV illumination while no orange or red autofluorescence from chlorophyll pigment under blue excitation light. A total of 50 randomly chosen fields of view under magnification at ×1000 were examined for each sample [71]. The abundance of the HNF was converted to the biomass using a factor of 4700 fg C cell–1 [72].
For prey abundance estimation, autotrophic and heterotrophic bacteria were counted, respectively. Paraformaldehyde-fixed water samples of 2 ml were enumerated by FACSAria flow cytometer (FACSAria, Becton Dickinson, USA). For each sample, a subsample of 1 ml was stained by SYBR Green (Molecular Probes Inc., Eugene, OR, USA) for 15 min in the dark for enumerating total bacteria [73] and the other unstained subsample (1 ml) was used for enumerating Prochlorococcus and Synochococcus (autotrophic bacteria) abundance [74]. The heterotrophic bacterial abundance was converted to the biomass using a conversion factor of 20 fg C cell–1 [75]. The abundance of Prochlorococcus and Synochococcus was converted to the biomass with a conversion factor of 60 fg C cell–1 and 178 fg C cell–1 [76], respectively, and the sum was used as the total autotrophic bacterial biomass.
We used the HNF/bacteria (autotrophic or heterotrophic bacteria) biomass ratio (predator/prey biomass ratio denoted as PPBR) as a proxy of trophic transfer efficiency between the HNF (predator) and the bacteria (prey). This usage was motivated by previous studies that used the biomass ratio of predators and prey as a proxy for trophic transfer efficiency [33, 44, 45]. We acknowledge that the predator-prey system we investigated is embedded in a larger food web: both predator and prey biomass and thus PPBR values may be influenced by organisms at other trophic levels that we did not estimate (for example, the grazing effect on nanoflagelaltes).
Statistical analysis
To test Hypotheses I–III (Fig. 1), we used a generalized linear mixed-effects model (GLMM) [77], with sampling season (spring: April, May, and June; summer: July, August, and September) as a random effect. To test Hypothesis I, predator biomass or PPBR was used as the dependent variable while prey diversity was used as the independent variable. For Hypothesis II, prey biomass or PPBR was used as the dependent variable while predator diversity as the independent variable. For Hypothesis I and II, GLMM was conducted with Gaussian distribution and a log-link function. Finally, for Hypothesis III, predator diversity was used as the dependent variable while prey diversity as the independent variable; here, the GLMM was analyzed with Gaussian distribution and an identity link function. Note that, we repeated the analyses for the four diversity indices and two functional groups of prey as explained above. We noted that for each analysis, we carried out multiple tests with various diversity indices; however, we did not correct the threshold α-value for multiple tests, because those diversity indices are correlated and did not represent independent tests; nevertheless, we reported the exact P-values.
To avoid possible spurious relationships between bacteria and HNF simply arising from shared environmental forces and to account for within trophic-level effects, we also evaluated the trophic interaction (prey diversity effect on prey biomass and predator diversity effect on prey biomass) using the partial least squares approach to structural equation modeling (PLS-SEM). PLS-SEM maximizes the amount of variance explained rather than fits a common factor model to the data when constructing the model (the relationships between observed data and latent variables, and between latent variables); it can accommodate the relatively small sample size, and is useful in identifying the key drivers in a complex network [78]. Our SEM model includes six variables: temperature (sea water temperature), nutrient (nitrite, nitrate, and phosphate with loadings of 0.95, 0.94, and 0.69, respectively), prey diversity (bacterial diversity), predator diversity (the HNF diversity), prey biomass (bacterial biomass), and predator biomass (the HNF biomass). Predator and prey biomass and PPBR were log10 transformed to achieve normality. PLS-SEM was run using 1000 bootstraps to validate the estimates of path coefficients and the coefficients of determination. These analyses were repeatedly performed for the two functional groups of prey (autotrophic and heterotrophic bacteria).
Computation
The diversity estimation was implemented using the iNEXT and iNextPD packages [68, 69], GLMM was conducted using the MASS package [77], and PLS-SEM was conducted using the plspm package [79] in the R program [80]. The R scripts of these analyses are provided in Supplementary Information.
Results
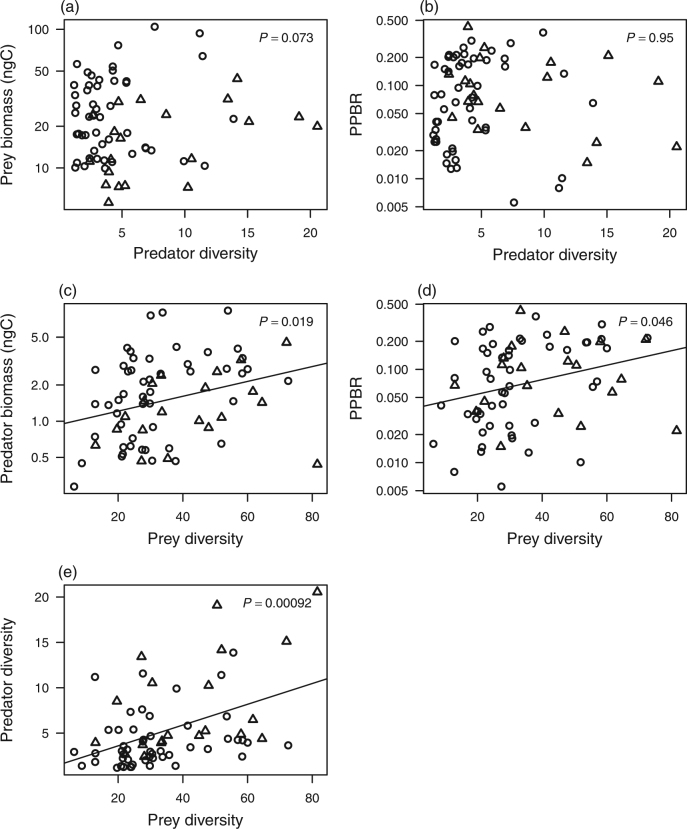
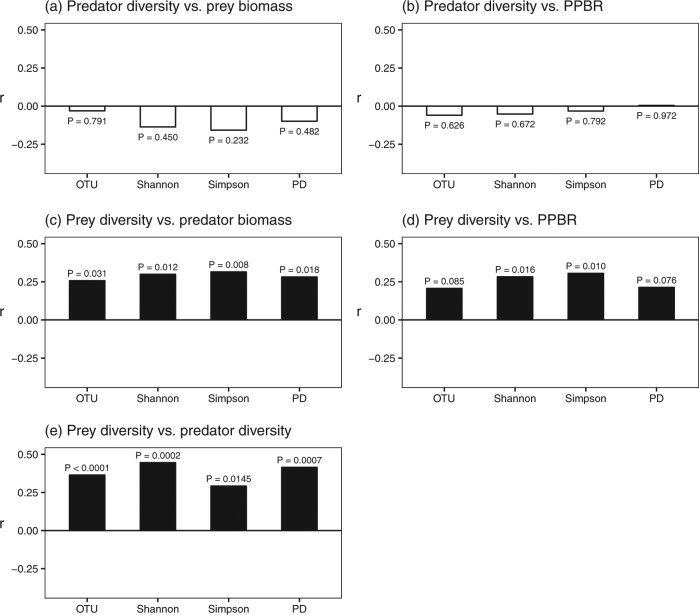
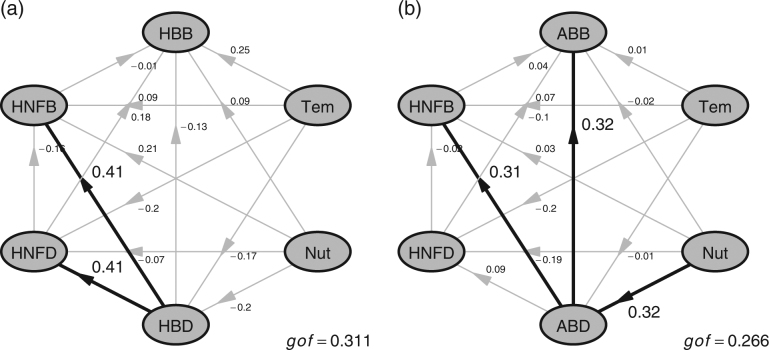
For Hypothesis I, we did not find a clear relationship between predator diversity and prey biomass (also PPBR), regardless of which different prey functional groups (autotrophic or heterotrophic bacteria), and diversity indices were examined (P > 0.40; Figs. 2a, b, 3a, b; Supplementary Figs. S2a, b and S3). The pattern based on PLS-SEM analysis also suggested non-significant effects of predator diversity on prey biomass (Fig. 4a, b). Thus, our results do not support either Hypothesis I-1 or I-2.
Fig. 2.
Relationships used to evaluate the aforementioned Hypotheses in Fig. 1. Hypothesis I: effects of predator diversity (Shannon diversity) on (a) prey (heterotrophic bacteria) biomass and (b) predator-prey biomass ratio (PPBR). Hypothesis II: effects of prey diversity on (c) predator biomass and (d) PPBR. Hypothesis III: (e) predator and prey diversity relationship. Note in a–d, the Y-axis is in log-scale with the base of 10. The circles and triangles represent the samples collected in the summer and spring seasons, respectively. The line indicates the best-fit linear regression line with significant (P < 0.05) relationship based on GLMM analysis with season as the random effect
Fig. 3.
Bar-plot summarizing the relative strength (Pearson’s correlation, r) and its P-value of biodiversity effect on trophic interaction based on different diversity indices: richness (OTU), Shannon, Simpson, and phylogenetic diversity (PD), for the heterotopic bacterial prey. Hypothesis I: effects of predator diversity on (a) prey biomass and (b) predator-prey biomass ratio (PPBR). Hypothesis II: effects of prey diversity on (c) predator biomass and (d) PPBR. Hypothesis III: (e) predator and prey diversity relationship. Filled and open bars indicate positive and negative effects, respectively
Fig. 4.
PLS-SEM analysis deciphering (a) the effect of Shannon diversity of heterotrophic nanoflagellates (HNFD, predator) and heterotrophic bacteria (HBD, prey) on their biomasses (HNFB and HBB), as well as (b) the effect of Shannon diversity of heterotrophic nanoflagellates (HNFD, predator) and autotrophic bacteria (ABD, prey) on their biomasses (HNFB and ABB). Both of the two analyses account for the effects of temperature (Tem), nutrient (Nut), and the interaction within individual trophic level (e.g., the relationship between HBD and HBB, as well as between HNFD and HNFB). Black and gray lines indicate the significant (P < 0.05), and non-significant (P > 0.05) relationships, respectively. The “gof” indicates the goodness of fit
For Hypothesis II, considering the prey diversity effect, the results for heterotrophic bacteria (prey) were supported, showing that increasing prey diversity enhanced predator biomass. This result was consistent for Shannon (Pearson’s correlation, r = 0.30, P = 0.012; GLMM, P = 0.0186) and Simpson diversity (Pearson’s correlation, r = 0.31, P = 0.008; GLMM, P = 0.046) (Figs. 2c and 3c; Supplementary Figs. S4a, c and e). The pattern remained consistent in PLS-SEM analyses, which accounted for the interaction within trophic levels and environmental factors (Fig. 4a). In addition, heterotrophic prey diversity promoted trophic transfer efficiency (Figs. 2d and 3d; Supplementary Figs. S4b, d and f). Whereas, for autotrophic bacteria (prey), positive prey diversity effect on predator biomass held only for the Shannon and Simpson diversity (Supplementary Figs. S2c). Furthermore, a significant positive relationship between autotrophic bacteria (prey) diversity and PPBR held only for phylogenetic diversity (Supplementary Figs. S2d).
Finally, considering the relationship between predator (the HNF) and heterotrophic prey (bacteria) diversity (Fig. 3e), our findings supported Hypothesis III, showing a positive predator-prey diversity relationship. The analyses based on multiple diversity indices showed consistent results for OTU richness (Pearson’s correlation, r = 0.36, P < 0.0001; GLMM, P = 0.002), Shannon (Pearson’s correlation, r = 0.45, P = 0.0002; GLMM, P = 0.00005) and phylogenetic diversity (Pearson’s correlation, r = 0.41, P = 0.0007; GLMM, P = 0.002) (Figs. 2e and 3e; Supplementary Fig. S5). On the contrary, the diversity of autotrophic prey (bacteria) did not show a clear relationship with predator (the HNF) diversity (Supplementary Fig. S2e). This pattern remained consistent after accounting for environmental effects and interaction within trophic levels in the PLS-SEM analysis (Fig. 4a, b).
We did not find biodiversity effects within the same trophic level. There was no clear relationship between diversity and biomass for predator or prey, regardless of different functional groups of prey and which diversity indices we examined (Fig. 4a, b; Supplementary Table S2). In the main text we focused on Faith’s phylogenetic diversity (PD), and noted that Faith’s PD does not consider relative abundance (Hill’s number q = 0) [68]; we further estimated the higher Hill’s number of phylogenetic diversity (q = 1 and 2) and presented the results in Supplementary Information (Supplementary Fig. S6).
Discussion
The effects of predator diversity on trophic interactions
We did not find any significant effect of predator diversity on prey biomass or PPBR, based on GLMM (Fig. 2a, b) or after accounting for environmental factors using the PLS-SEM analysis (Fig. 4); therefore, our results supported neither Hypothesis I-1 nor -2. This conclusion contradicts many previous marine studies that have emphasized the importance of ecosystem responses to predator diversity loss in multi-trophic levels (e.g., refs. [4, 34, 35, 81]). This unexpected result may be explained in several ways: First, previous studies which demonstrated significant impacts of predator diversity on prey biomass were often conducted in closed systems (e.g., refs. [12, 82]), which precluded the possibility of immigration. In contrast to those studies, ours was an open system study allowing for movement of organisms, which may weaken predator effects on the prey communities by compensating the predator-induced reduction in prey biomass [11, 83]. Furthermore, in the context of marine microorganisms, the bacterial growth rate is nearly equal to the mortality rate imposed by their grazers [38, 47, 84]. As such, fast bacterial growth may allow bacteria to recover their biomasses from losses due to predation, and dampen the effects of predator grazing on prey biomass. Another possibility is that we only considered the effect between two trophic levels. The effects of higher trophic-level grazing cannot be estimated in this study but may weaken or even eliminate the nanoflagellates top-down control on bacteria community. In addition to biotic effects, environmental factors possibly alter bacterial abundance and community structure, which may override the effect of trophic interaction in the field. In the ECS, many studies have demonstrated significant relationships between environmental factors and bacterial assemblages (e.g., refs. [36, 46, 85]). Indeed, in this study, environmental effects on heterotrophic bacterial biomass were also found, and it was stronger than that of top-down control: heterotrophic bacteria biomass was controlled primarily by temperature rather than the diversity or biomass of predators (Fig. 3a).
The effects of prey diversity on trophic interactions
We found that prey diversity significantly enhanced predator biomass and trophic transfer efficiency (Fig. 2c, d), supporting Hypothesis II-1, instead of Hypothesis II-2. These findings are in keeping with a previous experimental study that demonstrated a positive effect of bacteria diversity on predator production [82]; whereas, our findings stand in contrast to many previous studies in aquatic systems showing that diverse prey assemblages hinder trophic transfer. For example, a negative effect of prey diversity on predator consumption has usually been found in aquatic systems with algal prey [31, 33, 34, 86], in which increasing algal diversity may be accompanied by an increasing size variety and enhanced proportion of defensive prey [25, 26, 87]. The difference between findings for bacterial versus algal prey may lie in the fact that bacterial prey have little ability to defend against predatory grazing via size-based or chemical-based inhibition, as the size of free-living marine bacteria is consistently small and chemical defense is less prevalent among planktonic bacteria (compared with biofilm bacteria; [88]). Overall, our findings are in line with two hypotheses: (i) more diverse prey assemblages may contain a greater variety of essential biomolecules, which benefit generalist predators, known as “the balance diet hypothesis” or “mixed-diet effect” [12, 24, 89], and (ii) higher prey diversity may provide a wider range of niche breath for specialist predators and promote predator consumption and production via niche partitioning effect (e.g., refs. [19–21, 89]). We acknowledge that our conclusion here is based on correlation analyses; thus, we cannot rule out the possibility that the positive relationship between prey diversity and predator biomass could be a result of predation-mediated species coexistence [90].
Interestingly, Simpson and Shannon diversity indices of the prey (heterotrophic bacteria) showed better correlation with predator biomass and PPBR than OTU richness (Fig. 3c, d). These results suggest a need to consider the effect of prey evenness on trophic interaction, a topic rarely discussed in previous studies [91]. To further address this idea, we computed prey (heterotrophic bacteria) evenness (i.e., inverse Simpson’s index divided by richness) and investigated the relationship between prey evenness versus predator biomass and PPBR and found that predator biomass and PPBR tended to increase with prey evenness (Supplementary Fig. S7). Our results elucidate the effect of prey evenness, in addition to richness, in trophic interaction.
Predator and prey diversity relationship
Our results support Hypothesis III, that predator and prey diversities were positively correlated when considering heterotrophic bacterial prey (Figs. 2e, 3e and 4a). Our finding agrees with the majority of studies in terrestrial systems (e.g., refs. [28–30, 92]). However, to the best of our knowledge, this is the first study demonstrating a significant “positive” predator–prey diversity relationship in marine ecosystems since most previous oceanic studies have focused mainly on zooplankton–phytoplankton interactions, and have presented a weak, neutral or even negative predator-prey diversity relationship [31–33, 86]. The lack of a significant positive predator–prey diversity relationship has been attributed to the reduced degree of specialization for zooplankton [31] or the involvement of a high proportion of defensive phytoplankton taxa [26]. Our finding suggests that these issues may not be critical for interactions of bacteria and nanoflagellates.
Considering different functional groups of prey
The heterotrophic bacteria showed a more consistent and stronger relationship with their predators than autotrophic bacteria. While heterotrophic bacterial diversity significantly promoted the predator diversity and trophic transfer efficiency, these relationships were absent when considering autotrophic bacteria as prey (comparing Fig. 4a, b; Supplementary Fig. 2e). This difference is probably related to the diet preference of predators. It is generally believed that heterotrophic prey are a more important energy source than autotrophic prey [66, 93–95] in spite of a few contradictory results [96]. Thus, our results underline an important (but often-omitted) consideration in empirical research that different functional groups of prey have different effects on trophic interactions among microorganisms in natural marine ecosystems. Nevertheless, we cannot rule out a simple alternative statistical explanation: the range of variation in the diversity of autotrophic bacteria (on average OTU = 3.0 ± 2.6 for each community) is much smaller than that of heterotrophic bacteria (on average OTU = 320.9 ± 141.3 for each community).
Considering taxonomic composition
As a final check, we also investigated the effects of predator and prey taxonomic compositions. Our results indicate that, while the predator biomass changed with prey taxonomic composition, no relationship was found between predator composition and prey biomass (Supplementary Figs. S8 and S9). Quantifying relative contributions of taxonomic composition and diversity in affecting ecosystem functioning is an important issue [1] but it is beyond the scope of this study.
Conclusion
For the first time, we show a biodiversity effect across trophic levels using the nanoflagellate-bacteria linkage in the ECS. We report that prey diversity has positive effects on the biomass (Fig. 2c) and diversity (Fig. 2e) of predators, as well as trophic transfer efficiency (Fig. 2d). However, predator diversity has little effect on prey biomass or trophic transfer efficiency (Fig. 2a, b); whereas, prey biomass was chiefly affected by environmental factors rather than top-down control (Fig. 4a). Overall, multiple trophic-level diversity effects on trophic interaction are more prominent than effects within trophic levels for microbial assemblages (Fig. 4 ad Supplementary Table 2) in the ECS. Nevertheless, the relationships we found in our data are not strong; this may reflect the complexity of natural systems where more than two trophic levels are actively interacting. Our findings corroborate the recent emphasis on multi-trophic biodiversity effects on ecosystem functioning in natural systems (e.g., refs. [4–8]).
Electronic supplementary material
Acknowledgements
We thank Szulung Huang, Ching-Wei Hsu, Yuching Lee, Fan-Sian Lin, and Yuchu Lin for sampling, Hon-Tsen Yu for providing facilities and advice on laboratory work, Chun-Wei Chang, Shao-Lun Liu, Hsiao-Pei Lu, Cheng-Han Tsai, and Yi-Chun Yeh for comments, and Akash Sastri for English editing. This work was supported by the National Center for Theoretical Sciences, Foundation for the Advancement of Outstanding Scholarship, and the Ministry of Science and Technology, Taiwan.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0111-3) contains supplementary material, which is available to authorized users.
References
- 1.Loreau M, Naeem S, Inchausti P, Bengtsson J, Grime JP, Hector A, et al. Biodiversity and ecosystem functioning: current knowledge and future challenges. Science. 2001;294:804–8. doi: 10.1126/science.1064088. [DOI] [PubMed] [Google Scholar]
- 2.Bracken MES, Friberg SE, Gonzalez-Dorantes CA, Williams SL. Functional consequences of realistic biodiversity changes in a marine ecosystem. Proc Natl Acad Sci USA. 2008;105:924–8. doi: 10.1073/pnas.0704103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tilman D, Isbell F, Cowles JM. Biodiversity and ecosystem functioning. Annu Rev Ecol Evol Syst. 2014;45:471–93. doi: 10.1146/annurev-ecolsys-120213-091917. [DOI] [Google Scholar]
- 4.Shurin JB, Borer ET, Seabloom EW, Anderson K, Blanchette CA, Broitman B, et al. A cross-ecosystem comparison of the strength of trophic cascades. Ecol Lett. 2002;5:785–91. doi: 10.1046/j.1461-0248.2002.00381.x. [DOI] [Google Scholar]
- 5.Estes JA, Terborgh J, Brashares JS, Power ME, Berger J, Bond WJ, et al. Trophic downgrading of planet Earth. Science. 2011;333:301–6. doi: 10.1126/science.1205106. [DOI] [PubMed] [Google Scholar]
- 6.Barnes AD, Jochum M, Mumme S, Haneda NF, Farajallah A, Widarto TH, et al. Consequences of tropical land use for multitrophic biodiversity and ecosystem functioning. Nat Commun. 2014;5:5351. doi: 10.1038/ncomms6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardinale BJ, Duffy JE, Gonzalez A, Hooper DU, Perrings C, Venail P, et al. Biodiversity loss and its impact on humanity. Nature. 2012;486:59–67. doi: 10.1038/nature11148. [DOI] [PubMed] [Google Scholar]
- 8.Brose U, Hillebrand H. Biodiversity and ecosystem functioning in dynamic landscapes. Philos Trans R Soc B. 2016;371:20150267. doi: 10.1098/rstb.2015.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin JN, Byrnes JEK, Cardinale BJ. Effects of predator richness on prey suppression: a meta-analysis. Ecology. 2013;94:2180–7. doi: 10.1890/13-0179.1. [DOI] [PubMed] [Google Scholar]
- 10.Katano I, Doi H, Eriksson BK, Hillebrand H. A cross-system meta-analysis reveals coupled predation effects on prey biomass and diversity. Oikos. 2015;124:1427–35. doi: 10.1111/oik.02430. [DOI] [Google Scholar]
- 11.Duffy JE, Cardinale BJ, France KE, McIntyre PB, Thébault E, Loreau M. The functional role of biodiversity in ecosystems: incorporating trophic complexity. Ecol Lett. 2007;10:522–38. doi: 10.1111/j.1461-0248.2007.01037.x. [DOI] [PubMed] [Google Scholar]
- 12.Gamfeldt L, Hillebrand H, Jonsson PR. Species richness changes across two trophic levels simultaneously affect prey and consumer biomass. Ecol Lett. 2005;8:696–703. doi: 10.1111/j.1461-0248.2005.00765.x. [DOI] [Google Scholar]
- 13.Hillebrand H, Shurin JB. Biodiversity and aquatic food webs. In: Belgrano A, Scharler UM, Dunne J, Ulanowicz RE, editors. Aquatic Food Webs – an Ecosystem Approach. Oxford, UK: Oxford University Press; 2005. pp. 184–197. [Google Scholar]
- 14.Schmitz OJ. Predatory diversity and trophic interactions. Ecology. 2007;88:2415–26. doi: 10.1890/06-0937.1. [DOI] [PubMed] [Google Scholar]
- 15.Thébault E, Loreau M. Food-web constraints on biodiversity-ecosystem functioning relationships. Proc Natl Acad Sci USA. 2003;100:14949–54. doi: 10.1073/pnas.2434847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruno JF, Cardinale BJ. Cascading effects of predator richness. Front Ecol Environ. 2008;6:539–46. doi: 10.1890/070136. [DOI] [Google Scholar]
- 17.Ives AR, Cardinale BJ, Snyder WE. A synthesis of subdisciplines: predator-prey interactions, and biodiversity and ecosystem functioning. Ecol Lett. 2005;8:102–16. doi: 10.1111/j.1461-0248.2004.00698.x. [DOI] [Google Scholar]
- 18.Behl S, Stibor H. Prey diversity and prey identity affect herbivore performance on different time scales in a long term aquatic food-web experiment. Oikos. 2015;124:1192–202. doi: 10.1111/oik.01463. [DOI] [Google Scholar]
- 19.Hooper DU, Chapin FS, Ewel JJ, Hector A, Inchausti P, Lavorel S, et al. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge and needs for future research. Ecol Monogr. 2005;75:3–35. doi: 10.1890/04-0922. [DOI] [Google Scholar]
- 20.Loreau M, Hector A. Partitioning selection and complementarity in biodiversity experiments. Nature. 2001;412:72–6. doi: 10.1038/35083573. [DOI] [PubMed] [Google Scholar]
- 21.Tilman D, Knops J, Wedin D, Reich PB, Ritchie M, Siemann E. The influence of functional diversity and composition on ecosystem processes. Science. 1997;277:1300–2. doi: 10.1126/science.277.5330.1300. [DOI] [Google Scholar]
- 22.Bruno JF, O’Connor MI. Cascading effects of predator diversity and omnivory in a marine food web. Ecol Lett. 2005;8:1048–56. doi: 10.1111/j.1461-0248.2005.00808.x. [DOI] [Google Scholar]
- 23.Finke DL, Denno RF. Predator diversity and the functioning of ecosystems: the role of intraguild predation in dampening trophic cascades. Ecol Lett. 2005;8:1299–306. doi: 10.1111/j.1461-0248.2005.00832.x. [DOI] [Google Scholar]
- 24.DeMott WR, Gulati RD, Siewertsen K. Effects of phosphorus-deficient diets on the carbon and phosphorus balance of Daphnia magna. Limnol Oceanogr. 1998;43:1147–61. doi: 10.4319/lo.1998.43.6.1147. [DOI] [Google Scholar]
- 25.Fox JW. Effects of algal and herbivore diversity on the partitioning of biomass within and among trophic levels. Ecology. 2004;85:549–59. doi: 10.1890/03-0095. [DOI] [Google Scholar]
- 26.Hillebrand H, Cardinale BJ. Consumer effects decline with prey diversity. Ecol Lett. 2004;7:192–201. doi: 10.1111/j.1461-0248.2004.00570.x. [DOI] [Google Scholar]
- 27.Steiner CF. The effects of prey heterogeneity and consumer identity on the limitation of trophic-level biomass. Ecology. 2001;82:2495–506. doi: 10.1890/0012-9658(2001)082[2495:TEOPHA]2.0.CO;2. [DOI] [Google Scholar]
- 28.Crutsinger GM, Collins MD, Fordyce JA, Gompert Z, Nice CC, Sanders NJ. Plant genotypic diversity predicts community structure and governs an ecosystem process. Science. 2006;313:966–8. doi: 10.1126/science.1128326. [DOI] [PubMed] [Google Scholar]
- 29.Hawkins BA, Porter EE. Does herbivore diversity depend on plant diversity? The case of California butterflies. Am Nat. 2003;161:40–9. doi: 10.1086/345479. [DOI] [PubMed] [Google Scholar]
- 30.Scherber C, Eisenhauer N, Weisser WW, Schmid B, Voigt W, Fischer M, et al. Bottom-up effects of plant diversity on multitrophic interactions in a biodiversity experiment. Nature. 2010;468:553–6. doi: 10.1038/nature09492. [DOI] [PubMed] [Google Scholar]
- 31.Irigoien X, Huisman J, Harris RP. Global biodiversity patterns of marine phytoplankton and zooplankton. Nature. 2004;429:863–7. doi: 10.1038/nature02593. [DOI] [PubMed] [Google Scholar]
- 32.Longmuir A, Shurin JB, Clasen JL. Independent gradients of producer, consumer, and microbial diversity in lake plankton. Ecology. 2007;88:1663–74. doi: 10.1890/06-1448.1. [DOI] [PubMed] [Google Scholar]
- 33.García-Comas C, Sastri AR, Ye L, Chang CY, Lin FS, Su MS, et al. Prey size diversity hinders biomass trophic transfer and predator size diversity promotes it in planktonic communities. Proc R Soc B. 2016;283:20152129. doi: 10.1098/rspb.2015.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duffy JE, Stachowicz JJ. Why biodiversity is important to oceanography: potential roles of genetic, species, and trophic diversity in pelagic ecosystem processes. Mar Ecol Prog Ser. 2006;311:179–89. doi: 10.3354/meps311179. [DOI] [Google Scholar]
- 35.Gamfeldt L, Lefcheck JS, Byrnes JEK, Cardinale BJ, Duffy JE, Griffin JN. Marine biodiversity and ecosystem functioning: what’s known and what’s next? Oikos. 2015;124:252–65. doi: 10.1111/oik.01549. [DOI] [Google Scholar]
- 36.Jiao N, Yang Y, Hong N, Ma Y, Harada S, Koshikawa H, et al. Dynamics of autotrophic picoplankton and heterotrophic bacteria in the East China Sea. Cont Shelf Res. 2005;25:1265–79. doi: 10.1016/j.csr.2005.01.002. [DOI] [Google Scholar]
- 37.Lin YC, Campbell T, Chung CC, Gong GC, Chiang KP, Worden AZ. Distribution patterns and phylogeny of marine stramenopiles in the North Pacific Ocean. Appl Environ Microbiol. 2012;78:3387–99. doi: 10.1128/AEM.06952-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai AY, Gong GC, Sanders RW, Huang JK. Contribution of viral lysis and nanoflagellate grazing to bacterial mortality in the inner and outer regions of the Changjiang River plume during summer. J Plankton Res. 2013;35:1283–93. doi: 10.1093/plankt/fbt074. [DOI] [Google Scholar]
- 39.Azam F, Fenchel T, Field JG, Gray JS, Meyer-Reil LA, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–63. doi: 10.3354/meps010257. [DOI] [Google Scholar]
- 40.Haas LW, Webb KL. Nutritional mode of several non-pigmented microflagellates from the York River estuary, Virginia. J Exp Mar Bio Ecol. 1979;39:125–34. doi: 10.1016/0022-0981(79)90009-1. [DOI] [Google Scholar]
- 41.Saleem M, Fetzer I, Harms H, Chatzinotas A. Trophic complexity in aqueous systems: bacterial species richness and protistan predation regulate dissolved organic carbon and dissolved total nitrogen removal. Proc R Soc B. 2016;283:20152724. doi: 10.1098/rspb.2015.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chow CE, Kim DY, Sachdeva R, Caron DA, Fuhrman JA. Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J. 2014;8:816–29. doi: 10.1038/ismej.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saleem M, Fetzer I, Dormann CF, Harms H, Chatzinotas A. Predator richness increases the effect of prey diversity on prey yield. Nat Commun. 2012;3:1305. doi: 10.1038/ncomms2287. [DOI] [PubMed] [Google Scholar]
- 44.Yvon-Durocher G, Montoya J, Trimmer M, Woodward G. Warming alters the size spectrum and shifts the distribution of biomass in aquatic ecosystems. Glob Change Biol. 2010;17:1681–94. doi: 10.1111/j.1365-2486.2010.02321.x. [DOI] [Google Scholar]
- 45.Jeppesen E, Jensen JP, Jensen C, Faafeng B, Hessen DO, Søndergaard M, et al. The impact of nutrient state and lake depth on top-down control in the pelagic zone of lakes: a study of 466 lakes from the temperate zone to theArctic. Ecosystems. 2003;6:313–25. doi: 10.1007/PL00021503. [DOI] [Google Scholar]
- 46.Chung CC, Gong GC, Huang CY, Lin JY, Lin YC. Changes in the Synechococcus assemblage composition at the surface of the East China Sea due to flooding of the Changjiang River. Microb Ecol. 2015;70:677–88. doi: 10.1007/s00248-015-0608-5. [DOI] [PubMed] [Google Scholar]
- 47.Christaki U, Van Wambeke F, Dolan JR. Nanoflagellates (mixotrophs, heterotrophs and autotrophs) in the oligotrophic eastern Mediterranean: standing stocks, bacterivory and relationships with bacterial production. Mar Ecol Prog Ser. 1999;181:297–307. doi: 10.3354/meps181297. [DOI] [Google Scholar]
- 48.Gong GC, Wen YH, Wang BW, Liu GJ. Seasonal variation of chlorophyll a concentration, primary production and environmental conditions in the subtropical East China Sea. Deep Sea Res Part II Top Stud Oceanogr. 2003;50:1219–36. doi: 10.1016/S0967-0645(03)00019-5. [DOI] [Google Scholar]
- 49.Stoeck T, Bass D, Nebel M, Christen R, Jones MDM, Breiner HW, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 50.Cai L, Ye L, Tong AHY, Lok S, Zhang T. Biased diversity metrics revealed by bacterial 16S pyrotags derived from different primer sets. PLoS ONE. 2013;8:e53649. doi: 10.1371/journal.pone.0053649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berry D, Mahfoudh KBen, Wagner M, Loy A. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol. 2011;77:7846–9. doi: 10.1128/AEM.05220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger AM, Lohse M, Usadel B. Genome analysis trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: PAired-eND Assembler for Illumina sequences. BMC Bioinform. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jackson MA, Bell JT, Spector TD, Steves CJ. A heritability-based comparison of methods used to cluster 16S rRNA gene sequences into operational taxonomic units. PeerJ. 2016;4:e2341. doi: 10.7717/peerj.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mercier C, Boyer F, Bonin A, Coissac E. SUMATRA and SUMACLUST: fast and exact comparison and clustering of sequences. Available at http://metabarcoding.org/sumatra (2013).
- 58.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 61.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKie-Krisberg ZM, Sanders RW. Phagotrophy by the picoeukaryotic green alga Micromonas: implications for Arctic Oceans. ISME J. 2014;8:1953–61. doi: 10.1038/ismej.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Princiotta SD, Sanders RW. Heterotrophic and mixotrophic nanoflagellates in a mesotrophic lake: abundance and grazing impacts across season and depth. Limnol Oceanogr. 2016;62:632–44. doi: 10.1002/lno.10450. [DOI] [Google Scholar]
- 64.Safi KA, Hall JA. Mixotrophic and heterotrophic nanoflagellate grazing in the convergence zone east of New Zealand. Aquat Microb Ecol. 1999;20:83–93. doi: 10.3354/ame020083. [DOI] [Google Scholar]
- 65.Unrein F, Gasol JM, Not F, Forn I, Massana R. Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J. 2014;8:164–76. doi: 10.1038/ismej.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monger BC, Landry MR, Brown SL. Feeding selection of heterotrophic marine nanoflagellates based on the surface hydrophobicity of their picoplankton prey. Limnol Oceanogr. 1999;44:1917–27. doi: 10.4319/lo.1999.44.8.1917. [DOI] [Google Scholar]
- 67.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. doi: 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- 68.Chao A, Chiu CH, Hsieh TC, Davis T, Nipperess DA, Faith DP. Rarefaction and extrapolation of phylogenetic diversity. Methods Ecol Evol. 2014;84:45–67. [Google Scholar]
- 69.Hsieh TC, Ma KH, Chao A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers) Methods Ecol Evol. 2016;7:1451–6. doi: 10.1111/2041-210X.12613. [DOI] [Google Scholar]
- 70.Caron DA. Technique for enumeration of heterotrophic and phototrophic nanoplankton, using epifluorescence microscopy, and comparison with other procedures. Appl Environ Microbiol. 1983;46:491. doi: 10.1128/aem.46.2.491-498.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai AY, Gong GC, Chiang KP, Tsai PJ, Chao CF. Picoplankton dynamics and their trophic roles in the microbial food-web processes in the southern East China Sea upwelling region during summer. Terr Atmos Ocean Sci. 2014;25:435–48. doi: 10.3319/TAO.2014.01.15.01(Oc). [DOI] [Google Scholar]
- 72.Chen CC, Chiang KP, Gong GC, Shiah FK, Tseng CM, Liu KK. Importance of planktonic community respiration on the carbon balance of the East China Sea in summer. Glob Biogeochem Cycles. 2006;20:GB4001. doi: 10.1029/2005GB002647. [DOI] [Google Scholar]
- 73.Marie D, Partensky F, Jacquet S, Biologique S, Universitté I, Curie M. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl Environ Microbiol. 1997;63:186–93. doi: 10.1128/aem.63.1.186-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu H, Suzuki K, Minami C, Saino T, Watanabe M. Picoplankton community structure in the subarctic Pacific Ocean and the Bering Sea during summer 1999. Mar Ecol Prog Ser. 2002;237:1–14. doi: 10.3354/meps237001. [DOI] [Google Scholar]
- 75.Lee S, Fuhrman JA. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl Environ Microbiol. 1987;53:1298–13033. doi: 10.1128/aem.53.6.1298-1303.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Charpy L, Blanchot J. Photosynthetic picoplankton in French Polynesian atoll lagoons: estimation of taxa contribution to biomass and production by flow cytometry. Mar Ecol Prog Ser. 1998;162:57–70. doi: 10.3354/meps162057. [DOI] [Google Scholar]
- 77.Venables WN, Ripley BD. Modern Applied Statistics with S. Fourth Edition. Springer, New York. ISBN 0-387-95457-0 (2002).
- 78.Reinartz WJ, Haenlein M, Henseler J. An empirical comparison of the efficacy of covariance-based and variance-based SEM. Int J Mark Res. 2009;26:332–44. doi: 10.1016/j.ijresmar.2009.08.001. [DOI] [Google Scholar]
- 79.Sanchez G. PLS path modeling with R. Available at www.gastonsanchez.com/PLS_Path_Modeling_with_R.pdf. Accessed 3 July 2013.
- 80.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/ (2015).
- 81.Byrnes J, Stachowicz JJ, Hultgren KM, Randall Hughes AR, Olyarnik SV, Thornber CS. Predator diversity strengthens trophic cascades in kelp forests by modifying herbivore behavior. Ecol Lett. 2006;9:61–71. doi: 10.1111/j.1461-0248.2005.00842.x. [DOI] [PubMed] [Google Scholar]
- 82.Saleem M, Fetzer I, Harms H, Chatzinotas A. Diversity of protists and bacteria determines predation performance and stability. ISME J. 2013;7:1912–21. doi: 10.1038/ismej.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Connor MI, Bruno JF. Predator richness has no effect in a diverse marine food web. J Anim Ecol. 2009;78:732–40. doi: 10.1111/j.1365-2656.2009.01533.x. [DOI] [PubMed] [Google Scholar]
- 84.Zubkov MV, Sleigh MA, Burkill PH, Leakey RJG. Bacterial growth and grazing loss in contrasting areas of North and South Atlantic. J Plankton Res. 2000;22:685–711. doi: 10.1093/plankt/22.4.685. [DOI] [Google Scholar]
- 85.Wu W, Lu HP, Sastri A, Yeh YC, Gong GC, et al. Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. ISME J. 2018;2:485–94. doi: 10.1038/ismej.2017.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Declerck P, Behets J, Delaedt Y, Margineanu A, Lammertyn E, Ollevier F. Impact of non-Legionella bacteria on the uptake and intracellular replication of Legionella pneumophila in Acanthamoeba castellanii and Naegleria lovaniensis. Microb Ecol. 2005;50:536–49. doi: 10.1007/s00248-005-0258-0. [DOI] [PubMed] [Google Scholar]
- 87.Sommer U, Adrian R, De Senerpont Domis L, Elser JJ, Gaedke U, Ibelings B, et al. Beyond the plankton ecology group (PEG) model: Mechanisms driving plankton succession. Annu Rev Ecol Evol Syst. 2012;43:429–48. doi: 10.1146/annurev-ecolsys-110411-160251. [DOI] [Google Scholar]
- 88.Matz C, Webb J, Schupp P, Phang S. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS ONE. 2008;3:e2744. doi: 10.1371/journal.pone.0002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Striebel M, Singer G, Stibor H, Andersen T. “Trophic overyielding”: phytoplankton diversity promotes zooplankton productivity. Ecology. 2012;93:2719–27. doi: 10.1890/12-0003.1. [DOI] [PubMed] [Google Scholar]
- 90.Caswell H. Predator mediated co-existence: a non-equilibrium model. Am Nat. 1978;112:127–54. doi: 10.1086/283257. [DOI] [Google Scholar]
- 91.Byrnes J, Cardinale BJ, Reed DC. Interactions between sea urchin grazing and prey diversity on temperate rocky reef communities. Ecology. 2013;94:1636–46. doi: 10.1890/11-2310.1. [DOI] [PubMed] [Google Scholar]
- 92.Hertzog LR, Meyer ST, Weisser WW, Ebeling A. Experimental manipulation of grassland plant diversity induces complex shifts in aboveground arthropod diversity. PLoS ONE. 2016;11:e0148768. doi: 10.1371/journal.pone.0148768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Christaki U, Giannakourou A, Van Wambeke F, Gregori G. Nanoflagellate predation on auto- and heterotrophic picoplankton in the oligotrophic Mediterranean Sea. J Plankton Res. 2001;23:1297–310. doi: 10.1093/plankt/23.11.1297. [DOI] [Google Scholar]
- 94.Nakagawa Y, Endo Y. Contributions of heterotrophic and autotrophic prey to the diet of euphausiid, Euphausia pacifica in the coastal waters off northeastern Japan. Polar Biosci. 2002;2:52–65. [Google Scholar]
- 95.Broglio E, Saiz E, Calbet A, Trepat I, Alcaraz M. Trophic impact and prey selection by crustacean zooplankton on the microbial communities of an oligotrophic coastal area (NW Mediterranean Sea) Aquat Microb Ecol. 2004;35:65–78. doi: 10.3354/ame035065. [DOI] [Google Scholar]
- 96.Broglio E, Jónasdóttir SH, Calbet A, Jakobsen HH, Saiz E. Effect of heterotrophic versus autotrophic food on feeding and reproduction of the calanoid copepod Acartia tonsa: relationship with prey fatty acid composition. Aquat Microb Ecol. 2003;31:267–78. doi: 10.3354/ame031267. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.