Figure 7. Autoinhibition and allosteric activation of the TEC kinases.
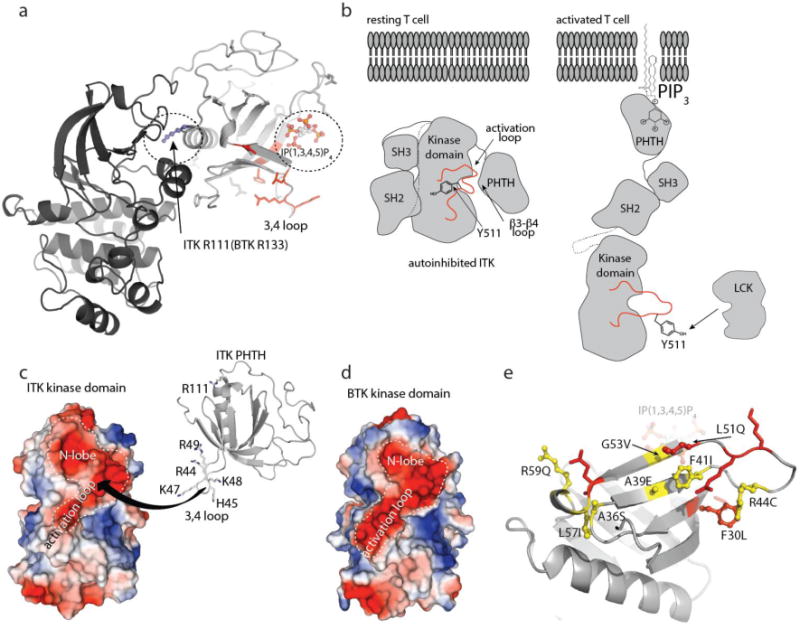
(a) The structure of tethered BTK PHTH-Kinase domain (PDB ID: 4Y93) where the ITK PHTH domain has been superimposed with the BTK PHTH domain (BTK PHTH not shown for clarity). R133 in the BTK PHTH domain makes contact with the BTK kinase domain. R111 is the corresponding residue in the ITK PHTH domain but mutation of R111 does not affect the ITK PHTH/Kinase interaction. The ITK PHTH interface residues located in and around the β3-β4 loop (colored red) are not located at the interface with the kinase domain defined in the crystal structure for the tethered BTK domains but are close to the IP(1,3,4,5)P4 binding site (circled). (b) left, Model of closed, autoinhibited ITK in a resting T cell (arrangement of the SH3 and SH2 domains is based on the BTK crystal structure of the SH3-SH2-Kinase fragment (4XI2)). Right, model of an open form of ITK (based on SAXS structure of full length BTK35 in the presence of membrane anchored PIP3 revealing Y511 for phosphorylation by LCK. (c) The ITK kinase domain presents a large negatively charged surface that spans the N-lobe and activation loop and may serve as a binding site for the positively charged region of ITK PHTH defined in this study. The highly basic stretch of residues on the β3-β4 loop are shown and labeled. (d) The BTK kinase domain shares the acidic patch across the N-lobe and activation loop. (e) Autoinhibitory ITK PHTH surface defined in this study. Residues shown in red are those side chains mutated in the current study that have an effect on the inhibitory interaction with the ITK kinase domain, those in yellow, ball and stick are ITK residues listed in the COSMIC database. F30 is shown in red with ball and stick to indicate that it both affects the PHTH/Kinase interface and is mutated in the database of somatic mutations in cancer.
