Abstract
Background
Residual feed intake (RFI) in dairy cattle typically calculated at peak lactation is a measure of feed efficiency independent of milk production level. The objective of this study was to evaluate differences in ruminal bacteria, biopolymer hydrolyzing enzyme activities, and overall performance between the most- and the least-efficient dairy cows during the peripartal period. Twenty multiparous Holstein dairy cows with daily ad libitum access to a total mixed ration from d − 10 to d 60 relative to the calving date were used. Cows were classified into most-efficient (i.e. with low RFI, n = 10) and least-efficient (i.e. with high RFI, n = 10) based on a linear regression model involving dry matter intake (DMI), fat-corrected milk (FCM), changes in body weight (BW), and metabolic BW.
Results
The most-efficient cows had ~ 2.6 kg/d lower DMI at wk 4, 6, 7, and 8 compared with the least-efficient cows. In addition, the most-efficient cows had greater relative abundance of total ruminal bacterial community during the peripartal period. Compared with the least-efficient cows, the most-efficient cows had 4-fold greater relative abundance of Succinivibrio dextrinosolvens at d − 10 and d 10 around parturition and tended to have greater abundance of Fibrobacter succinogenes and Megaspheara elsdenii. In contrast, the relative abundance of Butyrivibrio proteoclasticus and Streptococcus bovis was lower and Succinimonas amylolytica and Prevotella bryantii tended to be lower in the most-efficient cows around calving. During the peripartal period, the most-efficient cows had lower enzymatic activities of cellulase, amylase, and protease compared with the least-efficient cows.
Conclusions
The results suggest that shifts in ruminal bacteria and digestive enzyme activities during the peripartal period could, at least in part, be part of the mechanism associated with better feed efficiency in dairy cows.
Electronic supplementary material
The online version of this article (10.1186/s40104-018-0258-9) contains supplementary material, which is available to authorized users.
Keywords: Dairy cows, Enzyme activity, Peripartal period, RFI, Rumen bacteria
Background
Improving feed efficiency in dairy cows has become increasingly important for the dairy industry since feed expenses are the most costly component of dairy systems. Therefore, identifying and selecting for dairy cows that use feed efficiently, i.e. require less feed for maintenance and for the same level of milk production, provides opportunities for reducing production costs and maximizing the economic returns for dairy producers. Residual feed intake (RFI) has been used in dairy cows to define feed efficiency independent of body size and milk production level [1, 2]. Residual feed intake is calculated as the difference between the actual and the predicted feed consumption of individual dairy cows after adjusting the DMI for the level of production through a linear regression model [3]. Hence, the most-efficient dairy cows, i.e. with a negative RFI, consume less feed than expected for their production level compared with the least-efficient dairy cows having a positive RFI [4].
Phenotypic variation in RFI between individual animals is directly related to variation in feed digestion, efficiency of nutrient use, and microbial protein production, all of which take place in the rumen, suggesting a vital role for the rumen in improving feed efficiency [5]. The rumen harbors a complex anaerobic microbial community, mainly bacteria, capable of producing various biopolymer hydrolyzing enzymes (e.g., amylase, xylanase, cellulase, and protease) that convert low-quality feed consumed into energy- and protein-rich compounds for the host [6]. Production of VFA and microbial protein can supply the ruminant with 70% and 50% of its daily energy and protein requirements, respectively [7].
To the best of our knowledge, the only available study on the association between rumen bacteria and RFI phenotypes in lactating dairy cows was conducted by Jewell et al. [8] who observed that ruminal bacteria profiles in the most-efficient lactating dairy cows differed from the least-efficient ones; for example, over the course of two lactations, the higher abundance of bacterial genera Anaerovibrio and Butyrivibrio were associated with the least-efficient cows. That study only focused on profiling the composition of rumen bacterial community between the most- and the least-efficient lactating dairy cows and did not provide information on the microbial enzyme activities in the rumen between the two RFI phenotypes. Therefore, it is reasonable to hypothesize that shifts in bacterial composition could be accompanied by changes in major microbial enzyme activities and impact RFI phenotypes.
The “peripartal” period in dairy cows is characterized by a marked negative energy and metabolizable protein balance at least in part due to the decrease in voluntary DMI and the high requirements for nutrients by the fetus and lactating mammary gland. Therefore, the peripartal period is challenging for dairy cows resulting in higher susceptibility for developing metabolic disorders [9–11]. Current nutritional management of peripartal cows encompasses the feeding of higher-concentrate diets postcalving to provide the rumen bacterial communities with a more readily-available source of energy. As a result, bacterial composition in the rumen changes relative to the dry period [12–14]. Although various studies were recently conducted to evaluate shifts in rumen bacterial communities during the peripartal period in dairy cows, little attention has been given to changes in microbial enzyme activities. Clearly, it is possible that changes in the bacterial community composition of the rumen during the peripartal period may contribute to differences in the major biopolymer hydrolyzing enzyme activities.
The current study aimed to evaluate abundance of selected ruminal bacterial species and activities of enzymes associated with protein and carbohydrate metabolism between cows classified as most-efficient (i.e., with negative RFI) and least-efficient (i.e., with positive RFI) using data collected from d − 10 to d 60 relative to the calving date. An important goal was to determine potential linkages between RFI phenotype, ruminal microorganisms, digestive enzyme activities, and overall performance.
Methods
All experimental procedures were approved by The Institutional Animal Care and Use Committee at the University of Illinois (protocol number 14270).
Animals, experimental design, and diets
A subset of 20 multiparous Holstein cows from a larger cohort were used [15]. Before calving, cows were housed in a freestall barn equipped with electronic recognition feeding system for each cow (American Calan Inc., Northwood, NH), while cows were housed in a tie-stall system during lactation. Cows were fed individually a TMR and allowed free access to feed and water at all times. The ration was formulated to meet cow predicted requirements according to NRC [16]. The feed ingredients for the close-up (from d − 28 to calving), fresh (from 1 to 30 DIM), and high-producing (from 31 to 60 DIM) TMR diets are shown in Table 1. Feed offered and refused were measured daily to calculate feed intake throughout the entire study. Weekly samples of the diets were collected to determine the DM content. Samples of the TMR and feed ingredients were stored frozen at − 20 °C and composited monthly for analyses of crude protein [[17]; method 990.03], NDF with heat-stable α-amylase and sodium sulfite [18], ADF [18], and ether extract [[17]; method 2003.05] by Cumberland Valley Analytical Services (Hagerstown, MD). The nutrient analysis of the diets is shown in Table 1. Body weight was recorded weekly during the entire feeding period.
Table 1.
Ingredient composition and nutrient analysis of close-up (from d − 28 to parturition), fresh (from 1 to 30 DIM), and high-producing (from 31 to 60 DIM) diets
| Item | Close-up | Fresh | High-producing |
|---|---|---|---|
| Ingredient composition, % of DM | |||
| Alfalfa haylage | 6.55 | 7.81 | 10.8 |
| Corn silage | 26.6 | 31.0 | 31.9 |
| Wheat straw | 26.5 | 3.25 | – |
| Corn grain, ground, dry | 12.6 | 22.21 | 20.7 |
| Cottonseed | – | 2.17 | 1.83 |
| Molasses, beet sugar | 4.03 | 5.50 | 4.51 |
| Soybean hulls | 3.46 | 4.25 | 9.96 |
| Soybean meal, 48% CP | 7.83 | 10.1 | 7.98 |
| Expeller soybean meala | 5.80 | 5.16 | 5.17 |
| Protein supplementb | 0.78 | 1.81 | 1.58 |
| Urea | 0.59 | 0.39 | 0.40 |
| Soychlorc | 1.23 | – | – |
| Saturated fat supplementd | – | 2.25 | 2.14 |
| Limestone | – | 1.41 | 0.96 |
| Salt | – | 0.02 | 0.04 |
| Dicalcium phosphate | 0.52 | 1.17 | 0.92 |
| Magnesium oxide | – | 0.08 | 0.04 |
| Magnesium sulfate | 2.08 | 0.02 | – |
| Sodium bicarbonate | – | 0.84 | 0.59 |
| Mineral vitamin mixe | 0.17 | 0.17 | 0.20 |
| Vitamin Af | 0.03 | 0.02 | 0.02 |
| Vitamin Dg | 0.03 | – | – |
| Vitamin Eh | 0.60 | – | – |
| Biotini | 0.70 | 0.42 | 0.32 |
| Nutrient analysis, % of DM | |||
| CP | 15.6 ± 0.32 | 17.7 ± 0.36 | 17.4 ± 0.36 |
| NDF | 40.8 ± 0.68 | 29.2 ± 0.59 | 31.4 ± 0.62 |
| ADF | 27.5 ± 0.50 | 19.5 ± 0.38 | 21.5 ± 0.48 |
| NFC | 34.9 ± 0.81 | 41.4 ± 0.55 | 40.7 ± 0.54 |
| Ether extract | 2.32 ± 0.05 | 5.12 ± 0.14 | 5.13 ± 0.14 |
aSoyPlus, West Central Soy (Ralston, IA)
bProVAAl AADvantage, Perdue AgriBusiness (Salisbury, MD)
cWest Central Soy
dEnergy Booster 100, Milk Specialties Global (Eden Prairie, MN)
eContained a minimum of 5% Mg, 10% S, 7.5% K, 2.0% Fe, 3.0% Zn, 3.0% Mn, 5,000 mg/kg Cu, 250 mg/kg I, 40 mg/kg Co, 150 mg/kg Se, 2,200 kIU/kg vitamin A, 660 kIU/kg vitamin D3, and 7,700 IU/kg vitamin E
fContained 30,000 kIU/kg
gContained 5,000 kIU/kg
hContained 44,000 kIU/kg
iADM Animal Nutrition (Quincy, IL)
Sample collection
Cows were automatically milked 3 times daily, and individual milk production recorded daily. Consecutive morning, midday, and evening milk samples were collected once a week, and stored at 4 °C for fat analysis by an infrared system (Dairy Lab Services, Dubuque, IA). The fat-corrected milk (FCM) yield for each cow was calculated according to NRC (2001) equations.
Although cannulation has been previously considered as the standard method collection of a representative sample of ruminal contents [19], performing surgical cannulations and its associated post-surgical costs limit its application in large groups of cows and often leads to reduced statistical power [20, 21]. The lack of significant differences in ruminal fermentation and microbiota composition between samples harvested via cannula or stomach tubing demonstrated that the latter is suitable for ruminal digesta sampling [20, 22–24]. Therefore, stomach tubing was deemed suitable and allowed us to use a greater number of cows. Four-hundred mL of mixed ruminal contents was collected from each cow 4 h postfeeding via stomach tubing at d − 10 before expected calving date and at 10, 30, and 60 DIM. Ruminal contents were immediately frozen in liquid nitrogen, transported to the laboratory and stored at − 80 °C for later analysis.
Residual feed intake calculation
The RFI (kg/d) for the 20 cows used from the bigger cohort in the experiment of Batistel et al. [15] was calculated using individual data from the entire period between − 28 d through 60 d around parturition. This number of cows gave us a complete set of samples across the chosen time points, i.e. for ruminal fluid and production. Thus, the calculated RFI for the 20 cows allowed us to divide them into two groups based on their divergent in feed efficiency: the most-efficient cows with desirable (i.e. more negative) RFI coefficient vs. the least-efficient cows with unfavorable (i.e. more positive) RFI coefficient. The RFI coefficients were computed as the difference between the actual and the predicted DMI, where the predicted DMI was computed through a linear regression model using the regression of actual DMI on FCM, metabolic BW, and ADG as described previously [3]. The RFI coefficients for the most-efficient (n = 10) and the least-efficient (n = 10) cows are depicted in Fig. 1a.
Fig. 1.
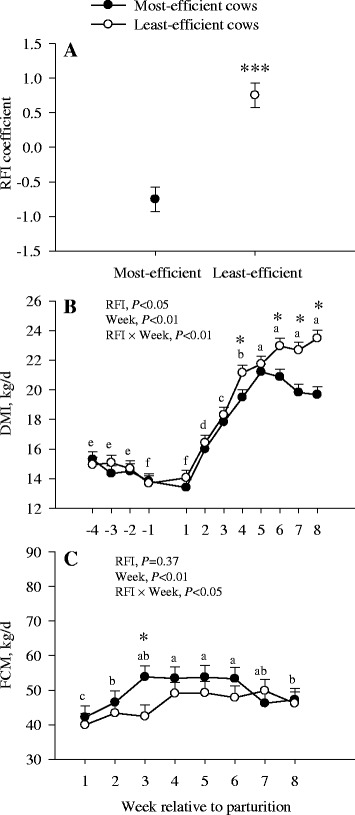
RFI coefficients (a), DMI (b), and fat-corrected milk (FCM) (c) in the most- and the least-efficient multiparous Holstein dairy cows during the peripartal period. Significant differences between RFI groups are denoted with an asterisk (*P < 0.05 or *** P < 0.0001). a-eDifferent letters indicate differences due to the main effect of time (P < 0.05)
Ruminal bacteria DNA extraction and RT-PCR amplification
The total genomic DNA was isolated using the repeated bead-beating plus column (RBB + C) purification method described by Yu and Morrison [25] for mechanical lysis of bacterial cell wall employing the QIAamp DNA mini kit (QIAGEN) for DNA purification. This method has been applied in several hundred studies to extract a high yield microbial DNA from rumen contents [21]. The DNA quantity and quality were checked using 0.8% (w/v) agarose gel electrophoresis and NanoDrop spectrophotometer (ND 1000, NanoDrop Technologies, Inc., Wilmington, DE, USA) at 260 nm. Extracted DNA was standardized to 8 ng/μL for RT-PCR.
Primers were selected to amplify 10 major ruminal bacteria species that play key roles in cellulose and hemicellulose digestion, xylan degradation, proteolysis, propionate production, lactate utilization and ruminal biohydrogenation. The chosen primers along with 3 universal primers are listed in Table 2. A total of 10 μL of RT-PCR mixture contained 4 μL sample DNA, 5 μL 1× SYBR Green with ROX (Quanta BioSciences, Gaithersburg, MD, USA), 0.4 μL each of 10 μmol/L forward and reverse primers, and 0.2 μL DNase/RNase free water in a MicroAmpTM Optical 384-Well Reaction Plate (Applied Biosystems, Foster City, CA, USA). Negative controls without template DNA, standards, and samples were run on the same plate in triplicate. The RT-PCR reactions were performed with QuantStudio-7 Real-Time PCR instrument (ThermoFisher Scientific, USA) using the following program: initial denaturation at 95 °C for 5 min, followed by 40 cycles of 1 s at 95 °C and 30 s annealing at 60 °C. A dissociation stage was performed to determine the specificity of the amplification. Relative abundance of bacterial species was calculated using the geometric mean of the universal primers eubacterial primer 1 and eubacterial primer 2 (Table 2) [26, 27] with the efficiency-corrected Δ−CT method [28]. The copy number of total bacterial 16S rRNA genes was measured to estimate the total bacterial population using RT-PCR analysis with eubacterial primer 3 (Table 2) [29] following the procedures described previously by Zhou et al. [30].
Table 2.
Species-specific primers for the quantification of selected rumen bacterial populations using a real-time qPCR assay
| Target bacterial species | Primer sequence (5` → 3`) | Reference | Efficiencya, % | |
|---|---|---|---|---|
| Anaerovibrio lipolytica | F:b R:c |
GAAATGGATTCTAGTGGCAAACG ACATCGGTCATGCGACCAA |
[13] | 96.06 |
| Butyrivibrio proteoclasticus | F: R: |
GGGCTTGCTTTGGAAACTGTT CCCACCGATGTTCCTCCTAA |
[13] | 100.00 |
| Eubacterium ruminantium | F: R: |
CTCCCGAGACTGAGGAAGCTTG GTCCATCTCACACCACCGGA |
[53] | 106.08 |
| Fibrobacter succinogenes | F: R: |
GCGGGTAGCAAACAGGATTAGA CCCCCGGACACCCAGTAT |
[53] | 100.67 |
| Megaspheara elsdenii | F: R: |
AGATGGGGACAACAGCTGGA CGAAAGCTCCGAAGAGCCT |
[53] | 101.35 |
| Prevotella bryantii | F: R: |
AGCGCAGGCCGTTTGG GCTTCCTGTGCACTCAAGTCTGAC |
[53] | 105.03 |
| Selenomonas ruminantium | F: R: |
CAATAAGCATTCCGCCTGGG TTCACTCAATGTCAAGCCCTGG |
[53] | 97.91 |
| Succinimonas amylolytica | F: R: |
CGTTGGGCGGTCATTTGAAAC CCTGAGCGTCAGTTACTATCCAGA |
[54] | 96.80 |
| Streptococcus bovis | F: R: |
TTCCTAGAGATAGGAAGTTTCTTCGG ATGATGGCAACTAACAATAGGGGT |
[53] | 103.89 |
| Succinivibrio dextrinosolvens | F: R: |
TAGGAGCTTGTGCGATAGTATGG CTCACTATGTCAAGGTCAGGTAAGG |
[54] | 96.80 |
| Eubacterial primer 1 | F: R: |
GGATTAGATACCCTGGTAGT CACGACACGAGCTGACG |
[27] | 95.26 |
| Eubacterial primer 2 | F: R: |
GTGSTGCAYGGYTGTCGTCA ACGTCRTCCMCACCTTCCTC |
[26] | 95.30 |
| Eubacterial primer 3 | F: | CCTACGGGAGGCAGCAG | [29] | 99.30 |
| R: | ATTACCGCGGCTGCTGG |
aMeasured efficiencies of the primers in the qPCR reactions
bForward primer
cReverse primer
Enzyme activities
Details of ruminal enzymatic assays for determining amylase, xylanase, cellulase, and protease enzymatic activities are reported in detail in the Additional file 1.
Statistical analysis
The MIXED procedure of SAS (SAS Institute Inc., Cary, NC) was used for repeated measures analysis of DMI, FCM, bacterial abundance, and enzyme activities. The fixed effects in the model were RFI and time (week or day), and the random effect was cow. Significance was determined at P ≤ 0.05, and tendencies were determined at P ≤ 0.10.
Results
Production
The most-efficient cows had lower (P = 0.04) DMI at wk 4, 6, 7, and 8 (average = 2.6 kg/d) (Fig. 1b). Dry matter intake in both groups decreased (P < 0.01) from − 4 wk to calving but progressively increased (P < 0.01) postpartum. No differences (P = 0.37) between RFI groups were observed for FCM (Fig. 1c). A RFI × week (P = 0.02) was observed for FCM yield due to a greater increase (P < 0.01) in the most-efficient cows at wk 3. Both groups of cows increased FCM yield (P < 0.01) after calving.
Relative abundance of bacteria
The relative abundance of target ruminal bacteria species between the most- and the least-efficient cows during the peripartal period are depicted in Fig. 2. Results indicate that Selenomonas ruminantium was the most-abundant bacteria among the 10 analyzed, averaging 0.8% of 16S rRNA copy number. There were no RFI, day or RFI × day effects observed for abundance of Anaerovibrio lipolytica. A RFI × week interaction (P = 0.04) was observed for Succinivibrio dextrinosolvens due to a greater relative abundance in the most-efficient compared with least-efficient cows of ~ 6-fold at d − 10 and ~ 4-fold at d 10. Similarly, the most-efficient cows tended (P = 0.09) to have a greater overall abundance of Fibrobacter succinogenes and Megaspheara elsdenii, whereas a tendency (P = 0.08) for RFI × day interaction in the abundance of Fibrobacter succinogenes was due to greater levels of Fibrobacter succinogenes in the most-efficient cows at all days except d 30. There was no RFI × day (P = 0.48) effect for Megaspheara elsdenii.
Fig. 2.
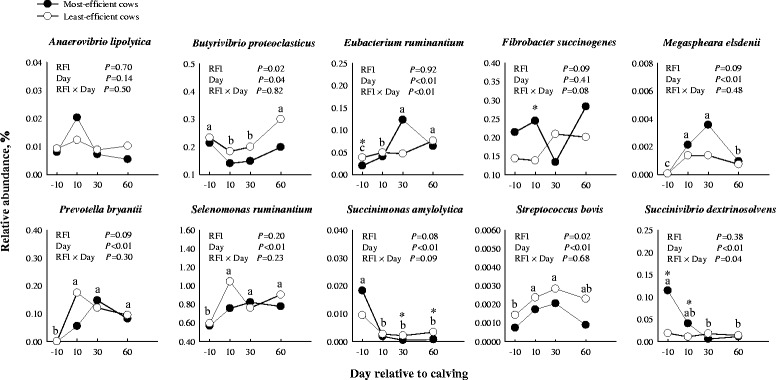
Relative abundance of 10 rumen bacterial species in the most- and the least-efficient multiparous Holstein dairy cows during the peripartal period. Significant differences (P < 0.05) between RFI groups at a given time point are denoted with an asterisk (*). a-cDifferent letters indicate differences due to the main effect of time (P < 0.05). Data were logit transformed to ensure normality of residuals
Compared with the least-efficient cows, the abundance of Butyrivibrio proteoclasticus and Streptococcus bovis was lower (P = 0.02) in the most-efficient cows during the peripartal period. In addition, the most-efficient cows had lower (P < 0.01) overall abundance of Eubacterium ruminantium only at − 10 d. In addition, the most-efficient cows tended to have a lower abundance of Succinimonas amylolytica (P = 0.08) and Prevotella bryantii (P = 0.09) during the peripartal period. A tendency for a RFI × day (P = 0.09) effect in Succinimonas amylolytica was due to the most-efficient cows having a greater relative abundance at d − 10 followed by a decrease at d 30 and d 60 postpartum in both RFI groups. No RFI × day interaction (P = 0.30) was observed for Prevotella bryantii.
Several shifts in the bacterial populations were observed over time. For example, the relative abundance of Eubacterium ruminantium, Prevotella bryantii, Selenomonas ruminantium, and Streptococcus bovis was higher (P < 0.01) in both RFI groups after parturition. In contrast, Megaspheara elsdenii abundance decreased (P < 0.01) at d 60 compared with d 10 and d 30. The relative abundance of Succinimonas amylolytica was lower (P < 0.01) postpartum in both RFI groups while Succinivibrio dextrinosolvens was lower (P < 0.01) in the most-efficient group postpartum. Butyrivibrio proteoclasticus had low relative abundance (P = 0.04) at d 10 and d 30 postpartum, and abundance returned to prepartum values at d 60.
The 16S rRNA copy numbers of the total ruminal bacterial community detected in the present study are shown in Fig. 3. The results indicate that the most-efficient cows had a greater (P = 0.04) relative abundance of bacteria compared with the least-efficient cows. However, there was a tendency for an RFI × day (P = 0.09) effect due to a greater bacterial community copy number in the rumen of the most-efficient cows at − 10 and 60 d, and a lower bacterial community copy number in the least-efficient cows at d 10.
Fig. 3.
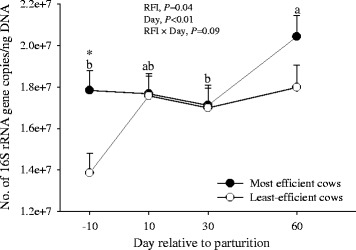
16S rRNA gene copy number of the total rumen bacterial community in rumen contents in the most- and the least-efficient multiparous Holstein dairy cows during the peripartal period. Significant differences (P < 0.05) between RFI groups at a given time point are denoted with an asterisk (*). a, bDifferent letters indicate differences due to the main effect of time (P < 0.05)
Digestive enzyme activities
The microbial enzyme activities in the rumen of the most- and the least-efficient cows during the peripartal period is shown in Fig. 4. Compared with the least-efficient cows, the results indicate that the most-efficient cows had lower overall activities of cellulase (P = 0.04), amylase (P = 0.02), and protease (P < 0.01). In addition, the most-efficient cows had lower (RFI × day, P = 0.04) xylanase activity at d 30 postpartum. No day or RFI × day effect (P > 0.10) was observed for amylase, cellulase, and protease activity. An RFI × day interaction for xylanase revealed that the most-efficient compared with least-efficient cows had lower (P = 0.04) activity at d 30. The interaction effect was also due to lower activity at d 30 relative to other time points.
Fig. 4.
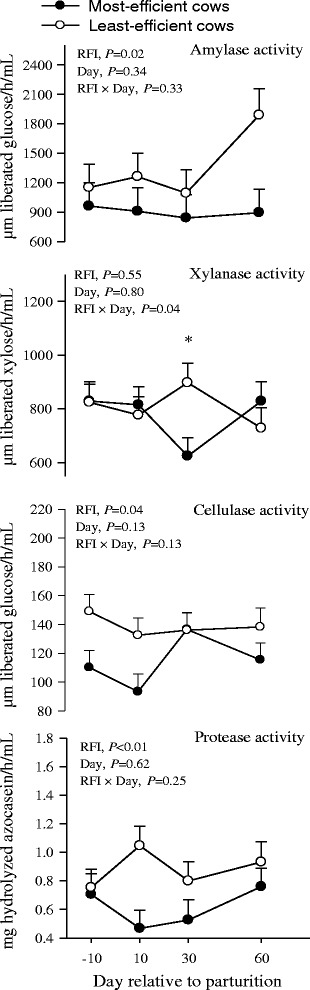
Activities of amylase, xylanase, cellulase, and protease in rumen contents from the most- and the least-efficient multiparous Holstein dairy cows during the peripartal period. Significant differences (P < 0.05) between RFI groups at a given time point are denoted with an asterisk (*)
Discussion
Although RFI calculations account for BW changes to determine individual feed efficiency independent of changes in BW during the feeding period, published RFI studies in dairy cows have been conducted during mid-lactation where minimal changes in BW occur [31]. Therefore, exploring the physiological differences between the most- and the least-efficient cows during the peripartal period and early lactation when changes in the physiology and metabolism of the dairy cow affect DMI, BW, and FCM appears warranted in the context of assessing the usefulness of RFI-based selection and performance during negative energy and protein balance.
The lack of overall effect of RFI on DMI between wk − 4 to 3 relative to calving or on FCM during the first 60 DIM suggests that RFI per se was not associated with measures of performance during a period when energy and protein balance are at a nadir. It could be possible that the metabolic and immune challenges cows experience during the peripartal period [9, 32] exert some level of control on DMI in the least-efficient cows while the most-efficient cows are able to maintain or decrease DMI within a narrow margin such that marked differences among groups are difficult to detect statistically. The fact that the most-efficient compared with least-efficient cows consumed on average 2.6 kg DMI/d less by wk 4 postpartum indicates that once “stressors” (e.g. proinflammatory cytokines, plasma free fatty acids, hydroxybutyrate) were not impinging on the cow’s ability to achieve their efficiency potential, the most-efficient cows restored the ability to utilize feed more efficiently. Succinivibrio dextrinosolvens and Megaspheara elsdenii bacteria are involved in propionate production in the rumen [33, 34]. It is well-known that propionate is vital for cow health and milk production since it serves as the main precursor for hepatic gluconeogenesis [35]. Therefore, although ruminal VFA concentration was not measured in this study, we speculate that the greater abundance of Succinivibrio dextrinosolvens and Megaspheara elsdenii in the most-efficient cows would have increased ruminal propionate production and its availability to the animal for productive purposes. This idea is supported by previous data with mid-lactation dairy cows [36] in which the most-efficient versus least-efficient cows had greater concentrations of propionate and higher propionate:acetate ratio in ruminal fluid. Megaspheara elsdenii is also known for its ability to remove lactate from the rumen environment, thus, Megaspheara elsdenii plays a vital role in preventing lactic acidosis [37]. In contrast, Streptococcus bovis is a lactate producer, and can stimulate lactic acidosis with potential negative effects on the ruminal epithelium [38]. The tendency for the most-efficient cows to have greater relative abundance of Megaspheara elsdenii and lower Streptococcus bovis from d − 10 to d 60 relative to parturition suggests that the most-efficient cows would have been at a lower risk of developing acidosis.
Fibrobacter succinogenes is equipped with various polysaccharide-degrading enzymes able to ferment cellulose primarily to succinic acid and to a lesser extent to acetic and formic acids, rendering this microorganism among the most-active and predominant bacteria involved in fiber degradation in the rumen [39, 40]. Thus, the tendency for greater abundance of this species in the most-efficient cows suggests that fiber digestion and feed utilization contribute to higher feed efficiency. This notion is further supported by data from recent studies [41, 42] indicating that the most-efficient bulls and heifers had higher NDF and DM digestibility compared with the least-efficient cattle.
Butyrivibrio proteoclasticus is a fibrolytic bacterium able to breakdown xylan, and produce butyrate [43]. Prevotella bryantii is a succinate producer that ferments hemicellulose, pectin, peptides, and amino acids [44–46], whereas Succinimonas amylolytica can ferment α-linked glucose molecules such as maltose, dextrin, and starch [47]. Thus, the lower abundance of Butyrivibrio proteoclasticus, Prevotella bryantii, and Succinimonas amylolytica coupled with greater abundance of propionate producers such as Succinivibrio dextrinosolvens and Megaspheara elsdenii in the most-efficient cows around parturition seems to support the notion that these cows were able to shift the ruminal fermentation pathways in a way that enhanced the production of glucogenic precursors and reduced dietary energy losses. The overall greater relative abundance of Eubacterium ruminantium, Megaspheara elsdenii, Prevotella bryantii, Selenomonas ruminantium, and Streptococcus bovis after parturition in both RFI groups could be explained in part by the gradual increase in DMI and the switch from a higher-forage diet prepartum to a higher-concentrate diet postpartum [13, 34].
The fact that there was a greater total copy number of the 16S rRNA gene, an indicator of total bacterial density in the rumen, in the most-efficient cows at d − 10 compared with postpartum times indicated that bacterial species other than the 10 evaluated likely proliferated in the most-efficient cows before calving. It is possible that such changes contributed to the improved feed efficiency, e.g., enhancing ruminal fermentation and fiber digestibility. This speculation is in line with recent findings reported by Bonilha et al. [41] who detected greater NDF and DM digestibility in the most-efficient bulls. It is also possible that the greater population of total ruminal bacteria observed in the most-efficient cows would have increased the production and outflow of total microbial mass from the rumen to the small intestine, allowing greater availability of amino acids for absorption and utilization, despite the lower DMI. This idea is supported by previous work demonstrating that stimulating bacterial growth in the rumen increases microbial protein synthesis [48, 49].
The lower activity of amylase, cellulase, and protease in the most-efficient cows from d − 10 to d 60 around parturition could be taken as indication that digestive function in the rumen might have been curtailed. However, several studies reported that efficient beef bulls and heifers had greater digestibility of dry matter, organic matter, NDF, protein, and total digestible nutrients [41, 42]. This apparent discrepancy seems to suggest that microbial digestive enzymes per se may not reflect the actual capacity for feed digestion in feed-efficient cattle. However, the shifts in ruminal bacteria and digestive enzymes observed in the most-efficient cows in the current study could be associated with the reduction in feed intake because decreasing DMI would slow down the rumen passage rate [50], allowing more time for microbes to degrade dietary fiber in the rumen which may explain the improvement in feed digestibility in feed-efficient cattle [41, 42]. Some studies reported that decreasing rumen passage rate is associated with increased energy costs of maintenance for rumen microbes [51, 52], hence, potentially decreasing the production of digestive enzymes.
Conclusions
Results indicate that better feed efficiency in dairy cows after calving could be attributed, at least in part, to shifts in ruminal bacteria and digestive enzyme activities during the peripartal period and early lactation. Future studies on the association between ruminal parameters such as feed retention time, passage rate, and microbial metabolic functions in cows divergent for RFI during the peripartal period are warranted.
Additional file
Preparation of rumen contents for enzyme activities. (DOCX 16 kb)
Acknowledgements
Ahmed A. Elolimy was supported by a scholarship from Missions Sector, Higher Education Ministry, Egypt. Fernanda Batistel was partially supported by a fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazilian Ministry of Education, Brasília, Brazil) and by Hatch funds under project ILLU-538-914, National Institute of Food and Agriculture (Washington, DC). We thank Perdue AgriBusiness (Salisbury, MD) for the donation of ProVAAl AADvantage during the course of the experiment.
Funding
Portions of the work were supported by Hatch funds under project ILLU-538-914, National Institute of Food and Agriculture (Washington, DC).
Availability of data and materials
The datasets during and/or analyzed during the current study available from the corresponding authors on reasonable request.
Abbreviations
- ADF
Acid detergent fiber
- BW
Body weight
- DMI
Dry matter intake
- FCM
Fat-corrected milk
- NDF
Neutral detergent fiber
- NFC
Non-fiber carbohydrates
- RBB + C
Repeated bead-beating plus column
Authors’ contributions
JJL designed the experiments; FB, AAE and JMA conceived the animal experiments; AAE and MAI conceived and performed the analyses; AAE wrote the manuscript. All authors read and approved the final version of the manuscript.
Authors’ information
Ahmed A. Elolimy is PhD candidate, University of Illinois, Urbana, Illinois, 61801, USA. José M. Arroyo is Assistant Professor, Departamento de Nutrición Animal, Instituto de Producción Animal, Facultad de Veterinaria, Universidad de la Republica, San José, Uruguay. Fernanda Batistel is PhD candidate, University of Illinois, Urbana, Illinois, 61801, USA. Michael A Iakiviak is PhD candidate, University of Illinois, Urbana, Illinois, 61801, USA. Juan J. Loor is Associate Professor in the Department of Animal Sciences, University of Illinois, Urbana, Illinois, 61801, USA.
Ethics approval
All procedures for this study (protocol no. 14270) were approved by the Institutional Animal Care and Use Committee of the University of Illinois.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40104-018-0258-9) contains supplementary material, which is available to authorized users.
Contributor Information
Ahmed A. Elolimy, Email: elolimy2@illinois.edu
José M. Arroyo, Email: chemaarroyo@gmail.com
Fernanda Batistel, Email: batiste2@illinois.edu.
Michael A. Iakiviak, Email: miakivi2@illinois.edu
Juan J. Loor, Email: jloor@illinois.edu
References
- 1.Shetty N, Lovendahl P, Lund MS, Buitenhuis AJ. Prediction and validation of residual feed intake and dry matter intake in Danish lactating dairy cows using mid-infrared spectroscopy of milk. J Dairy Sci. 2017;100:253–264. doi: 10.3168/jds.2016-11609. [DOI] [PubMed] [Google Scholar]
- 2.Rathbun FM, Pralle RS, Bertics SJ, Armentano LE, Cho K, Do C. Relationships between body condition score change, prior mid-lactation phenotypic residual feed intake, and hyperketonemia onset in transition dairy cows. J Dairy Sci. 2017;100:3685–3696. doi: 10.3168/jds.2016-12085. [DOI] [PubMed] [Google Scholar]
- 3.Xi YM, Wu F, Zhao DQ, Yang Z, Li L, Han ZY. Biological mechanisms related to differences in residual feed intake in dairy cows. Animal. 2016;10:1311–1318. doi: 10.1017/S1751731116000343. [DOI] [PubMed] [Google Scholar]
- 4.Potts SB, Boerman JP, Lock AL, Allen MS, VandeHaar MJ. Relationship between residual feed intake and digestibility for lactating Holstein cows fed high and low starch diets. J Dairy Sci. 2017;100:265–278. doi: 10.3168/jds.2016-11079. [DOI] [PubMed] [Google Scholar]
- 5.Herd RM, Arthur PF. Physiological basis for residual feed intake. J Anim Sci. 2009;87:E64–E71. doi: 10.2527/jas.2008-1345. [DOI] [PubMed] [Google Scholar]
- 6.Pitta DW, Kumar S, Vecchiarelli B, Shirley DJ, Bittinger K, Baker LD, et al. Temporal dynamics in the ruminal microbiome of dairy cows during the transition period. J Anim Sci. 2014;92:4014–4022. doi: 10.2527/jas.2014-7621. [DOI] [PubMed] [Google Scholar]
- 7.Yeoman CJ, White BA. Gastrointestinal tract microbiota and probiotics in production animals. Annu Rev Anim Biosci. 2014;2:469–486. doi: 10.1146/annurev-animal-022513-114149. [DOI] [PubMed] [Google Scholar]
- 8.Jewell KA, McCormick CA, Odt CL, Weimer PJ, Suen G. Ruminal Bacterial Community Composition in Dairy Cows Is Dynamic over the Course of Two Lactations and Correlates with Feed Efficiency. Appl Environ Microbiol. 2015;81:4697–4710. doi: 10.1128/AEM.00720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batistel F, Osorio JS, Tariq MR, Li C, Caputo J, Socha MT, et al. Peripheral leukocyte and endometrium molecular biomarkers of inflammation and oxidative stress are altered in peripartal dairy cows supplemented with Zn, Mn, and Cu from amino acid complexes and Co from Co glucoheptonate. J Anim Sci Biotechnol. 2017;8:33. doi: 10.1186/s40104-017-0163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertoni G, Trevisi E, Han X, Bionaz M. Effects of inflammatory conditions on liver activity in puerperium period and consequences for performance in dairy cows. J Dairy Sci. 2008;91:3300–3310. doi: 10.3168/jds.2008-0995. [DOI] [PubMed] [Google Scholar]
- 11.Trevisi E, Amadori M, Cogrossi S, Razzuoli E, Bertoni G. Metabolic stress and inflammatory response in high-yielding, periparturient dairy cows. Res Vet Sci. 2012;93:695–704. doi: 10.1016/j.rvsc.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Derakhshani H, Tun HM, Cardoso FC, Plaizier JC, Khafipour E, Loor JJ. Linking Peripartal Dynamics of Ruminal Microbiota to Dietary Changes and Production Parameters. Front Microbiol. 2016;7:2143. doi: 10.3389/fmicb.2016.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minuti A, Palladino A, Khan MJ, Alqarni S, Agrawal A, Piccioli-Capelli F, et al. Abundance of ruminal bacteria, epithelial gene expression, and systemic biomarkers of metabolism and inflammation are altered during the peripartal period in dairy cows. J Dairy Sci. 2015;98:8940–8951. doi: 10.3168/jds.2015-9722. [DOI] [PubMed] [Google Scholar]
- 14.Scharen M, Kiri K, Riede S, Gardener M, Meyer U, Hummel J, et al. Alterations in the Rumen Liquid-, Particle- and Epithelium-Associated Microbiota of Dairy Cows during the Transition from a Silage- and Concentrate-Based Ration to Pasture in Spring. Front Microbiol. 2017;8:744. doi: 10.3389/fmicb.2017.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batistel F, Arroyo JM, Bellingeri A, Wang L, Saremi B, Parys C, et al. Ethyl-cellulose rumen-protected methionine enhances performance during the periparturient period and early lactation in Holstein dairy cows. J Dairy Sci. 2017;100:7455–7467. doi: 10.3168/jds.2017-12689. [DOI] [PubMed] [Google Scholar]
- 16.NRC. Nutrient requirements of beef cattle. 7th rev. ed. . National Academy Press, Washington, DC; 2000.
- 17.AOAC International . Official Methods of Analysis., 17 edition. Association of Official Analytical Chemists: Arlington, VA; 2000. [Google Scholar]
- 18.Van Soest PJ, Robertson JB, Lewis BA. Methods for Dietary Fiber, Neutral Detergent Fiber, and Nonstarch Polysaccharides in Relation to Animal Nutrition. J Dairy Sci. 1991;74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- 19.Nocek JE. Bovine acidosis: implications on laminitis. J Dairy Sci. 1997;80:1005–1028. doi: 10.3168/jds.S0022-0302(97)76026-0. [DOI] [PubMed] [Google Scholar]
- 20.Paz HA, Anderson CL, Muller MJ, Kononoff PJ, Fernando SC. Rumen Bacterial Community Composition in Holstein and Jersey Cows Is Different under Same Dietary Condition and Is Not Affected by Sampling Method. Front Microbiol. 2016;7:1206. doi: 10.3389/fmicb.2016.01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M, Park T, Yu Z. Metagenomic investigation of gastrointestinal microbiome in cattle. Asian-Australas J Anim Sci. 2017;30:1515–1528. doi: 10.5713/ajas.17.0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen JS, Chai Z, Song LJ, Liu JX, Wu YM. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J Dairy Sci. 2012;95:5978–5984. doi: 10.3168/jds.2012-5499. [DOI] [PubMed] [Google Scholar]
- 23.Terre M, Castells L, Fabregas F, Bach A. Short communication: Comparison of pH, volatile fatty acids, and microbiome of rumen samples from preweaned calves obtained via cannula or stomach tube. J Dairy Sci. 2013;96:5290–5294. doi: 10.3168/jds.2012-5921. [DOI] [PubMed] [Google Scholar]
- 24.Ramos-Morales E, Arco-Pérez A, Martín-García AI, Yáñez-Ruiz DR, Frutos P, Hervás G. Use of stomach tubing as an alternative to rumen cannulation to study ruminal fermentation and microbiota in sheep and goats. Anim Feed Sci Technol. 2014;198:57–66. doi: 10.1016/j.anifeedsci.2014.09.016. [DOI] [Google Scholar]
- 25.Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques. 2004;36:808–812. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]
- 26.Maeda H, Fujimoto C, Haruki Y, Maeda T, Kokeguchi S, Petelin M, et al. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol. 2003;39:81–86. doi: 10.1016/S0928-8244(03)00224-4. [DOI] [PubMed] [Google Scholar]
- 27.Fliegerova K, Tapio I, Bonin A, Mrazek J, Callegari ML, Bani P, et al. Effect of DNA extraction and sample preservation method on rumen bacterial population. Anaerobe. 2014;29:80–84. doi: 10.1016/j.anaerobe.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 28.Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. 2009;101:541–550. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- 29.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou M, Hernandez-Sanabria E, Guan LL.Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl Environ Microbiol. 2009, 75:6524-33. [DOI] [PMC free article] [PubMed]
- 31.Tempelman RJ, Spurlock DM, Coffey M, Veerkamp RF, Armentano LE, Weigel KA, et al. Heterogeneity in genetic and nongenetic variation and energy sink relationships for residual feed intake across research stations and countries. J Dairy Sci. 2015;98:2013–2026. doi: 10.3168/jds.2014.8510. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z, Bulgari O, Vailati-Riboni M, Trevisi E, Ballou MA, Cardoso FC, et al. Rumen-protected methionine compared with rumen-protected choline improves immunometabolic status in dairy cows during the peripartal period. J Dairy Sci. 2016;99:8956–8969. doi: 10.3168/jds.2016-10986. [DOI] [PubMed] [Google Scholar]
- 33.Bryant MP, Small N. Characteristics of two new genera of anaerobic curved rods isolated from the rumen of cattle. J Bacteriol. 1956;72:22–26. doi: 10.1128/jb.72.1.22-26.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernando SC, Purvis HT, 2nd, Najar FZ, Sukharnikov LO, Krehbiel CR, Nagaraja TG, et al. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl Environ Microbiol. 2010;76:7482–7490. doi: 10.1128/AEM.00388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell JB, Wilson DB. Why are ruminal cellulolytic bacteria unable to digest cellulose at low pH? J Dairy Sci. 1996;79:1503–1509. doi: 10.3168/jds.S0022-0302(96)76510-4. [DOI] [PubMed] [Google Scholar]
- 36.Shabat SK, Sasson G, Doron-Faigenboim A, Durman T, Yaacoby S, Berg Miller ME, et al. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016;10:2958–2972. doi: 10.1038/ismej.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meissner HH, Henning PH, Horn CH, Leeuw KL, Hagg FM, Fouche G. Ruminal acidosis: A review with detailed reference to the controlling agent Megasphaera elsdenii NCIMB 41125. S Afr J Anim Sci. 2010;40:79–100. doi: 10.4314/sajas.v40i2.57275. [DOI] [Google Scholar]
- 38.Aslan V, Thamsborg SM, Jorgensen RJ, Basse A. Induced acute ruminal acidosis in goats treated with yeast (Saccharomyces cerevisiae) and bicarbonate. Acta Vet Scand. 1995;36:65–77. doi: 10.1186/BF03547703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ransom-Jones E, Jones DL, McCarthy AJ, McDonald JE. The Fibrobacteres: an important phylum of cellulose-degrading bacteria. Microb Ecol. 2012;63:267–281. doi: 10.1007/s00248-011-9998-1. [DOI] [PubMed] [Google Scholar]
- 40.Neumann AP, McCormick CA, Suen G. Fibrobacter communities in the gastrointestinal tracts of diverse hindgut-fermenting herbivores are distinct from those of the rumen. Environ Microbiol. 2017;19(9):3768-3783 [DOI] [PMC free article] [PubMed]
- 41.Bonilha SF, Branco RH, Mercadante ME, JN DSGC, Monteiro FM, Ribeiro EG. Digestion and metabolism of low and high residual feed intake Nellore bulls. Trop Anim Health Prod. 2017;49:529–535. doi: 10.1007/s11250-017-1224-9. [DOI] [PubMed] [Google Scholar]
- 42.McDonnell RP, Hart KJ, Boland TM, Kelly AK, McGee M, Kenny DA. Effect of divergence in phenotypic residual feed intake on methane emissions, ruminal fermentation, and apparent whole-tract digestibility of beef heifers across three contrasting diets. J Anim Sci. 2016;94:1179–1193. doi: 10.2527/jas.2015-0080. [DOI] [PubMed] [Google Scholar]
- 43.Moon CD, Pacheco DM, Kelly WJ, Leahy SC, Li D, Kopecny J, et al. Reclassification of Clostridium proteoclasticum as Butyrivibrio proteoclasticus comb. nov., a butyrate-producing ruminal bacterium. Int J Syst Evol Microbiol. 2008;58:2041–2045. doi: 10.1099/ijs.0.65845-0. [DOI] [PubMed] [Google Scholar]
- 44.Matsui H, Ogata K, Tajima K, Nakamura M, Nagamine T, Aminov RI, et al. Phenotypic characterization of polysaccharidases produced by four Prevotella type strains. Curr Microbiol. 2000;41:45–49. doi: 10.1007/s002840010089. [DOI] [PubMed] [Google Scholar]
- 45.Miyazaki K, Martin JC, Marinsek-Logar R, Flint HJ. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B(1)4. Anaerobe. 1997;3:373–381. doi: 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- 46.Ling JR, Armstead IP. The in vitro uptake and metabolism of peptides and amino acids by five species of rumen bacteria. J Appl Bacteriol. 1995;78:116–124. doi: 10.1111/j.1365-2672.1995.tb02831.x. [DOI] [PubMed] [Google Scholar]
- 47.Li F, Cao Y, Liu N, Yang X, Yao J, Yan D. Subacute ruminal acidosis challenge changed in situ degradability of feedstuffs in dairy goats. J Dairy Sci. 2014;97:5101–5109. doi: 10.3168/jds.2013-7676. [DOI] [PubMed] [Google Scholar]
- 48.Phesatcha K, Wanapat M. Tropical legume supplementation influences microbial protein synthesis and rumen ecology. J Anim Physiol Anim Nutr (Berl). 2017;101:552–562. doi: 10.1111/jpn.12458. [DOI] [PubMed] [Google Scholar]
- 49.Clark JH, Klusmeyer TH, Cameron MR. Microbial Protein Synthesis and Flows of Nitrogen Fractions to the Duodenum of Dairy Cows1. J Dairy Sci. 1992;75:2304–2323. doi: 10.3168/jds.S0022-0302(92)77992-2. [DOI] [PubMed] [Google Scholar]
- 50.Colucci PE, Chase LE, Van Soest PJ. Feed Intake, Apparent Diet Digestibility, and Rate of Particulate Passage in Dairy Cattle. J Dairy Sci. 1982;65:1445–1456. doi: 10.3168/jds.S0022-0302(82)82367-9. [DOI] [Google Scholar]
- 51.Firkins JL. Maximizing microbial protein synthesis in the rumen. J Nutr. 1996;126:1347s–1354s. doi: 10.1093/jn/126.suppl_4.1347S. [DOI] [PubMed] [Google Scholar]
- 52.Sniffen CJ, Robinson PH. Protein and fiber digestion, passage, and utilization in lactating cows. Microbial growth and flow as influenced by dietary manipulations. J Dairy Sci. 1987;70:425–441. doi: 10.3168/jds.S0022-0302(87)80027-9. [DOI] [PubMed] [Google Scholar]
- 53.Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol. 2007;75:165–174. doi: 10.1007/s00253-006-0802-y. [DOI] [PubMed] [Google Scholar]
- 54.Khafipour E, Li S, Plaizier JC, Krause DO. Rumen Microbiome Composition Determined Using Two Nutritional Models of Subacute Ruminal Acidosis. Appl Environ Microbiol. 2009;75:7115–7124. doi: 10.1128/AEM.00739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Preparation of rumen contents for enzyme activities. (DOCX 16 kb)
Data Availability Statement
The datasets during and/or analyzed during the current study available from the corresponding authors on reasonable request.


