Abstract
Targeted therapy against actionable gene mutations shows a significantly higher response rate as well as longer survival compared to conventional chemotherapy, and has become a standard therapy for many cancers. Recent progress in next-generation sequencing (NGS) has enabled to identify huge number of genetic aberrations. Based on sequencing results, patients recommend to undergo targeted therapy or immunotherapy. In cases where there are no available approved drugs for the genetic mutations detected in the patients, it is recommended to be facilitate the registration for the clinical trials. For that purpose, a NGS-based sequencing panel that can simultaneously target multiple genes in a single investigation has been used in daily clinical practice. To date, various types of sequencing panels have been developed to investigate genetic aberrations with tumor somatic genome variants (gain-of-function or loss-of-function mutations, high-level copy number alterations, and gene fusions) through comprehensive bioinformatics. Because sequencing panels are efficient and cost-effective, they are quickly being adopted outside the lab, in hospitals and clinics, in order to identify personal targeted therapy for individual cancer patients.
Keywords: next-generation sequencing, clinical sequencing, gene sequencing panel, personalized medicine
The importance of genetic testing for cancer therapy
Recent progress in next-generation sequencing (NGS) has enabled the performance of whole genome sequencing (WGS), whole exome sequencing (WES), or RNA sequencing (RNA-Seq), as well as the identification of huge number of genetic aberrations1). Using NGS method, several large-scale investigations, such as The Cancer Genome Atlas (TCGA), have revealed genome profiles in many cancers, including gastric (GC), colorectal (CRC), breast, gynecological, and non-small cell lung (NSCLC) cancer2-9). After determining the disease-specific distributions of mutational frequencies10,11), clinically actionable gene mutations are focused on among various gene aberrations12). Several gene mutations that may be predictive biomarkers for targeted therapy and prognostic biomarkers have also been identified.
Currently, targeted therapy is being developed based on the identification of actionable gene mutations and the development of targeted drugs. For patients with GC, anti-vascular endothelial growth factor (VEGF)- and anti-human epidermal growth factor receptor 2 (HER2)-targeted therapies have become a standard therapeutic regimen. In addition, HER2-targeted therapy for patients with HER2-positive GC has been reported to have a better therapeutic outcome than conventional chemotherapy, demonstrating that HER2-targeted therapy is a significant step forward to achieve personalized therapy. Therefore, the National Comprehensive Cancer Network (NCCN) guidelines recommend the assessment of HER2 overexpression. Genetic testing for HER2 can also assess HER2 amplification, providing the adaptation of HER2-targeted therapy.
For patients with CRC, NCCN guidelines recommend investigating RAS (KRAS and NRAS) mutation status in advance for the use of inhibitors for epidermal growth factor receptor (EGFR) mutations. These guidelines also recommend investigating BRAF mutation status in patients with CRC, because CRC patients with BRAF mutations might not benefit from anti-EGFR therapy. Because KRAS is a downstream component of the EGFR signaling network, EGFR mutations can not be a therapeutic target in CRC with KRAS mutations.
EGFR and KRAS mutations are considered to be oncogenic driver mutations and are mutually exclusive in lung adenocarcinoma (LADC) (Fig. 1)19). The EGFR exon 19 mutation and exon 21 L858R mutation activate the tyrosine kinase domain, accelerating tumor development. Therefore, targeted therapies using EGFR tyrosine kinase inhibitors (TKIs) against tumors with EGFR mutations have remarkable therapeutic effects13). Similar to EGFR mutations, anaplastic lymphoma kinase (ALK) and ROS1 fusions are treated using specific TKIs that are currently used as standard therapy (Fig. 2)14,15). The current NCCN guidelines recommend genetic testing for EGFR mutations, ALK fusions, and ROS1 fusions to determine first-line therapy16). The therapeutic effects on other actionable gene mutations, such as BRAF, ERBB2, MET, and RET, are being investigated in several clinical trials13). After confirming the therapeutic effects of emerging targeted therapies, the NCCN guidelines will be updated the consensus of genetic testing and treatment strategy in the future.
Fig. 1.
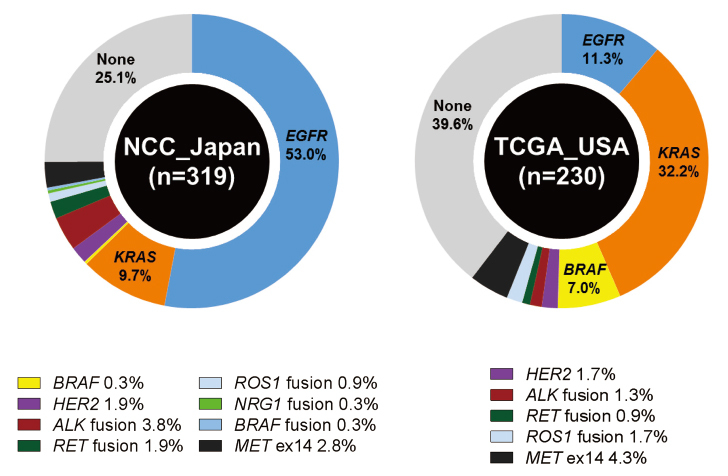
Frequencies of driver gene aberrations in lung adenocarcinoma (LADC). The frequencies are shown for mutations in EGFR, KRAS, BRAF, and HER2, fusions involving ALK, RET, ROS1, NRG1, and BRAF and skipping of MET exon 14. The data were obtained from a Japanese cohort (319 cases from National Cancer Center Hospital) and a US cohort (230 cases from the TCGA study). This figure was modified from our previous reports13,19).
Fig. 2.
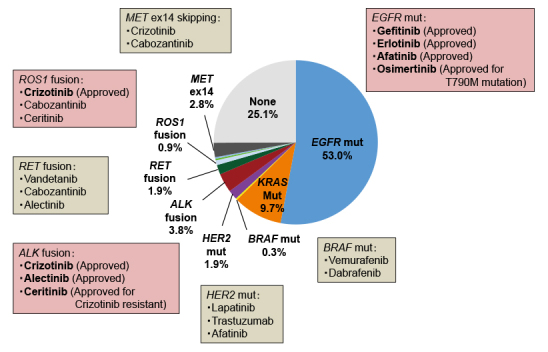
Drugs approved in Japan or candidate targeted drugs against driver gene aberrations in lung adenocarcinoma (LADC). Driver gene aberrations in EGFR, KRAS, HER2, and BRAF; driver fusions involving ALK, RET, and ROS1; and skipping of MET exon 14 were examined in 319 LADC patients who had undergone surgical resection at the National Cancer Center Hospital13)
Clinical sequencing panel
Sequencing panels can simultaneously target multiple genes efficiently, quickly and accurately to maximize the available information from a single investigation. In addition, these panels use clinical samples of both tumor and normal tissue, or tumor tissue only, and reveal gene mutations, copy number variation and gene fusions in tumors through comprehensive bioinformatics.
The predesigned panel covers the most commonly mutated genes or candidate actionable genes in various cancers. On the other hand, custom panels or tumor-specific predesigned panels are developed to investigate the genes that are specifically focused on or found in tumor-specific mutation. While TP53 mutations are broadly identified in various cancers, most actionable gene mutations are identified differently among different cancers. For CRC, genetic testing of KRAS, NRAS, and BRAF mutations is necessary, while on the other hand, for LADC, that of EGFR, KRAS, BRAF and HER2 mutations and ALK, RET, and ROS1 fusions is necessary. We and others have reported that gene mutation profiles differ even in different histological cancer subtypes17,18). Therefore, it is necessary to select the appropriate sequencing panel for each patient, in order to determine actionable gene mutations to perform personalized targeted therapy.
Several cancer centers all over the world have developed their own in-house platforms. For example, the Memorial Sloan Kettering (MSK) Cancer Center developed MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets), a hybridization capture-based NGS panel that can detect all protein-coding mutations, copy number alterations, selected promoter mutations, and structural rearrangement in 468 cancer-associated genes. In 2017, the MSK-IMPACT test was approved by the US Food and Drug Administration (FDA) for in vitro diagnostic test for tumor profiling. To date, MSK-IMPACT has sequenced tumors from more than 20,000 patients with advanced cancer. According to a recent study that sequenced tumors from more than 10,000 patients using MSK-IMPACT, nearly 37% of patients had at least one actionable gene mutation and 11% were able to participate in clinical trials of treatments that directly targeted their genetic alterations20). The FDA has also approved the Oncomine Dx Target test, which targets gene mutations in NSCLC. This test is used as a companion diagnosis to aid in the selection of specific drugs for individual NSCLC patients with EGFR mutations, BRAF mutations, or ROS1 fusions.
The most impressive feature of MSK-IMPACT is that this panel can be used to analyzes both tumor and matched normal tissue and blood and identifies both somatic and germline mutations20). Therefore, MSK-IMPACT results in accurate somatic mutation calls due to eliminate germline variants. MSK-IMPACT also accurately determines mutational signatures to reveal multiple mutational processes and tumor mutation burden to identify patients who mostly can receive the most benefit from immunotherapy. In addition, germline variants can provide therapeutic opportunity as well as cancer susceptibility21-23). The FDA approved a poly ADP ribose polymerase (PARP) inhibitors for the germline BRCA1/2-mutant ovarian cancer24). Furthermore, the PARP inhibitor is also approved for maintenance therapy in both germline and somatic BRCA-mutant ovarian cancer.
Clinical sequencing in Japan
The use of clinical sequencing using NGS-based multiplex gene panels has recently been started in Japan. To date, the MSK-IMPACT test is available at Juntendo University Hospital, Tohoku University Hospital, and Yokohama City University Hospital in Japan. Tumor and non-tumor specimens are sent from these hospitals to the Memorial Sloan Kettering Cancer Center in the U.S. After being sequenced and annotated, the analyzed results are delivered to their respective hospitals in Japan. Based on the identified actionable gene mutations, each patient is recommended to undergo appropriate targeted therapies or clinical trials. Another sequencing panel, the OncoPrime test, targets 232 cancer-related genes and is used at the Kyoto University Hospital, Okayama University Hospital, and Hokkaido University Hospital. However, to date, neither MSK-IMPACT nor OncoPrime tests are covered by social insurance in Japan. Similar to other cancer centers in the world, the National Cancer Center (NCC) in Japan has developed an original sequencing panel, the NCC oncopanel, which targets 90 gene mutations/amplifications and 12 gene fusions, with the aim to identify actionable gene mutations and select suitable molecular targeted therapies25).
NCC Japan launched SCRUM-Japan (Cancer Genome Screening Project for Individualized Medicine in Japan), a nation-wide genome screening consortium, in March 2015. The aim of this project is to investigate the frequency of oncogenic genome alterations in Japanese patients and to facilitate the registration of clinical trials for targeted therapies. In this project, the Oncomine Cancer Research Panel (Oncomine Comprehensive Assay) is used to detect gene mutation, copy number variation and gene rearrangement26). The 143-gene Oncomine Comprehensive Assay is also used in the US National Cancer Institute-Molecular Analysis for Therapy Choice (NCI-MATCH) trial27). The NCI-MATCH trial is a phase II basket study which aims to identify actionable gene mutations.
Conclusion
The recent advances of NGS technologies have enabled the performance of gene sequencing panels for cancer patients in daily clinical practice. Personalized therapy against actionable gene mutations shows positive efficacy; therefore, we believe that the demand for this therapy will increase in all cancer subtypes.
Acknowledgment
This work was supported by JSPS KAKENHI Grant Number 15K10275.
References
- 1.Ashworth A, Hudson TJ. Genomics: Comparisons across cancers. Nature, 502: 306-307, 2013. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487: 330-337, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature, 490: 61-70, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature, 474: 609-615, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature, 489: 519-525, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature, 499: 43-49, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature, 511: 543-550, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature, 513: 202-209, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research, N, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, Yau C, Laird PW, Ding L, Zhang W, Mills GB, Kucherlapati R, Mardis ER, Levine DA. Integrated genomic characterization of endometrial carcinoma. Nature, 497: 67-73, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD M, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12 major cancer types. Nature, 502: 333-339, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet, 45: 1127-1133, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dancey JE, Bedard PL, Onetto N, Hudson TJ. The genetic basis for cancer treatment decisions. Cell, 148: 409-420, 2012. [DOI] [PubMed] [Google Scholar]
- 13.Saito M, Suzuki H, Kono K, Takenoshita S, Kohno T. Treatment of lung adenocarcinoma by molecular-targeted therapy and immunotherapy. Surg Today, 48: 1-8, 2018. [DOI] [PubMed] [Google Scholar]
- 14. Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, De Pas, T, Besse B, Solomon BJ, Blackhall F, Wu YL, Thomas M, O’Byrne KJ, Moro-Sibilot D, Camidge DR, Mok T, Hirsh V, Riely GJ, Iyer S, Tassell V, Polli A, Wilner KD, Janne PA. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med, 368: 2385-2394, 2013. [DOI] [PubMed] [Google Scholar]
- 15. Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, Riely GJ, Varella-Garcia M, Shapiro GI, Costa DB, Doebele RC, Le LP, Zheng Z, Tan W, Stephenson P, Shreeve SM, Tye LM, Christensen J G, Wilner KD, Clark JW, Iafrate AJ. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med, 371: 1963-1971, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman J, Chirieac LR, D’Amico TA, DeCamp MM, Dilling TJ, Dobelbower M, Doebele RC, Govindan R, Gubens MA, Hennon M, Horn L, Komaki R, Lackner RP, Lanuti M, Leal TA, Leisch LJ, Lilenbaum R, Lin J, Loo BW Jr., Martins R, Otterson GA, Reckamp K, Riely GJ, Schild SE, Shapiro TA, Stevenson J, Swanson SJ, Tauer K, Yang SC, Gregory K, Hughes M. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw, 15: 504-535, 2017. [DOI] [PubMed] [Google Scholar]
- 17. Takenaka M, Saito M, Iwakawa R, Yanaihara N, Saito M, Kato M, Ichikawa H, Shibata T, Yokota J, Okamoto A, Kohno T. Profiling of actionable gene alterations in ovarian cancer by targeted deep sequencing. Int J Oncol, 46: 2389-2398, 2015. [DOI] [PubMed] [Google Scholar]
- 18.Saito M, Takenoshita S, Kohno T. Advances in targeted therapy and immunotherapy for treatment of lung cancer. Ann Cancer Res Ther 24: 1-6, 2016. [Google Scholar]
- 19.Saito M, Shiraishi K, Kunitoh H, Takenoshita S, Yokota J, Kohno T. Gene aberrations for precision medicine against lung adenocarcinoma. Cancer Sci, 107: 713-720, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, Hellmann MD, Barron DA, Schram AM, Hameed M, Dogan S, Ross DS, Hechtman JF, DeLair DF, Yao J, Mandelker DL, Cheng DT, Chandramohan R, Mohanty AS, Ptashkin RN, Jayakumaran G, Prasad M, Syed MH, Rema AB, Liu ZY, Nafa K, Borsu L, Sadowska J, Casanova J, Bacares R, Kiecka IJ, Razumova A, Son JB, Stewart L, Baldi T, Mullaney KA, Al-Ahmadie H, Vakiani E, Abeshouse AA, Penson AV, Jonsson P, Camacho N, Chang MT, Won HH, Gross BE, Kundra R, Heins ZJ, Chen HW, Phillips S, Zhang H, Wang J, Ochoa A, Wills J, Eubank M, Thomas SB, Gardos SM, Reales DN, Galle J, Durany R, Cambria R, Abida W, Cercek A, Feldman DR, Gounder MM, Hakimi AA, Harding JJ, Iyer G, Janjigian YY, Jordan EJ, Kelly CM, Lowery MA, Morris LGT, Omuro AM, Raj N, Razavi P, Shoushtari AN, Shukla N, Soumerai TE, Varghese AM, Yaeger R, Coleman J, Bochner B, Riely GJ, Saltz LB, Scher HI, Sabbatini PJ, Robson ME, Klimstra DS, Taylor BS, Baselga J, Schultz N, Hyman DM, Arcila ME, Solit DB, Ladanyi M, Berger MF. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med, 23: 703-713, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shiraishi K, Okada Y, Takahashi A, Kamatani Y, Momozawa Y, Ashikawa K, Kunitoh H, Matsumoto S, Takano A, Shimizu K, Goto A, Tsuta K, Watanabe S, Ohe Y, Watanabe Y, Goto Y, Nokihara H, Furuta K, Yoshida A, Goto K, Hishida T, Tsuboi M, Tsuchihara K, Miyagi Y, Nakayama H, Yokose T, Tanaka K, Nagashima T, Ohtaki Y, Maeda D, Imai K, Minamiya Y, Sakamoto H, Saito A, Shimada Y, Sunami K, Saito M, Inazawa J, Nakamura Y, Yoshida T, Yokota J, Matsuda F, Matsuo K, Daigo Y, Kubo M, Kohno T. Association of variations in HLA class II and other loci with susceptibility to EGFR-mutated lung adenocarcinoma. Nat Commun, 7: 12451, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shiraishi K, Kohno T, Tanai C, Goto Y, Kuchiba A, Yamamoto S, Tsuta K, Nokihara H, Yamamoto N, Sekine I, Ohe Y, Tamura T, Yokota J, Kunitoh H. Association of DNA repair gene polymorphisms with response to platinum-based doublet chemotherapy in patients with non-small-cell lung cancer. J Clin Oncol, 28: 4945-4952, 2010. [DOI] [PubMed] [Google Scholar]
- 23. Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, Ohnami S, Shimada Y, Ashikawa K, Saito A, Watanabe S, Tsuta K, Kamatani N, Yoshida T, Nakamura Y, Yokota J, Kubo M, Kohno T. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet, 44: 900-903, 2012. [DOI] [PubMed] [Google Scholar]
- 24.Maxwell KN, Domchek SM. Cancer treatment according to BRCA1 and BRCA2 mutations. Nat Rev Clin Oncol, 9: 520-528, 2012. [DOI] [PubMed] [Google Scholar]
- 25. Asano N, Yoshida A, Mitani S, Kobayashi E, Shiotani B, Komiyama M, Fujimoto H, Chuman H, Morioka H, Matsumoto M, Nakamura M, Kubo T, Kato M, Kohno T, Kawai A, Kondo T, Ichikawa H. Frequent amplification of receptor tyrosine kinase genes in welldifferentiated/ dedifferentiated liposarcoma. Oncotarget, 8: 12941-12952, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hovelson DH, McDaniel AS, Cani AK, Johnson B, Rhodes K, Williams PD, Bandla S, Bien G, Choppa P, Hyland F, Gottimukkala R, Liu G, Manivannan M, Schageman J, Ballesteros-Villagrana E, Grasso CS, Quist MJ, Yadati V, Amin A, Siddiqui J, Betz BL, Knudsen KE, Cooney KA, Feng FY, Roh MH, Nelson PS, Liu CJ, Beer DG, Wyngaard P, Chinnaiyan AM, Sadis S, Rhodes DR, Tomlins SA. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia, 17: 385-399, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullard A. NCI-MATCH trial pushes cancer umbrella trial paradigm. Nat Rev Drug Discov, 14: 513-515, 2015. [DOI] [PubMed] [Google Scholar]


