Abstract
The structural basis of the interaction between the CD4 coreceptor and a class II major histocompatibility complex (MHC) is described. The crystal structure of a complex containing the human CD4 N-terminal two-domain fragment and the murine I-Ak class II MHC molecule with associated peptide (pMHCII) shows that only the “top corner” of the CD4 molecule directly contacts pMHCII. The CD4 Phe-43 side chain extends into a hydrophobic concavity formed by MHC residues from both α2 and β2 domains. A ternary model of the CD4-pMHCII-T-cell receptor (TCR) reveals that the complex appears V-shaped with the membrane-proximal pMHCII at the apex. This configuration excludes a direct TCR–CD4 interaction and suggests how TCR and CD4 signaling is coordinated around the antigenic pMHCII complex. Human CD4 binds to HIV gp120 in a manner strikingly similar to the way in which CD4 interacts with pMHCII. Additional contacts between gp120 and CD4 give the CD4–gp120 complex a greater affinity. Thus, ligation of the viral envelope glycoprotein to CD4 occludes the pMHCII-binding site on CD4, contributing to immunodeficiency.
The adaptive immune response depends on the specific recognition by a T-cell receptor (TCR) of an antigenic peptide bound to a major histocompatibility complex (MHC) molecule (pMHC), as well as interaction of this same pMHC with a CD8 or CD4 coreceptor (1–4). The structural basis of the TCR-pMHCI, TCR-pMHCII, and CD8αα–pMHCI interactions has been crystallographically defined (1, 5–8), whereas the structural nature of CD4-pMHCII is not yet known. The extracellular rod-like CD4 segment consists of four concatamerized Ig-like domains (D1–D4) (9–11). Mutagenesis studies have suggested that residues in the membrane distal D1-D2 module bind to predominantly nonpolymorphic residues of MHC class II molecules (12–14). During pMHCII recognition, CD4 and TCR colocalize to interact with the same pMHCII (2). Hence, CD4 brings the p56lck that associates with the short CD4 cytoplasmic tail to the site of immune recognition (15). In fact, such tyrosine kinase recruitment, rather than any major contribution to binding energy, appears to be the major function of CD4 (16).
Structural information about the CD4 receptor–ligand interaction would be of particular importance, given the expression of CD4 on the surface of all helper T cells and the essential role of this protein in their activation. Once triggered, helper T cells stimulate B-cell antibody production and cytolytic T-cell generation, thereby regulating the induction of the immune response. Development of methods to block CD4–pMHCII interaction might lead to an important new class of immunosuppressive compounds, depending on structural data. With this goal in mind, we have determined the crystal structure of a CD4–pMHCII complex. As reported below, it is now possible to compare the nature of coreceptor binding to pMHC for both CD4 and CD8 T-cell subsets as well as to examine how CD4 is ligated by its physiologic pMHCII ligand versus the pathologic HIV1 gp120 ligand.
Materials and Methods
Crystallization and Data Collection.
The human CD4 N-terminal two-domain fragment (amino acid residues 1–183) and the murine class II molecule I-Ak with a covalently linked 13-residue hen egg conalbumin (CA) peptide (residues 134–146) were prepared as previously reported (10, 17). Crystals were grown by using the hanging-drop vapor-diffusion method. CD4 and CA/I-Ak were stoichiometrically mixed to a concentration of 20 mg/ml. The complex solution was further mixed with reservoir crystallization buffer in equal volume to 2 μl. The reservoir contains 1 ml of 17% polyethylene glycol 4,000/0.2 M Li2SO4/0.1 M Tris at pH 8.5. The rod-like crystals of 0.25 × 0.05 × 0.05 mm grew readily at room temperature in a few days. They belong to space group P4322 with unit cell dimension: a = b = 145.2 Å and c = 103.8 Å. There is one complex in the asymmetric unit with a solvent content of ≈68%. These crystals diffract very anisotropically (3.5 Å along the c-axis, the rod direction, and only around 4.5–5 Å in the perpendicular direction) and are extremely sensitive to any changes in harvest condition.
Data were collected at the SBC-CAT 19ID beamline of the Advanced Photon Source at Argonne National Laboratories. The best data set was obtained from a single prefrozen crystal that was directly picked up from the crystallization droplet with a loop, “soaked” in mineral oil for half a second, and then dipped into liquid nitrogen for freezing. Data with an oscillation range covering 160° were collected to ensure a high degree of redundancy. The data were processed by using the HKL2000 suite (18). For the overall data within 30–4.3 Å, the I/σ = 12.0, Rmerge = 15.2%; completeness = 83% (100% completeness to 5 Å); redundancy = 9.6; whereas at the outer resolution bin (4.45–4.3 Å), the corresponding figures are 1.9, 61%, 56%, and 7.8, respectively.
Structure Determination.
The structure was determined with the molecular replacement method. The search for I-Ak (without a peptide) was first carried out within a data range of 10–5 Å with the program amore (19) by using the pdb file 1iak (20) as a search model. The solution was straightforward, giving an outstanding rotation function peak with a correlation coefficient of 6.3 versus 3.3 for the next peak. After the fitting step, the correlation coefficient reached a value of 35.1, and the R factor was 49.8% at 5 Å. The CD4 molecule was located by using an evolutionary six-dimensional search algorithm, implemented in the program epmr (21), also at 5 Å resolution. The properly positioned MHC molecule was added as a constraint such that the centers of gravity of the CD4 molecule and the MHC molecule are 12 Å apart. The best solution has a correlation coefficient of 60.3 versus 51.2 for the second highest. In addition, fffear (22) was run to perform a reciprocal space search with data to 4.3 Å to position the CD4 molecule by using phases from the properly placed MHC molecule. The rms deviation between the models derived from fffear and epmr was 1.5 Å for all Cα atoms. Further rigid body refinement, including all measured reflections in the range 20–4.3 Å with refmac (23), gave an R factor of 42% (48% from 4.5 to 4.3 Å) and an Rfree of 45% (48% from 4.5 to 4.3 Å) by using bulk solvent correction and anisotropic scaling. The rigid bodies consisted of each of the four domains of the class II MHC molecule, the peptide, and each of the two domains of the CD4 D1-D2 molecule. No major domain movements were observed on rigid body refinement. TLS refinement was performed on the same rigid bodies to get an estimate for the atomic displacements of the domains. We obtained relatively low B values for the class II MHC molecule (57 Å2 for α1, 52 Å2 for α2, 63 Å2 for β1, and 47 Å2 for β2) and slightly higher values for the peptide (82 Å2) and the respective CD4 domains (73 Å2 for D1 and 100 Å2 for D2). Molecular dynamics were performed on the side chains of Phe-43 and Arg-59 with cns (24) to ascertain their optimal position, whereas the remainder of the model was not adjusted further. The structure coordinates arrived at after rigid body refinement, as well as the structure factor amplitudes, have been deposited in the Protein Data Base under code 1JL4.
Mutagenesis, Transfection, and Binding Assay.
Mutagenesis of human CD4 by using single-stranded DNA and oligonucleotides (18–22 bases) was carried out as described (12). Mutant CD4 was subcloned into the XbaI site of the CDM8 vector. COS7 cells were plated into each well of Falcon six-well dishes and were transiently transfected with 5 μg of wild-type or mutant CD4 DNA by the calcium phosphate/chloroquine method. Two days after transfection, binding of MHC class II+ human B lymphoblastoid Raji cells to CD4-transfected COS7 cells was assayed as described (12). Briefly, 1 × 107 Raji cells were added onto the COS7 cell monolayer and incubated at 37°C for 1 h. Unbound Raji cells were removed by aspiration and further washing, and binding of CD4 to class II MHC was enumerated by counting the number of COS7-cell-B-cell rosettes in 10 random optical fields under a microscope. Cells transfected with CDM8 vector alone served as negative controls for rosette formation. Cell-surface CD4 expression was determined by using mAbs and indirect immunofluorescence, as described (13, 14).
Results and Discussions
CD4–pMHCII Complex.
Prior studies established that human CD4 interacts with murine as well as human pMHCII (25–27). To examine the CD4–pMHCII interaction structurally, the human CD4 two domain fragment (amino acids 1–183) (10) and the murine class II molecule I-Ak (17) were stoichiometrically mixed and cocrystallized. The tetragonal crystals diffract anisotropically, to beyond 3.5 Å in the best direction and to within 4.5–5 Å in the worst direction. The structure was determined by using the molecular replacement method and rigid-body refined to 4.3 Å with an R factor reaching 42%. Although the complex structure has a limited resolution, the result can be interpreted with confidence in light of known high-resolution structures of each of the two molecular components. This information includes the general binding topology and interacting structural elements involved. However, at this relatively low resolution, any description of detailed interaction is tentative.
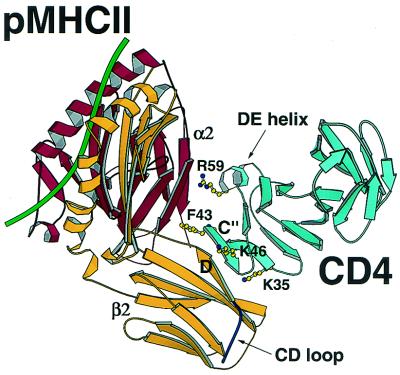
The complex structure shows that the CD4 N-terminal immunoglobulin variable region-like domain is directed toward and reaching into the two membrane-proximal domains of the MHC class II molecule. The C terminus of the CD4 molecule points away from pMHCII (Fig. 1). Consequently, domain 2 of the CD4 fragment makes no contact with the pMHCII molecule. In other words, the long axes of the two molecules are neither antiparallel, as expected, nor perpendicular, as in the case of the CD8–pMHCI interaction (5, 8). There are three major binding elements on the N-terminal domain of CD4: Phe-43, the C" strand, and the short α-helical segment between β strands D and E. Collectively, these structural elements are located at the “top corner” of the domain, encompassing what has been previously referred to as the C′C" ridge (10).
Figure 1.
Ribbon diagram of the CD4–pMHCII complex. The murine I-Ak MHC class II molecule with a CA peptide (green) bound to the antigen-presenting platform interacts with hCD4 (cyan) through both the α2 (red) and the β2 (yellow) domains of pMHCII and domain 1 (D1) of hCD4. Residues Lys-35, Phe-43, Lys-46, and Arg-59 on CD4 D1 essential for binding are highlighted. The CD loop (delineated with an arrow) on the β2 domain of the I-Ak molecule is shown to have no direct interactions with CD4. Note also how CD4 D2 (unlabeled, cyan) makes no contact to pMHCII. All the figures were prepared with molscript (46) and RASTER 3D (47).
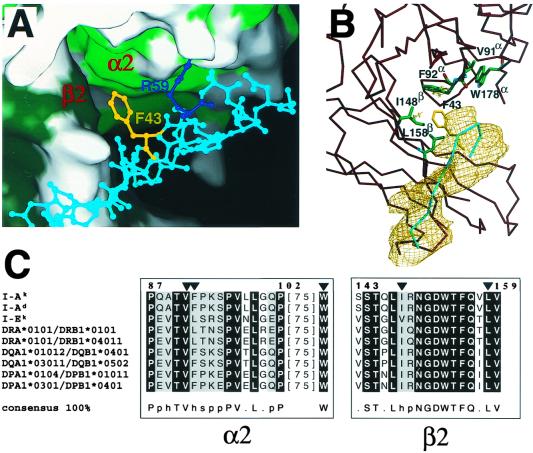
The most striking feature of the interactions is that the aromatic ring of Phe-43 at the beginning of the C" strand is inserted into a site surrounded by a conserved group of hydrophobic pMHCII residues, appearing to include Val-91, Phe-92, and Trp-178 from the α2 domain as well as Ile-148 and Leu-158 from the β2 domain. Fig. 2A is a local view of this area and depicts how α2 and β2 domains of the pMHCII molecule together form the concavity for CD4 binding. Fig. 2B shows the position of these hydrophobic MHC class II residues relative to the CD4 C" strand. Superimposed is an omit map showing a local region where the C" strand lies. The rather striking conservation of residues in this region between human and mouse alleles and across loci is evident from the Fig. 2C sequence alignment.
Figure 2.
The conserved hydrophobic pocket formed by class II MHC α2 and β2 domain residues into which CD4 Phe-43 inserts. (A) Class II MHC is depicted in surface representation form and adjacent CD4 residues in ball-and-stick format. Hydrophobic residues from domain α2 (light green) and domain β2 (dark green) of the I-Ak molecule are shown. The positions of the side chains of Phe-43 (yellow) and Arg-59 (blue) have been optimized by molecular dynamics by using cns (24). The surface representation was prepared with spock. (B) The same region is presented with residues that contribute to the hydrophobic pocket shown, including Val-91, Phe-92, and Trp-178 from the α2 domain and Ile-148 and Leu-158 from the β2 domain of the MHC class II molecule in green. The Cα trace of the MHC class II molecule in this region is depicted in dark red. The Cα trace of the C" strand of the CD4 molecule is depicted in cyan, and the Phe-43 residue is shown in yellow. A portion of an omit map at a contour level of 1σ is shown in gold for the C" strand of CD4, which was obtained by rigid body refinement with this segment excluded from the calculation. (C) Sequence alignment of the highly conserved contact regions in MHC class II for different murine (I-Ak, I-Ad, and I-Ek) and human (DRA*0101/DRB1*0101, DRA*0101/DRB1*04011, DQA1*01012/DQB1*0401, DQA1*03011/DQB1*0502, DPA1*0104/DPB1*01011, DPA1*0301/DPB1*0401) alleles by using clustalx (48). Residues Val-91, Phe-92, and Trp-178 of the α2 domain and Ile-148 and Leu-158 of the β2 domain are indicated by a triangle. The consensus is defined so that entirely conserved residues are depicted by their amino acid code, whereas p indicates conservation of a polar residue, h conservation of a hydrophobic residue, and s conservation of a small side-chain-containing residue.
The second major CD4–MHC class II interaction is between the middle portions of the CD4 C" strand and the pMHCII β2 domain D strand. The backbones of CD4 Lys-46–Leu-44 and pMHCII Ser-144–Gln-146 segments meet to form a much twisted, antiparallel mini-β-sheet. This mini-β-sheet helps bring the Phe-43 at the beginning of the C" strand into the contact site to perform its key binding function.
The third interaction site involves the helical region between β strands D and E of the CD4 N-terminal domain and approximated residues of the pMHCII α2 domain. There is a group of charged and other hydrophilic residues in the contact area for both interacting partners that could potentially form a series of salt bridges and hydrogen bonds between the two molecules. These hydrophilic contacts may include Ser-60 and Asp-63 from CD4 and Gln-88α and Lys-176α of pMHCII, respectively. In addition, other hydrophilic interactions might involve CD4 residues Lys-35, Lys-46, and Arg-59 that are within reach of Glu-162β and Gln-146β as well as Gln-88α and Asp-110α of pMHCII. As suggested by analysis of other surface proteins involved in cell–cell interactions and their “hot spots,” a hydrophobic residue such as Phe-43 may provide the major binding energy, whereas surrounding charged residues facilitate specificity (1). A role for both β2 and α2 pMHCII domains in the CD4 interaction was anticipated by domain swap and mutagenesis studies, although the functional disruption from those pMHCII point mutations is secondary to internal structural rearrangements (28–30).
Although the CD8αα homodimer also uses the N-terminal portions of its paired variable-like domains to contact a pMHCI molecule, it does so differently, in several respects, from the way in which CD4 binds to a pMHCII (5, 8). The major CD8-binding site on pMHCI is the CD loop of the α3 domain, the counterpart of the β2 domain in pMHCII. By contrast, the major CD4-binding site on pMHCII involves the junction of α2 and β2 domains (the D and E strands of β2 and the A and G strands of α2). There are no direct contacts between CD4 and the CD loop of the pMHCII β2 domain. CD8αα clamps onto this CD loop of the pMHCI α3 domain in an antibody-like manner with all six CDRs (three from each V-like domain) across the top of the two CD8 subunits. On the other hand, CD4 interacts with pMHCII by using only the “top corner.” Finally, CD8 binding causes the α3 domain to swing relative to the rest of the pMHCI molecule (5, 8), whereas CD4 binding does not seem to induce significant pMHCII domain movements.
The structure agrees well with CD4 mutational data if one considers those mutations that do not alter CD4 expression (≥90% of wild-type CD4 levels) yet significantly impair class II MHC binding (≤30% rosetting), as determined by adhesion assays between hCD4 transfected COS cell and class II MHC expressing human B lymphoblastoid cells and, in the case of Phe-43→Ile, analysis of transgene mice (13, 14, 31). On the other hand, mutations that more modestly impair cell binding and/or are associated with reduced CD4 cell surface expression in the functional adhesion assay do not correspond to CD4-pMHCII contact sites observed crystallographically. The precise geometry of the C′C" ridge relative to the rest of CD4 D1 is undoubtedly important because internal alterations that shift or eliminate hydrogen bonds, for example, mutations of Trp-62→Tyr or Ser-49→Val, strongly disrupt pMHCII binding. Only CD4 Lys-72 and Thr-81 residues fail to make direct contacts to pMHCII in the crystallographic structure yet virtually eliminate pMHCII binding when mutated to alanine. We did not find crystal contacts between symmetry-related molecules in the unit cell involving Lys-72 or Thr-81 to suggest that they represent points of CD4-CD4 oligomerization. Thus, these two residues are neither sites of pMHC ligand contacts nor oligomer contacts. However, effects of those two residues may be secondary such that, for example, the Thr-81→Ala mutation impacts on a hydrogen bonding network involving Gln-94 near the CD4 D1–D2 junction.
Ternary Model of pMHCII, TCR, and CD4.
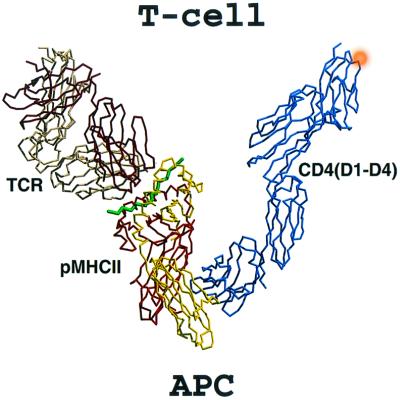
To date, there has been no structural report on a ternary complex that contains a pMHCII, a TCR, and an intact CD4 molecule. Because a binary complex structure of the TCR–CA/I-Ak complex (17) and a structure of the entire extracellular four-domain fragment of hCD4 (CD4 D1-D4) (11) are available, a ternary complex model can be constructed in conjunction with the current binary complex structure of the hCD4 D1D2-CA/I-Ak. We first superimposed the I-Ak components of the CD4 D1D2-CA/I-Ak structure and our recently published structure of the D10 TCR–CA/I-Ak complex (17). The D10-CA/I-Ak structure contains only the V-module of the TCR. Because all TCRs share a common C module that is structurally similar in human and mouse, and the V modules of D10 and a class I-restricted human TCR B7 (32) can be superimposed onto each other extremely well (17), the C module of D10 TCR can thus be modeled from the B7 structure. We then applied the transform derived from the superposition of the CD4 D1D2 components of the current hCD4 D1D2-CA/I-Ak structure and the hCD4 D1-D4 structure (11) to incorporate the C-terminal D3D4 module of hCD4. The final ternary complex model (Fig. 3) gives a view of how an MHC class II-restricted TCR and an intact CD4 molecule derived from a T-helper cell might simultaneously bind to the same pMHCII on an opposing antigen-presenting cell.
Figure 3.
Model of the ternary CD4–pMHCII–TCR complex. The assembly of several crystal structures reveals a ternary complex composed of the hCD4 D1-D4, pMHCII, and TCR having a V-shaped configuration. The D10 TCR–CA/I-Ak complex consists of the α (bright red) and β (yellow) subunits of the I-Ak molecule, the CA peptide (green), and the two TCR subunits α (dark red) and β (beige). The hCD4 D1-D4 is shown in blue, with the approximate position of the D4 glycan given as an orange ball. The assembly is oriented with the T-cell membrane on top and the antigen-presenting cell membrane on the bottom.
From the perspective of the T-cell surface membrane (Fig. 3 Upper), both TCR and CD4 are tilted rather than oriented vertically. Assuming the insertion point of the CD4 ectodomain into lipid rafts where CD4 is a resident protein (33) and that of the non-raft resident TCR proteins are at equivalent vertical positions in the cell membrane, this “tilt” is likely because of the intrinsic features of the TCR complex components. The D1–D2 junction and D3–D4 junction of the CD4 molecule are quite rigid, with the D2–D3 junction itself having only limited flexibility (11). Thus, the total length of the concatenated four-domain CD4 rod is ≈120 Å, with virtually no capacity to extend further. The length of the binary TCR–pMHCII complex is around 130–140 Å, slightly longer than CD4. On the basis of prior analysis of TCR–pMHCII interactions (17, 34), it is likely that the variation of TCR docking topology onto pMHCII is limited, making the D10–CA/I-Ak complex a reasonable surrogate for all TCR–pMHCII interactions. The stalk regions connecting the membrane proximal domain and transmembrane-spanning segment of CD4, TCR Cα, and TCR Cβ are, respectively, 9, 19, and 13 amino acids in length. If the TCR–pMHCII binary complex oriented vertically to the cell membrane, then the CD4 molecule would not be of sufficient length to reach out from the T-cell membrane to maintain the V-shaped binding mode. Movement at the CD4 D2–D3 junction or alteration of the relative TCR and CD4 insertion points and intervening membrane may modify the picture to some degree. However, such variation is unlikely to change the overall binding topology of the components.
Several interesting features about the ternary complex model shown in Fig. 3 can be noted. There are no direct contacts between CD4 and the TCR αβ heterodimer, consistent with recent Biacore studies (16). In fact, the membrane-proximal ends of these two molecules are as far as 100 Å apart. On the basis of mAb epitope mapping and chain association studies (35, 36), CD3ɛ and CD3δ subunits may sit within or near this interval, but the ample space makes direct CD4-CD3 contact unlikely. The antigen-binding groove of pMHCII is no longer parallel to the cell surface but makes about a 45° angle with the membrane. Despite the overall predicted V shape for the ternary complex, the membrane-proximal domains of each of the components are all roughly vertical, including domain 4 of CD4, the α2 and β2 domains of pMHCII, as well as the Cβ domain of TCR. The only exception is the unique Cα domain, which hangs almost parallel above the membrane with its relatively lengthy stalk region bridging the space. Human CD4 has one glycan attached to Asn-300, a residue located on the AB loop of domain 4 and pointing toward the membrane and away from TCR-pMHCII. This glycan may help preconfigure CD4 to assume a favorably tilted orientation for ligand binding in a role analogous to that observed in the structure of intercellular adhesion molecule-2 (37). Overall, this unexpected V-shaped binding mode raises intriguing questions as to how the cytoplasmic portions of the TCR subunits, including their immune receptor tyrosine activation motifs and the CD4-associated p56lck, coordinate in signaling, the precise disposition of the CD3 components within the TCR machinery, and whether this ectodomain model represents a pre- and/or post-activation complex.
In vitro data have suggested that functional CD4-pMHCII binding may depend on CD4 oligomerization (29, 38). Dimerization of the CD4 D1-D4 via D4 in several crystal lattices has been interpreted as consistent with this view (11). Assuming this is the case, then the V shape of the ternary complex modeled herein could become a “W” shape by crosslinking of two adjacent V-shaped CD4–pMHC–TCR complexes. However, this is unlikely to occur, because the two protomers within the CD4 D1-D4 dimer have a very large opening angle. Consequently, the vertical distance from the apex of the CD4 D1-D4 dimer interface to the other end of each CD4 molecule is only ≈80 Å. In comparison, excluding their respective stalks, the ternary complex spans 120 Å between the two opposing cell membranes. Thus, the different geometries make it difficult to imagine how CD4 dimers could create an oligomer array between the T cell and the antigen-presenting cell. In our current crystal structure, two CD4 D1D2 molecules are related by a crystal dyad in a parallel fashion. Both D1 and D2 are so closely juxtaposed to their symmetry mates that CD4 D3D4 cannot be modeled onto these CD4 D1D2 fragments without serious molecular collision. We have also examined molecular packing modes in published crystal structures that contain CD4 fragments (CD4 D1D2, CD4 D3D4, CD4 D1-D4), and pMHCII and relevant TCR molecules (9–11, 17, 39, 40). In fact, the CD4-binding site on the class II MHC molecule is nearly coincident with the “dimer of class II MHC heterodimers” interface previously proposed (40). From this analysis, we conclude that dimerization of the ternary complex is not likely to occur via the ectodomains of components of the complex.
Mutagenesis of CD4-D4.
Nevertheless, to directly assess the role of potential CD4-D4 oligomerization in pMHCII binding, a series of CD4-D4 mutations was made and examined for their capacity to alter CD4-pMHCII-dependent cell conjugate formation. On the basis of the dimer interaction in the CD4 D1-D4 crystals, Glu-356, Gln-344, and Lys-318 residues of one CD4 molecule interact with Lys-318, Gln-344, and Glu-356 residues, respectively, of the pseudodyad related molecules (11). As shown in Table 1, mutations at these positions separately or collectively are without significant effect on CD4-pMHCII binding, excluding this CD4 dimerization mode as essential for physiologic pMHCII interaction. Likewise, a potential tetramer model involving the D4 CC′ loop of one CD4 molecule and the C′ strand of a neighboring protomer as necessary for pMHCII binding is implausible. Note that earlier human–mouse species shuffle mutations at CD4 residues 328–330, which demonstrated significant reduction in pMHCII binding (41), altered surface charge; alanine substitutions of these same residues are without functional effect. Although not shown, mutations on the BC and FG loops of CD4 D4 also provide no evidence for tetramerization via these D4 elements. From these results, the ternary model in Fig. 3, molecular packing within our CD4 D1D2-CA/I-Ak crystal and related structural data (9–11, 17, 39), physiologic CD4 oligomerization seems unlikely despite the indirect functional suggestions to the contrary. Perhaps the dominant negative effect of the Phe-43→Ile CD4 variant in blocking wild-type CD4–pMHCII interaction (38) is a consequence of local clustering of variant and wild-type CD4, which are both compartmentalized in lipid rafts, rather than because of a molecular oligomerization.
Table 1.
CD4 D4 mutations are without effect on CD4–MHC class II interaction
| Mutation | Residue location | CD4 level | Rosette formation |
|---|---|---|---|
| Arg-293→Ala | A | I | ++ |
| Lys-318→Alaa | C | II | + |
| Asn-321→Ala | CC′ | I | ++ |
| Lys-322→Ala | CC′ | I | ++ |
| Glu-323→Ala | CC′ | I | ++ |
| Glu-320→Lys/Asn-321→Ala/Glu-323→Ala | CC′ | I–II | + |
| Lys-328→Ala/Arg-329→Ala/Glu-330→Ala | C′,C′E | I | ++ |
| Lys-328→Glu/Arg-329→Glu/Glu-330→Glnb | C′,C′E | I–II | ± |
| Lys-328→Glu/Arg-329→Glub | C′,C′E | III | − |
| Lys-328→Glu/Glu-330→Glnb | C′,C′E | II | ++ |
| Arg-329→Glu/Glu-330→Glnb | C′E | I | ++ |
| Gln-344→Alaa | F | I | ++ |
| Gln-344→Ala/Glu-356→Lysa | F,G | I | ++ |
| Glu-356→Alaa | G | I | ++ |
Mutations are designated by the name of the residue (three-letter amino acid code) and the position in the sequence followed by the name of the substituting residue.
, mutation of the residues observed in the study of Wu et al. (11) to be involved in CD4 dimer contacts in different crystals of hCD4 D1–D4.
, mutation of human CD4 residues to those of murine CD4. Relative levels of CD4 expression were defined by groups I–III, as follows: I, comparable to wild-type CD4; II, reduced to 31–60% of wild-type CD4; III, reduced to 10–30% of wild-type CD4. Rosette formation between CD4-expressing COS7 cells and MHC class II-expressing Raji B cells were measured and categorized as follows: ++, comparable to the rosette formation of wild-type CD4; +, reduced (31–60%) of wild-type CD4; ±, poor (10–30%) of wild-type CD4; −, <10% rosette formation. Results are representative of three independent experiments.
Comparison of CD4 Binding to pMHCII and HIV1 gp120.
CD4 is the primary receptor for the HIV virus. The mature HIV gp160 envelope spike protein consists of the CD4-binding gp120 portion and the noncovalently associated gp41. CD4–gp120 interaction constitutes the first step of virus attachment, followed by cellular chemokine receptor binding (ref. 42 and refs. therein). The virus envelope gp160 spike protein then undergoes dramatic conformational changes, exposing its fusogenic gp41 portion, which coalesces viral and host cell membranes for viral genome entry. The binding of CD4-gp120 is orders of magnitude stronger than that of CD4-pMHCII (16, 43). Although some studies map the HIV- and pMHCII-binding sites to the same region of CD4, others have mapped the two ligands to opposite sides (12, 13, 44, 45). Having obtained the current CD4-CA/I-Ak structure, the direct three-dimensional–structural comparison with the previously published CD4-gp120-Fab structure (42) becomes possible.
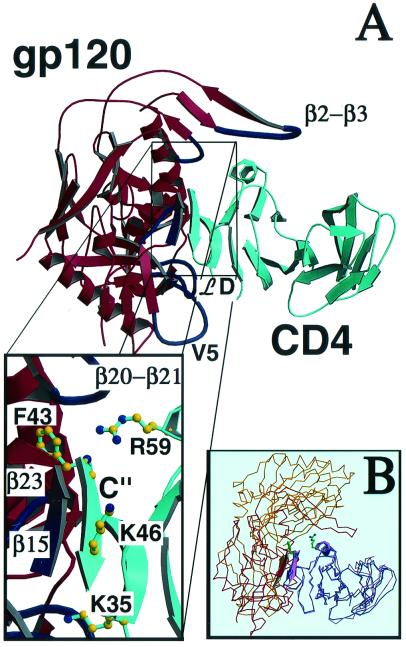
Fig. 4A is a picture of the HIV gp120–CD4 complex with a view such that CD4 is in a similar orientation to CD4 in the Fig. 1 CD4–pMHCII complex. Strikingly, CD4 uses the same “top corner” for both pMHCII and gp120 binding. The key CD4 residue Phe-43 inserts into the junction of gp120's outer and inner domains as well as the bridging sheet (42). The other two CD4 D1-binding regions associated with pMHCII interaction, the mini-antiparallel β sheet involving CD4 segment Lys-46–Leu-44 on the C" strand and the small DE helix, also play a very similar role in gp120 binding. For example, gp120 uses its β15 strand Gly-366–Asp-368 segment to pair with the CD4 C" segment Lys-46–Leu-44 in an identical manner as the MHC class II molecule does to help position the CD4 Phe-43 residue (Fig. 4A and Insert). The Phe-43-binding environment seems more favorable in the CD4–pMHCII complex than in the CD4–gp120 complex, because the gp120-binding pocket contains a negatively charged glutamic acid and includes a large cavity. The gp120 also makes less favorable interactions with the CD4 DE helix. However, gp120 binding to CD4 is accompanied by significant conformational changes that may be energetically favorable (43) and involves a broader area than CD4-pMHCII binding. Details of the way in which the six segments of gp120 bind to CD4 have already been described (42). The buried surface area for CD4-pMHCII and CD4-gp120 is 850 Å2 and 1,880 Å2, respectively (using probe radius = 1.7 Å and density = 4). From Fig. 4, it is evident that the gp120 loop between β2 and β3 strands (VIV2), the £D loop, and the V5 loop may all contribute to greater binding. Because the current CD4-gp120-Fab structure comprises the core part of gp120, the intact molecule may make additional contacts with CD4 and/or further kinetically facilitate its binding. Superposition of CD4 D1D2-CA/I-Ak and CD4 D1D2-gp120 complexes on the basis of hCD4 D1 further emphasizes the common CD4-binding mode (Fig. 4B).
Figure 4.
Molecular mimicry of gp120 binding to CD4 relative to MHC class II. (A) The CD4 D1D2–gp120 complex (ref. 42, with Fab not included for clarity) is shown in similar orientation to the complex of CD4 D1D2/I-Ak in Fig. 1. The interactions that are in common are displayed in the box, which shows the Phe-43 inserted in a hydrophobic pocket involving gp120 β15 and β23, the formation of an antiparallel mini-β-sheet between the C" strand of CD4 and β15 from gp120 as well as interactions between CD4 Arg-59 and the loop connecting β20 to β21 in gp120. Besides these core interactions, gp120 has additional contacts through the V1/V2 loop connecting the β2 and β3 strands, the ℒD loop, and the V5 loop. (B) Overlay of CD4–CA/I-Ak and CD4–gp120 (42) complexes based on hCD4 D1 (blue and pink, respectively) with I-Ak in yellow and gp120 in red. Phe-43 and Arg-59 side chains are shown.
In conclusion, the crystal structure of the CD4-pMHCII presented here argues for a V-shaped CD4–pMHCII–TCR ternary complex as the basis of helper T-cell activation. It also demonstrates that gp120 binds to the identical structural elements of CD4 used by pMHCII, as well as to additional sites. Hence the virus has evolved to mimic the normal pMHCII interaction with CD4 but, in so doing, has augmented its interaction to bolster binding affinity. In this way, the viral protein usurps the normal CD4-pMHCII binding necessary for helper T-cell function (12), hence causing the human immunodeficiency. Whether some other viral envelope proteins morph themselves to mimic the normal ligands of cellular receptors as well remains to be seen.
Abbreviations
- MHC
major histocompatibility complex
- pMHCII
peptide bound to a MHC class II molecule
- pMHCII
peptide bound to a MHC class I molecule
- TCR
T-cell receptor
- CA
conalbumin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code IJL4).
References
- 1.Wang J-H, Reinherz E L. Curr Opin Struct Biol. 2000;10:656–661. doi: 10.1016/s0959-440x(00)00150-0. [DOI] [PubMed] [Google Scholar]
- 2.Janeway C A., Jr Annu Rev Immunol. 1992;10:645–674. doi: 10.1146/annurev.iy.10.040192.003241. [DOI] [PubMed] [Google Scholar]
- 3.Kruisbeek A M, Mond J J, Fowlkes B J, Carmen J A, Bridges S, Longo D L. J Exp Med. 1985;161:1029–1047. doi: 10.1084/jem.161.5.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marrack P, Kappler J. Adv Immunol. 1986;38:1–30. doi: 10.1016/s0065-2776(08)60005-x. [DOI] [PubMed] [Google Scholar]
- 5.Gao G F, Tormo J, Gerth U C, Wyer J R, McMichael A J, Stuart D I, Bell J I, Jones E Y, Jakobsen B K. Nature (London) 1997;387:630–634. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- 6.Garcia K C, Teyton L, Wilson I A. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 7.Hennecke J, Wiley D C. Cell. 2001;104:1–4. doi: 10.1016/s0092-8674(01)00185-4. [DOI] [PubMed] [Google Scholar]
- 8.Kern P, Teng M-K, Smolyar A, Liu J-H, Liu J, Hussey R E, Chang H-C, Reinherz E L, Wang J-H. Immunity. 1998;9:519–530. doi: 10.1016/s1074-7613(00)80635-4. [DOI] [PubMed] [Google Scholar]
- 9.Ryu S E, Kwong P D, Truneh A, Porter T G, Arthos J, Rosenberg M, Dai X, Xuong N, Axel R, Sweet R W, Hendrickson W A. Nature (London) 1990;348:419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J-H, Yan Y, Garrett T P J, Liu J, Rodgers D W, Garlick R L, Tarr G E, Husain Y, Reinherz E L, Harrison S C. Nature (London) 1990;348:411–418. doi: 10.1038/348411a0. [DOI] [PubMed] [Google Scholar]
- 11.Wu H, Kwong P D, Hendrickson W A. Nature (London) 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 12.Clayton L K, Sieh M, Pious D A, Reinherz E L. Nature (London) 1989;339:548–551. doi: 10.1038/339548a0. [DOI] [PubMed] [Google Scholar]
- 13.Moebius U, Clayton L K, Abraham S, Diener A, Yunis J J, Harrison S C, Reinherz E L. Proc Natl Acad Sci USA. 1992;89:12008–12012. doi: 10.1073/pnas.89.24.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moebius U, Pallai P, Harrison S C, Reinherz E L. Proc Natl Acad Sci USA. 1993;90:8259–8263. doi: 10.1073/pnas.90.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H, Littman D R. Cell. 1993;74:633–643. doi: 10.1016/0092-8674(93)90511-n. [DOI] [PubMed] [Google Scholar]
- 16.Xiong Y, Kern P, Chang H-C, Reinherz E L. J Biol Chem. 2001;276:5659–5667. doi: 10.1074/jbc.M009580200. [DOI] [PubMed] [Google Scholar]
- 17.Reinherz E L, Tan K, Tang L, Kern P, Liu J-H, Xiong Y, Hussey R E, Smolyar A, Hare B, Zhang R, et al. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 20.Fremont D H, Monnaie D, Nelson C A, Hendrickson W A, Unanue E R. Immunity. 1998;8:305–317. doi: 10.1016/s1074-7613(00)80536-1. [DOI] [PubMed] [Google Scholar]
- 21.Kissinger C R, Gehlhaar D K, Fogel D B. Acta Crystallogr D. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 22.Cowtan K. Acta Crystallogr D. 1998;54:750–756. doi: 10.1107/s0907444997016247. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov G N, Vagin A A, Lebedev A, Wilson K S, Dodson E J. Acta Crystallogr D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 24.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Killeen N, Sawada S, Littman D R. EMBO J. 1993;12:1547–1553. doi: 10.1002/j.1460-2075.1993.tb05798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law Y M, Yeung R S M, Mamalaki C, Kioussis D, Mak T W, Flavell R A. J Exp Med. 1994;179:1233–1242. doi: 10.1084/jem.179.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Hoegen P, Miceli M C, Tourvieille B, Schilham M, Parnes J R. J Exp Med. 1989;170:1879–1886. doi: 10.1084/jem.170.6.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.König R, Huang L-Y, Germain R N. Nature (London) 1992;356:796–798. doi: 10.1038/356796a0. [DOI] [PubMed] [Google Scholar]
- 29.König R, Shen X, Germain R N. J Exp Med. 1995;182:779–787. doi: 10.1084/jem.182.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vignali D A A, Moreno J, Schiller D, Hämmerling G J. J Exp Med. 1992;175:925–932. doi: 10.1084/jem.175.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakihama T, Hunsicker M E, Hussey R E, Reinherz E L. Eur J Immunol. 2000;30:279–290. doi: 10.1002/1521-4141(200001)30:1<279::AID-IMMU279>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 32.Ding Y H, Baker B M, Garboczi D N, Biddison W E, Wiley D C. Immunity. 1999;11:45–56. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- 33.Xavier R, Brennan T, Li Q, McCormack C, Seed B. Immunity. 1998;8:723–732. doi: 10.1016/s1074-7613(00)80577-4. [DOI] [PubMed] [Google Scholar]
- 34.Hennecke J, Carfi A, Wiley D C. EMBO J. 2000;19:5611–5624. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghendler Y, Smolyar A, Chang H-C, Reinherz E L. J Exp Med. 1998;187:1529–1536. doi: 10.1084/jem.187.9.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manolios N, Kemp O, Li Z G. Eur J Immunol. 1994;24:84–92. doi: 10.1002/eji.1830240114. [DOI] [PubMed] [Google Scholar]
- 37.Casasnovas J M, Springer T A, Liu J-H, Harrison S C, Wang J-H. Nature (London) 1997;387:312–315. doi: 10.1038/387312a0. [DOI] [PubMed] [Google Scholar]
- 38.Sakihama T, Smolyar A, Reinherz E L. Proc Natl Acad Sci USA. 1995;92:6444–6448. doi: 10.1073/pnas.92.14.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brady R L, Dodson E J, Dodson G G, Lange G, Davis S J, Williams A F, Barclay A N. Science. 1993;260:979–983. doi: 10.1126/science.8493535. [DOI] [PubMed] [Google Scholar]
- 40.Brown J H, Jardetzky T S, Gorga J C, Stern L J, Urban R G, Strominger J L, Wiley D C. Nature (London) 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 41.Houlgatte R, Scarmato P, el Marhomy S, Martin M, Ostankovitch M, Lafosse S, Vervisch A, Auffray C, Platier-Tonneau D. J Immunol. 1994;152:4475–4488. [PubMed] [Google Scholar]
- 42.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Nature (London) 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myszka D G, Sweet R W, Hensley P, Brigham-Burke M, Kwong P D, Hendrickson W A, Wyatt R, Sodroski J, Doyle M L. Proc Natl Acad Sci USA. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fleury S, Lamarre D, Meloche S, Ryu S E, Cantin C, Hendrickson W A, Sekaly P. Cell. 1991;66:1037–1049. doi: 10.1016/0092-8674(91)90447-7. [DOI] [PubMed] [Google Scholar]
- 45.Piatier-Tonneau D, Gastinel L-N, Moussy G, Benichou B, Amblard F, Vaigot P, Auffray C. Proc Natl Acad Sci USA. 1991;88:6858–6862. doi: 10.1073/pnas.88.15.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 47.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 48.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]