The title coordination polymer was obtained by combining an aqueous solution of copper(II) dichloride with the ligand {tert-butylmethyl[4-(6-{[4-(pyridin-2-yl-)1H-1,2,3-triazol-1-yl]methyl}-1,3-benzothiazol-2-yl)phenyl]carbamate in acetonitrile.
Keywords: crystal structure, pyridine–triazole, Alzheimer’s disease, copper(II) complex, hydrogen bonding, C—H⋯π interactions, offset π–π interactions
Abstract
In the title coordination polymer, {[CuCl2(C27H26N6O2S)]·CH3CN}n, the copper(II) ion is fivefold coordinated, with an almost perfect square-pyramidal coordination sphere. In the equatorial plane, it is ligated to a pyridine N atom and an N atom of the triazole unit and to two Cl− ions, while the apical position is occupied by the carbonyl O atom of the tert-butyl carbamate group. In the crystal, the polymer chains propagate in the [11-1] direction, with the acetonitrile solvent molecules linked to the chain by C—H⋯N hydrogen bonds. The chains are linked by C—H⋯Cl hydrogen bonds forming sheets parallel to the plane (011). The crystal packing is further consolidated by C—H⋯π interactions and offset π–π stacking interactions [intercentroid distance = 3.6805 (15) Å], forming a three-dimensional supramolecular structure.
Chemical context
Alzheimer’s Disease (AD) is a neurodegenerative disease characterized by aggregation of amyloid peptide and extensive inflammation related to a strong oxidative stress (Cheignon et al., 2018 ▸). Metals are known to play a key role in this oxidative stress and also to be associated with peptide aggregation, at the core of the pathology (Faller et al., 2013 ▸; Viles, 2012 ▸). More specifically, CuII has been found to form a complex with the amyloid peptide for which aggregation is one of the major hallmarks of AD (Eury et al., 2011 ▸; Faller et al., 2014 ▸). This has triggered significant ongoing interest in the development of chelators able to interact with metals in the context of AD (Santos et al., 2016 ▸; Conte-Daban et al., 2017 ▸).
In the course of our studies on the development of bifunctional molecules able to target amyloid fibrils, for example via a 2-arylbenzothiazole core (Noel et al., 2013 ▸), and interact with copper ions found within the senile plaques, we have designed and synthesized a benzothiazole moiety decorated with a triazole-pyridine subunit, viz. tert-butyl methyl[4-(6-{[4-(pyridin-2-yl)-1H-1,2,3-triazol-1-yl]methyl} benzo[d]thiazol-2-yl]phenyl}carbamate (L). Indeed integrating the N-binding from the triazole moiety in the binding site of a chelator has been shown to be a successful approach (Jones et al., 2012 ▸, 2017 ▸). Compared to these seminal works, the additional aryl-benzothiazole moiety in compound L is expected to enhance the ability of the chelator to interact with amyloid aggregates and thus to retrieve deleterious CuII ions from Aβ fibrils. Investigation of the ability to chelate CuII ions, by studying the reaction of L with CuCl2, led to the formation of the title coordination polymer whose synthesis and molecular and crystal structures are described herein.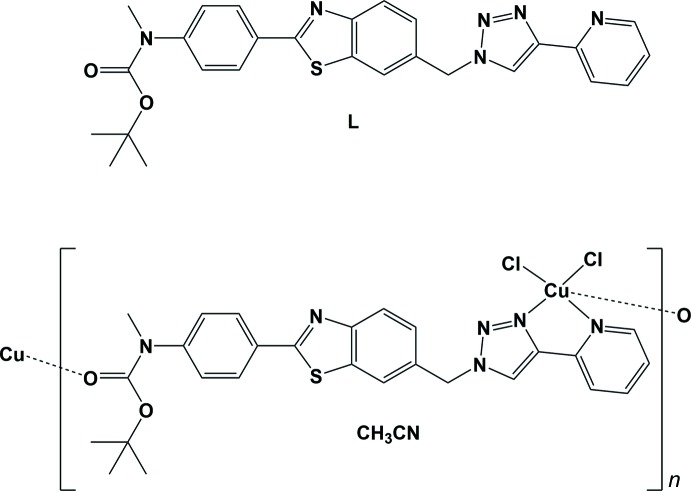
Structural commentary
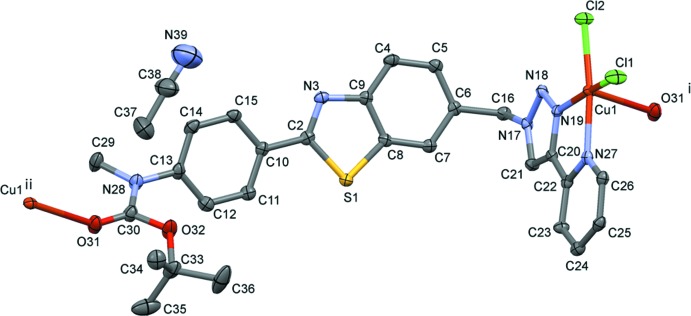
The molecular structure of the asymmetric unit of the title coordination polymer is shown in Fig. 1 ▸. Selected bond lengths and bond angles are given in Table 1 ▸. The ligand is L-shaped with the benzothiazole ring system (S1/N3/C2/C4–C9; r.m.s. deviation = 0.01 Å) being inclined to the triazole ring (N17-N197C20/C21) by 79.54 (12)°. The benzene ring is inclined to the benzothiazole ring system by 12.27 (11)°, while the pyridine ring is inclined to the triazole ring by 4.07 (14)°. The copper(II) ion is fivefold coordinate with an almost perfect square-pyramidal coordination sphere. In the equatorial plane, the copper(II) ion coordinates the pyridine N atom N27 and atom N19 of the triazole unit and two Cl− anions, while the apical position is occupied by the carbonyl O atom, O31, of the tert-butyloxycarbamate group. The τ5 descriptor for the fivefold coordination sphere is 0.08 (τ5 = 0 for an ideal square-pyramidal coordination sphere, and = 1 for an ideal trigonal–pyramidal coordination sphere; Addison et al., 1984 ▸). The triazole ring (N17–N19/C20/C21) exhibits a slightly shorter Cu1—N19 bond length [2.004 (2) Å] than the pyridine Cu1—-N27 bond length [2.054 (2) Å], yet no trans effect is observed as the two Cu—-Cl bond lengths are very close [2.2344 (7) and 2.2380 (7) Å]. These bond lengths are similar to those observed for a related complex, viz. dichloro-(4-{2-[4-(pyridin-2-yl)-1H-1,2,3-triazol-1-yl]ethyl}morpholine)copper(II) (Jones et al., 2012 ▸).
Figure 1.
The molecular structure of the asymmetric unit of the title coordination polymer, with atom labelling. Displacement ellipsoids are drawn at the 50% probability level. The H atoms have been omitted for clarity. [Symmetry codes: (i) x − 1, y − 1, z + 1; (ii) x + 1, y + 1, z − 1.]
Table 1. Selected geometric parameters (Å, °).
| Cu1—O31i | 2.508 (2) | Cu1—Cl1 | 2.2344 (7) |
| Cu1—N19 | 2.004 (2) | Cu1—Cl2 | 2.2380 (7) |
| Cu1—N27 | 2.054 (2) | ||
| Cl1—Cu1—N19 | 168.01 (7) | Cl2—Cu1—N27 | 172.70 (6) |
Symmetry code: (i) 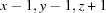 .
.
Supramolecular features
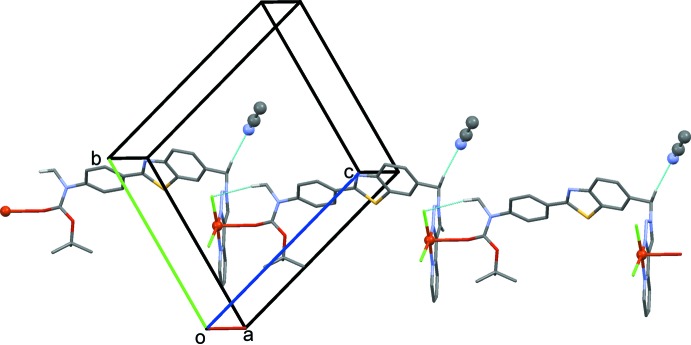
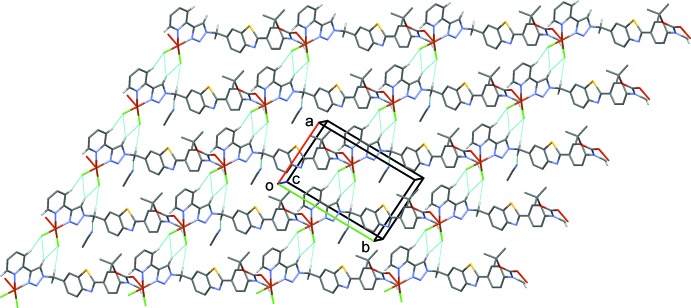
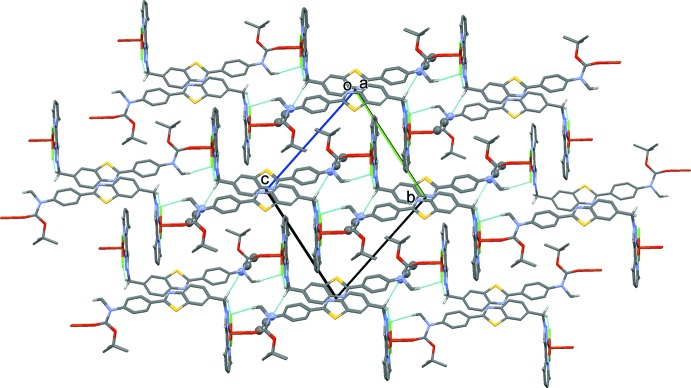
In the crystal, the polymer chains propagate in the [11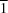 ] direction (Fig. 2 ▸). They are linked by C—H⋯Cl hydrogen bonds, forming sheets parallel to (011); see Fig. 3 ▸ and Table 2 ▸. The acetonitrile solvent molecules are linked to the polymer chains within the network by C—H⋯N hydrogen bonds (Figs. 2 ▸ and 3 ▸; Table 2 ▸). The crystal packing is further consolidated by C—H⋯π interactions (Table 2 ▸) and offset π–π stacking interactions, forming a three-dimensional supramolecular structure (Fig. 4 ▸). The offset π–π interactions involve inversion-related triazole and pyridine rings with interplanar distances of 3.3848 (11) and 3.300 (1) Å [Cg3⋯Cg4i = 3.6805 (15) Å, α = 4.07 (14)°, slippages are 1.63 and 1.45 Å; Cg3 and Cg4 are the centroids of rings N17–N19/C20/C21 and N27/C22–C26, respectively; symmetry code: (i) −x, −y − 1, −z + 2].
] direction (Fig. 2 ▸). They are linked by C—H⋯Cl hydrogen bonds, forming sheets parallel to (011); see Fig. 3 ▸ and Table 2 ▸. The acetonitrile solvent molecules are linked to the polymer chains within the network by C—H⋯N hydrogen bonds (Figs. 2 ▸ and 3 ▸; Table 2 ▸). The crystal packing is further consolidated by C—H⋯π interactions (Table 2 ▸) and offset π–π stacking interactions, forming a three-dimensional supramolecular structure (Fig. 4 ▸). The offset π–π interactions involve inversion-related triazole and pyridine rings with interplanar distances of 3.3848 (11) and 3.300 (1) Å [Cg3⋯Cg4i = 3.6805 (15) Å, α = 4.07 (14)°, slippages are 1.63 and 1.45 Å; Cg3 and Cg4 are the centroids of rings N17–N19/C20/C21 and N27/C22–C26, respectively; symmetry code: (i) −x, −y − 1, −z + 2].
Figure 2.
A view along the a axis of the acetonitrile solvent molecules (ball and stick) linked to the polymer chains, that propagate along direction [11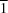 ], via a C—H⋯N hydrogen bond (see Table 2 ▸ for details). Other H atoms have been omitted for clarity.
], via a C—H⋯N hydrogen bond (see Table 2 ▸ for details). Other H atoms have been omitted for clarity.
Figure 3.
A view along the c axis of the crystal packing of the title compound, showing the hydrogen bonds (dashed lines; see Table 2 ▸ for details) forming sheets parallel to (011). H atoms not involved in these interactions have been omitted.
Table 2. Hydrogen-bond geometry (Å, °).
Cg is the centroid of the C4–C9 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C16—H162⋯N39ii | 0.97 | 2.52 | 3.451 (6) | 161 |
| C16—H161⋯Cl2iii | 0.97 | 2.72 | 3.606 (3) | 152 |
| C21—H211⋯Cl1iii | 0.94 | 2.81 | 3.633 (3) | 147 |
| C23—H231⋯Cl1iii | 0.94 | 2.62 | 3.494 (3) | 155 |
| C26—H261⋯Cl1 | 0.94 | 2.55 | 3.154 (3) | 122 |
| C29—H291⋯Cl2iv | 0.95 | 2.80 | 3.741 (3) | 172 |
| C25—H251⋯Cg v | 0.94 | 2.85 | 3.583 (3) | 135 |
Symmetry codes: (ii) 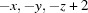 ; (iii)
; (iii) 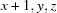 ; (iv)
; (iv) 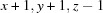 ; (v)
; (v) 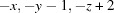 .
.
Figure 4.
A view along the a axis of the crystal packing of the title compound, showing the hydrogen bonds as dashed lines (see Table 2 ▸ for details). H atoms not involved in these interactions have been omitted.
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.38, update May 2017; Groom et al., 2016 ▸) for pyridine-triazole copper(II) dichloride complexes gave seven hits. Two of these compounds have a similar geometry involving the copper(II) atom, viz. dichloro-(4-{2-[4-(pyridin-2-yl)-1H-1,2,3-triazol-1-yl]ethyl}morpholine)copper(II) (CSD refcode MEHHEO; Jones et al., 2012 ▸) and bis(μ-chloro)dichloro-bis(2-{[4-(pyridin-2-yl)-1H-1,2,3-triazol-1-yl]methyl}benzonitrile)di-copper (UMIYEW; Bai et al., 2016 ▸). As in the title compound (see Table 1 ▸), the CuII ions have fivefold coordination spheres with a square-pyramidal geometry. In addition, the Cu—Npyridine bond lengths [2.063 (3) and 2.075 (2) Å, respectively] are slightly longer than the Cu—Ntriazole bond lengths [2.024 (3) and 2.005 (3) Å, respectively], while the Cu—Cl bonds lengths are very similar in both complexes [2.265 (1) and 2.242 (1) Å in MEHHEO, and 2.246 (1) and 2.264 (1) Å in UMIYEW]. However, both of these compounds are binuclear complexes, possessing inversion symmetry, with bis(μ-chloro) Cl− anions bridging the metal ions.
Synthesis and crystallization
The synthesis of the ligand, tert-butyl methyl[4-(6-{[4-(pyridin-2-yl)-1H-1,2,3-triazol-1-yl]methyl}benzo[d]thiazol-2-yl)phenyl]carbamate (L), was performed according to literature precedents (Noel et al., 2013 ▸; Jones et al., 2012 ▸). A mixture of 15 mg of L dissolved in 1 ml of acetonitrile, and 1.1 equiv. of CuCl2 dissolved in 10 ml of a mixture acetonitrile/H2O (6/3) was heated to 353 K. The mixture was cooled at room temperature, allowing a precipitate to form. The supernatant was removed and the precipitate was dissolved with a minimum volume of hot acetonitrile, filtered and left at room temperature in a closed vessel producing overnight pale-green plate-like crystals.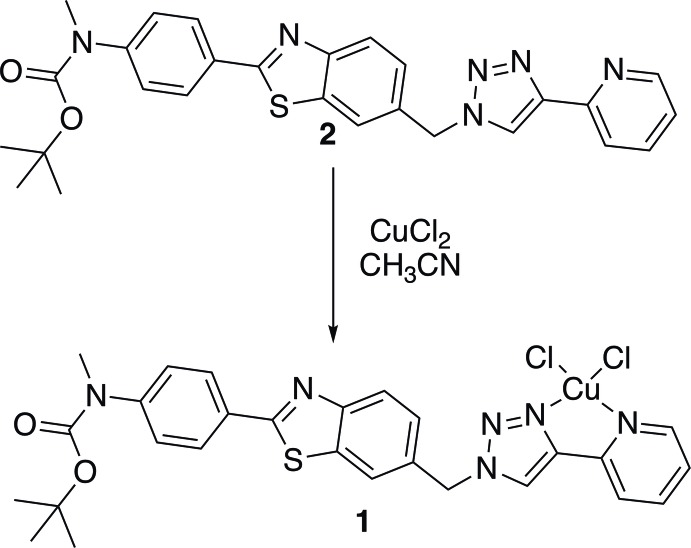
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The H atoms were all located in difference-Fourier maps, but those attached to carbon atoms were repositioned geometrically. The H atoms were initially refined with soft restraints on the bond lengths and angles to regularize their geometry [C—H = 0.93–0.98 Å with U iso(H) = 1.5U eq(C-methyl) and 1.2U eq(C) for other H atoms], after which the positions were refined with riding constraints (Cooper et al., 2010 ▸).
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | [CuCl2(C27H26N6O2S)]·CH3CN |
| M r | 674.11 |
| Crystal system, space group | Triclinic, P
|
| Temperature (K) | 100 |
| a, b, c (Å) | 8.6374 (7), 13.1553 (10), 14.2243 (11) |
| α, β, γ (°) | 73.755 (3), 73.863 (3), 84.226 (3) |
| V (Å3) | 1490.1 (2) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 1.02 |
| Crystal size (mm) | 0.12 × 0.09 × 0.02 |
| Data collection | |
| Diffractometer | Bruker Kappa APEXII |
| Absorption correction | Multi-scan (SADABS; Bruker, 2006 ▸) |
| T min, T max | 0.91, 0.98 |
| No. of measured, independent and observed [I > 2.0σ(I)] reflections | 26982, 5475, 4358 |
| R int | 0.053 |
| (sin θ/λ)max (Å−1) | 0.603 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.037, 0.036, 1.05 |
| No. of reflections | 4062 |
| No. of parameters | 379 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.45, −0.36 |
Supplementary Material
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S2056989018000488/su5417sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018000488/su5417Isup2.hkl
CCDC reference: 1815501
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| [CuCl2(C27H26N6O2S)]·CH3CN | Z = 2 |
| Mr = 674.11 | F(000) = 694 |
| Triclinic, P1 | Dx = 1.502 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.6374 (7) Å | Cell parameters from 7700 reflections |
| b = 13.1553 (10) Å | θ = 2–25° |
| c = 14.2243 (11) Å | µ = 1.02 mm−1 |
| α = 73.755 (3)° | T = 100 K |
| β = 73.863 (3)° | Plate, pale green |
| γ = 84.226 (3)° | 0.12 × 0.09 × 0.02 mm |
| V = 1490.1 (2) Å3 |
Data collection
| Bruker Kappa APEXII diffractometer | 4358 reflections with I > 2.0σ(I) |
| Graphite monochromator | Rint = 0.053 |
| φ & ω scans | θmax = 25.4°, θmin = 1.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2006) | h = −10→8 |
| Tmin = 0.91, Tmax = 0.98 | k = −15→15 |
| 26982 measured reflections | l = −17→17 |
| 5475 independent reflections |
Refinement
| Refinement on F | Primary atom site location: other |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.036 | H-atom parameters constrained |
| S = 1.05 | Method, part 1, Chebychev polynomial, (Watkin, 1994; Prince, 1982) [weight] = 1.0/[A0*T0(x) + A1*T1(x) ··· + An-1]*Tn-1(x)] where Ai are the Chebychev coefficients listed below and x = F /Fmax Method = Robust Weighting (Prince, 1982) W = [weight] * [1-(deltaF/6*sigmaF)2]2 Ai are: 0.270 0.160 0.128 |
| 4062 reflections | (Δ/σ)max = 0.001 |
| 379 parameters | Δρmax = 0.45 e Å−3 |
| 0 restraints | Δρmin = −0.36 e Å−3 |
Special details
| Experimental. The crystal was placed in the cold stream of an Oxford Cryosystems open-flow nitrogen cryostat (Cosier & Glazer, 1986) with a nominal stability of 0.1 K. Cosier, J. & Glazer, A.M., 1986. J. Appl. Cryst. 105-107. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.49721 (8) | −0.10533 (5) | 0.91493 (5) | 0.0178 | |
| Cu1 | −0.26343 (4) | −0.55390 (3) | 1.17008 (3) | 0.0153 | |
| Cl1 | −0.43826 (8) | −0.62080 (6) | 1.11470 (6) | 0.0283 | |
| C2 | 0.4057 (3) | 0.0195 (2) | 0.8788 (2) | 0.0166 | |
| Cl2 | −0.45219 (8) | −0.45258 (6) | 1.24664 (5) | 0.0241 | |
| N3 | 0.2719 (3) | 0.03835 (18) | 0.94172 (17) | 0.0182 | |
| C4 | 0.0978 (3) | −0.0538 (2) | 1.1089 (2) | 0.0198 | |
| C5 | 0.0743 (3) | −0.1456 (2) | 1.1862 (2) | 0.0184 | |
| C6 | 0.1821 (3) | −0.2326 (2) | 1.18434 (19) | 0.0154 | |
| C7 | 0.3190 (3) | −0.2270 (2) | 1.1035 (2) | 0.0168 | |
| C8 | 0.3417 (3) | −0.1348 (2) | 1.0253 (2) | 0.0157 | |
| C9 | 0.2337 (3) | −0.0477 (2) | 1.0261 (2) | 0.0164 | |
| C10 | 0.4785 (3) | 0.0928 (2) | 0.7807 (2) | 0.0167 | |
| C11 | 0.6322 (3) | 0.0724 (2) | 0.7241 (2) | 0.0196 | |
| C12 | 0.6982 (3) | 0.1394 (2) | 0.6295 (2) | 0.0218 | |
| C13 | 0.6083 (3) | 0.2264 (2) | 0.5891 (2) | 0.0205 | |
| C14 | 0.4567 (4) | 0.2492 (2) | 0.6463 (2) | 0.0226 | |
| C15 | 0.3921 (3) | 0.1834 (2) | 0.7420 (2) | 0.0197 | |
| C16 | 0.1480 (3) | −0.3355 (2) | 1.2670 (2) | 0.0176 | |
| N17 | 0.0898 (3) | −0.41369 (17) | 1.22896 (16) | 0.0142 | |
| N18 | −0.0676 (3) | −0.41805 (17) | 1.23796 (16) | 0.0156 | |
| N19 | −0.0798 (2) | −0.48723 (16) | 1.18938 (17) | 0.0149 | |
| C20 | 0.0688 (3) | −0.5257 (2) | 1.14907 (19) | 0.0143 | |
| C21 | 0.1801 (3) | −0.4783 (2) | 1.17465 (19) | 0.0165 | |
| C22 | 0.0753 (3) | −0.6034 (2) | 1.09231 (19) | 0.0153 | |
| C23 | 0.2169 (3) | −0.6451 (2) | 1.0407 (2) | 0.0177 | |
| C24 | 0.2065 (3) | −0.7194 (2) | 0.9903 (2) | 0.0206 | |
| C25 | 0.0551 (3) | −0.7488 (2) | 0.9927 (2) | 0.0191 | |
| C26 | −0.0798 (3) | −0.7017 (2) | 1.0431 (2) | 0.0171 | |
| N27 | −0.0723 (3) | −0.63013 (17) | 1.09261 (16) | 0.0145 | |
| N28 | 0.6716 (3) | 0.29292 (18) | 0.48943 (17) | 0.0230 | |
| C29 | 0.6889 (4) | 0.4067 (2) | 0.4767 (2) | 0.0320 | |
| C30 | 0.7287 (4) | 0.2548 (2) | 0.4066 (2) | 0.0228 | |
| O31 | 0.7877 (3) | 0.30981 (16) | 0.32214 (15) | 0.0289 | |
| O32 | 0.7071 (3) | 0.15044 (16) | 0.43036 (15) | 0.0304 | |
| C33 | 0.7874 (4) | 0.0873 (2) | 0.3586 (2) | 0.0286 | |
| C34 | 0.7093 (4) | 0.1108 (3) | 0.2724 (2) | 0.0303 | |
| C35 | 0.9671 (4) | 0.1073 (3) | 0.3221 (3) | 0.0478 | |
| C36 | 0.7533 (6) | −0.0256 (3) | 0.4247 (3) | 0.0546 | |
| C37 | 0.3253 (6) | 0.2890 (4) | 0.3651 (4) | 0.0763 | |
| C38 | 0.1812 (5) | 0.2908 (3) | 0.4432 (3) | 0.0502 | |
| N39 | 0.0648 (6) | 0.2875 (4) | 0.5048 (4) | 0.0862 | |
| H41 | 0.0246 | 0.0045 | 1.1119 | 0.0245* | |
| H51 | −0.0172 | −0.1504 | 1.2437 | 0.0226* | |
| H71 | 0.3946 | −0.2857 | 1.1026 | 0.0214* | |
| H111 | 0.6940 | 0.0122 | 0.7511 | 0.0246* | |
| H121 | 0.8046 | 0.1270 | 0.5925 | 0.0270* | |
| H141 | 0.3970 | 0.3095 | 0.6193 | 0.0274* | |
| H151 | 0.2892 | 0.2001 | 0.7807 | 0.0247* | |
| H161 | 0.2462 | −0.3640 | 1.2865 | 0.0221* | |
| H162 | 0.0655 | −0.3234 | 1.3253 | 0.0219* | |
| H211 | 0.2925 | −0.4869 | 1.1601 | 0.0196* | |
| H231 | 0.3174 | −0.6224 | 1.0400 | 0.0224* | |
| H241 | 0.2997 | −0.7503 | 0.9546 | 0.0264* | |
| H251 | 0.0439 | −0.7998 | 0.9599 | 0.0235* | |
| H261 | −0.1829 | −0.7191 | 1.0427 | 0.0217* | |
| H291 | 0.6493 | 0.4483 | 0.4217 | 0.0499* | |
| H292 | 0.6300 | 0.4275 | 0.5370 | 0.0497* | |
| H293 | 0.8010 | 0.4230 | 0.4655 | 0.0508* | |
| H341 | 0.7619 | 0.0674 | 0.2266 | 0.0456* | |
| H342 | 0.5970 | 0.0936 | 0.2989 | 0.0468* | |
| H343 | 0.7196 | 0.1844 | 0.2362 | 0.0455* | |
| H351 | 1.0195 | 0.0571 | 0.2860 | 0.0706* | |
| H352 | 1.0120 | 0.0978 | 0.3794 | 0.0711* | |
| H353 | 0.9903 | 0.1784 | 0.2784 | 0.0709* | |
| H361 | 0.7925 | −0.0372 | 0.4838 | 0.0841* | |
| H362 | 0.8064 | −0.0739 | 0.3855 | 0.0839* | |
| H363 | 0.6376 | −0.0356 | 0.4445 | 0.0840* | |
| H371 | 0.3941 | 0.2312 | 0.3908 | 0.1152* | |
| H372 | 0.3774 | 0.3554 | 0.3457 | 0.1152* | |
| H373 | 0.2956 | 0.2769 | 0.3077 | 0.1154* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0155 (3) | 0.0172 (3) | 0.0189 (3) | 0.0004 (2) | −0.0021 (3) | −0.0046 (3) |
| Cu1 | 0.01124 (16) | 0.01627 (17) | 0.02098 (18) | −0.00100 (12) | −0.00448 (12) | −0.00854 (13) |
| Cl1 | 0.0169 (3) | 0.0348 (4) | 0.0440 (4) | 0.0014 (3) | −0.0124 (3) | −0.0236 (3) |
| C2 | 0.0181 (13) | 0.0180 (13) | 0.0193 (13) | −0.0008 (10) | −0.0095 (11) | −0.0091 (11) |
| Cl2 | 0.0161 (3) | 0.0283 (4) | 0.0327 (4) | 0.0033 (3) | −0.0062 (3) | −0.0174 (3) |
| N3 | 0.0194 (11) | 0.0178 (11) | 0.0196 (12) | 0.0015 (9) | −0.0055 (9) | −0.0086 (10) |
| C4 | 0.0200 (13) | 0.0200 (14) | 0.0228 (14) | 0.0015 (11) | −0.0054 (11) | −0.0121 (12) |
| C5 | 0.0191 (13) | 0.0198 (14) | 0.0191 (13) | −0.0021 (10) | −0.0042 (11) | −0.0098 (11) |
| C6 | 0.0170 (12) | 0.0169 (13) | 0.0166 (13) | −0.0047 (10) | −0.0069 (10) | −0.0073 (11) |
| C7 | 0.0160 (12) | 0.0177 (13) | 0.0206 (14) | −0.0011 (10) | −0.0064 (11) | −0.0094 (11) |
| C8 | 0.0130 (12) | 0.0187 (13) | 0.0191 (13) | −0.0032 (10) | −0.0049 (10) | −0.0090 (11) |
| C9 | 0.0169 (12) | 0.0175 (13) | 0.0187 (13) | −0.0023 (10) | −0.0063 (10) | −0.0087 (11) |
| C10 | 0.0201 (13) | 0.0158 (13) | 0.0184 (13) | −0.0029 (10) | −0.0073 (11) | −0.0077 (11) |
| C11 | 0.0207 (13) | 0.0217 (14) | 0.0188 (14) | 0.0003 (11) | −0.0075 (11) | −0.0071 (11) |
| C12 | 0.0218 (14) | 0.0259 (15) | 0.0192 (14) | −0.0016 (11) | −0.0054 (11) | −0.0082 (12) |
| C13 | 0.0286 (15) | 0.0157 (13) | 0.0188 (14) | −0.0040 (11) | −0.0058 (11) | −0.0064 (11) |
| C14 | 0.0311 (15) | 0.0185 (14) | 0.0210 (14) | 0.0044 (12) | −0.0105 (12) | −0.0081 (12) |
| C15 | 0.0224 (14) | 0.0198 (14) | 0.0192 (14) | 0.0009 (11) | −0.0042 (11) | −0.0106 (11) |
| C16 | 0.0187 (13) | 0.0189 (13) | 0.0182 (13) | −0.0012 (10) | −0.0053 (11) | −0.0088 (11) |
| N17 | 0.0147 (10) | 0.0149 (11) | 0.0140 (11) | −0.0031 (8) | −0.0031 (8) | −0.0050 (9) |
| N18 | 0.0148 (10) | 0.0149 (11) | 0.0172 (11) | −0.0030 (8) | −0.0029 (9) | −0.0046 (9) |
| N19 | 0.0137 (10) | 0.0113 (10) | 0.0194 (11) | −0.0010 (8) | −0.0043 (9) | −0.0031 (9) |
| C20 | 0.0131 (12) | 0.0139 (12) | 0.0147 (12) | −0.0010 (10) | −0.0024 (10) | −0.0029 (10) |
| C21 | 0.0166 (12) | 0.0180 (13) | 0.0161 (13) | −0.0004 (10) | −0.0037 (10) | −0.0072 (11) |
| C22 | 0.0163 (12) | 0.0146 (13) | 0.0159 (13) | −0.0012 (10) | −0.0074 (10) | −0.0019 (10) |
| C23 | 0.0154 (12) | 0.0202 (13) | 0.0205 (14) | 0.0019 (10) | −0.0069 (11) | −0.0087 (11) |
| C24 | 0.0219 (14) | 0.0192 (14) | 0.0215 (14) | 0.0025 (11) | −0.0064 (11) | −0.0068 (11) |
| C25 | 0.0266 (14) | 0.0140 (13) | 0.0187 (14) | 0.0005 (11) | −0.0074 (11) | −0.0062 (11) |
| C26 | 0.0202 (13) | 0.0152 (12) | 0.0176 (13) | −0.0030 (10) | −0.0069 (11) | −0.0044 (11) |
| N27 | 0.0169 (11) | 0.0148 (11) | 0.0125 (11) | −0.0016 (9) | −0.0057 (9) | −0.0025 (9) |
| N28 | 0.0349 (14) | 0.0170 (12) | 0.0162 (12) | −0.0019 (10) | −0.0036 (10) | −0.0052 (10) |
| C29 | 0.053 (2) | 0.0167 (14) | 0.0221 (15) | −0.0005 (14) | −0.0055 (14) | −0.0035 (12) |
| C30 | 0.0293 (15) | 0.0183 (14) | 0.0213 (15) | −0.0010 (12) | −0.0069 (12) | −0.0057 (12) |
| O31 | 0.0433 (13) | 0.0218 (11) | 0.0165 (10) | −0.0017 (9) | −0.0011 (9) | −0.0033 (9) |
| O32 | 0.0522 (14) | 0.0183 (10) | 0.0181 (10) | −0.0036 (9) | −0.0013 (9) | −0.0078 (8) |
| C33 | 0.0434 (18) | 0.0222 (15) | 0.0241 (16) | 0.0037 (13) | −0.0105 (14) | −0.0125 (13) |
| C34 | 0.0380 (17) | 0.0315 (17) | 0.0265 (16) | −0.0045 (14) | −0.0108 (14) | −0.0125 (13) |
| C35 | 0.040 (2) | 0.054 (2) | 0.068 (3) | 0.0172 (17) | −0.0253 (19) | −0.042 (2) |
| C36 | 0.110 (4) | 0.0202 (17) | 0.037 (2) | 0.0028 (19) | −0.023 (2) | −0.0102 (15) |
| C37 | 0.051 (3) | 0.056 (3) | 0.084 (4) | −0.005 (2) | 0.021 (2) | 0.004 (3) |
| C38 | 0.038 (2) | 0.058 (3) | 0.049 (2) | 0.0078 (18) | −0.0059 (19) | −0.015 (2) |
| N39 | 0.073 (3) | 0.099 (4) | 0.072 (3) | 0.013 (3) | 0.005 (2) | −0.029 (3) |
Geometric parameters (Å, º)
| S1—C2 | 1.754 (3) | C20—C21 | 1.373 (4) |
| S1—C8 | 1.736 (3) | C20—C22 | 1.458 (4) |
| Cu1—O31i | 2.508 (2) | C21—H211 | 0.937 |
| Cu1—N19 | 2.004 (2) | C22—C23 | 1.388 (4) |
| Cu1—N27 | 2.054 (2) | C22—N27 | 1.355 (3) |
| Cu1—Cl1 | 2.2344 (7) | C23—C24 | 1.386 (4) |
| Cu1—Cl2 | 2.2380 (7) | C23—H231 | 0.943 |
| C2—N3 | 1.300 (3) | C24—C25 | 1.390 (4) |
| C2—C10 | 1.468 (4) | C24—H241 | 0.946 |
| N3—C9 | 1.387 (4) | C25—C26 | 1.377 (4) |
| C4—C5 | 1.375 (4) | C25—H251 | 0.943 |
| C4—C9 | 1.403 (4) | C26—N27 | 1.339 (3) |
| C4—H41 | 0.947 | C26—H261 | 0.944 |
| C5—C6 | 1.402 (4) | N28—C29 | 1.474 (4) |
| C5—H51 | 0.959 | N28—C30 | 1.356 (4) |
| C6—C7 | 1.393 (4) | C29—H291 | 0.949 |
| C6—C16 | 1.516 (4) | C29—H292 | 0.964 |
| C7—C8 | 1.385 (4) | C29—H293 | 0.973 |
| C7—H71 | 0.961 | C30—O31 | 1.214 (3) |
| C8—C9 | 1.403 (4) | C30—O32 | 1.338 (3) |
| C10—C11 | 1.392 (4) | O32—C33 | 1.476 (3) |
| C10—C15 | 1.401 (4) | C33—C34 | 1.503 (4) |
| C11—C12 | 1.386 (4) | C33—C35 | 1.518 (5) |
| C11—H111 | 0.959 | C33—C36 | 1.526 (5) |
| C12—C13 | 1.395 (4) | C34—H341 | 0.974 |
| C12—H121 | 0.948 | C34—H342 | 0.963 |
| C13—C14 | 1.390 (4) | C34—H343 | 0.962 |
| C13—N28 | 1.431 (4) | C35—H351 | 0.952 |
| C14—C15 | 1.389 (4) | C35—H352 | 0.969 |
| C14—H141 | 0.948 | C35—H353 | 0.972 |
| C15—H151 | 0.948 | C36—H361 | 0.960 |
| C16—N17 | 1.473 (3) | C36—H362 | 0.961 |
| C16—H161 | 0.973 | C36—H363 | 0.972 |
| C16—H162 | 0.971 | C37—C38 | 1.422 (6) |
| N17—N18 | 1.336 (3) | C37—H371 | 0.970 |
| N17—C21 | 1.352 (3) | C37—H372 | 0.957 |
| N18—N19 | 1.315 (3) | C37—H373 | 0.977 |
| N19—C20 | 1.362 (3) | C38—N39 | 1.131 (6) |
| C2—S1—C8 | 89.01 (13) | C20—C21—N17 | 103.8 (2) |
| O31i—Cu1—Cl1 | 107.68 (6) | C20—C21—H211 | 130.3 |
| O31i—Cu1—Cl2 | 100.13 (5) | N17—C21—H211 | 125.8 |
| Cl1—Cu1—Cl2 | 93.31 (3) | C20—C22—C23 | 124.4 (2) |
| O31i—Cu1—N19 | 80.21 (8) | C20—C22—N27 | 113.2 (2) |
| Cl1—Cu1—N19 | 168.01 (7) | C23—C22—N27 | 122.4 (2) |
| Cl2—Cu1—N19 | 94.14 (6) | C22—C23—C24 | 118.6 (2) |
| O31i—Cu1—N27 | 83.26 (8) | C22—C23—H231 | 119.9 |
| Cl1—Cu1—N27 | 91.79 (6) | C24—C23—H231 | 121.5 |
| Cl2—Cu1—N27 | 172.70 (6) | C23—C24—C25 | 119.0 (2) |
| N19—Cu1—N27 | 80.00 (8) | C23—C24—H241 | 121.6 |
| S1—C2—N3 | 115.9 (2) | C25—C24—H241 | 119.5 |
| S1—C2—C10 | 119.49 (19) | C24—C25—C26 | 118.9 (2) |
| N3—C2—C10 | 124.6 (2) | C24—C25—H251 | 121.0 |
| C2—N3—C9 | 110.5 (2) | C26—C25—H251 | 120.0 |
| C5—C4—C9 | 118.8 (3) | C25—C26—N27 | 123.0 (2) |
| C5—C4—H41 | 120.6 | C25—C26—H261 | 119.4 |
| C9—C4—H41 | 120.6 | N27—C26—H261 | 117.6 |
| C4—C5—C6 | 121.8 (3) | C22—N27—C26 | 118.0 (2) |
| C4—C5—H51 | 119.6 | C22—N27—Cu1 | 115.25 (17) |
| C6—C5—H51 | 118.6 | C26—N27—Cu1 | 126.77 (18) |
| C5—C6—C7 | 120.1 (2) | C13—N28—C29 | 118.9 (2) |
| C5—C6—C16 | 120.9 (2) | C13—N28—C30 | 122.9 (2) |
| C7—C6—C16 | 119.0 (2) | C29—N28—C30 | 118.0 (2) |
| C6—C7—C8 | 118.0 (2) | N28—C29—H291 | 110.7 |
| C6—C7—H71 | 120.5 | N28—C29—H292 | 111.1 |
| C8—C7—H71 | 121.5 | H291—C29—H292 | 108.1 |
| S1—C8—C7 | 128.5 (2) | N28—C29—H293 | 110.9 |
| S1—C8—C9 | 109.2 (2) | H291—C29—H293 | 109.6 |
| C7—C8—C9 | 122.3 (2) | H292—C29—H293 | 106.3 |
| C8—C9—C4 | 119.0 (2) | N28—C30—O31 | 123.7 (3) |
| C8—C9—N3 | 115.5 (2) | N28—C30—O32 | 111.1 (2) |
| C4—C9—N3 | 125.5 (2) | O31—C30—O32 | 125.3 (3) |
| C2—C10—C11 | 120.7 (2) | Cu1ii—O31—C30 | 146.2 (2) |
| C2—C10—C15 | 120.1 (2) | C30—O32—C33 | 121.0 (2) |
| C11—C10—C15 | 119.2 (2) | O32—C33—C34 | 109.9 (3) |
| C10—C11—C12 | 120.6 (3) | O32—C33—C35 | 110.4 (2) |
| C10—C11—H111 | 119.8 | C34—C33—C35 | 112.3 (3) |
| C12—C11—H111 | 119.5 | O32—C33—C36 | 101.8 (2) |
| C11—C12—C13 | 119.9 (3) | C34—C33—C36 | 110.6 (3) |
| C11—C12—H121 | 120.5 | C35—C33—C36 | 111.4 (3) |
| C13—C12—H121 | 119.7 | C33—C34—H341 | 109.0 |
| C12—C13—C14 | 119.8 (3) | C33—C34—H342 | 109.2 |
| C12—C13—N28 | 120.4 (2) | H341—C34—H342 | 109.1 |
| C14—C13—N28 | 119.7 (3) | C33—C34—H343 | 110.2 |
| C13—C14—C15 | 120.2 (3) | H341—C34—H343 | 109.5 |
| C13—C14—H141 | 119.7 | H342—C34—H343 | 109.8 |
| C15—C14—H141 | 120.1 | C33—C35—H351 | 109.2 |
| C10—C15—C14 | 120.1 (3) | C33—C35—H352 | 110.3 |
| C10—C15—H151 | 120.3 | H351—C35—H352 | 107.5 |
| C14—C15—H151 | 119.6 | C33—C35—H353 | 112.0 |
| C6—C16—N17 | 109.6 (2) | H351—C35—H353 | 109.2 |
| C6—C16—H161 | 110.1 | H352—C35—H353 | 108.5 |
| N17—C16—H161 | 108.3 | C33—C36—H361 | 110.0 |
| C6—C16—H162 | 109.9 | C33—C36—H362 | 108.5 |
| N17—C16—H162 | 108.7 | H361—C36—H362 | 110.0 |
| H161—C16—H162 | 110.3 | C33—C36—H363 | 108.6 |
| C16—N17—N18 | 119.7 (2) | H361—C36—H363 | 110.1 |
| C16—N17—C21 | 127.2 (2) | H362—C36—H363 | 109.6 |
| N18—N17—C21 | 112.7 (2) | C38—C37—H371 | 108.2 |
| N17—N18—N19 | 105.46 (19) | C38—C37—H372 | 109.3 |
| N18—N19—Cu1 | 134.95 (17) | H371—C37—H372 | 110.9 |
| N18—N19—C20 | 110.4 (2) | C38—C37—H373 | 107.7 |
| Cu1—N19—C20 | 114.51 (16) | H371—C37—H373 | 109.9 |
| N19—C20—C21 | 107.6 (2) | H372—C37—H373 | 110.6 |
| N19—C20—C22 | 116.9 (2) | C37—C38—N39 | 176.6 (5) |
| C21—C20—C22 | 135.4 (2) |
Symmetry codes: (i) x−1, y−1, z+1; (ii) x+1, y+1, z−1.
Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C4–C9 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C16—H162···N39iii | 0.97 | 2.52 | 3.451 (6) | 161 |
| C16—H161···Cl2iv | 0.97 | 2.72 | 3.606 (3) | 152 |
| C21—H211···Cl1iv | 0.94 | 2.81 | 3.633 (3) | 147 |
| C23—H231···Cl1iv | 0.94 | 2.62 | 3.494 (3) | 155 |
| C26—H261···Cl1 | 0.94 | 2.55 | 3.154 (3) | 122 |
| C29—H291···Cl2ii | 0.95 | 2.80 | 3.741 (3) | 172 |
| C25—H251···Cgv | 0.94 | 2.85 | 3.583 (3) | 135 |
Symmetry codes: (ii) x+1, y+1, z−1; (iii) −x, −y, −z+2; (iv) x+1, y, z; (v) −x, −y−1, −z+2.
Funding Statement
This work was funded by Alzheimer Association of France grant .
References
- Addison, A. W., Rao, T. N., Reedijk, J., van Rijn, J. & Verschoor, G. C. (1984). J. Chem. Soc. Dalton Trans. pp. 1349–1356.
- Bai, S.-Q., Jiang, L., Young, D. J. & Hor, T. S. A. (2016). Aust. J. Chem. 69, 372–378.
- Betteridge, P. W., Carruthers, J. R., Cooper, R. I., Prout, K. & Watkin, D. J. (2003). J. Appl. Cryst. 36, 1487.
- Bruker (2006). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Cheignon, C., Tomas, M., Bonnefont-Rousselot, D., Faller, P., Hureau, C. & Collin, F. (2018). Redox Biology. 14, 450–464. [DOI] [PMC free article] [PubMed]
- Conte-Daban, A., Boff, B., Candido Matias, A., Aparicio, C. N. M., Gateau, C., Lebrun, C., Cerchiaro, G., Kieffer, I., Sayen, S., Guillon, E., Delangle, P. & Hureau, C. (2017). Chem. Eur. J. 23, 17078–17088. [DOI] [PMC free article] [PubMed]
- Cooper, R. I., Thompson, A. L. & Watkin, D. J. (2010). J. Appl. Cryst. 43, 1100–1107.
- Eury, H., Bijani, C., Faller, P. & Hureau, C. (2011). Angew. Chem. Int. Ed. 50, 901–905. [DOI] [PubMed]
- Faller, P., Hureau, C. & Berthoumieu, O. (2013). Inorg. Chem. 52, 12193–12206. [DOI] [PubMed]
- Faller, P., Hureau, C. & La Penna, G. (2014). Acc. Chem. Res. 47, 2252–2259. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Jones, M. R., Mathieu, E., Dyrager, C., Faissner, S., Vaillancourt, Z., Korshavn, K. J., Lim, M. H., Ramamoorthy, A., Wee Yong, V., Tsutsui, S., Stys, P. K. & Storr, T. (2017). Chem. Sci. 8, 5636–5643. [DOI] [PMC free article] [PubMed]
- Jones, M. R., Service, E. L., Thompson, J. R., Wang, M. C., Kimsey, I. J., DeToma, A. S., Ramamoorthy, A., Lim, M. H. & Storr, T. (2012). Metallomics, 4, 910–920. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Noël, S., Cadet, S., Gras, E. & Hureau, C. (2013). Chem. Soc. Rev. 42, 7747–7762. [DOI] [PubMed]
- Palatinus, L. & Chapuis, G. (2007). J. Appl. Cryst. 40, 786–790.
- Prince, E. (1982). Mathematical Techniques in Crystallography and Materials Science, pp. 96–106. New York: Springer-Verlag.
- Santos, M. A., Chand, K. & Chaves, S. (2016). Coord. Chem. Rev. 327–328, 287–303.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Viles, J. H. (2012). Coord. Chem. Rev. 256, 2271–2284.
- Watkin, D. (1994). Acta Cryst. A50, 411–437.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S2056989018000488/su5417sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018000488/su5417Isup2.hkl
CCDC reference: 1815501
Additional supporting information: crystallographic information; 3D view; checkCIF report