Abstract
Rationale: Twist is a key transcription factor for induction of epithelial-mesenchymal transition (EMT), which promotes cell migration, invasion, and cancer metastasis, confers cancer cells with stem cell-like characteristics, and provides therapeutic resistance. However, the functional roles and targeted genes of Twist in EMT and cancer progression remain elusive.
Methods: The potential targeted genes of Twist were identified from the global transcriptomes of T47D/Twist cells by microarray analysis. EMT phenotype was detected by western blotting and immunofluorescence of marker proteins. The dual-luciferase reporter and chromatin immunoprecipitation assays were employed to observe the direct transcriptional induction of ROR1 by Twist. A lung metastasis model was used to study the pro-metastatic role of Twist and ROR1 by injecting MDA-MB-231 cells into tail vein of nude mice. Bio-informatics analysis was utilized to measure the metastasis-free survival of breast cancer patients.
Results: Twist protein was proved to directly activate the transcription of ROR1 gene, a receptor of Wnt5a in non-canonical WNT signaling pathway. Silencing of ROR1 inhibited EMT process, cell migration, invasion, and cancer metastasis of basal-like breast cancer (BLBC) cells. Knockdown of ROR1 also ameliorated the pro-metastatic effect of Twist. Furthermore, analyses of clinical specimens indicated that high expression of both ROR1 and Twist tightly correlates with poor metastasis-free survival of breast cancer patients.
Conclusion: ROR1 is a targeted gene of Twist. Twist/ROR1 signaling is critical for invasion and metastasis of BLBC cells.
Keywords: ROR1, Twist, EMT, tumor metastasis, basal-like breast cancer (BLBC)
Introduction
Breast cancer is the most common malignancy in women worldwide; about 90% of breast cancer deaths are the result of metastasis 1. Breast cancer is a heterogeneous disease in terms of tumor histology, clinical presentation, and response to therapy. There are four major subtypes based on gene expression profiling: luminal A, luminal B, ErbB2, and basal-like 1, 2. BLBC cells are characterized by the lack of expression of estrogen receptor (ER), progesterone receptor (PR), and epidermal growth factor receptor 2 (HER2) and positive expression of basal markers (Cytokeratin 5/6 (CK5/6) and CK14) 3. BLBC cells undergo epithelial-mesenchymal transition (EMT) and highly express several basal markers 4. Several transcriptional factors have been reported to be involved in the regulation of EMT, and Twist and Snail are two major transcriptional factors that contribute to the induction of EMT process 5-8. During mesoderm development in Drosophila, Snail functions as a transcriptional repressor to prevent expression of genes that belong to ectoderm, whereas Twist serves as a transcriptional activator to induce mesodermal gene expression 9, 10. These results suggest that Snail and Twist work synergistically to induce EMT. A previous study has reported the mechanism underlying gene transcriptional activation by Twist 2; however, the functional roles of Twist in EMT and cancer progression are still not fully characterized.
Receptor tyrosine kinases (RTKs) are crucial to cell differentiation, proliferation, and migration. The aberrant expression of RTKs has been observed in different types of cancer cells 11-14. Therefore, the identification and subsequent targeting of specific RTKs that overexpress uniquely in malignant cells, but not in normal cells, is crucial to anticancer therapies 15. Receptor tyrosine kinase-like orphan receptors (RORs), as members of the RTKs family, have been recently brought into focus in the scientific community for the diagnosis and treatment of different cancer types 4, 13, 15, 16. In humans, the expression of ROR1 is predominantly limited to embryo and fetus and it is not observed in adult tissues; however, the expression profile of ROR1 in cancer is quite different 15. Similar to altered expression and activity of RTKs in different cancer types, the overexpression of ROR1 has been found in malignant cancers, such as chronic lymphocytic leukemia (CLL) 15, lung adenocarcinoma 13, and ovarian cancer 17. High expression of ROR1 in tumor cells is associated with enhanced cell migration, EMT, increased risk of tumor relapse and metastasis, as well as poor prognosis 18-20. Recently, ROR1 has been identified as a receptor for Wnt5a, which induces non-canonical Wnt signaling, potentially leading to enhanced tumor cell survival, cell movement, and polarity during development. Silencing of ROR1 in metastatic breast cancer cell lines attenuated expression of EMT markers, and impaired their migration/invasion capacity as well as metastatic potential 21-23. Therefore, ROR1 may serve as a potential therapeutic target for patients with aggressive tumor 24. However, its manner of transcriptional regulation and how tumor cells maintain high levels of ROR1 still remain elusive.
In this study, we identified that Twist serves as a key factor for the transcriptional activation of ROR1 in BLBC cells by occupying the ROR1 gene promoter, and demonstrated that increased ROR1 expression is responsible for Twist-mediated EMT and breast cancer metastasis.
Methods
Microarray data analysis
Microarray data of Twist expression in T-47D cells was deposited in the Gene expression Omnibus database with the accession number GSE53222. Raw data were normalized by Affymetrix Expression Console Software.
Cell culture
T47D, MDA-MB231, Hs578T, and 293T cells were obtained from ATCC. T47D cell line was cultured in RPMI-1640 (Life Technologies) with 10% FBS (Life Technologies) and 0.2 U/mL bovine insulin (Sigma). Hs578T was maintained in DMEM (Life Technologies) supplemented with 10% FBS (Life Technologies) and 0.01 mg/mL bovine insulin (Sigma). MDA-MB231 and 293T were cultured in DMEM (Life Technologies) with 10% FBS (Sigma). All cell lines were incubated in a humidified incubator at 37 °C with 5% CO2.
Plasmids construction
The ROR1 promoter was purchased from Yingrun Biotechnology Inc. (Changsha, China). The sequences used for cloning ROR1 promoter mutants are shown in Supplementary Material. All plasmids above were verified by DNA sequencing. shRNA targeting ROR1 and Twist were purchased from Sigma.
shROR1#4,5'-CCGGGCACCGTCTATATGGAGTCTTCTCGAGAAGACTCCATATAGACGGTGCTTTTT-3'; shROR1#5,5'-CCGGCTTTACTAGGAGACGCCAATACTCGAGTATTGGCGTCTCCTAGTAAAGTTTTT-3'; shROR1#8,5'-CCGGCGGAGAGCAACTTCATGTAAACTCGAGTTTACATGAAGTTGCTCTCCGTTTTT-3'; shTwist#1,5'-CCGGCCTGAGCAACAGCGAGGAAGACTCGAGTCTTCCTCGCTGTTGCTCAGGTTTTT-3'; shTwist#2,5'-CCGGTCCGCAGTCTTACGAGGAGCTCTCGAGAGCTCCTCGTAAGACTGCGGATTTTT-3'.
RNA extraction and qRT-PCR
Total RNA isolation, qRT-PCR, and the quantification of target gene expression were performed as previously described 25. Briefly, total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized using the Revert AidTM First Strand cDNA Synthesis Kit (MBI Fermentas). The primers used to amplify the indicated genes are shown in Supplementary Material.
Immunofluorescence
Cells seeded in confocal dishes were fixed with 4% paraformaldehyde and then kept stable in 0.2% Triton X-100 for 10 min to rupture the cell membranes. Following three PBS washings, non-specific antigen-binding sites were blocked by 2% BSA for 30 min. Cells were then incubated with anti-Twist (Merck Millipore, ABD29,1:200), anti-ROR1 (Santa cruz, sc-130867, 1;100) and anti-E-cadherin (BD, 610181, 1:100) antibodies overnight at 4 °C. After washing, cells were incubated with secondary antibody (Invitrogen) for 60 min and the nuclei were stained with DAPI (Invitrogen) for 2 min, which was washed with PBS later. Cells were protected from light before observation with a fluorescence microscope.
Western blotting
Western blotting was performed as previously described 25. Cells were collected and lysed by RIPA buffer (150 mM NaCl, 0.5% EDTA, 50 mM Tris, 0.5% NP40) and centrifuged for 20 min at 14,000 x g and 4 °C. Fifty micrograms of harvested total protein was loaded and separated on the 8% SDS-polyacrylamide gradient gel.
The proteins were then transferred onto polyvinylidene difluoride membranes and blocked with 5% non-fat milk for 2 h at room temperature. The membranes were incubated with primary antibody and horseradish peroxidase-conjugated secondary antibody, and proteins were then detected using an ECL chemiluminescence system (Pierce). Primary antibodies used were as follows: Twist (Abcam, ab50581, 1:1000), ROR1 (Santa cruz, sc-130867, 1:1000), E-Cadherin (CST, 3195, 1:1000), N-Cadherin (CST, 13116, 1:1000), Vimentin (CST, 5741,1:1000), ZO-1 (CST, 8193, 1:1000) and β-Actin (CST, 4970, 1:1000).
Transwell migration and invasion assay
Cells were seeded into Boyden chambers containing 24-well Transwell plates with a pore size of 8 µm (BD Bioscience). The upper chamber was either left uncoated for the migration assay or precoated with 50 µL of 1:8 diluted Matrigel (BD Bioscience) for the invasion assay. For the migration assay, the Hs578T (4×104) and MDA-MB231 (5×104) cells were seeded into the upper chamber; for the invasion assay, 8×104 Hs578T and 1×105 MDA-MB231 cells were seeded into the upper chamber. The upper chamber was filled with 200 µL serum-free specified medium whereas the lower chamber was filled with specified medium containing 10% FBS. Following incubation for 16 h or 24 h at 37 °C, cells that had invaded into the lower chamber were fixed with 4% paraformaldehyde and stained with crystal violet for 1 h at room temperature, and counted in five randomly-selected microscopic fields. All these experiments were performed in triplicate.
Dual-luciferase reporter assay
Cells were seeded in 12-well plates until growth to 80% confluence. Then, flag-Twist as well as its controls were co-transfected with ROR1, ROR1 mutants' promoter constructs and internal control pRK-TK into the indicated cells. After transaction for 48 h, the luciferase activity was measured using a Dual-luciferase Assay kit (Promega). Reporter luciferase activity was normalized to Renilla luciferase activity.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation assay was performed as described by the ChIP kit (Millipore, 17-10085 & 17-10086) 25. Briefly, each cell line was seeded in a 15 cm plate and allowed to grow to 70-80% confluence. To fix cells, complete cell fixative solution (10% of the volume of the growth medium) was added to the existing culture medium. The fixation reaction was stopped by adding stop solution (5% of the volume of the growth medium) to the existing culture medium. The cells were collected by centrifugation, and the nuclear pellet was resuspended in chromatin immunoprecipitation (ChIP) buffer. The cell lysate was subjected to sonication and then incubated with 5 mg of antibodies overnight, followed by incubation with the protein A/G agarose beads overnight at 4 ℃. Bound DNA-protein complexes were eluted, and cross-links were reversed after a series of washes. The purified DNA was resuspended in TE buffer for qRT-PCR. The primers for the indicated promoters are shown in Supplementary Material.
Immunohistochemical staining
Human tissue specimens (HBre-Duc140Sur-01) was purchased from Shanghai Outdo Biotech CO. The primary antibodies against Twist (Merck Millipore, ABD29) or ROR1 (Santa cruz, sc-130867) were diluted 1:3000 or 1:800, respectively, and then incubated at 4 °C overnight in a humidified container. Following washing three times with PBS, the section was treated with a non-biotin horseradish peroxidase detection system according to the manufacturer's instructions (Dako). The tissue samples were examined by two pathologists who were blinded to the pathological information, and immunoreactivity. For Twist and ROR1, each tissue sample was scored according to its staining intensity (0, none; 1, weak; 2, moderate; 3, strong), and the percentage of stained cells (0, 0%; 1, 1-24%; 2, 25-49%; 3, 50-74%; 4, 75-100%). The score for each tissue was calculated by multiplying the staining intensity and the percentage of stained cells, yielding a value between 0 and 12. Then, the ROC curve analysis was used to determine the immunohistochemical cut-off for high or low expression. Cancers with scores above the cut-off value were considered to have high expression of the indicated molecule and vice versa.
Statistical analysis
Data were analyzed using SPSS software version 16.0 (SPSS Inc.). The significance of differences was assessed using two-tailed Student's t-test or a χ2 test, as appropriate. The relationship between Twist expression, ROR1 expression, and the clinical pathologic parameters was examined using Pearson χ2 test. The correlations between Twist expression, ROR1 expression, and overall survival curves were assessed using Kaplan-Meier plots and compared with the log-rank test. Univariate and multivariate Cox regression analyses were used to evaluate survival data. Differences were considered significant when the p-values were < 0.05.
Animal experiments
All animal experiments were performed in accordance with protocols approved by the Research Animal Resource Center of Sun Yat-sen University. The animal ethics number of this project is L102012016050I. All animals were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). A lung metastasis model was used. Indicated cells were harvested and washed twice with PBS. Approximately 2×106 cells resuspended in 150 µL PBS were injected into the tail veins of five-week-old female athymic mice. All mice were sacrificed 10 weeks after the injection, and the lungs were harvested. The metastatic nodules in each lung were counted.
Results
Twist induces ROR1 expression during EMT
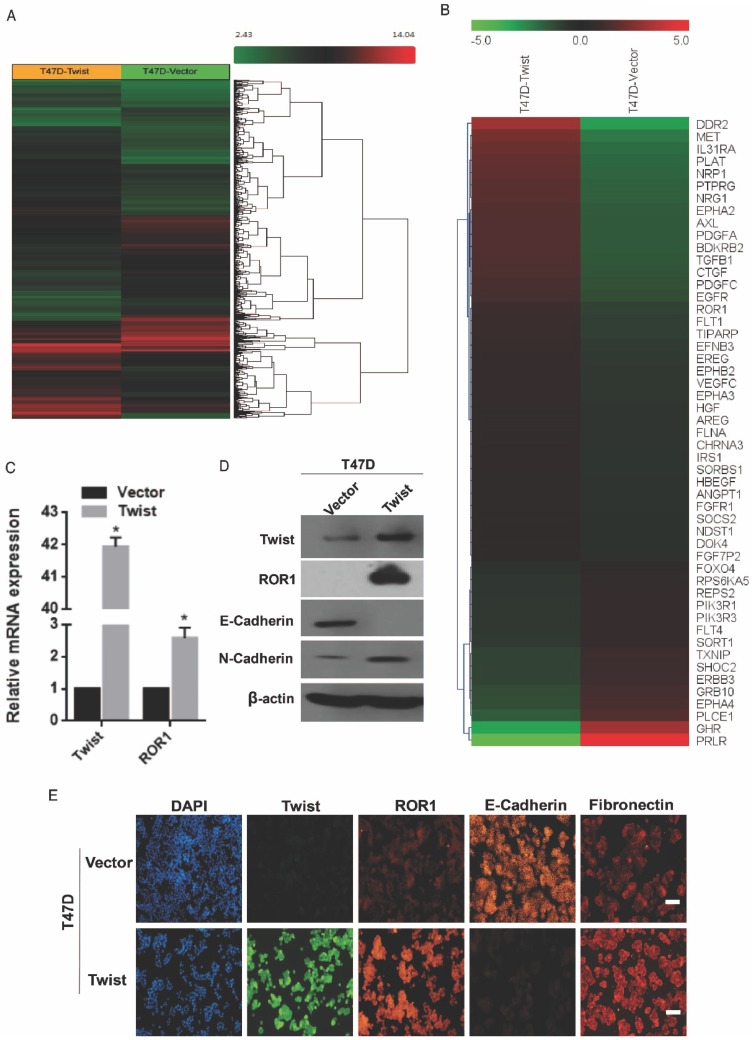
To investigate the potential targets of Twist, the global transcriptomes were analyzed in T47D cells in which Twist was overexpressed exogenously, compared with those cells transfected with vector control (Figure 1A). Intriguingly, we found expression changes of nearly 2000 genes in T47D/Twist cells that were more than 2-fold higher than those of vector control cells. Among them, 54 genes encoded transmembrane proteins. ROR1, which was initially identified as a member of the receptor tyrosine kinase family, was revealed to be one of the most altered molecules (Figure 1B). As shown in Figure 1C-D, overexpression of Twist in T47D cells promoted ROR1 expression at both mRNA and protein levels. In addition, immunofluorescence staining also indicated a positive correlation between Twist and ROR1 expression (Figure 1E and Figure S1A). Taken together, these data indicate that Twist can induce ROR1 expression by promoting its transcription during EMT in breast cancer cells.
Figure 1.
Twist promotes the expression of ROR1. (A-B) Representative heatmaps from global comparative transcriptome analysis indicating genes that are up-regulated upon Twist overexpression. (C) The relative mRNA levels of the indicated genes were normalized to the GAPDH level in the T47D cells stably transfected with pLenti6.3/V5-vector or pLenti6.3/V5-Twist as determined by qRT-PCR. The results are expressed as the mean ± SD of three independent experiments. * P < 0.05 using Student's t-test. (D) The indicated proteins were analyzed by Western blotting in T47D cells stably expressing pLenti6.3/V5-vector or pLenti6.3/V5-Twist, as indicated. (E) T47D cells stably expressing pLenti6.3/V5-vector or pLenti6.3/V5-Twist were analyzed by immunofluorescence as indicated. Scale bars, 50μm.
ROR1 is a direct targeted gene of Twist
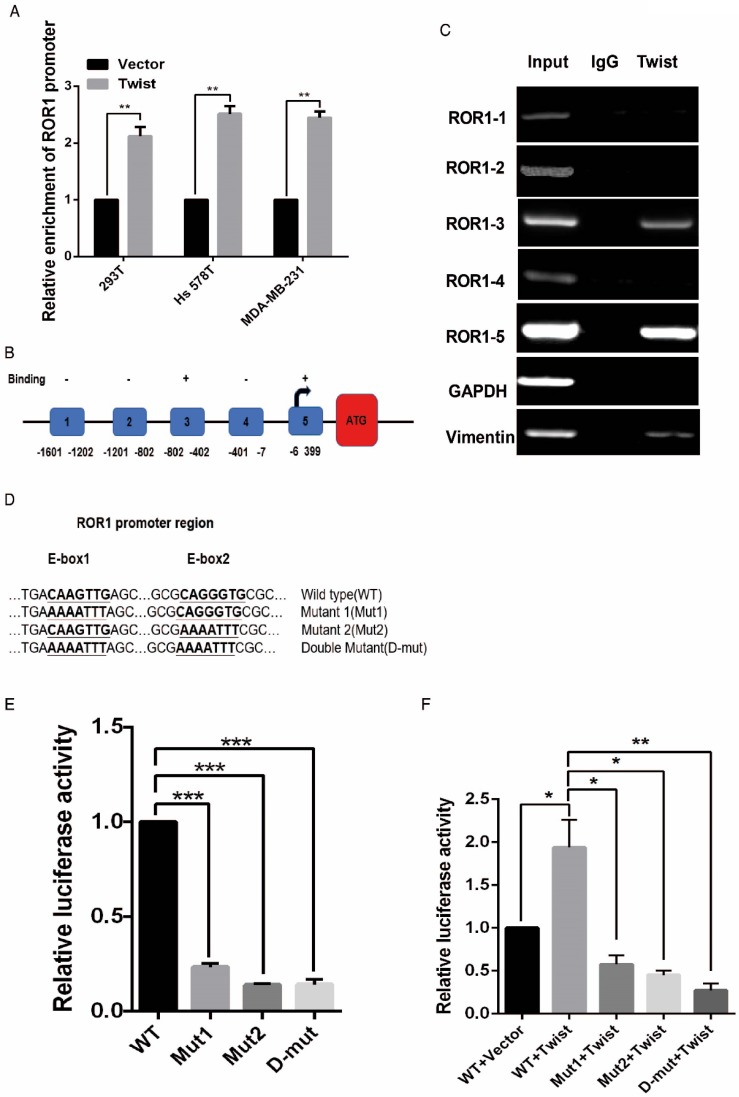
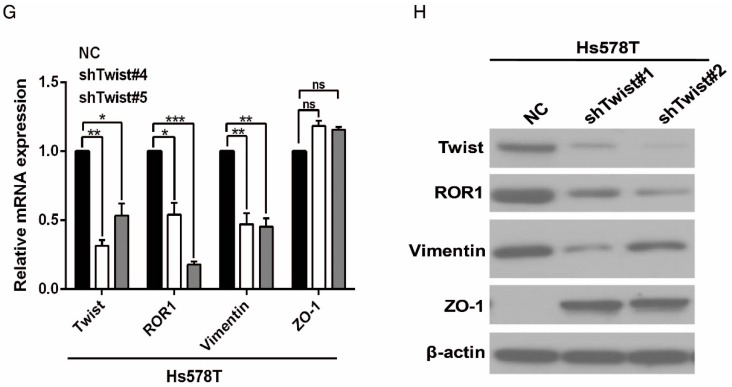
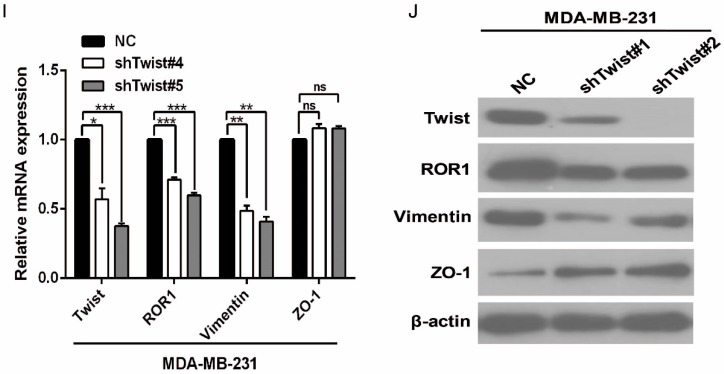
After screening a panel of breast cancer cells, only in basal-like subtypes we find high Twist and ROR1 expression (Figure S2A). To investigate whether ROR1 is directly regulated by Twist, luciferase assays were performed to measure the effect of Twist on the ROR1 promoter reporter, which was cloned into a pGL-3 basic vector. As shown in Figure 2A, Twist promotes ROR1 promoter luciferase activity in HEK293T, Hs578T, and MDA-MB231 cells. In addition, ChIP results showed that Twist binds to the region of ROR1 promoter from -802 to -402 (fragment 3) and from -6 to 399 (fragment 5); the Vimentin and GAPDH promoter were used as positive and negative controls, respectively (Figure 2B-C). We found that there was one E-box motif in fragment 3 (Figure S3A) and two E-box elongated motifs 26, 27 in fragment 5 (EL1 and EL2) (Figure 2D). Intriguingly, the promoter activity of ROR1 marginally affected by mutating E-box in fragment 3 (Figure S3B); however, its activity was greatly reduced by mutating either one or both of the two elongated E-box motifs in fragment 5 (Figure 2E). Furthermore, the elevation of ROR1 promoter activity by Twist was dependent on these two elongated E-box motifs (EL1 and EL2); as such, the effect of Twist was abolished by mutating either one of them (Figure 2F). Consistently, knockdown of Twist, which led to the expression of ZO-1 and the suppression of Vimentin, also decreased the mRNA and protein level of ROR1 in Hs578T and MDA-MB231 cell lines (Figure 2G-J). These results demonstrate that Twist can promote ROR1 transcription by occupying its promoter in BLBC cells.
Figure 2.
Twist directly upregulates ROR1 by occupying its promoter. (A) The indicated cells overexpressing empty vector or Twist were transfected with ROR1 promoter linked to luciferase, and, after 48 h, luciferase activity was assayed. (B) Illustration of the ROR1 promoter region with the indicated Twist-binding capability. "+" indicates binding and "-" indicates no binding based on the results shown in (C). (C) MDA-MB-231 cells were analyzed by ChIP using anti-Twist antibody. (D) Schematic illustration of wild-type ROR1 promoter and its mutants. (E) ROR1 promoters (wild-type or mutant) linked to luciferase were transfected into 293T cells, and, after 48 h, luciferase activity was assayed. (F) The indicated cells overexpressing empty vector or Twist were transfected with ROR1 promoter (wild-type or mutant) linked to luciferase, and, after 48 h, luciferase activity was assayed. (G, I) The indicated cells stably transfected with control or Twist shRNA were used to analyze the mRNA level of the indicated molecules by qRT-PCR. (H, J) The indicated cells stably transfected with control or Twist shRNA were used to analyze the protein levels by Western blot.
ROR1 positively regulates cancer metastasis in BLBC
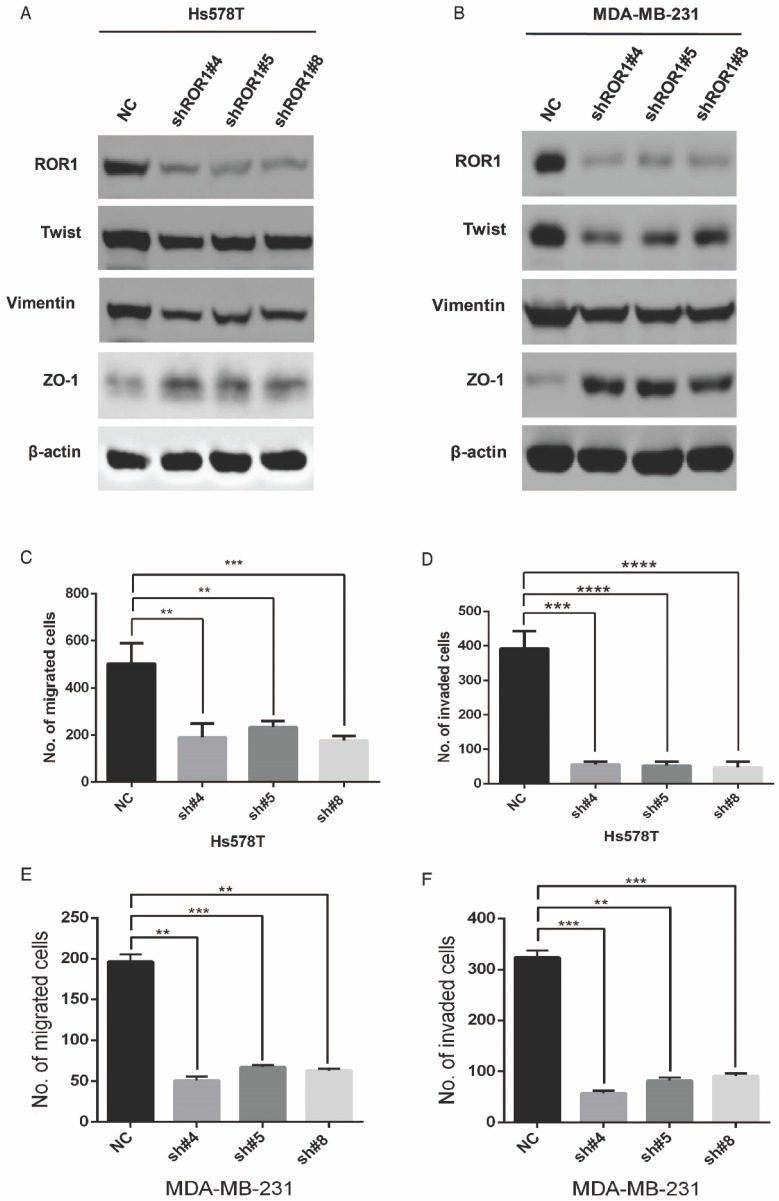
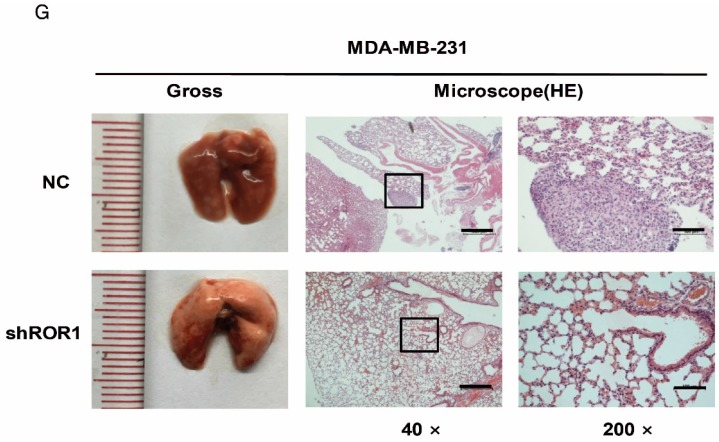
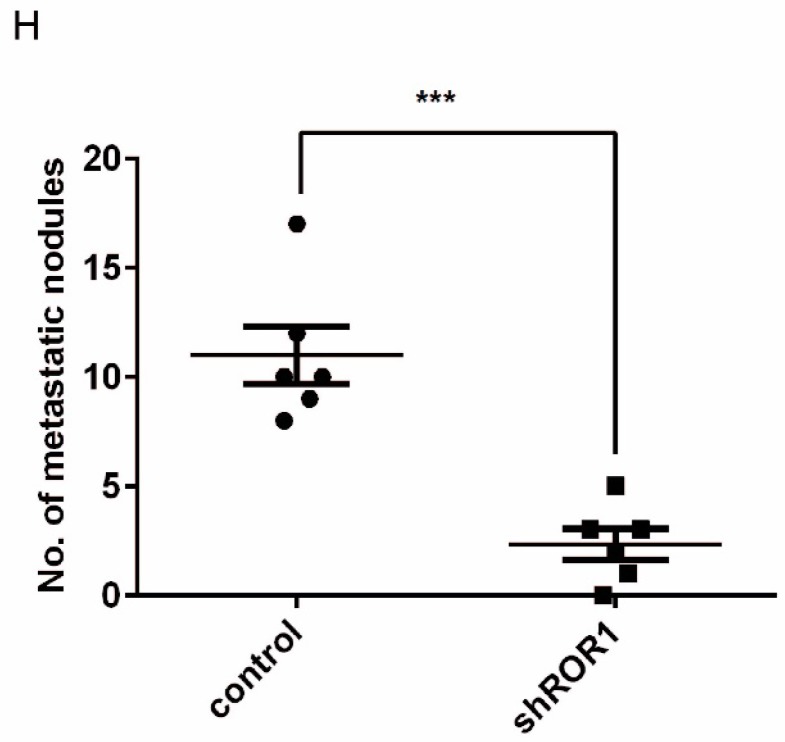
A previous study proved that ROR1 plays an important role in cell proliferation and is associated with enhanced tumor cell growth 28 (Figure S4A-B). To further determine the roles of ROR1 in BLBC, five pairs of short hairpin RNAs (shRNA) were used to knockdown endogenous ROR1 expression. As shown in Figure S5A, the shROR1#4, shROR1#5 and shROR1#8 had the best knockdown efficiency in Hs578T cells. As has been reported, knockdown of ROR1 reduced Vimentin expression, and increased ZO-1 expression, in Hs578T and MDA-MB-231cells (Figure 3A-B), suggesting that suppression of ROR1 is sufficient to inhibit the EMT process in breast cancer cells 21. Furthermore, silencing of ROR1 significantly inhibited the migration and invasion of both Hs578T and MDA-MB231 cells (Figure 3C-F and Figure S5B-C). Moreover, knockdown of ROR1 also reduced the number of metastatic nodules when MDA-MB-231 cells were injected into the tail veins of mice (Figure 3G-H). Taken together, ROR1 positively regulates cell migration, invasion, and cancer metastasis in BLBC.
Figure 3.
ROR1 positively regulates migration, invasion and cancer metastasis in BLBC. (A-B) The indicated molecules were analyzed by Western blot in the Hs578T and MDA-MB-231 cells with stable ROR1 knockdown. (C-F) Cell migration and invasion were determined in the indicated stable cell lines. These experiments were repeated at least three times. The results are expressed as the mean ± SD. * p < 0.05 and ** p < 0.01, *** p < 0.001 using Student's t-test. (G) MDA-MB-231 cells stably transfected with control or shROR1 were injected into the lateral tail vein of nude mice. The left panel presents macroscopic appearances of metastatic lung tumors; central and right panels present the H&E staining. Scale bars, 40×,500μm; 200×,100μm. (H) Illustration of the statistical results (n=6). The results are expressed as the mean ± SD. ** P < 0.01 using Student's t test.
ROR1 mediates Twist-induced breast cancer metastasis
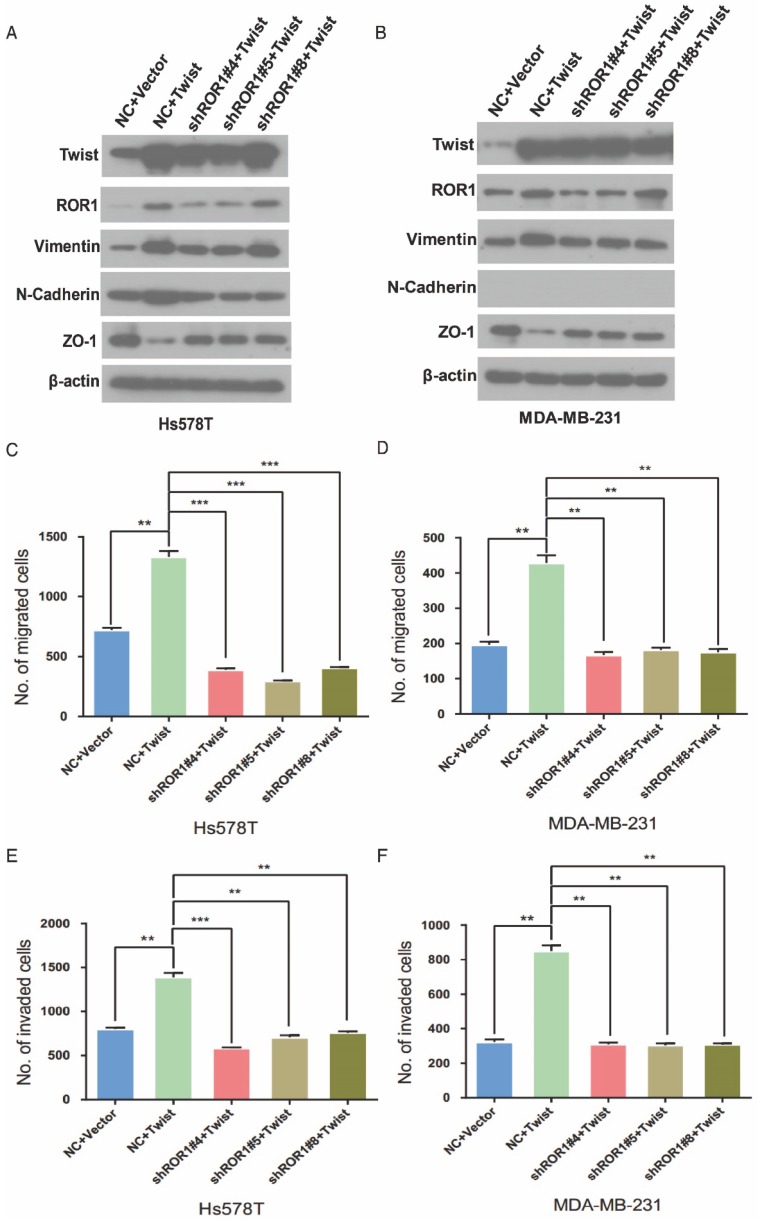
It is known that Twist can induce the EMT process in breast cancer cells 1. To investigate the relationship between Twist and ROR1 during EMT process, we tested the dependence of increased EMT and metastasis due to overexpression of Twist on ROR1. As expected, silencing of ROR1 abolished the EMT markers alteration induced by Twist in both Hs578T and MDA-MB231 cells (Figure 4A-B). Consistently, silencing ROR1 also inhibited the increased migration, invasion, and cancer metastasis mediated by Twist in both Hs578T and MDA-MB-231 cells (Figure 4C-H and Figure S6A-B). These results demonstrate that Twist promotes EMT process and metastasis by inducing ROR1 expression.
Figure 4.
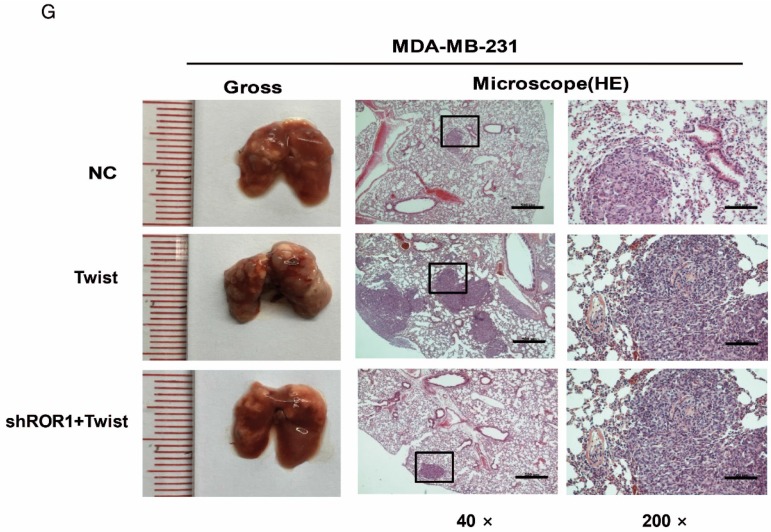
The promotion of cell migration, invasion and cancer metastasis after Twist overexpression primarily depends on ROR1. (A-B) The indicated molecules were analyzed by Western blot in the Hs578T and MDA-MB-231 cells stably expressing control, ROR1 shRNA, Twist, or both, as indicated. (C-F) Cell migration and invasion were determined in the indicated stable cell lines. The results are expressed as the mean ± SD of three independent experiments. * P < 0.05, ** P < 0.01 and *** P < 0.001 using Student's t-test. (G-H) An in vivo lung metastasis model was established in nude mice using MDA-MB-231 cells stably expressing control, ROR1 shRNA, Twist, or both, as indicated. (G) Representative results of gross and H&E staining (middle scale: 40×; right scale: 200×) of metastatic lung nodules. Scale bars, 40×,500μm; 200×,100μm. (H) Illustration of the statistical results (n = 6). The results are expressed as the mean ± SD. * P < 0.05, *** P < 0.001 using Student's t-test.
Twist and ROR1 expression are positively correlated in breast cancer tissues
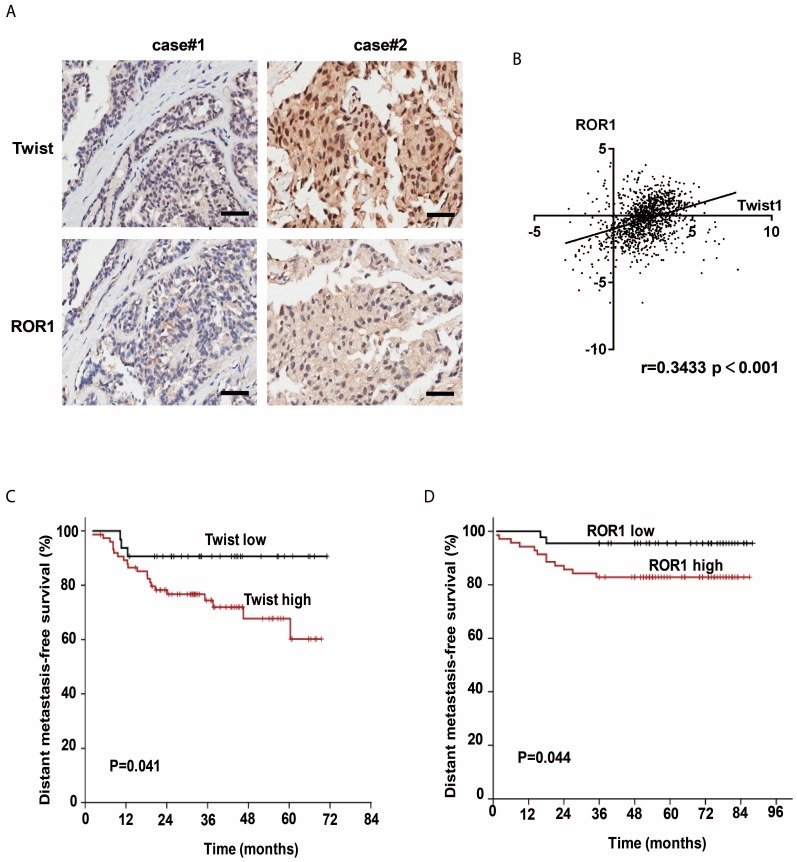
The clinical significance of the regulation of ROR1 by Twist was then evaluated in 134 breast cancer patient tissues by immunohistochemical (IHC) staining. Using both anti-Twist and anti-ROR1 antibodies, 23 of 134 tissues were shown to co-express high levels of Twist, whereas 60 of 134 tissues were shown to co-express high levels of ROR1. Moreover, the level of these two proteins was positively correlated (Figure 5A and Figure S7A). In addition, both Twist and ROR1 were associated with poor overall survival in breast cancer patients (Figure S7B-C). The chi-squared test revealed that the ROR1 level is related to pathology grade (p=0.001) and vital status (p=0.017) (Table 1), and the univariate and multivariate analyses revealed that the ROR1 expression level (p=0.021), with a HR of 2.024 and a 95% CI of 1.106-3.706, is an independent prognostic factor in patients with breast cancer (Table 2). We also found that ROR1 mRNA expression is positively correlated with that of Twist1 in breast cancer patient samples from The Cancer Genome Atlas [TCGA] database after analyzing by ChIPBase v2.0 (Figure 5B) 29, 30. We analyzed 220 patients with breast cancer in the GEO database. After separating patients into 4 groups based upon their relative expression of Twist and ROR1, we found that both high Twist (Figure 5C) and ROR1 (Figure 5D) expression predict shorter DMFS in these breast cancer tissue samples. These data suggest that the Twist/ROR1 signaling is a new biomarker to predict clinical outcomes in breast cancer patients.
Figure 5.
The expression of Twist and ROR1 correlates with clinical prognosis in breast cancer. (A) Representative immunohistochemical staining of Twist and ROR1 from 134 paraffin-embedded breast cancer tissues. Scale bars, 100μm. (B) Correlation of the expression of ROR1 and Twist in TCGA breast cancer datasets. (C-D) Graphs were derived from published data available through the PubMed GEO database (GSE16446 and GSE19615). Kaplan-Meier curves depict the prognostic impact of Twist (C) and ROR1 (D) expression on distant metastasis-free survival (DMFS), respectively. Statistical differences were determined by log-rank test.
Table 1.
Correlation between ROR1 expression and clinicopathologic characteristics of breast cancer patients
| ROR1 expression | Chi-square test | ||||
|---|---|---|---|---|---|
| Characteristics | n | Low or none No. cases (n) |
High No. cases(n) |
p value | χ2 |
| Age | 0.385 | 0.756 | |||
| ≤50 | 63 | 41 | 22 | ||
| >50 | 71 | 41 | 30 | ||
| Clinical Stage | 0.380 | 1.936 | |||
| TNM1 | 11 | 6 | 5 | ||
| TNM2 | 77 | 51 | 26 | ||
| TNM3 | 46 | 25 | 21 | ||
| T classification | 0.682 | 0.764 | |||
| T1 | 26 | 15 | 11 | ||
| T2 | 94 | 57 | 37 | ||
| T3 | 14 | 10 | 4 | ||
| N classification | 0.083 | 6.667 | |||
| N0 | 51 | 30 | 21 | ||
| N1 | 40 | 30 | 10 | ||
| N2 | 34 | 19 | 15 | ||
| N3 | 9 | 3 | 6 | ||
| Tumor Size | |||||
| ≤5 | 109 | 65 | 44 | 0.459 | 0.600 |
| >5 | 25 | 17 | 8 | ||
| Pathology grade | 0.001* | 13.653 | |||
| I | 9 | 10 | 1 | ||
| II | 117 | 71 | 43 | ||
| III | 8 | 1 | 8 | ||
| Vital status (at follow-up) | 0.017* | 5.748 | |||
| Alive | 91 | 62 | 29 | ||
| Death | 43 | 20 | 23 | ||
*P<0.05.
Table 2.
Univariate and multivariate analysis for overall survival (Cox proportional hazards regression model)
| Variable | Univariate analysis | Mutivariate analysis | |||
|---|---|---|---|---|---|
| No. | p value | HR | 95% CI | p value | |
| ROR1 expression | 0.021* | 2.024 | 1.106-3.706 | 0.022* | |
| low expression | 63 | ||||
| high expression | 71 | ||||
| Age | 0.309 | 1.683 | 0.871-3.250 | 0.121 | |
| ≤50 | 63 | ||||
| >50 | 71 | ||||
| T classification | 0.204 | 1.305 | 0.360-4.728 | 0.685 | |
| T1+ T2 | 120 | ||||
| T3 | 14 | ||||
| N classification | 0.088 | 1.571 | 0.787-3.136 | 0.204 | |
| N0 | 51 | ||||
| N1+ N2+ N3 | 83 | ||||
| Tumor size | 0.168 | 1.691 | 0.578-4.944 | 0.338 | |
| ≤5 | 109 | ||||
| >5 | 25 | ||||
Discussion
Accumulating evidences indicate that Twist is a key transcriptional regulator that endows cells with malignant traits, such as invasion, migration and therapeutic resistance. However, identification of new Twist-targeted genes and how these genes function during these events are required to fully understand the molecular mechanism of EMT and metastasis. In this report, we have demonstrated that Twist functions as a transcriptional activator for ROR1 gene expression in EMT and breast cancer, and Twist-induced EMT and breast cancer metastasis partially depends on ROR1.
In previous studies, several RTKs were observed to be significantly deregulated in cancers including vascular endothelial growth factor (VEGF) 31, epidermal growth factor receptor (EGFR) 32, fibroblast growth factor receptor (FGFR) 33, and RTK-like orphan receptor 1 (ROR1) 34. These deregulated RTKs offer promising drug targets for cancer treatment with specific targeting by monoclonal antibodies. Recently, ROR1 has been identified as the receptor of Wnt5a 18, 22, 35, which activates non-canonical WNT signaling to promote breast cancer metastasis 21. Almost a third of human breast cancer cases express high levels of ROR1, which is associated with activation of AKT, cAMP response element-binding protein (CREB), and enhanced tumor cell growth 28, 36. Moreover, the expression of ROR1 is particular to BLBC cell lines that have high metastatic potential 21. Intriguingly, we showed previously that Twist can recruit BRD4 to direct WNT5A gene expression in BLBC by binding to WNT5A gene promoter and enhancer; therefore, Twist may contribute to activation of non-canonical WNT signaling 2. Herein, we established a T47D derivative cell line stably expressing Twist, and this cell line displayed a mesenchymal cell phenotype. In this process, Twist activates ROR1 gene transcription in a luminal breast cancer cell line, which contains little expression of endogenous ROR1. In the other hand, silencing ROR1 in BLBC cell lines, Hs578T and MDA-MB231, inhibited Twist-induced EMT, cell migration, invasion, and cancer metastasis. Furthermore, a positive correlation between Twist and ROR1 was observed in breast cancer patient tissues; this may become a new marker to predict DMFS and overall survival of patients with breast cancer. Mechanistically, we discovered that although the -802 to -402 (fragment 3) region of ROR1 promoter has Twist-binding affinity, mutation of the E-box in this region did not affect ROR1 promoter reporter activity. However, mutations on either one or both of two elongated E-box motifs in the -6 to 399 (fragment 5) region could abrogate the Twist-induced ROR1 promoter reporter activity. Previous studies identified a non-canonical E-box motif, which is called E-box elongated CANNNTG, that may also serve as a potential DNA-binding site 26, 27. Although there is no report on whether Twist can recognize this E-box elongated motif, our evidences showed that mutations in this motif can abrogated Twist-induced ROR1-promoter activity, suggesting that Twist may recognize this E-box elongated motif on the gene promoter.
In summary, our findings suggest that not only Wnt5a but also ROR1 is a transcription target of Twist. We revealed cross-talk between Twist-mediated EMT and non-canonical WNT signaling in metastatic breast cancer, because both Wnt5a and ROR1 are involved in this pathway. Therefore, Twist functions as an important transcriptional factor to activate non-canonical WNT signaling, and thus promote EMT, cell migration, invasion, and cancer metastasis in BLBC. Moreover, ROR1 may serve as a better therapeutic target for metastatic breast cancer compared with Wnt5a, because either Wnt5a or ROR1-induced non-canonical WNT signaling can be blocked by ROR1 inhibitor. The involvement of Twist and ROR1 in a pathway regulating EMT, as shown here, may inform the design of new drugs or treatments for patients with BLBC.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This work was supported by grants from Science and Technology Program of Guangzhou, China (201508020102) to Tiebang Kang; National Natural Science Foundation of China (81672629), and Science and Technology Program of Guangzhou, China (201707010331) to Jian Shi.
Authors' Contributions
Kang T, Zhou BP and Shi J conceived the idea. Cao J and Wang X performed the experiments. Cao J, Wang X, Dai T, Cao R, Wu Y, Zhang M, Zhang R, Jiang R, Wang G, and Shi J analyzed the data. Kang T, Zhou BP, Shi J and Cao J wrote the manuscript. All co-authors have seen and approved this version of the manuscript.
Abbreviations
- EMT
epithelial-mesenchymal transition
- BLBC
basal-like breast cancer
- RTK
receptor tyrosine kinases.
References
- 1.Wang Y, Liu J, Ying X, Lin PC, Zhou BP. Twist-mediated epithelial-mesenchymal transition promotes breast tumor cell invasion via inhibition of hippo pathway. Sci Rep. 2016;6:24606. doi: 10.1038/srep24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q. et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–25. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–81. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 4.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–97. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 5.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C. et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Villarejo A, Cortes-Cabrera A, Molina-Ortiz P, Portillo F, Cano A. Differential role of Snail1 and Snail2 zinc fingers in E-cadherin repression and epithelial to mesenchymal transition. J Biol Chem. 2014;289:930–41. doi: 10.1074/jbc.M113.528026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 1991;5:1568–76. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- 10.Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA. et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–90. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XY, Zhang PY. Receptor tyrosine kinases in carcinogenesis. Oncol Lett. 2016;12:3679–82. doi: 10.3892/ol.2016.5200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Green JL, Kuntz SG, Sternberg PW. Ror receptor tyrosine kinases: orphans no more. Trends Cell Bio. 2008;18:536–44. doi: 10.1016/j.tcb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi T, Yanagisawa K, Sugiyama R, Hosono Y, Shimada Y, Arima C. et al. NKX2-1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell. 2012;21:348–61. doi: 10.1016/j.ccr.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda T, Chen L, Endo T, Tang L, Lu D, Castro JE. et al. Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc Natl Acad Sci U S A. 2008;105:3047–52. doi: 10.1073/pnas.0712148105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aghebati-Maleki L, Shabani M, Baradaran B, Motallebnezhad M, Majidi J, Yousefi M. Receptor tyrosine kinase-like orphan receptor 1 (ROR-1): An emerging target for diagnosis and therapy of chronic lymphocytic leukemia. Biomed Pharmacother. 2017;88:814–22. doi: 10.1016/j.biopha.2017.01.070. [DOI] [PubMed] [Google Scholar]
- 16.Balakrishnan A, Goodpaster T, Randolph-Habecker J, Hoffstrom BG, Jalikis FG, Koch LK. et al. Analysis of ROR1 Protein Expression in Human Cancer and Normal Tissues. Clin Cancer Res. 2017;23:3061–3071. doi: 10.1158/1078-0432.CCR-16-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S, Cui B, Lai H, Liu G, Ghia EM, Widhopf GF 2nd. et al. Ovarian cancer stem cells express ROR1, which can be targeted for anti-cancer-stem-cell therapy. Proc Natl Acad Sci U S A. 2014;111:17266–71. doi: 10.1073/pnas.1419599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Chen L, Cui B, Widhopf GF 2nd, Shen Z, Wu R. et al. Wnt5a induces ROR1/ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. J Clin Invest. 2016;126:585–98. doi: 10.1172/JCI83535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Wang S, Xing Z, Lin A, Liang K, Song J. et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19:106–19. doi: 10.1038/ncb3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bleckmann A, Conradi LC, Menck K, Schmick NA, Schubert A, Rietkotter E. et al. beta-catenin-independent WNT signaling and Ki67 in contrast to the estrogen receptor status are prognostic and associated with poor prognosis in breast cancer liver metastases. Clin Exp Metastasis. 2016;33:309–23. doi: 10.1007/s10585-016-9780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui B, Zhang S, Chen L, Yu J, Widhopf GF 2nd, Fecteau JF. et al. Targeting ROR1 inhibits epithelial-mesenchymal transition and metastasis. Cancer Res. 2013;73:3649–60. doi: 10.1158/0008-5472.CAN-12-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klemm F, Bleckmann A, Siam L, Chuang HN, Rietkotter E, Behme D. et al. beta-catenin-independent WNT signaling in basal-like breast cancer and brain metastasis. Carcinogenesis. 2011;32:434–42. doi: 10.1093/carcin/bgq269. [DOI] [PubMed] [Google Scholar]
- 23.Chien HP, Ueng SH, Chen SC, Chang YS, Lin YC, Lo YF. et al. Expression of ROR1 has prognostic significance in triple negative breast cancer. Virchows Arch. 2016;468:589–95. doi: 10.1007/s00428-016-1911-3. [DOI] [PubMed] [Google Scholar]
- 24.Hojjat-Farsangi M, Moshfegh A, Daneshmanesh AH, Khan AS, Mikaelsson E, Osterborg A. et al. The receptor tyrosine kinase ROR1-an oncofetal antigen for targeted cancer therapy. Semin Cancer Biol. 2014;29:21–31. doi: 10.1016/j.semcancer.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Li L, Wu Y, Zhang R, Zhang M, Liao D. et al. CBX4 Suppresses Metastasis via Recruitment of HDAC3 to the Runx2 Promoter in Colorectal Carcinoma. Cancer Res. 2016;76:7277–89. doi: 10.1158/0008-5472.CAN-16-2100. [DOI] [PubMed] [Google Scholar]
- 26.Rabissi A, Vilela B, Lumbreras V, Ludevid D, Culianez-Macia FA, Pages M. Molecular characterization of maize bHLH transcription factor (ZmKS), a new ZmOST1 kinase substrate. Plant Sci. 2016;253:1–12. doi: 10.1016/j.plantsci.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Jee SH, Kim GE, Hong SH, Seo SB, Shim JK, Park SC. et al. Characterization of EamaT1, a member of maT family of transposable elements from the earthworm Eisenia andrei (Annelida, Oligochaeta) Mol Genet Genomics. 2007;278:479–86. doi: 10.1007/s00438-007-0266-5. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S, Chen L, Cui B, Chuang HY, Yu J, Wang-Rodriguez J. et al. ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PloS one. 2012;7:e31127. doi: 10.1371/journal.pone.0031127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang JH, Li JH, Jiang S, Zhou H, Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res. 2013;41:D177–87. doi: 10.1093/nar/gks1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou KR, Liu S, Sun WJ, Zheng LL, Zhou H, Yang JH. et al. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017;45:D43–D50. doi: 10.1093/nar/gkw965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular-specific growth factors and blood vessel formation. Nature. 2000;407:242–8. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 32.Hayman MJ, Enrietto PJ. Cell transformation by the epidermal growth factor receptor and v-erbB. Cancer Cells. 1991;3:302–7. [PubMed] [Google Scholar]
- 33.Touat M, Ileana E, Postel-Vinay S, Andre F, Soria JC. Targeting FGFR Signaling in Cancer. Clin Cancer Res. 2015;21:2684–94. doi: 10.1158/1078-0432.CCR-14-2329. [DOI] [PubMed] [Google Scholar]
- 34.Borcherding N, Kusner D, Liu GH, Zhang W. ROR1, an embryonic protein with an emerging role in cancer biology. Protein Cell. 2014;5:496–502. doi: 10.1007/s13238-014-0059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho HY, Susman MW, Bikoff JB, Ryu YK, Jonas AM, Hu L. et al. Wnt5a-Ror-Dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc Natl Acad Sci U S A. 2012;109:4044–51. doi: 10.1073/pnas.1200421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Chen L, Wang-Rodriguez J, Zhang L, Cui B, Frankel W. et al. The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am J Pathol. 2012;181:1903–10. doi: 10.1016/j.ajpath.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.