Abstract
Purpose
To investigate the association between novel PAX6 mutations to bilateral anterior pyramidal congenital cataracts (APyC), complete and intact irides, and nystagmus.
Observations
This is a retrospective observational case series in a multi-center setting with genetic testing. Three female patients were diagnosed with bilateral APyC, intact irides and nystagmus. Genetic testing identified the three patients had novel missense mutations in PAX6 – c.128C > T; p.Ser43Phe (S43F), c. 197T > A; p.Ile66Asn (I66N) and c.781C > G; p.Arg261Gly (R261G).
Conclusions and importance
This study demonstrates a novel phenotype of bilateral APyC, intact irides, and nystagmus in whom genetic testing for PAX6 identified novel missense mutations (S43F, I66N, R261G) in highly conserved DNA-binding domains. Implications of understanding why the iris is present in these cases is discussed.
Keywords: Pyramidal cataracts, Intact irides, Nystagmus, Novel PAX6 missense mutations
1. Introduction
Pax6, a 422-amino-acid highly-conserved transcription factor, plays critical roles in the development of the eye, brain, olfactory system, pancreas, pituitary gland and spinal cord.1 Human PAX6 is located on chromosome 11p13, spanning 14 exons over 22 kb. PAX6 heterozygous mutations are commonly associated with an ocular phenotype involving complete or partial absence of iris, anterior polar cataract (APoC), aniridia-associated keratopathy, foveal and/or optic nerve hypoplasia, nystagmus, and glaucoma.2, 3, 4 Other ocular phenotypes reported with PAX6 heterozygous mutations include isolated foveal hypoplasia,5 Peters' anomaly,6,7 autosomal dominant keratitis,8 microphthalmia,9, 10, 11, 12 and certain optic nerve defects.13 PAX6 homozygous mutations are extremely rare, as they generate debilitating central nervous system defects that lead to post-natal loss of life.14 Thus, PAX6 mutations producing primarily ocular phenotypes are likely inherited in an autosomal dominant pattern.
We herein describe three cases of bilateral anterior pyramidal cataracts (APyC), intact irides, and nystagmus due to novel missense mutations in PAX6. A thorough review of the literature suggests, to the best of our knowledge, that we are describing a novel phenotype. We speculate as to why this phenotype occurs with these mutations using molecular modeling and analysis of the PAX6 mutation database.
2. Findings
A total of three female patients with a mean age of 96.5 months (M) (Median = 88.5M; range = 5 months-17 years) with bilateral APyC, intact iris, and nystagmus were included.
2.1. Case 1
A 9-month old girl presented with fine vertical nystagmus. Her BCVA was 20/136 in the right eye and 20/200 in the left with a refraction of +2.5DS-3.0DC × 10° and +3.5DS-3.5DC × 180° D in the right and left eye, respectively (Table 1). On anterior segment evaluation she was noted to have bilateral APyC (Fig. 1A, B, C). The iris was intact and IOP was within normal limits. While posterior segment examination showed no evidence of foveal hypoplasia, given the nystagmus it was difficult to be sure. OCT was not successfully performed in this child. At 11 years, she underwent cataract extraction and posterior chamber intraocular lens (IOL) implantation in the left eye. At the last follow-up visit (12 months after cataract extraction), her BCVA was 20/125 in the right eye and 20/200 in the left.
Table 1.
Clinical characteristics and genetic testing results.
| Cases |
1 |
2 |
3 |
||||
|---|---|---|---|---|---|---|---|
| Age at presentation |
9 months |
5 months |
17 years |
||||
| OD | OS | OD | OS | OD | OS | ||
| Vision (BCVA) | 20/136 | 20/200 | 20/1200 | 20/1200 | 20/80 | 20/200 | |
| Refraction | +2.50 −3.00 ×10° |
+3.50 −3.50 ×180° |
+7.50 −2.00 ×45° |
+7.00 −1.00 ×170° |
−12.25 −4.75 ×87° |
−12.25 −4.50 ×105° |
|
| IOP | normal | normal | 18 mmHg | 19 mmHg | |||
| Foveal hypoplasia | no | yes (PVEP abnormal) |
yes | ||||
| Nystagmus | fine vertical | yes | high frequency, low amplitude pendular | ||||
| Iris present | yes | yes | yes | yes | yes | yes | |
| Pyramidal cataract | yes | yes | yes | yes | yes | yes | |
| Other observations and clinical notes | Performed cataract extraction and posterior chamber IOL. Vision after surgery was 20/125 OD and 20/200 OS | persistent pupillary membranes | inferior corneal pannus, persistent pupillary membranes, small optic nerves | ||||
| PAX6 mutation | Nucleotide Transcript variant 1, mRNA NCBI Reference Sequence: NM_000280.4 |
c.128C > T exon 5 |
c. 197T > A exon 6 |
c. 781C > G exon 10 |
|||
| Protein | p.Ser43Phe (S43F) | p.Ile66Asn (I66N) | p.Arg261Gly (R261G) | ||||
| Structural location (refer to Fig. 4) | PAI - α2 | linker 1 between PAI and RED sub-domains | HD - α9 | ||||
| Impact on helical structure and DNA-binding stability | undisrupted | undisrupted | undisrupted | ||||
Abbreviations: BCVA, best-corrected visual acuity; HD, homeodomain DNA-binding domain; IOL, intraocular lens; IOP, intraocular pressure; NCBI, National Center for Biotechnology Information; PAI, PAI DNA-binding subdomain; PVEP, pattern visual evoked potential; RED, RED DNA-binding subdomain.
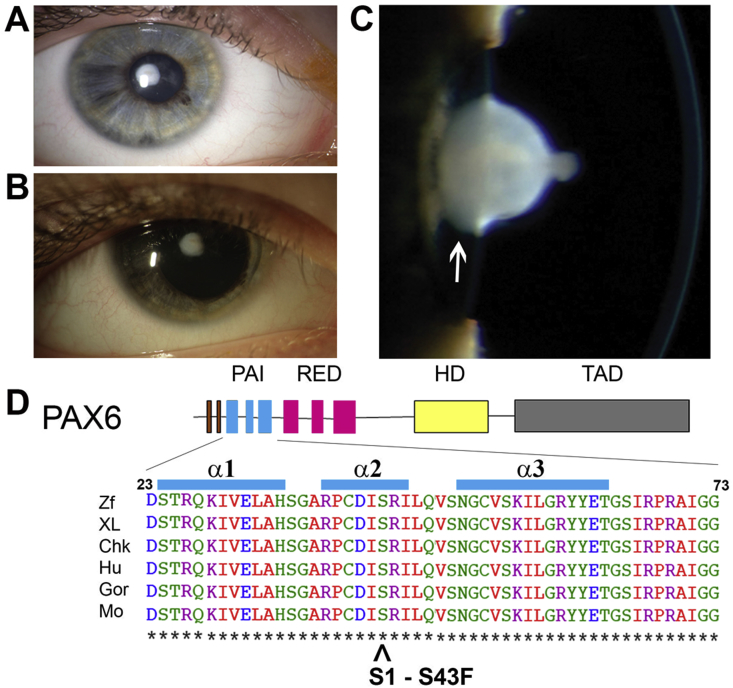
Fig. 1.
Clinical examination and genetic testing of case 1. A: APyC through the undilated pupil with intact iris. B: APyC through the dilated pupil. C: Slit-lamp biomicroscopy clearly demonstrating the morphology of APyC with conical protrusion into the anterior chamber and some amount of posterior cortical opacification. No evidence of corneal opacity. D: Missense mutation, p.Ser43Phe (S43F; denoted by ˆ), is in the α2 helix of the DNA-binding PAI domain. The protein structure of human PAX6 is shown at the top. Amino acid sequence alignments from several vertebrate species in the region of the missense mutation is at the bottom. The numbers refer to amino acid positions for human PAX6. This shows that the serine which in this mutation has been replaced by a phenylalanine amino acid, is highly conserved across six species, including human. Abbreviations: APyC, Anterior Pyramidal Cataracts; PAI, first subunit of the PAIRED DNA-binding domain; RED, second subunit of the PAIRED DNA-binding subunit; HD; homeodomain DNA-binding domain; TAD, transactivation domain; Chk, chicken; Gor, gorilla; Hu, human; M, mouse; XL, Xenopus leavis (frog); Zf, zebrafish.
Genetic testing revealed a novel PAX6 missense mutation for this young patient. The proband was heterozygous for a c.128C > T mutation in exon 5 that changes the amino acid at codon 43 from serine to phenylalanine (p.Ser43Phe). (Fig. 1D). No other family members were affected and, therefore, genetic testing was not carried out on the parents.
2.2. Case 2
A 5 months old girl was presented with vision of 20/1200 both eyes open, with no fixation preference, and nystagmus. Her cycloplegic refraction was +7.50DS-2.00DC × 45° in the right and +7.00DS −1.00DC × 170° in the left eye (Table 1). Prior to dilation, the child had mid-dilated pupils revealing APyC in both eyes (Fig. 2A). On EUA, upon pupil dilation, the APyC became more apparent and she was noted to have persistent pupillary membrane (Fig. 2B). The iris was slightly hypoplastic. IOP was 13 mmHg in both eyes with applanation tonometry. Ultrasound biomicroscopy (UBM) of the right eye revealed a more accurate morphology of the cataracts, where a conical shape was observed arising from a break in the lens capsule (Fig. 2C). Fundoscopy revealed good optic nerve size in both eyes and the possibility of foveal hypoplasia. The electrophysiological evaluation using flash electroretinogram (ERG) showed normal retinal responses, but the pattern evoked pattern visual evoked potentials (PVEP) indicated macular pathway dysfunction confirming the foveal hypoplasia suspected on fundus examination.
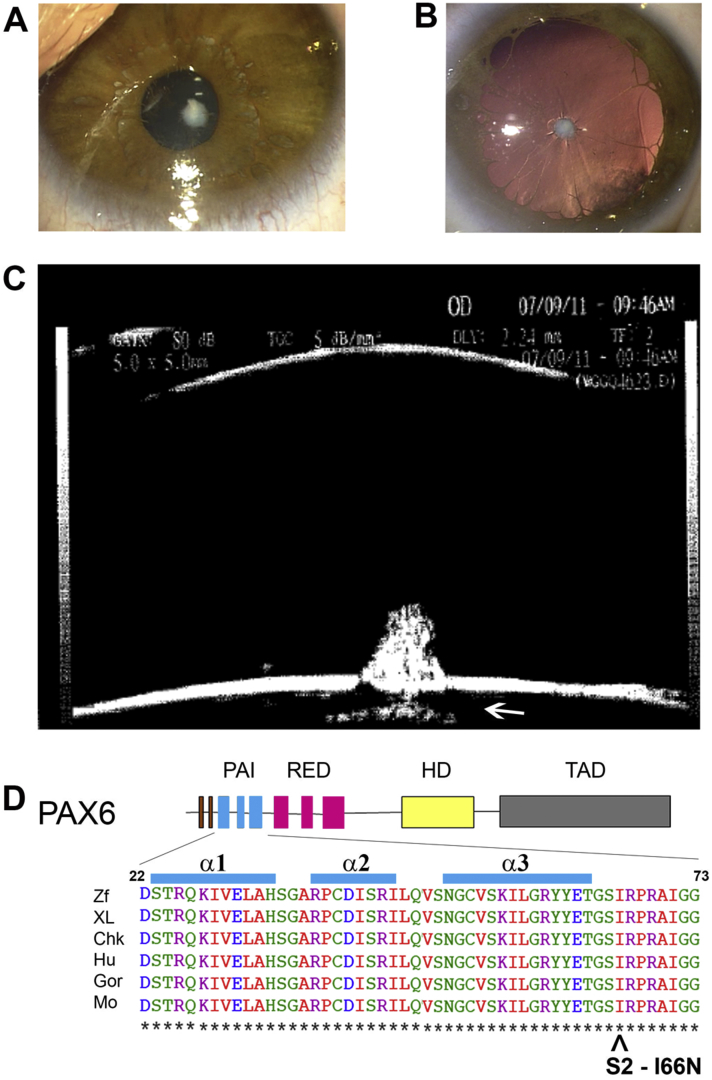
Fig. 2.
Clinical examination and genetic testing of case 2. A: APyC through the undilated pupil with intact iris. B: Dilated eye exam showing a persistent pupillary membrane. C: Ultrasound biomicroscopy demonstrating the morphology of the APyC. D. Missense mutation, p.Ile66Asn (I66N; denoted by ˆ), in the region after the α3 helix of the PAI domain. The isoleucine at this position, which in this mutation is replaced by asparagine, is highly conserved across 6 species, including human. Amino acid sequence alignments from several vertebrate species in the region of the missense mutation is at the bottom. The numbers refer to amino acid positions for human PAX6. Abbreviations: APyC, Anterior Pyramidal Cataracts; PAI, first subunit of the PAIRED DNA-binding domain; Chk, chicken; Gor, gorilla; Hu, human; M, mouse; XL, Xenopus leavis (frog); Zf, zebrafish.
Genetic testing revealed a novel missense PAX6 mutation. She was heterozygous for a c.197T > A mutation in exon 6 that changes the amino acid at codon 80 from isoleucine to asparagine (p.Ile66Asn) (Fig. 2D). In order to determine the pathogenicity of this novel amino acid substitution, 100 controls were tested for this sequence variation and shown to be negative suggesting that this could be the disease-causing mutation. The proband's parents and maternal uncle, who were all clinically unaffected, were negative for the mutation in blood-derived genomic DNA.
2.3. Case 3
A 17 year-old girl presented with bilateral APyC and nystagmus. Her BCVA was 20/80 with −12.25 DS/+4.75 DC in the right eye and 20/200 with −12.25 DS/+4.50 DC in the left eye. She exhibited high frequency, low amplitude pendular nystagmus (Table 1). On examination of the anterior segment, APyC, inferior corneal pannus and persistent pupillary membrane was noted in both eyes. The iris pattern was intact. Pneumotonometry was 18 mmHg in the right eye and 19 mmHg in the left eye. On fundus examination, she had small optic discs in both eyes suggestive of optic nerve hypoplasia. There was no history of consanguinity, but the family history was significant for cataract, nystagmus and iris coloboma in the father and paternal aunt. There was a history of nystagmus and coloboma in the paternal uncle. The maternal family history was negative for significant ocular disease. The girl's clinical features and family history led us to suspect a PAX6 mutation.
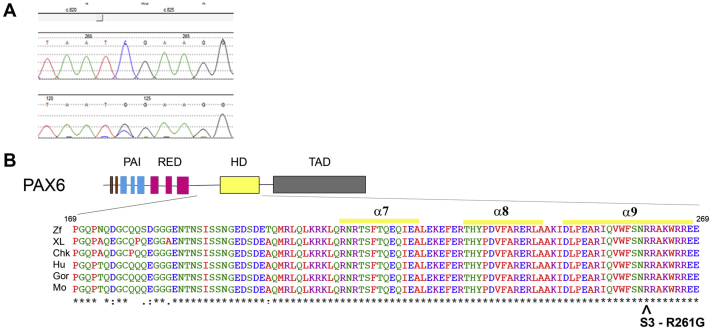
On genetic testing, she was found to be heterozygous for a novel variation in PAX6, namely c.781C > G (Fig. 3A) resulting in the amino acid substitution at position 261 of arginine for glycine, p.Arg261Gly (Fig. 3B). No other family members were affected and, therefore, genetic testing was not carried out on the parents.
Fig. 3.
Genetic testing of case 3 A: Chromatogram of the patient showing the c.781C > G missense mutation in PAX6. B: Missense mutation, p.Arg261Gly (R261G; denoted by ˆ), in the α9 helix of the human PAX6 DNA-binding homeodomain (HD). Amino acid sequence alignments from several vertebrate species in the region of the missense mutation is at the bottom. The numbers refer to amino acid positions for human PAX6. Abbreviations: Chk, chicken; Gor, gorilla; Hu, human; M, mouse; XL, Xenopus leavis (frog); Zf, zebrafish.
3. Discussion
We report three unrelated patients who shared a common phenotype consisting of APyC, intact irides and nystagmus from novel heterozygous missense mutations in PAX6, specifically at S43F, I66N and R261G.
To understand why the three novel but different mutations caused the iris to remain intact, we must briefly understand: (1) the structure of the PAX6 protein, (2) type of mutations in this protein, (3) the location of previous missense mutations and their effect on iris formation, and (4) protein stability as a result of the mutation. Structurally, the PAX6 protein is composed of three domains - two DNA-binding domains and an activation domain. The two DNA-binding domains are the PAIRED domain and the homeodomain, where the former is split into 2 subunits, the PAI and RED subdomains. The most common types of mutations seen in PAX6 are deletions, frameshifts, splicing errors, and formation of nonsense codons, all of which result in premature translational termination (PTT) with loss of normal protein function and, in the vast majority, total or near total absence of development of the iris.15 The three novel mutations we describe do not result in PTT and, instead, are all missense mutations resulting in protein structure changes.
Correlation of a database of all reported PAX6 missense mutations to date and location of our mutations in the PAX6 DNA-binding sites with the phenotype seen (Fig. 4 and Table 2) reveals that some missense mutations in the DNA-binding site lead to drastic protein structure changes. Such structural events prevent correct binding to DNA and, therefore, no activation of the protein causing absence of iris. However, some missense mutations lead to subtle protein changes that do not affect binding to DNA and enable intact iris formation. Two of the three missense mutations in our patients (I66N in case 2 and R261G in case 3) are indeed in those sites where intact irides have been reported (Fig. 4), in addition to symptoms of typical aniridia (e.g. autosomal dominant keratitis and foveal hypoplasia only). However, one mutation (S43F) in case 1 was in a site where you would expect total aniridia. To explain why the iris was intact in case 1, we looked at protein stability through three independent molecular modeling simulations (Fig. 5). The results from these simulations demonstrate that iris formation is dependent not only on the region of mutation with respect to more/less sensitivity for iris formation (Fig. 4), but also to disruption of the DNA-binding specificity because of reduced protein stability that leads to significant reduction in protein function. While the S43F mutation (S1) is in a highly sensitive region for iris formation that would be expected to result in a no iris phenotype, its effect on secondary and tertiary protein structure is minimal resulting in a stable protein with decreased DNA-binding but no loss of DNA-binding specificity and, therefore, protein function (Fig. 5). We speculate this result explains the intact irides phenotype. Therefore, to have an intact iris certain regions of DNA-binding domain within the PAX6 protein must be intact and the DNA-binding protein domains must be stable.
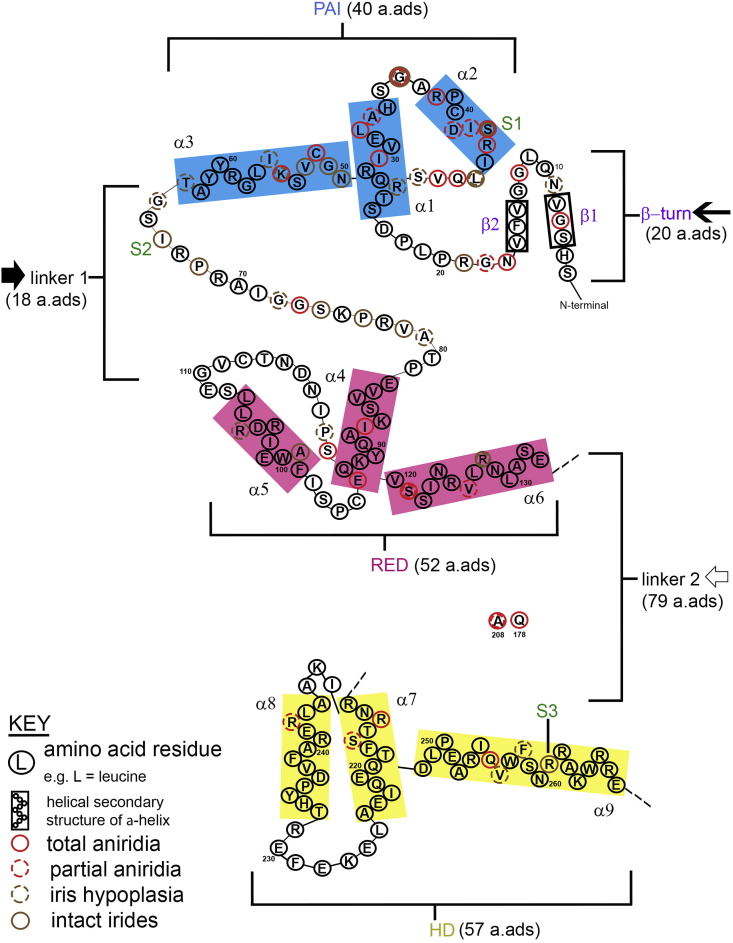
Fig. 4.
Schematic of PAX6 DNA-binding domains with all reported human missense mutations to date resulting in varying degrees of iris hypoplasia shown at their positions and the resulting iris phenotypes. The β-turn (purple designation comprising of 20 amino acids; 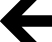 ) consists of two β-turn motifs (β1, β2) at the N-terminus of the protein. The PAI subdomain secondary structure consists of three α-helices (α1-, α2-and α3-; blue rectangles, 40 amino acids). The RED subdomain secondary structure also consists of three α-helices (α4-, α5-and α6-; pink rectangles, 52 amino acids). Linker 1 connects the PAI subdomain to the RED subdomain (
) consists of two β-turn motifs (β1, β2) at the N-terminus of the protein. The PAI subdomain secondary structure consists of three α-helices (α1-, α2-and α3-; blue rectangles, 40 amino acids). The RED subdomain secondary structure also consists of three α-helices (α4-, α5-and α6-; pink rectangles, 52 amino acids). Linker 1 connects the PAI subdomain to the RED subdomain ( ) and linker 2 connects the RED-subdomain to the HD (
) and linker 2 connects the RED-subdomain to the HD ( ). The HD is shown following the linker region after the RED subdomain and consists of 3 α-helices (α7-, α8-and α9; yellow rectangles, 57 amino acids). Amino acid residues are denoted by their standard letter abbreviations (i.e. L = leucine; see Key). Secondary structure of the α-helices is represented by residue positioning in the α-helix boxes (blue, pink, yellow rectangles; see Key). The complete amino acid residue chain for linker 2 (
). The HD is shown following the linker region after the RED subdomain and consists of 3 α-helices (α7-, α8-and α9; yellow rectangles, 57 amino acids). Amino acid residues are denoted by their standard letter abbreviations (i.e. L = leucine; see Key). Secondary structure of the α-helices is represented by residue positioning in the α-helix boxes (blue, pink, yellow rectangles; see Key). The complete amino acid residue chain for linker 2 ( ) are not shown due to space limitations, but the iris hypoplasia phenotypes for the only missense mutations in this region are shown as residues Gln178 (Q178) and Arg208 (R208).
) are not shown due to space limitations, but the iris hypoplasia phenotypes for the only missense mutations in this region are shown as residues Gln178 (Q178) and Arg208 (R208).
Data from Fig. 4 and Table 2 shows that two of our novel missense mutations (I66N (S2) and R261G (S3)) are in DNA-binding regions that are less critical to iris formation. This is because missense mutations in these regions lead more often to intact irides/intact irides with iris hypoplasia (brown circles/broken brown circles) than total aniridia (red circles). For example, linker 1 (where the S2 mutant lies) has 5 mutations resulting in intact irides, 3 to intact irides with iris hypoplasia and only 1 with total aniridia (see Table 2), therefore the intact irides phenotype seen for S2 would be expected in linker 1. However, one of our missense mutations (S43F (S1)) is in a critical region to iris formation (PAI:α2-helix), where a mutation would be expected to give a total aniridia phenotype. This is clearly observed in Table 2 where we observe six missense mutations in PAI:α2-helix that lead to total aniridia compared to 3 missense mutations leading to intact irides. An explanation for this unexpected phenotype is given in Fig. 5. Abbreviations: PAI, first subunit of the PAIRED DNA-binding domain; RED, second subunit of the PAIRED DNA-binding subunit; HD; homeodomain DNA-binding domain. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Compendium of PAX6 missense mutations in DNA-binding domains resulting in various iris phenotypes. Mutations, expressed as amino acid substitutions, and associated phenotypes extracted from the PAX6 mutation database (http://lsdb.hgu.mrc.ac.uk/home.php? select_db = PAX6).30
| Mutation | Iris phenotype | Mutation | Iris phenotype | Mutation | Iris phenotype |
|---|---|---|---|---|---|
| β-turn | Gly51Arg | II | linker 2 | ||
| Gly7Arg | TAN | Cys52Arg | TAN | Gln178His | TAN |
| Asn9Ile | IH | Val53Leu | II | Ala208Gln | TAN |
| Gly12Arg | TAN | Lys55Thr | TAN | Ala208Trp | PAN |
| Asn17Ser | TAN | Lys55Arg | IH | HD:α7-helix | |
| Gly18Trp | PAN | Ile56Thr | IH | Arg214Gly | TAN |
| Arg19Pro | II | Thr63Pro | IH | Arg214Ser | TAN |
| PAI:α1-helix | linker 1 | Ser216Pro | PAN | ||
| Arg26Pro | IH | Gly64Val | IH | HD:α8-helix | |
| Ile29Ser | TAN | Ile66Asn | II | Arg242Thr | PAN |
| Ile29Val | TAN | Pro68Ser | II | HD:α9-helix | |
| Leu32Val | TAN | Gly72Ser | IH | Val256Ala | IH |
| Ala33Pro | PAN | Gly73Asp | TAN | Phe258Ser | IH |
| Gly36Trp | TAN | Ser74Gly | II | Arg261Gly | II |
| Gly36Arg | IH | Pro76Arg | II | ||
| Gly36Ala | II | Val78Ala | II | ||
| PAI:α2-helix | Ala79Glu | IH | |||
| Arg38Trp | TAN | RED:α4-helix | |||
| Asp41Gly | PAN | Ile87Arg | TAN | ||
| Ile42Ser | PAN | Ile87Lys | TAN | ||
| Ser43Pro | TAN | Glu93Lys | TAN | ||
| Ser43Phe | II | RED:α5-helix | |||
| Arg44Gln | TAN | Ala99Pro | II | ||
| Leu46Arg | IH | Arg105Ser | IH | ||
| Leu46Pro | II | Pro118Arg | IH | ||
| Leu46Val | II | Pro118Gln | IH | ||
| Gln47Arg | TAN | Ser119Arg | TAN | ||
| Gln47Glu | TAN | RED:α6-helix | |||
| Val48Gln | TAN | Ser121Pro | PAN | ||
| Ser49Phe | IH | Ser121Leu | TAN | ||
| PAI:α3-helix | Val126Asp | PAN | |||
| Asn50Lys | II | Arg128Cys | II | ||
| Gly51Ala | II | Arg128Pro | IH |
Abbreviations: TAN, total aniridia; PAN, partial aniridia; IH, intact irides with some iris hypoplasia; II, intact irides; PAI, PAI DNA-binding subdomain; RED, RED DNA-binding subdomain; HD, homeodomain DNA-binding domain.
Terms in bold refer to the structural regions of the PAX6 protein (see Fig. 4).
Fig. 5.
Molecular model simulations to understand differences in phenotype between the intact irides-S43F mutant (S1) and the total aniridia-S43P mutant. Three results of molecular stability studies from three independent simulations are shown in this figure. All show that the mutant S43F (our mutation) is more stable than the aniridic S43P mutant, thereby generating complete intact irides in the former mutant and total aniridia in the latter mutant. A: Heat map of the percentage helicity (stability of structure) of each amino acid residue within the PAI domain in the wild-type (Wt, normal), S43F mutation (S1, intact irides) and S43P mutation (classic aniridia with no iris). Darker shading indicates higher percentage helicity for the amino acid residue. Therefore, by looking at this simulation, particularly the α2-helix chain, helicity is highest for the wild-type allele, followed by the S1 mutant, and then the S43P total aniridia mutant. B: Average and standard deviation of the Root Mean-Square Deviations (RMSD) for the alpha-carbon (in the carboxyl chain of an amino acid residue). This is a measure of each alpha-carbon distance for an amino acid residue between alignments of the PAI domain helical motif (from the crystal structure of PAX6) for PAI domain wild-type (blue), S43F mutation (S1, green) and S43P mutation (red) simulations. Cartoon of the secondary structure of the PAI domain of PAX6 is shown below the RMSD values. This demonstrates that S1 (green; intact irides) and Wt (blue) are aligned tightly, but the S43P (total aniridia) is not (red). C: Backbone RMSD of the helical motifs of the PAI domain (tertiary structure) in simulations of the Wt (blue), S43F mutation (S1, green) and S43P mutation (red). Backbone RMSD considers all the atoms over the protein chain backbone, not particular atoms in an amino acid (like alpha-carbon atom above in B). This shows disruption of the protein backbone in the S43P mutant resulting in total aniridia, but not in S1 (irides intact) or in the Wt (normal). Abbreviations: PAI, first subunit of the PAIRED DNA-binding domain. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
While APyC have been considered morphologically a subgroup of anterior polar cataracts (APoC), certain characteristics make considering APyC a separate entity a reasonable proposal. These characteristics include: asymmetry, association with surrounding cortical opacification necessitating removal, and extension anteriorly into the anterior chamber with corneal fusion and rarely detachment into the anterior chamber. In the largest series of 15 patients with APyC by Wheeler and colleagues, 9 were found to have bilateral APyC with only 2 patients having nystagmus with poor vision. Surprisingly they did not find irregular astigmatism in any of the patients. The authors also did not describe the iris details nor have they reported any genetic analysis related to these patients.16 While we are describing a specific genotype-phenotype correlation we feel that the phenotype may have been incompletely described in the previous two reports, but we cannot be sure.
A review of the literature suggests that previous causes of APyC have been considered to be acquired or congenital with acquired causes being varied, including corneal ulcer.16, 17, 18 Congenitally, the morphology of the APyC suggests that they could arise from remnants of prior attachment to the presumptive cornea during lens vesicle separation from the surface ectoderm in the fifth week of embryonic development. This could possibly explain the irregular astigmatism seen in our patients.19 Furthermore, even though refractive errors in aniridia do vary, anterior congenital cataracts are usually associated with variable astigmatism.20
While the association of congenital cataracts with nystagmus has been previously reported,21 they were not noted to be APyC and no genetic analysis was done. In a study by Hingorani et al., in a cohort of 43 patients with identified PAX6 mutations, only 3 patients with identified PAX6 mutations were found to have intact iris, cataract, irregular astigmatism and nystagmus (2 with foveal hypoplasia).22 It is possible that these three cases represent a similar phenotype seen in our study, though no lenticular phenotype was mentioned.
If pyramidal cataracts arise from abnormal detachment of the lens vesicle to the overlying presumptive cornea, then our results suggest that PAX6 may have a role in lens vesicle detachment. This has been documented before in cases of Peters' anomaly.6 In mouse models, genes downstream from Pax6 have been identified that have a specific role in lens vesicle separation defect when mutated.23, 24, 25, 26, 27, 28, 29 We speculate that Pax6 has a role in the expression of the above genes and the missense mutations reported in this study mildly affect their expression leading to APyC.
4. Conclusions
We define a novel phenotype with bilateral anterior pyramidal cataract, intact irides, and nystagmus due to missense mutations in PAX6. To the best of our knowledge this precise genotype-phenotype has not been recognized as one seen in PAX6 mutations previously, and we show for the first time a molecular explanation at the protein level as to why iris may remain intact despite mutations or deletions in PAX6.
5. Patient consent
The Institutional Review Board approval was waived as this study was considered a retrospective case series.
Genetic testing was performed after the informed consent from the parents. The patients provided oral consent for publication of this case series.
Funding
This work is supported by the National Institutes of Health CORE Grant P30 EY008098, Eye and Ear Foundation of Pittsburgh, Pennsylvania, USA and an unrestricted grant from Research to Prevent Blindness, New York, New York, USA.
Authorship
All authors attest that they meet the current ICMJE criteria for authorship.
Conflicts of interest
No financial disclosures.
Acknowledgements and disclosures
None.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ajoc.2018.02.021.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Simpson T.I., Price D.J. Pax6; a pleiotropic player in development. Bioessays. 2002;24:1041–1051. doi: 10.1002/bies.10174. [DOI] [PubMed] [Google Scholar]
- 2.Khan A.O., Aldahmesh M.A., Alkuraya F.S. Genetic and genomic analysis of classic aniridia in Saudi Arabia. Mol Vis. 2011;17:708–714. [PMC free article] [PubMed] [Google Scholar]
- 3.Khan A.O., Aldahmesh M.A. PAX6 analysis of two unrelated families from the Arabian Peninsula with classic hereditary aniridia. Ophthalmic Genet. 2008;29:145–148. doi: 10.1080/13816810802078195. [DOI] [PubMed] [Google Scholar]
- 4.Neethirajan G., Krishnadas S.R., Vijayalakshmi P., Shashikant S., Sundaresan P. PAX6 gene variations associated with aniridia in south India. BMC Med Genet. 2004;5:9. doi: 10.1186/1471-2350-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azuma N., Nishina S., Yanagisawa H., Okuyama T., Yamada M. PAX6 missense mutation in isolated foveal hypoplasia. Nat Genet. 1996;13:141–142. doi: 10.1038/ng0696-141. [DOI] [PubMed] [Google Scholar]
- 6.Hanson I.M., Fletcher J.M., Jordan T. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters' anomaly. Nat Genet. 1994;6:168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- 7.Churchill A.J., Yeung A. A compound heterozygous change found in Peters' anomaly. Mol Vis. 2005;11:66–70. [PubMed] [Google Scholar]
- 8.Mirzayans F., Pearce W.G., MacDonald I.M., Walter M.A. Mutation of the PAX6 gene in patients with autosomal dominant keratitis. Am J Hum Genet. 1995;57:539–548. [PMC free article] [PubMed] [Google Scholar]
- 9.Deml B., Reis L.M., Lemyre E., Clark R.D., Kariminejad A., Semina E.V. Novel mutations in PAX6, OTX2 and NDP in anophthalmia, microphthalmia and coloboma. Eur J Hum Genet. 2016;24:535–541. doi: 10.1038/ejhg.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mauri L., Franzoni A., Scarcello M. SOX2, OTX2 and PAX6 analysis in subjects with anophthalmia and microphthalmia. Eur J Med Genet. 2015;58:66–70. doi: 10.1016/j.ejmg.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Xiao X., Li S., Zhang Q. Microphthalmia, late onset keratitis, and iris coloboma/aniridia in a family with a novel PAX6 mutation. Ophthalmic Genet. 2012;33:119–121. doi: 10.3109/13816810.2011.642452. [DOI] [PubMed] [Google Scholar]
- 12.Solomon B.D., Pineda-Alvarez D.E., Balog J.Z. Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am J Med Genet A. 2009;149A:2543–2546. doi: 10.1002/ajmg.a.33081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azuma N., Yamaguchi Y., Handa H. Mutations of the PAX6 gene detected in patients with a variety of optic-nerve malformations. Am J Hum Genet. 2003;72:1565–1570. doi: 10.1086/375555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser T., Jepeal L., Edwards J.G., Young S.R., Favor J., Maas R.L. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 15.Robinson D.O., Howarth R.J., Williamson K.A., van Heyningen V., Beal S.J., Crolla J.A. Genetic analysis of chromosome 11p13 and the PAX6 gene in a series of 125 cases referred with aniridia. Am J Med Genet A. 2008;146A:558–569. doi: 10.1002/ajmg.a.32209. [DOI] [PubMed] [Google Scholar]
- 16.Collins E.T. Diseases of the lens. 1. On the minute anatomy of pyramidal cataract. Trans Ophthalmol Soc U K. 1892;12:89–105. [Google Scholar]
- 17.Brown G.C., Shields J.A., Oglesby R.B. Anterior polar cataracts associated with bilateral retinoblastoma. Am J Ophthalmol. 1979;87:276. doi: 10.1016/0002-9394(79)90061-8. [DOI] [PubMed] [Google Scholar]
- 18.Kostick A.M., Romanchuck K.G., Beebe D.C. Anterior pyramidal cataracts in Ehlers-Danlos syndrome. Can J Ophthalmol. 1996;31:133–136. [PubMed] [Google Scholar]
- 19.Wheeler D.T., Mullaney P.B., Awad A., Zwaan J. Pyramidal anterior polar cataracts. Ophthalmology. 1999;106:2362–2367. doi: 10.1016/S0161-6420(99)90540-7. [DOI] [PubMed] [Google Scholar]
- 20.Bouzas A.G. Anterior polar congenital cataract and corneal astigmatism. J Pediatr Ophthalmol Strabismus. 1992;29:210–212. doi: 10.3928/0191-3913-19920701-05. [DOI] [PubMed] [Google Scholar]
- 21.Yan N., Zhao Y., Wang Y. Molecular genetics of familial nystagmus complicated with cataract and iris anomalies. Mol Vis. 2011;17:2612–2617. [PMC free article] [PubMed] [Google Scholar]
- 22.Hingorani M., Williamson K.A., Moore A.T., van Heyningen V. Detailed ophthalmologic evaluation of 43 individuals with PAX6 mutations. Invest Ophthalmol Vis Sci. 2009;50:2581–2590. doi: 10.1167/iovs.08-2827. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y., Doughman Y.Q., Gu S. Cited2 is required for the proper formation of the hyaloid vasculature and for lens morphogenesis. Development. 2008;135:2939–2948. doi: 10.1242/dev.021097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blixt A., Mahlapuu M., Aitola M., Pelto-Huikko M., Enerback S., Carlsson P. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 2000;14:245–254. [PMC free article] [PubMed] [Google Scholar]
- 25.Wiley L.A., Dattilo L.K., Kang K.B., Giovannini M., Beebe D.C. The tumor suppressor merlin is required for cell cycle exit, terminal differentiation, and cell polarity in the developing murine lens. Invest Ophthalmol Vis Sci. 2010;51:3611–3618. doi: 10.1167/iovs.09-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semina E.V., Murray J.C., Reiter R., Hrstka R.F., Graw J. Deletion in the promoter region and altered expression of Pitx3 homeobox gene in aphakia mice. Hum Mol Genet. 2000;9:1575–1585. doi: 10.1093/hmg/9.11.1575. [DOI] [PubMed] [Google Scholar]
- 27.Pontoriero G.F., Smith A.N., Miller L.A., Radice G.L., West-Mays J.A., Lang R.A. Co-operative roles for E-cadherin and N-cadherin during lens vesicle separation and lens epithelial cell survival. Dev Biol. 2009;326:403–417. doi: 10.1016/j.ydbio.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuracha M.R., Burgess D., Siefker E. Spry1 and Spry2 are necessary for lens vesicle separation and corneal differentiation. Invest Ophthalmol Vis Sci. 2011;52:6887–6897. doi: 10.1167/iovs.11-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le T.T., Conley K.W., Mead T.J., Rowan S., Yutzey K.E., Brown N.L. Requirements for Jag1-Rbpj mediated Notch signaling during early mouse lens development. Dev Dyn. 2012;241:493–504. doi: 10.1002/dvdy.23739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown A., McKie M., van Heyningen V., Prosser J. The human PAX6 mutation database. Nucleic Acids Res. 1998;26:259–264. doi: 10.1093/nar/26.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.