Abstract
Background
Staphylococcus aureus is a major bacterial pathogen capable of causing a range of infections in humans from gastrointestinal disease, skin and soft tissue infections, to severe outcomes such as sepsis. Staphylococcal infections in humans can be frequent and recurring, with treatments becoming less effective due to the growing persistence of antibiotic resistant S. aureus strains. Due to the prevalence of antibiotic resistance, and the current limitations on antibiotic development, an active and highly promising avenue of research has been to develop strategies to specifically inhibit the activity of virulence factors produced S. aureus as an alternative means to treat disease.
Objective
In this review we specifically highlight several major virulence factors produced by S. aureus for which recent advances in antivirulence approaches may hold promise as an alternative means to treating diseases caused by this pathogen. Strategies to inhibit virulence factors can range from small molecule inhibitors, to antibodies, to mutant and toxoid forms of the virulence proteins.
Conclusion
The major prevalence of antibiotic resistant strains of S. aureus combined with the lack of new antibiotic discoveries highlight the need for vigorous research into alternative strategies to combat diseases caused by this highly successful pathogen. Current efforts to develop specific antivirulence strategies, vaccine approaches, and alternative therapies for treating severe disease caused by S. aureus have the potential to stem the tide against the limitations that we face in the post-antibiotic era.
Keywords: Staphylococcus aureus, virulence, antivirulence, MRSA, review, pathogenesis
1. INTRODUCTION
Staphylococcus aureus (S. aureus) is a Gram-positive, non-motile bacterium that can reside as a common carrier in humans, although its host range can extend to animals such as dogs, cats, sheep, cattle, and poultry (Fig. 1) [1, 2]. It is estimated that up to 30% of the human population can be colonized with S. aureus strains, with resident sites being the nares, skin, and the gastrointestinal (GI) tract [3]. Skin and soft tissue pathology comprise one of the most common manifestations of S. aureus infections, which include folliculitis, impetigo, and scalded skin syndrome [4–6]. More invasive infections can lead to outcomes such as endocarditis, bone and joint infections, bacteremia, and toxic shock syndrome. GI infections have also been widely reported, and have been associated with outbreaks of food poisoning [7–11]. Additionally, infections due to surgery wounds or prostheses have been reported, which are often associated with catheters, medical implantation, dialysis, and other procedures [12]. In addition to patients undergoing surgical procedures, other high risk groups for S. aureus infection include individuals undergoing immunosuppressive or cancer therapy, along with low birth weight infants and young children.
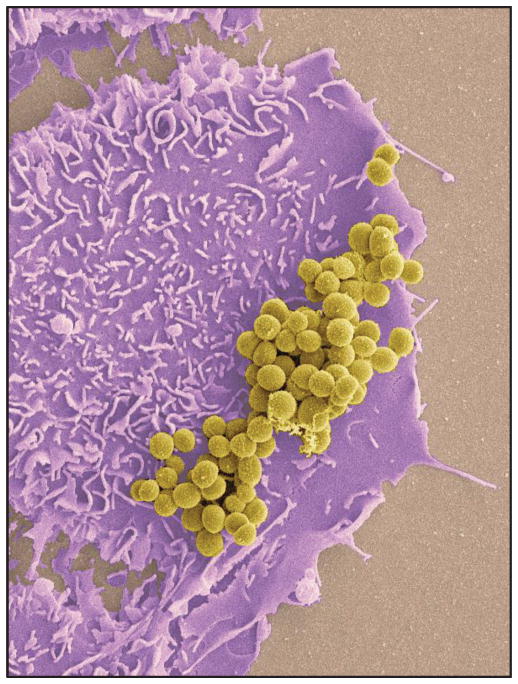
Fig. 1. Staphylococcus aureus on host keratinocytes.
Scanning electron micrograph image of S. aureus on HaCat eukaryotic cells. Image colorized using Adobe Photoshop.
Prior to the development of antibiotics, S. aureus mortality exceeded 80% in bacteremia cases [13]. The use of penicillin in the 1940s dramatically decreased infection mortality; however, resistant strains were observed as early as 1941 [14]. The improved β-lactam antibiotic Methicillin (trade name Celbenin) was developed and first used in 1959, but was rapidly followed by reports of resistance occurring in individuals [15]. This report first heralded the rise of what has now become known as MRSA, or methicillin-resistant S. aureus. MRSA strains are now prevalent worldwide, with estimates of over 50 million people colonized with MRSA strains at any given time, making efforts to limit bacterial spread difficult [16].
Currently, MRSA has become an inclusive common term used to describe S. aureus strains that are typified by resistance to most β-lactam antibiotics, with the exception of some modern cephalosporin classes of β-lactam compounds [17, 18]. Mechanisms of β-lactam resistance by MRSA have been extensively studied, with 1. acquisition of a penicillin-binding protein (MecA) that exhibits lower affinity for β-lactam compounds (methicillin-resistance), and 2. proteolytic inactivation of β-lactams by the expression of specific β-lactamase, being the prevalent means of resistance in MRSA strains [19, 20]. MRSA strains can also be classified as multiply resistant, with resistance to Vancomycin being a particular recent concern [21, 22]. MRSA is listed by the World Health Organization (WHO) as one of the nine bacteria of international concern due to its high level of antibiotic resistance (WHO 2014). 20–80% of all S. aureus infections worldwide can be attributed to MRSA strains, depending on the country reporting (WHO 2014). The WHO further notes that MRSA infections in general result in more hospital days to resolve the infection, an increase in sepsis outcomes and increased duration in intensive care units (WHO 2014).
MRSA strains have been historically separated on the basis of where the infection was acquired, with MRSA infections originating in community settings, such as daycares, prisons, dorm rooms, or locker rooms, termed community-acquired MRSA (CA-MRSA). Infections acquired in health care settings, including in-patient hospital stays, surgical procedures, dialysis, or catheters, are termed hospital acquired infections (HA-MRSA). These MRSA distinctions are becoming blurred as strains that are traditionally acquired in the community (such as USA300) are starting to gain footholds in health care settings [23–25]. In the last twenty years, particularly virulent strains of CA-MRSA have emerged in hospital settings and healthcare facilities [12, 26–28]. In the United States alone, present MRSA infections number around 75,000 per year, with over 20,000 deaths [29].
Along with evolving several mechanisms which confer resistance to antibiotic compounds, S. aureus produces a myriad array of individual virulence factors that manipulate host cell responses for its overall survival [30–33]. Virulence factors produced by S. aureus are manifold, and have the ability to not only cause lysis of host cells, and promote tissue invasion and destruction, but many specific virulence factors produced by S. aureus have the ability to specifically manipulate both innate and adaptive immune responses, including inhibition of complement activation, prevention of neutrophil function and recruitment, and inhibition of phagocyte function [34–42]. Excellent reviews have summarized recent scientific findings on virulence factors produced by S. aureus and host cell manipulation by S. aureus in the context of human disease [41, 43–46]. In light of vigorous research into mechanisms of S. aureus virulence, important avenues have been investigated with regard to emerging alternative therapies for treating S. aureus infections. In spite of the fact that antibiotic research and development has steadily decreased in the last twenty years, with only seven new antibiotics launched from 2009–2012, a range of approved antibiotics to treat MRSA infections have been implemented only recently [47–49]. These include established classes such as fluoroquinolones and cephalosporins, but also novel compound classes such as peptide mimics, oxadiazoles, and diaminopyrimidines [49–51]. Additionally, owing to the widespread knowledge of multi-drug resistance in global rates of S. aureus infections, efforts to develop non-drug alternatives to combat MRSA have been emerging. In particular, anti-virulence strategies have been heavily investigated not only for bacterial pathogens such as MRSA, but for many pathogens for which there are clear mechanistic links between the virulence mechanism and pathogenesis. Quorum molecule mimics, pilicides, active site inhibitors of specific toxins, secretion system inhibitors, and phage therapies, have all been extensively investigated as antivirulence strategies to combat mechanisms of bacterial pathogenesis by specific microbes [52–56]. In this review, we specifically highlight several major virulence factors produced by S. aureus for which recent advances in antivirulence approaches may hold promise as an alternative means to treating diseases caused by this pathogen (Fig. 2 and Table 1).
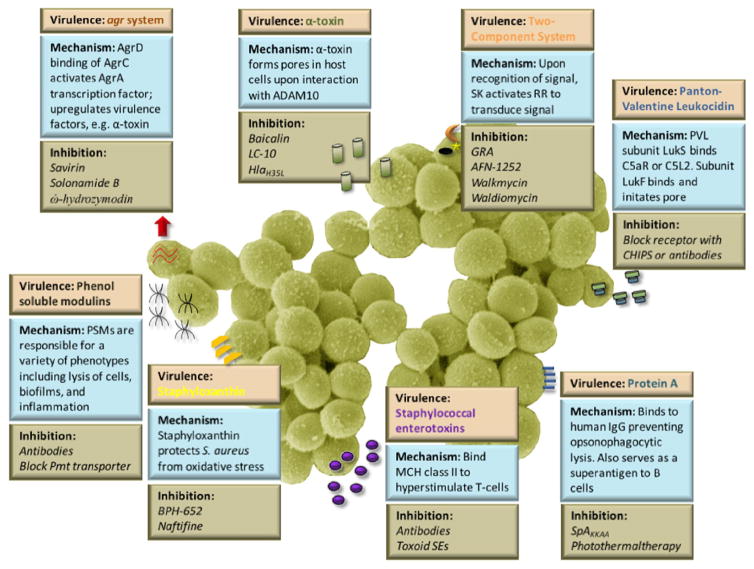
Fig. 2. Depiction of virulence factors (tan box), mechanism of action (blue box) and strategies to inhibit the virulence factors (green box) as discussed in this review.
All of the virulence factors discussed in this review are secreted with the exception of the agr system which serves as a regulator for many other virulence factors including α-toxin, phenol soluble modulins, and protein A.
Table 1.
Detailed list of the virulence factors discussed in this review including mechanism of action and inhibition approaches. Summary of the virulence factors discussed in this review as well as details on virulence factor mechanisms and specific inhibition approaches.
| Virulence Factor | Mechanism of Action | Inhibition Mechanism |
|---|---|---|
| agr system | agr is a quorum sensing system that is dependent on cell density. AgrD binds to AgrC, which results in activation of AgrA the transcription factor. AgrA is then able to increase transcription of SE, hemolysins, proteases, and capsular polysaccharides. |
|
| α-toxin | Binds to ADAM10 on eukaryotic cells and oligomerizes, forming pores and lysing the cell. |
|
| Phenol Soluble Modulins (PSM) | Family of 7 α helical peptides, split into PSMα and PSMβ. PSMα are able to lyse a variety of cells including leukocytes and erythrocytes. PSMβ are implicated in the spread of biofilms. |
|
| Protein A | Cell wall anchored protein that binds to the FC region of IgG antibodies and to the Fab of Variable Heavy 3 (VH3) idiotype B-cell receptors. |
|
| Panton-Valentine Leukocidin (PVL) | PVL consists of two subunits, LukS and LukF. LukS binds to human complement receptors C5aR and C5L2 and allows docking of LukF. The Luk subunits oligomerizes and form a pore lysing the cell. |
|
| Staphylococcal Enterotoxins (SE) | There are 21 known SEs. SEs bind MHC class II receptors on T-cells and hyperstimulate the T-cells. |
|
| Two-Component System (TCS) | Consist of a sensor kinase which senses the environmental signal and the response regulator which is activated and is able to bind DNA to affect transcription. |
|
| Staphyloxanthin | Pigment responsible for some strains of S. aureus golden color. Protects the bacterium from oxidative stress. |
|
We also highlight recent developments in vaccine research into MRSA, along with alternative anti-MRSA approaches such as phage-based treatment. Given the widespread rates of antibiotic resistant strains of S. aureus, active research into alternative strategies to combat diseases caused by this highly successful pathogen remains a paramount priority.
2. agr OPERON
The accessory gene regulator system (agr) operon was first discovered in 1986 by Recsei et al. using a transposon insertion [57]. They observed a large decrease in alpha, beta, and delta hemolysins, but an increase in the protein A production, and they termed this gene cluster agr for accessory gene regulator [57]. Decreases in hemolysins alpha, beta, and delta were determined by growth on blood agar plates, while protein A production was verified by western blot [57]. The agr cluster encodes two RNAs, RNAII and RNAIII [58, 59]. RNAII codes for the AgrD precursor which is eventually converted to AgrD, the extracellular signaling peptide [58]. The operon also encodes AgrB, the maturation and export protein for AgrD, as well as a two-component regulatory system consisting of AgrC, a sensor histidine kinase, and AgrA, the response regulator. Later in this review, two component systems will be discussed in detail. AgrA is activated when a quorum sensing molecule binds to AgrC. AgrA then binds to promoter regions in the agr operon, resulting in a positive feedback loop [58]. RNAIII serves as a transcription factor responsible for the change in gene expression [60]. One of the primary functions upon the transcription of RNAIII is the binding of RNAIII to the mRNA of the repressor of toxins (ROT) mRNA [61]. Antisense binding of the RNAIII to the mRNA of ROT results in the cleavage of ROT mRNA by RNase III as shown by northern blot [61, 62]. Specific roles of agr during the course of infection are complicated due to varying infections, disease models, and patient populations [63]. In acute mice infection models including intracranial abscesses and necrotizing pneumonia, there is positive evidence that agr is important for the virulence [64–67]. In humans, there has been a link between persistent colonization in nosocomial infections with S. aureus and agr dysfunction [68–71]. Other studies have found no link between agr deficiency and poor clinical outcomes [72]. In a case study of 75 patients with MRSA pneumonia, increased death was associated with dysfunctional agr; however, these data lacked statistical significance due to low sampling [72]. Further studies are needed to establish a clear link between the agr operon and specific pathogenic outcomes, as current studies have shown varied conclusions, which may be the result of strain-specific differences. There are at least four alleles of agr among the various strains of S. aureus, and each group is inhibitory to the other three agr [73–75]. Despite the differences in host pathologies implicated with agr regulation, the agr operon has been shown to be responsible for upregulating a variety of known virulence genes including SEs, hemolysins, capsular polysaccharides, and proteases [57, 59]. Given the known virulence factors that agr has been shown to regulate, strategies to target agr may serve as an attractive target for antivirulence approaches. Experiments where agr has been knocked out have been shown to exhibit decreased virulence in murine models of S. aureus pneumonia and dermonecrosis [67]. This decreased virulence was shown to coincide with reduced transcription of Panton-Valentine leukocidin and α-toxin [67]. These findings demonstrate that at least in the context of an acute infection mouse model, toxins such as hemolysins or PVL, that are regulated by agr during this type of infection, could play an important role in the progression of disease [57, 67].
A study by Sully et al. was recently published where the researchers were able to identify a small molecule inhibitor targeting AgrA of the agr operon they dubbed savirin (S. aureus virulence inhibitor) [76]. Savirin was identified using a high throughput screen designed to identify inhibitors of S. aureus agr, but not inhibitors of the closely related bacterium S. epidermidis [76]. By using a mouse model of skin and soft tissue infection, the authors were able to show that there was decreased tissue injury, and reduced bacterial burden upon treatment of infections with savirin [76]. In order to address a common concern, resistance, the authors also verified that resistance did not arise after serial passage of bacteria with savirin [76]. Using an Affymetrix GeneChip containing all open reading frames from S. aureus strain USA300 LAC, they were also able to show that virulence associated genes such as PVL, Hla, PSM, and serine proteases were down regulated upon treatment with savirin [76]. Treatment with savirin did not disrupt quorum sensing in the related skin commensal Staphylococcus epidermidis indicating that this could be a specific quorum sensing inhibitor to S. aureus [76].
Using a naturally isolated cyclodepsipeptide from Photobacterium halotolerans named Solonamide B, the Ingmer group was able to inhibit the action of the agr cluster [77]. Upon application to cultures of USA300, bacterial growth was not impaired, but transcription of PSMα and agrA were decreased as shown by Northern blot [77]. Decreases in hemolysis and human neutrophil lysis were shown via rabbit hemolysis assays and LDH release assay, respectively [77]. Follow up work regarding Solonamide B and derivatives examined toxicity on eukaryotic immune cells [78]. The authors were able to show no increase in cytotoxicity for human peripheral blood mononuclear cells, proliferated T-cells or murine dendritic cells [78].
An additional inhibitor of the agr gene cluster has been developed and chronicled by Daly et al. They have shown that ω-hydroxyemodin (OHM), a small compound isolated from Penicillium restrictum is able to inhibit the quorum sensing function of the agr operon by binding to the AgrA promoter protein and preventing interaction with DNA [79]. During the course of mouse infections with the LAC strain of S. aureus, treatment with OHM reduced lesion size as well as reducing the CFU of the injected bacteria [79]. Importantly, using cell culture systems they were also able to show effective agr inhibition using concentrations of OHM that were not toxic to eukaryotic cells [79]. At a concentration of roughly 16 μM, bacterial cell growth was uninhibited, while agr was inhibited by ~40%, and human kidney, alveolar, and hepatocyte cells were unaffected [79].
Targeting a system such as agr could prove to be very useful as it could disrupt a wide variety of virulence factors, rather than targeting each virulence factor individually [80]. Targeting the virulence regulator as opposed to simply trying to kill the microbe has several advantages, including: 1. a wider variety of targets, 2. preservation of commensal bacteria, and 3. less selective pressure towards resistance, as in most cases the virulence factor is not required for the overall survival of the bacterium [81]. The findings by Sully et al. emphasize the key advantage of targeting a single, specific gene cluster in that inactivation of virulence regulation genes such as agr can impact the expression of several virulence factors and thus serve as an efficient means of reducing virulence in model systems.
3. α-TOXIN
α-toxin is a secreted 33 kDa protein that is able to lyse cells by forming pores on the membrane of target host cells via interaction with the specific host component ADAM10 [82, 83]. ADAM10 is a cellular metalloprotease responsible for a variety of functions including E-cadherin shedding and endothelial permeability [84, 85]. α-toxin is repressed by ROT (Repressor of Toxins) and stimulated by the agr system [57, 86]. A recent study by Caballero et al. examined the role this toxin plays in rabbit models of Staphylococcal ocular infections. The authors note that S. aureus is the primary cause of eye infections in North America [82, 87, 88]. The researchers attempted to limit the damage caused in the course of infection to the eye by utilizing a high affinity monoclonal Fab fragment antibody to α-toxin. Fragment antibodies were used due to concerns that the full antibody could have limited penetration into the corneal tissue and thus would be unable to effectively neutralize α-toxin [82]. Using the LTM14 Fab antibody alone resulted in a significant decrease in corneal erosion in the rabbit infection model. Erosion areas without LTM14 Fab treatment averaged ~30 mm2, with LTM14 Fab treatment averaged ~17 mm2, and combination treatment with LTM14 Fab and BAK (a compound used to increase ocular permeability to drugs), resulted in average erosion size of ~11 mm2 [82].
An additional method of disrupting α-toxin activity is to prevent the formation of the toxin pore complex on the surface of the host cell. Baicalin (BAI), a naturally occurring compound from Scutellaria baicalensis georgi, a traditional Chinese medicinal herb, has been shown to be effective in disrupting α-toxin activity [89]. BAI was predicted to disrupt the formation of pores using molecular dynamics simulation studies [89]. The inability of the toxin to form pores was confirmed in biochemical studies using purified α-toxin treated with increasing levels of BAI. Upon treatment with BAI, the appearance of higher molecular weight bands of α-toxin corresponding to the pore complex were decreased in a dose-dependent manner, suggesting BAI treatment could disrupt the oligomerization of α-toxin [89]. Disruption of the pore assembly was also shown to reduce in vitro hemolysis, and treatment with BAI increased overall mice survival using a S. aureus pneumonia model [89].
In a very similar vein as the Caballero paper, a study was published by Hua et al. using α-toxin antibody LC10 in a mouse pneumonia model. Hua and collaborators were able to show that mice were better able to survive when given the LC10 α-toxin antibody [90]. In combination with LC10 and vancomycin or linezolid treatment, the mice had improved survival rates as compared to treatment with either treatment alone [90]. Data generated by Hua suggest that combination treatments incorporating virulence targeting and traditional antibiotics could both increase survival and reduce the duration of antibiotic treatment, thus possibly reducing antibiotic resistance [90]. LC10 is in clinical trials under the name MEDI4893 for high risk nosocomial pneumonia patients [91]. As patients facing S. aureus pneumonia may be immunocompromised, the ability of the LC10 to be efficacious in this patient population is important. In this manner, Hua et al. showed similar results of increased mice survival with LC10 using immunocompromised mice [91].
A study by Kennedy et al. was able to show that immunization to α-toxin using antisera or a non-toxic form of α-toxin (HlaH35L) resulted in a reduction of skin lesions and dermonecrosis in a murine model of S. aureus infection [92]. HlaH35L was demonstrated to reduce α-toxin oligomerization as shown by liposomal-membrane binding study [93, 94]. The Nagy group was able to develop a cross-reactive antibody that was capable of binding conserved conformational epitopes of α-toxin, γ-hemolysin, and Panton-Valentine leucocidin [95]. They were able to use this antibody to increase survival in murine challenges both intranasally and intravenously delivered MRSA strain USA300 [95]. Targeting multiple virulence factors using a single antibody may be a very effective and promising means of treatment with further development. Currently, there is at least one antibody targeting α-toxin in current phase two clinical trials, AstraZeneca compound MEDI4893. It is a modified form of the LC10 antibody from the Hua et al. studies above [48, 91, 96].
α-toxin is an extremely important virulence factor for S. aureus, and recent research has shown that inhibitors of this toxin could result in improved outcomes during the course of bacterial infection. Hyperexpression of α-toxin has been shown to be correlated to the increased virulence of the CA-MRSA strain ST93, underscoring the importance of approaches to inhibit this virulence factor for therapies against S. aureus infection [97].
4. PHENOL-SOLUBLE MODULINS
Phenol-soluble modulins (PSMs) comprise a family of amphipathic, α-helical peptides with diverse roles in Staphylococcal pathogenesis [98]. The peptides were named for their ability to be soluble in the phenol layer rather than the aqueous layer during hot phenol extraction, a characteristic attributed to the presence of the amphipathic α-helix, and were originally isolated from the concentrated supernatant of Staphylococcus epidermidis. Three forms of PSMs were identified during the course of the S. epidermidis study: PSMα, PSMβ, and PSMγ. PSMα and PSMβ did not exhibit strong homology to known staphylococcal toxins, but PSMγ was identified as the delta toxin of S. epidermidis [99]. PSMs were identified in S. aureus using a combination of reversed-phase HPLC/electrospray mass spectrometry and N-terminal peptide sequencing, which resulted in a total of seven PSMs: PSMα1-4, PSMβ1-2, and S. aureus δ-toxin, related to PSMα [100]. PSMα and PSMβ are grouped by size and charge; α-PSMs are shorter, 20–25 amino acids long and primarily have a net neutral or positive charge, while the β-PSMs are larger, with 43–45 amino acids, and primarily have a negative charge [98]. Unlike many of the virulence factors produced by S. aureus which are encoded by mobile genetic elements, PSMs are encoded in the core genome and so are present in nearly all strains of S. aureus [97]. One exception is the recently characterized PSM-mec, encoded on the mobile genetic element (MGE) staphylococcal cassette chromosome mec (SCCmec). This MGE also encodes resistance to methicillin, and may contribute to horizontal gene transfer of both virulence factors and antibiotic resistance in S. aureus strains [101]. Nevertheless, the ubiquitous nature of PSMα and PSMβ in S. aureus strains makes them suitable potential targets for anti-virulence therapy.
In addition to different size and charge, the two general categories of PSMs have differing roles in both S. aureus pathogenicity and initial colonization of human epithelial surfaces. PSMα peptides are cytolytic, with the ability to lyse a variety of human cells, including leukocytes and erythrocytes. Interestingly, while PSMαs have been show to lyse neutrophils, they can also trigger inflammatory responses through recruitment and activation of neutrophils prior to lysis [100]. This highlights the importance of stringent PSM regulation by bacteria to maintain a balance between immune evasion and immune cell recruitment. PSMβ peptides have been implicated in the spread of biofilms but have less cytolytic activity [100, 102, 103]. This alternative role of PSMs has been more thoroughly studied in S. epidermidis, where PSMβs were found at higher levels relative to other PSM peptides in biofilms compared to planktonic growth [104]. In S. aureus, all PSM classes have been implicated in both biofilm formation and detachment, including structuring of channels and dissemination from biofilms in an in vivo infection model [105]. With respect to overall virulence, it has been proposed that the increased virulence exhibited by CA-MRSA can be attributed at least in part to higher PSM expression levels in CA-MRSA strains compared to HA-MRSA strains [100, 106]. In a murine skin infection model of S. aureus, the Kahlenberg group showed that infection the wild type LAC strain resulted in massive lesions on mice, while the strain with both PSMα and PSMβ genes knocked out did not result in any significant lesion [107]. Additional deletion studies further underscore the differences between PSMα and PSMβ contributions to virulence. A study of the USA300 LAC strain with knockouts for either PSMα or β genes showed significant reduction in mice skin lesions only when the α-PSMs were knocked out. When applied directly to human neutrophils, a similar trend was confirmed, with α-PSMs demonstrating greater cytotoxicity than the β-PSM [100]. In particular, the PSMα3 was shown to be the most virulent, as a plasmid containing only PSMα3 was able to restore near wild type lysis levels in a global α-PSM knockout background [100].
Studies by Kaito et al. reported that the differences in PSM expression, and therefore virulence, between HA-MRSA and CA-MRSA could also be attributed to the mobile genetic element SCCmec mentioned previously [108, 109]. While SCCmec is present in both HA- and CA-MRSA, HA-MRSA alone contains the F-region. This region encodes the psm-mec and fudoh genes. It was found that transformation of the F-region into CA-MRSA strains that previously lacked the region resulted in suppression of PSMα production, reduction of colony spreading activity, and decreased virulence in murine models [108]. Interestingly, presence of the F-region also promoted the formation of biofilms, which helps explain the presence of HA-MRSA biofilms on medical equipment such as catheters. In a later study, the authors found that psm-mec RNA, acting as a regulatory RNA form, directly inhibited translation of agrA, interrupting the agr feedback loop and reducing overall PSMα expression. Additionally, HA-MRSA isolates with mutated or absent psm-mec had higher PSMα levels and increased virulence [109].
PSMs are tightly regulated by the quorum sensing agr operon described previously in this review, and it is therefore important to note for targeting purposes that PSM production is highly dependent on cell density. PSMα1-4 and PSMα-like δ-toxin are encoded in the psmα and RNAIII loci respectively (δ-toxin is encoded by the hld gene within RNAIII), and the two PSMβ peptides are encoded in the psmβ locus [100, 110]. Interestingly, PSM regulation involves direct binding of the agr response regulator AgrA to the psmα and psmβ promoters, and is independent of the agr transcription factor RNAIII [58]. Recent work has supported an additional role of PSMs: to aid S. aureus escape after phagocytosis by leukocytes [111]. Upon addition of 1% human serum to neutrophils, the authors discovered that the bacterial supernatant or purified PSMs were unable to activate, attract, or lyse neutrophils. This PSM inhibition was attributed to lipoproteins in the serum, which can sequester both PSM and components of the agr quorum sensing system to reduce production and pro-inflammatory activities of PSM. The authors conclude that based on these data, the PSMs are more likely to function intracellularly rather than extracellularly [111]. Another study examined the stringent response, a bacterial stress response characterized by the production of the alar-mone (p)ppGpp, as this response was found to be induced in S. aureus following phagocytosis by polymorphonuclear neutrophils (PMNs) [112]. Genes encoding PSMs were found to be positively regulated during the stringent response, thought to contribute to survival of the pathogen post-phagocytosis [112]. Interestingly, activation of the stringent response was found to be independent of AgrA, and the mechanism by which (p)ppGpp activates PSM production is unknown. These data suggest that both agr locus regulation and responses to intracellular environments may contribute to PSM production and bacterial survival in the phagosome prior to escape [98].
The conserved nature of PSMs makes them advantageous targets in anti-toxin therapy for S. aureus infections. It has been previously established that PSMs are immunogenic, and patients with lower antibody levels to S. aureus toxins, including δ-toxin and PSMα3, had higher incidence of sepsis in invasive infection [113]. To that end, it has been proposed that a targeting strategy for S. aureus disease would be the use of antibodies against PSMs. In a murine model of S. epidermidis infection, PSMβ1 and 2 were highly immunogenic and elicited a strong IgG response. In animals treated with anti-PSMβ, the researchers found reduced dissemination to organs including the liver, spleen, and lymph nodes from a biofilm-associated infection on a catheter [104]. However, immunization against the more cytolytic PSMα-type, which may be more protective in acute infection, requires further investigation. PSMα-like δ-toxin has long been established as immunogenic, but a study in rabbits and guinea pigs found that immunization with δ-toxin required Freund adjuvant for successful antibody production as opposed to the toxin alone, and that the induced antibody did not neutralize the hemolytic activity of the toxin [114]. A second targeting strategy involves blocking PSM export through the phenol-soluble modulin transporter, or Pmt, an ABC transporter. This strategy is advantageous for broad PSM inhibition, as Pmt is the dedicated transporter for all PSM types and is encoded in the core genome [115]. Importantly, functional Pmt is necessary for S. aureus growth. In the absence of Pmt, blocked export of PSMs results in their accumulation in the cytosol, causing cytoplasmic membrane damage and abnormal cell division thus inhibiting growth of the bacterium. Another reason to target Pmt in S. aureus is its ability to protect the organism from self-produced PSMs as well as non-self PSMs of S. epidermidis [115]. Finally, PSMs are under the control of the agr system, so inhibiting the agr operon as discussed above would result in reduced levels of PSM associated virulence. There may be opportunities to take advantage of the agrA suppression properties of psm-mec as an indirect method of inhibition for αPSMs.
To conclude, the core genome-encoded phenol-soluble modulins contribute to S. aureus pathogenesis in multiple ways including host cell lysis, biofilm formation, and the triggering of inflammatory responses. Inhibition of these proteins could help to increase survivability of MRSA infections. There are several promising avenues for targeting PSMs, both directly through the use of antibodies and indirectly through targeting of export and regulatory pathways. Due to their complex and varied nature, successful inhibition of PSM-associated virulence will likely occur through a multi-faceted approach.
5. PROTEIN A
Protein A (SpA) is a cell wall anchored protein with a mass of approximately 42kDa, released during bacterial growth and encoded by the spa gene [116–119]. As with many S. aureus virulence factors, protein A is regulated by the agr system, and expression of protein A is specifically repressed by RNAIII. The 3′ end of RNAIII binds to the complementary portion of the 5′ end of spa mRNA. This binding both blocks the the mRNA from translational machinery and allows for recruitment of double strand specific RNase III to degrade the mRNA [120]. Protein A is composed of two regions with clear structural and functional differences: Region X and the immunoglobulin binding domains. Region X contains two parts: the repetitive region (Xr) consisting of repeating octapeptide units, and the C-terminal domain (Xc) with a unique amino acid sequence. Region X is responsible for the cell wall attachment function of protein A [117]. The N-terminal region of protein A contains five immunoglobulin-binding domains (E, D, A, B, C) that can bind the Fc of IgG antibodies and the Fab of Variable Heavy 3 (VH3) idiotype B-cell receptors [121–123]. In fact, protein A binds to human IgG at these five sites so effectively that it is used as a column substrate to purify antibodies [124–126]. Production of protein A contributes to S. aureus pathogenesis by evasion and suppression of host immune system components. IgG binding activity protects the pathogen from opsonophagocytic killing [127, 128], and the protein acts as a superantigen against B-cells, inducing rapid activation and expansion followed by apoptotic death [129].
There have been several recent studies on developing therapeutics to S. aureus infection based on targeting protein A. It was established by Kim et al. that infection with a spa deletion mutant of S. aureus reduced mortality from 60% to 25% in a mouse model. The researchers then introduced amino acid substitutions into the immunoglobulin-binding domain (IgBD) D, forming the mutant SpA-DKKAA. This mutant had reduced immunoglobulin binding activity and B cell superantigen activity. Additionally, antibodies generated against this mutant had a protective effect against MRSA infections in mice [130]. The investigators then generated SpAKKAA that included all five IgBDs with four amino acid substitutions in each domain, and in which interactions between the Fcγ and Fab domains were disrupted [29]. The Fab domain is the part of the antibody responsible for binding to antigens and FCγ is the receptor on the leukocyte responsible for recognizing the constant Fc domain of the antibody. Immunization with this mutant, referred to as nontoxigenic SpA, promoted an antibody response against S. aureus USA300 and clearing of staphylococci [130]. Multiple studies used this mutated protein A to purify mice and human monoclonal antibodies [130–132]. Falugi et al. attempted to parse out the role that protein A plays using a mouse model of abscess formation. The researchers were able to show that the SpAKKAA variant of protein A was similar in virulence by renal abscess and total CFU load after infection to the spa knockout mutant [29]. Notably, pretreating mice with the SpAKKAA strain prior to infection with wild type USA300 led to a marked increase in IgG antibodies against clumping factor A, fibronectin-binding protein B, iron-regulated surface determinant B, coagulase, α-toxin, and SpA [29]. Survival was also significantly increased upon challenging the mice with lethal dose of USA300 following pretreatment with this IgG antibody [29]. Use of this antibody was also able to confer protection in a murine neonate model of S. aureus infection [131]. Kim et al. were able to raise an antibody specifically to one of the five IgG domains of protein A (domain E), but antibody to this domain alone failed to provide increased protection to the murine host [133]. Overall, these findings demonstrate promising results using an immunization strategy with nontoxigenic SpAKKAA. One potential caveat with any such vaccine approach is the difficulty in translating results in animal models to meaningful human outcomes [134–136].
As protein A is a cell wall anchored protein, using an antibody targeting protein A and containing a gold nanorod, the Zanjani group was able to target S. aureus for selective killing by photothermal therapy [137]. The antibody with its conjugated nanoparticle targets the bacterial cell, followed by irradiation of the gold nanorod. The subsequent energy transfer introduces heat and other physical damage to the bacterial cell. While this method had been used to kill bacteria in previous in vitro studies, Shokri et al. demonstrated its promise in an in vivo mouse model, and observed a 73% reduction in bacterial cell viability in a MRSA infection [137]. This bacterial targeting approach is highly specific with the use of conjugated antibodies to specific factors; here protein A, and the gold nanorods have low levels of toxicity in mammalian systems [138]. This method may be used in conjunction with or entirely replace antibiotics for bactericidal-based treatment of bacterial infections. While resistance in the form of mutations to the binding site of the antibody is likely to occur over time, the combination of the photothermal therapy and standard antibiotic treatment could prove to be an innovative and novel approach to treat bacterial infections.
Current research demonstrates that protein A could be a viable vaccine candidate as well as serving as a S. aureus specific marker for potential selective photothermal killing of the bacterium. Antibody approaches to neutralize virulence factors in S. aureus have reached phase two clinical trials in the case of α-toxin, so similar success could be seen with SpA antibodies in the future.
6. PANTON-VALENTINE LEUKOCIDIN
Panton-Valentine Leukocidin was first described by Van deVelde in 1894 when he noticed its ability to lyse leukocytes [139, 140]. Later work on this toxin was done by Panton and Valentine who were able to associate the toxin with skin and tissue infections, and show that the hemolytic ability was distinct and not caused by the S. aureus leukocidin [139, 141]. Panton and Valentine were able to show that these phenotypes were distinct by isolating the toxin from S. aureus, then subjecting the purified toxin to rabbit blood or leukocytes isolated from human blood [141, 142]. The toxin consists of two distinct subunits that are both needed for its function, named LukS-PV and LukF-PV, with sizes of 38kDa and 32 kDa respectively [143, 144]. Upon column separation of the two subunits, macrophage lysis was found to be abolished, but lytic activity was restored upon combination of the two subunit fractions [143, 144]. Binding of the LukS subunit of the toxin to the neutrophil occurs first, and thus allows the binding of the LukF subunit to form the fully active toxin [145]. Binding order was determined by using 125I-labeled PVL subunits which were then incubated with PMNs for a given time, then washed and counted in a gamma counter [145]. Binding of LukF was only observed when PMNs were pretreated with LukS [145]. LukS binds to the human complement receptors C5aR and C5L2 which are abundant on human neutrophils, and protection is conferred using the C5aR inhibitor CHIPS [146]. Specific binding was determined by allowing the proteins to bind to neutrophils, then western blotting for the C5aR receptor [146]. An octamer of four LukF and four LukS subunits then forms the open pore in the cell [147]. In low concentrations, PVL localizes to the mitochondrial membrane and induces apoptosis [148]. Upon incubation of isolated mitochondria with rPVL, release of apoptotic proteins cytochrome c and Smac/DIABLO can be observed via western blot [148]. Genes encoding PVL are nearly universally found in strains of CA-MRSA, and thus there is a strong link between severe infection outcomes and the PVL genes in S. aureus strains [139, 149].
Despite this close correlation between severe disease outcomes and the presence of PVL genes, establishing the role that PVL plays during the course of infection remains controversial, with some studies stressing the importance of PVL, and others refuting the notion that PVL is important [150–152]. Some of the dispute regarding the importance of PVL is likely due to the differing susceptibilities of various eukaryotic cell types to PVL toxin. PVL is active against human leukocytes and rabbit leukocytes [153, 154] but not against mouse leukocytes [146, 153, 155].
Work by Tseng and others were able to generate humanized mice using CD24+ cells from umbilical blood that require one-two log less inoculum of S. aureus to generate lesions [156]. This decreased inoculum brings the CFU count of S. aureus down to the estimated human range of infection of 105 or 106 CFU [156–158]. Using the humanized mice they were also able to see significantly increased lesion size in wild type S. aureus infections as compared to infection with an isogenic PVL knockout strain, indicating that humanized mice models could more accurately represent the results observed in human cellular phenotypes [156]. Interesting findings reported by Otto et al. showed that expression of PVL and other virulence factors such as SpA could be inhibited by using sublethal concentrations of antibiotics [159]. Although this is an interesting finding, using subinhibitory concentrations of antibiotics for treatment might exacerbate antibiotic resistance and therefore not be a suitable approach [160–163]. In a study by Laventie et al., the class of compounds known as SCns (p-sulfonato-calix [n]arenes) were shown to be able to prevent the PVL LukS subunit from binding to the eukaryotic cell membrane using flow cytometry studies [164]. In rabbit models of infection, treatment with SCns was shown to be effective in decreasing ocular inflammation associated with purified PVL protein [164].
Further work is needed to clarify the importance of PVL in host pathogenesis by S. aureus, with consideration to its variable activity against diverse eukaryotic cell types. Assuming that PVL does indeed play a significant role in human infection, blocking its activity via a receptor inhibitor such as CHIPS or antibody could confer important protection during infection. The downside of using PVL inhibitors is that because the majority of HA-MRSA strains lack this gene, inhibitors to PVL would be ineffective against many HA-MRSA strains. Additionally, there are also virulent CA-MRSA strains that lack PVL genes [165]. Therefore, strain typing prior to treatment may be an approach with such selective antivirulence therapeutics.
7. STAPHYLOCOCCAL ENTEROTOXINS
Staphylococcal enterotoxins (SE) are also dubbed ‘superantigens’ for their ability to bind to human T-cells and induce hyperstimulation [166]. SEs specifically bind to MHC class II T-cell receptors [167, 168]. X-ray crystallography was used to image T-cell receptors coupled to SE [167]. Staphylococcal enterotoxins are the primary cause of staphylococcal food poisoning [169, 170]. SEs are very robust and are resistant to heating, and proteolysis [171, 172]. Biological activity of SE has been shown to be recalcitrant to heating to 60°C for up to 16 hours, or to digestion with trypsin, chymotrypsin, rennin, or papain [171]. Although SEs are a member of the streptococcal superantigen family, only toxins that cause emesis in primate models are designated SEs [173]. Currently there are twenty-one known SEs, designated SE-A to SE-U [172, 174, 175]. Due to the expense of testing emesis in primates and the limited number of labs capable of testing in primates, toxins similar to SEs but unable to be validated in primate models are designated as staphylococcal enterotoxin-like (SEls) [169, 173]. As little as 10 μg of SE-B administered intragastrically has been shown to be sufficient to cause disease symptoms in primate models [176]. There are limited data regarding human exposure to various purities of SEB as a result of lab accidents, and symptoms such as ocular swelling and discharge can be observed upon exposure to as little as 50 μg of toxin [177]. Due to the small quantity of protein needed for symptoms, the ease of spread, and the robust nature of SEs, they are listed as a Category B select agent by the CDC (CDC, [172]. Strains of S. aureus produce vastly different amounts of SE toxin, ranging from more than 1 μg/mL culture to less than 10 ng/mL [178]. SEs do not appear to be fully regulated using the agr system, and higher production of SE toxins appears to be linked with prophage induction [178, 179]. Transposon insertions in the agr allele was not found to affect overall levels of SE in S. aureus strains [179]. In strains of S. aureus that naturally produce high levels of SE-A (>1000 ng/mL measured via ELISA), induction of prophages using mitomycin C resulted in further increases of up to 10 fold [178].
Work by Bavari et al. found that immunizing mice with an antibody towards one SE would elicit cross-immunity against the other SE toxins [180]. Mice were challenged with recombinant SEs, and mice that had been immunized with antibodies raised against one particular SE received partial protection against challenge with a different SE isoform. This promising finding demonstrates the potential of developing an antibody species that would offer protection against many SE isoforms. Additional findings by Tiahun et al. showed that a mouse antibody raised against SE-B was readily able to inhibit the T-cell activation associated with SE-B, and this protective effect was found to extend to human PBMCs as well [181]. Mice antibodies raised to SE-B were also shown to be effective in human donor T-cells by Varsheny et al. 2011 [182]. T-cell activation from human donors treated with antibodies created to SE-B was up 50% compared to those that did not receive treatment [182]. The authors did not extend the test to other forms of SEs, but it would be interesting to test with multiple SEs given the results from Bavari et al. Similarly, human antibodies developed against SE-B showed increased survival in mice models of SE-B induced toxic shock syndrome [183]. Mice that were given antibodies raised to SE-B had a 68% increased survival as compared to mice that did not receive any antibodies [183]. Use of a mutated SE-B subunit as a vaccine approach also lead to increased immunity against SE-B toxicity [184]. Piglets immunized with the non-toxic form of SE-B resulted in increased levels of IgG and IgA levels that was shownt to cross react with wild type SE-B, indicating that this method may be a viable immunization strategy [184]. Recently, Reddy and colleagues were able to generate a recombinant peptide consisting of parts of SE-A and toxic shock syndrome toxin 1 in order to generate an immune response to these native toxins [185]. Immunization of the mice prior to cell-free toxin challenge resulted in a 50–80% survival rate when confronted with a lethal dose of SE-A [185]. Aptamers have also been shown to be effective in inhibiting SE function [186]. Aptamers created to bind specifically to SE-A were used in a mouse challenge model, where a 60% survival increase was observed when mice were pretreated with aptamers prior to toxin challenge (10 ug SE-A) vs. no pretreatment.
Given the promising findings regarding immunity generated towards SEs using antibodies, aptamers, or mutated forms of the SE protein itself, antivirulence and vaccine-based approaches against these potent Staphylococcal enterotoxins may offer significant potential for the treatment of severe S. aureus disease.
8. TWO-COMPONENT SYSTEMS
Bacterial two-component systems (TCS) are one of the primary means bacteria have to sense their environment [187, 188]. As the name suggests, TCSs consist of two proteins, a sensor kinase which responds to the signal and a response regulator that binds to DNA [187, 188]. In S. aureus, these TCS can affect the virulence of the bacterium. agr is an important regulator of virulence and is also a TCS consisting of the response regulator AgrA and the kinase AgrC. The SaeR/S TCS in essential to the evasion of the innate immune system by reducing reactive oxygen species, aiding survival in the face of host defense peptides, reducing expression of IL-8, and increasing the expression of leukocidins [32, 40, 42, 189, 190]. SaeRS can also regulate the expression of virulence genes including hla [191].
Work has shown that SaeRS can be inhibited chemically in order to reduce the expression of virulence genes. Using a natural product isolated from Glycyrrhiza, dubbed 18β-Glycyrrhetinic acid (GRA), the Voyich group was able to show that at high concentrations the compound was able to kill S. aureus, and at sub-lethal concentrations, virulence factor genes including saeR were inhibited [192]. The authors determined that during the course of an in vivo infection where the mice were treated with MRSA and GRA, the CFUs recovered were not significantly different [192]. While the CFUs were unchanged, the lesion size of the mice were significantly reduced, implying that the GRA treatment was having an anti-virulence effect as opposed to an antibiotic effect [192]. To further explore this phenomenon, the authors measured the change in gene expression upon treatment with GRA. Of the five genes examined, only saeR was significantly different with a 14 fold reduction in transcription [192]. This paper illustrates a possible ideal in treatment of bacterial infections. At high doses the compound is able to kill the S. aureus, but even at lower doses, the virulence of the bacteria is inhibited.
Using a small molecular inhibitor of staphylococcal type 2 fatty acid synthesis called AFN-1252, Parsons et al. were able to show that in treatment of S. aureus with AFN-1252 led to a reduction in the expression of the sae genes [193]. Using an Affymetrix array, the authors were able to observe a ~2 fold reduction in the saeS and saeR genes [193]. When treatment was translated to an in vivo model, there was a significant reduction in the amount of recoverable CFUs of more than three log after 72 hours following a single dose of AFN-1252 [193]. Upon multiple doses of AFN-1252, the recoverable CFU was reduced nearly 5 log [193]. The authors noted that the timing of treatment also had a significant impact on the level of sae gene transcription as cells that were actively growing upon time of treatment had a much more robust reduction in sae levels as compared to when the bacteria were treated at stationary phase [193]. This paper has again reinforced the idea that targeting these two component systems can have a positive impact on infections, however in this case the bacterial growth dynamics can have a large effect on the efficacy of the treatment.
TCS inhibiton has also been shown to cause rapid bactericidal effects [194–196]. Specifically targeting the WalK/WalR (YycG/YycF) two component system which is essential to cell wall metabolism in S. aureus results in death of the bacteria [194–196]. Work by Okada et al. isolated a natural product from Streptomyces sp. MK632-100F11 dubbed walkmycin B which exhibited antibacterial activity against B. subtilis [195]. The compound also exhibited antibacterial characteristics against both a MRSA strain and a MSSA strain [195]. Walkmycin was determined to be active against the WalK TCS protein component by observing a decrease in autophosphorylation of WalK when purified WalK was exposed to the drug treatment [195]. This interaction between walkmycin B and the TCS protein was confirmed by surface plasmon resonance using the B. subtilis WalK protein [195].
Similar research was also undertaken by Igarashi et al. The authors were also able to isolate a novel species of Streptomyces dubbed MK844-mF10 that produced an active compound against the WalK protein from B. subtilis [196]. The compound, dubbed waldiomycin bears structural similarity to diozamycin and was also active against several strains of S. aureus including two different MRSA strains with a MIC of 16 μg/mL [196]. Waldiomycin was determined to prevent the autophosphorylation of WalK for both the B. subtilis and S. aureus purified protein product [196].
Screening of targets to inhibit TCS can be done with high throughput fragment screening or with structure-based virtual screening [197]. A recent publication utilized both approaches in order to find new compounds to inhibit TCS. The authors used an in silico approach to screen 600,000 compounds from a commercially available drug database, and 898 compound fragment library [197]. Ten final compounds from the in silico approach were tested experimentally, and 25 compounds from the fragment library were tested [197]. Of the ten compounds selected from the fragment library, two hits yielded effective inhibition of S. aureus, compounds F1 and F2 [197]. Along with F1 and F2, derivatives of one of the in silico compounds were tested against eight strains of S. aureus, with the in silico derived compound S1.13 able to inhibit all 8 strains between 8 μg/mL and 16 μg/mL [197]. This paper was able to show the benefit of using in silico and fragment based approaches to rapidly screen large numbers of compounds to generate leads which can then be modified to obtain very effective antibacterial agents.
Two component systems remain a viable area of research to either inhibit the virulence of the S. aureus bacterium in the case of SaeR and SaeS inhibition or as an antibacterial target. Dual approaches to screening compounds in the form of in silico or fragment library screening can generate positive hits from an initially massive library, which will ideally result in more rapid drug development times.
9. STAPHYLOXANTHIN
Staphyloxanthin is the pigment that S. aureus produces that is primarily responsible for the characteristic golden color that some strains of S. aureus exhibit [198]. This compound was originally isolated from S. aureus by Marshall and Wilmoth in 1981. Using a warm methanol extraction method followed by chromatography separation, they were able to obtain four fractions, and fraction three contained the staphyloxanthin [198]. Subsequent work using gene deletions in the staphyloxanthin biosynthesis pathway confirmed that staphyloxanthin functions as a virulence factor by serving as an antioxidant protecting the bacterium from neutrophil oxidative burst [199, 200].
The first step in the synthesis of staphyloxanthin is catalyzed by the dehydrosqualene synthase (CrtM) enzyme [199–201]. Liu et al. noticed that the structure of dehydrosqualene is similar to squalene in human cholesterol biosynthesis, and thus they hypothesized that inhibitors to human cholesterol synthesis may be effective against S. aureus producing staphyloxanthin [201]. Out of eight compounds tested, three phosphonosulfonates were found to be effective against pigment formation in vitro [201]. One of the compounds, BPH-652, is a derivative of S-BPH-652 which is a compound in preclinical trials as a human cholesterol lowering drug, and for these reasons, the authors chose to test BPH-652 against eukaryotic cell toxicity, the ability of phagocytes to kill S. aureus, and in vivo infections [201]. Researchers found no evidence of eukaryotic toxicity, and upon treatment of S. aureus with BPH-652, the bacteria were more susceptible to being killed in whole blood assays by a factor of 4, and were more susceptible to killing by hydrogen peroxide [201]. During an in vivo mice infection, the number of colonies recovered from kidneys were reduced by ~98% [201].
Follow up work from the same Oldfield group focused on synthesizing derivatives of BHP-652 and characterizing the compounds [202–204]. They were able to develop a computer model to more accurately predict compounds that were active against CrtM and also compounds that would have low affinity for the human analogue SQS [202]. From this paper the researchers found that two compounds had increased selectivity for CrtM as compared to SQS while maintaining similar inhibitory kinetics on CrtM [202]. One of the most potent inhibitors to CrtM the authors were able to obtain x-ray crystallographic structures of the inhibitor bound to the CrtM enzyme [203]. Interestingly, they noted that the inhibitor spanned both of the substrate biding sites on CrtM, unlike other compounds that selectively bound either one or the other substrate sites [203]. Later work by the Oldfield group noticed that inhibiting the SQS enzyme in neutrophils resulted in an increase in NET production [204, 205]. Based on this observation, the authors decided to create a compound that would be highly effective against both the human SQS and the bacterial CrtM [204]. Extensive in silico screening and crystallization with CrtM led the authors to finding a compound dubbed 16, which stimulated NET production, and also inhibited S. aureus growth [204].
Screening for staphyloxanthin inhibition can be accomplished by treating S. aureus with the compound of interest, and then observing the decrease in golden color [206]. Recently Chen and colleagues used that approach to screen 412 drugs to find drugs which inhibited staphyloxanthin biosynthesis [206]. They were able to find three compounds that resulted in the loss of pigmentation in S. aureus Newman, ibandronate, terbinafine and naftifine the last of which inhibited staphyloxanthin production most significantly [206]. Bacteria treated with naftifine exhibited increased sensitivity to hydrogen peroxide, and were 20 fold decreased in survival in human whole blood [206]. In a sepsis model of mice infection, 70% of mice that received naftifine treatment survived 12 days where all the mock treated mice were dead after 24 hours [206]. Using E. coli expression methods, the authors were able to show that naftifine is able to inhibit the activity of CrtN [206]. While these results are very promising, the authors do note that not all strains of S. aureus produce staphyloxanthin, and thus the treatment may be of limited utility in non-staphyloxanthin producing strains [206].
Despite the fact that not all strains of S. aureus produce the staphyloxanthin pigment, in the cases where it is present in the bacterium, inhibition of the molecule can be highly effective. Taken together, these findings indicate that inhibiting the synthesis of staphyloxanthin could be a viable anti-virulence strategy and that the use of computer inhibition models can help to inform compound synthesis for the modification of existing compounds.
10. PHAGE THERAPY
Bacteriophages have also been explored as a possible treatment against MRSA infection. In a recent case study, a woman who had been infected with a persistent vancomycin resistant S. aureus infection exhibited colonization for over 11 years [207]. Despite constant and varied treatment including cyclosporine, pristanamycine, and mupirocin, the infection never successfully cleared [207]. The patent decided to seek treatment at the phage therapy center in Tbilisi Georgia. Upon treatment with a bacteriophage ATCC PTA-9476 over the course of four weeks, the vancomycin resistant S. aureus infection was successfully cleared [207]. Although such examples highlight the promise of phage therapies, bacterophages are generally highly specific for bacterial strains and thus can exhibit lower efficiency within strains of the same species. In one report where researchers attempted to isolate additional strains of bacteriophage active against 12 different strains of MRSA using phage isolated from sewage sources, livestock areas, a river, and a water lock from a room that treats MRSA patients, only 5 different phages could be found to be active against MRSA strains [208]. The isolated phages also exhibited very poor cross-reactivity against additional MRSA strains [208]. Out of 12 tested MRSA strains, only two of the bacteriophages exhibited activity in 5/12 MRSA strains, and the other 3 phage were limited to 1/12 tested MRSA strains [208]. Using a commercially available S. aureus bacteriophage ATCC PTA-9476, researchers tested the ability of a phage to inhibit three different strains of S. aureus, USA300 LAC, USA100 71080, and USA100 626 [209]. The authors found that while the bacteriophage was able to inhibit the growth of USA300 and the USA100 626 strain, it was ineffective against the USA100 71080 strain [209]. When used specifically against the USA300 LAC strain, the phage highly effective in reducing the infection against this S. aureus strain, and this protection was extended to several routes of infection [209]. Phage treatment has also been proposed to prevent infections during implanted medical devices [210]. S. aureus has also been a major concern in causing hospital associated infections especially during surgeries where devices are implanted [211–213]. Wires impregnated with a biodegradable polymer containing a specific bacteriophage in combination with an antibiotic compound were implanted into mice, followed by infection with the S. aureus MRSA strain 43300 [210]. Mice that were implanted with the wire containing both the phage and antibiotic treatments exhibited significantly lower CFU counts [210]. Using a functional measure of how well the mice healed post-surgery, mice who received both bacteriophage and antibiotic treatment exhibited faster recovery than mice who received untreated wires or mono phage or antibiotic treatment [210].
Bacteriophage treatment of bacterial infection remains a promising area of research in the context of S. aureus pathogenesis. The studies highlighted above demonstrate the potential that phage therapy may offer, especially as an alternative to classical antibiotic treatment or in combination with available therapies to increase the effective of treatment strategies. One limitation to phage therapy has been the high level of strain specificity of phages against particular S. aureus isolates, making it difficult to develop a strain of phage that would act broadly against all pathogenic strains. Regulatory approval will also likely remain an area of challenge as there are currently no FDA approved phages for treatment of S. aureus infections. Resistance to phage could be slower to arise as the phages are a biological system and thus can co-evolve along with resistance by the susceptible bacteria.
11. VACCINE DEVELOPMENT FOR S. AUREUS
Vaccine approaches to Staphylococcus aureus have been thus far proven unsuccessful when progressed to clinical trials [135, 136]. As discussed in this review, S. aureus has a wide range of potential vaccine targets, including capsular, cell wall anchored proteins, and secreted factors, but a vaccine specific to only one of these factors would likely not elicit total protective immunity [25]. Similar to many of the anti-virulence approaches discussed herein, it may be more feasible to utilize vaccine-based strategies to limit the severity of infection while administering other treatments such as antibiotics to reduce the severity and duration of the overall infection [214]. Despite the current lack of a universal vaccine against S. aureus, significant research into Staph-based vaccines are ongoing. A recent four-component vaccine consisting of two capsular polysaccharides conjugated to tetanus toxoid, mutated α-toxin, and clumping factor A has been developed for S. aureus [215, 216]. This vaccine has recently completed phase one clinical trials with no safety concerns and with humoral immune responses noted.
Methods of vaccine delivery also play a pivotal role in the efficacy of vaccine treatment. Gomes and colleagues compared the efficiencies of a S. aureus whole antigen delivered either intranasally or intramuscularly [217]. Mice vaccinated intranasally with the whole antigen exhibited significantly increased production of IgG2a as compared to mice who were vaccinated the intramuscular route as measured via serum ELISA [217]. Importantly, the authors observed no detrimental effects on the mice during intranasal vaccination whereas the mice vaccinated intramuscularly exhibited redness and lameness near the injection site [217]. Upon challenge with 108 CFU of S. aureus intraperitoneally, mice who had been vaccinated intranasally exhibited significantly increased survival (95% survival after 72 hrs as compared to 60%) and significantly lower CFU counts in liver and spleen [217]. In another study by Selle et al., the route of vaccination was shown to have a significant effect on the disease outcome of S. aureus challenge [217]. When challenged with fluorescently labeled S. aureus the mice that were vaccinated I.V. exhibited significantly lowered luminescence and ~100 fold fewer CFUs than those vaccinated using an intramuscular route [217]. Interestingly, the route of vaccine also affected the antibody profiles of the mice, with I.V. vaccinated mice exhibiting greater levels of antibody to PVL, gamma toxin, and beta hemolysin whereas mice vaccinated intramuscularly exhibited greater antibody titers to intracellular antigens GreA and Tuf [218].
CONCLUSION
S. aureus is a versatile bacterial pathogen as demonstrated both by its ability to acquire rapid resistance to new antibiotics as well as its use of a wide arsenal of virulence factors that have evolved to specifically survive host immune responses for disease spread. Due to the prevalence of MRSA, and the current limitations on antibiotic development, an active and highly promising avenue of research has been to develop strategies to specifically inhibit the activity of virulence factors produced S. aureus as an alternative means to treat disease. This review has highlighted several virulence factors where mechanistic studies elucidating the structural and functional properties of these virulence factors have paved the way for exciting new strategies to inhibit or otherwise inactivate these virulence factors and ameliorate pathogenic outcomes. Strategies to inhibit virulence factors can range from small molecule inhibitors, to antibodies, to mutant and toxoid forms of the virulence proteins. Of particular interest are those inhibitors that have been shown to have cross reactivity to multiple virulence factor forms, as well as inhibitors that target global regulators such as the agr operon, which have multi-target activities. Current efforts to develop specific antivirulence strategies, vaccine approaches, and alternative therapies such as phages for treating severe disease caused by S. aureus have the potential to stem the tide against the limitations that we face in the post-antibiotic era.
Acknowledgments
Trevor Kane is supported by a fellowship from the Eck Institute of Global Health, University of Notre Dame. Katelyn Carothers is supported by a teaching fellowship from the Eck Institute of Global Health, University of Notre Dame. Shaun Lee is supported by NIH Grant 1DP2OD008468-01.
Footnotes
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Weese JS, van Duijkeren E. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet Microbiol. 2010;140(3–4):418–429. doi: 10.1016/j.vetmic.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 2.Katakweba AS, Muhairwa AP, Espinosa-Gongora C, Guardabassi L, Mtambo MMA, Olsen JE. Spa typing and antimicrobial resistance of staphylococcus aureus from healthy humans, pigs and dogs in Tanzania. J Infect Dev Ctries. 2016;10(2):143–148. doi: 10.3855/jidc.6790. [DOI] [PubMed] [Google Scholar]
- 3.van Belkum A, Melles DC, Nouwen J, et al. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol. 2009;9(1):32–47. doi: 10.1016/j.meegid.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Benson PF, Rankin GLS, Rippey JJ. an Outbreak of Exfoliative Dermatitis of the Newborn (Ritter’S Disease) Lancet. 1962;279(7237):999–1002. doi: 10.1016/s0140-6736(62)92036-6. [DOI] [PubMed] [Google Scholar]
- 5.Parker MT, Tomlinson AJ, Williams RE. Impetigo contagiosa; the association of certain types of Staphylococcus aureus and of Streptococcus pyogenes with superficial skin infections. J Hyg (Lond) 1955 Dec;53(4):458–73. doi: 10.1017/s0022172400000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MITCHELL-HEGGS GB, CROW KD. Folliculitis decalvans associated with penicillin-resistant Staph. aureus infection. Proc R Soc Med. 1946 Sep;39(11):686. [PubMed] [Google Scholar]
- 7.Archer GL. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis. 1998;26(5):1179–81. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 8.Ogston A. Micrococcus Poisoning. J Anat Physiol. 1882;17(Pt 1):24–58. [PMC free article] [PubMed] [Google Scholar]
- 9.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998 Aug 20;339(8):520–32. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki Y, Matsushita S, Kubota H, et al. Identification and functional activity of a staphylocoagulase type XI variant originating from staphylococcal food poisoning isolates. Lett Appl Microbiol. 2016;63(3):172–7. doi: 10.1111/lam.12595. [DOI] [PubMed] [Google Scholar]
- 11.Corpening A, Foxhall EP. Outbreak of Food Poisoning, Probably Due to Staphylococcus Aureus. Am J Public Health Nations Health. 1935;25(8):938–40. doi: 10.2105/ajph.25.8.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616–87. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skinner DKC. Significance of bacteremia caused by staphylococcus aureus. Arch Intern Med. 1941;68(5):851. [Google Scholar]
- 14.Abraham EP, Chain E, Fletcher CM, et al. Further observations on penicillin. Lancet. 1941;238(6155):177–189. [Google Scholar]
- 15.Jevons P. “Celbenin”-resistant Staphylococci. Br Med J. 1961;1(5219):124–125. [PMC free article] [PubMed] [Google Scholar]
- 16.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368(9538):874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 17.Saravolatz LD, Stein GE, Johnson LB. Ceftaroline: A novel cephalosporin with activity against methicillin-resistant staphylococcus aureus. Clin Infect Dis. 2011;52(9):1156–63. doi: 10.1093/cid/cir147. [DOI] [PubMed] [Google Scholar]
- 18.Barbour A, Schmidt S, Rand KH, Derendorf H. Ceftobiprole: a novel cephalosporin with activity against Gram-positive and Gram-negative pathogens, including methicillin-resistant Staphylococcus aureus (MRSA) Int J Antimicrob Agents. 2009;34(1):1–7. doi: 10.1016/j.ijantimicag.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Peacock SJ, Paterson GK. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu Rev Biochem. 2015;84(1):577–601. doi: 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- 20.Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008;8(6):747–763. doi: 10.1016/j.meegid.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Stryjewski ME, Corey GR. Methicillin-resistant staphylococcus aureus: An evolving pathogen. Clin Infect Dis. 2014;58(SUPPL 1):10–19. doi: 10.1093/cid/cit613. [DOI] [PubMed] [Google Scholar]
- 22.Kos VN, Desjardins CA, Griggs A, et al. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with Methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio. 2012;3(3):1–9. doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298(15):1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 24.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787–794. doi: 10.1086/528716. [DOI] [PubMed] [Google Scholar]
- 25.Daum RS, Spellberg B. Progress toward a Staphylococcus aureus vaccine. Clin Infect Dis. 2012;54(4):560–7. doi: 10.1093/cid/cir828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowy F. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111(9):1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu Rev Microbiol. 2010;64:143–62. doi: 10.1146/annurev.micro.112408.134309. [DOI] [PubMed] [Google Scholar]
- 28.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375(9725):1557–68. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falugi F, Kim HK, Missiakas DM, Schneewind O. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. MBio. 2013;4(5):e00575–13. doi: 10.1128/mBio.00575-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bröker B, Mrochen D, Péton V. The T Cell Response to Staphylococcus aureus. Pathogens. 2016;5(1):31. doi: 10.3390/pathogens5010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaili S, Cheung AL, Bayer AS, et al. The GraS sensor in Staphylococcus aureus mediates resistance to host defense peptides differing in mechanisms of action. Infect Immun. 2015;84(2):459–466. doi: 10.1128/IAI.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen MT, Kraft B, Wenqi Y, et al. Correction: The νSaα Specific Lipoprotein Like Cluster (lpl) of S. aureus USA300 Contributes to Immune Stimulation and Invasion in Human Cells (PLoS Pathog, 11-6, (2015), 10.1371/journal.ppat.1004984) PLoS Pathog. 2015;11(9):1–27. doi: 10.1371/journal.ppat.1004984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Haas CJC, Veldkamp KE, Peschel A, et al. Chemotaxis Inhibitory Protein of Staphylococcus aureus, a Bacterial Antiinflammatory Agent. J Exp Med. 2004;3687900(5):687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rooijakkers SH, Ruyken M, Roos A, et al. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat Immunol. 2005;6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 35.Haupt K, Reuter M, Van Den Elsen J, et al. The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with host complement factor H and C3b. PLoS Pathog. 2008;4(12) doi: 10.1371/journal.ppat.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stapels DA, Ramyar KX, Bischoff M, et al. Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors. Proc Natl Acad Sci USA. 2014;111(36):13187–92. doi: 10.1073/pnas.1407616111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilke GA, Bubeck J. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus α-hemolysin - mediated cellular injury. Proc Natl Acad Sci USA. 2010;107(30):13473–8. doi: 10.1073/pnas.1001815107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thammavongsa V, Missiakas D, Schneewind O. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science. 2013;342(6160):863–6. doi: 10.1126/science.1242255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerra FE, Addison CB, de Jong NWM, et al. Staphylococcus aureus SaeR/S-regulated factors reduce human neutrophil reactive oxygen species production. J Leukoc Biol. 2016 Nov;100:1–6. doi: 10.1189/jlb.4VMAB0316-100RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGuinness W, Kobayashi S, DeLeo F. Evasion of Neutrophil Killing by Staphylococcus aureus. Pathogens. 2016;5(1):32. doi: 10.3390/pathogens5010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zurek OW, Pallister KB, Voyich JM. Staphylococcus aureus Inhibits Neutrophil-derived IL-8 to Promote Cell Death. J Infect Dis. 2015;212(6):934–8. doi: 10.1093/infdis/jiv124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thammavongsa V, Kim HK, Missiakas D, Schneewind O. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol. 2015;13(9):529–43. doi: 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spaan AN, Surewaard BGJ, Nijland R, van Strijp JA. Neutrophils Versus Staphylococcus aureus: A Biological Tug of War. Annu Rev Microbiol. 2013;67:629–50. doi: 10.1146/annurev-micro-092412-155746. [DOI] [PubMed] [Google Scholar]
- 44.Lacey KA, Geoghegan JA, McLoughlin RM. The Role of Staphylococcus aureus Virulence Factors in Skin Infection and Their Potential as Vaccine Antigens. Pathog (Basel, Switzerland) 2016;5(1):22. doi: 10.3390/pathogens5010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28(3):603–61. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Projan SJ. Whither antibacterial drug discovery? Drug Discov Today. 2008;13(7–8):279–80. doi: 10.1016/j.drudis.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 47.Schäberle TF, Hack IM. Overcoming the current deadlock in antibiotic research. Trends Microbiol. 2014;22(4):165–7. doi: 10.1016/j.tim.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Magana M, Ioannidis A, Magiorkinis E, et al. Therapeutic Options and Emerging Alternatives for Multidrug Resistant Staphylococcal Infections. 2015:2058–2072. doi: 10.2174/1381612821666150310101851. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez-Ruiz A, Seaton RA, Hamed K. Daptomycin: an evidence-based review of its role in the treatment of Gram-positive infections. Infect Drug Resist. 2016;9(10):47–58. doi: 10.2147/IDR.S99046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russo A, Concia E, Cristini F, et al. Current and future trends in antibiotic therapy of acute bacterial skin and skin-structure infections. Clin Microbiol Infect. 2016;22(Suppl 2):S27–36. doi: 10.1016/S1198-743X(16)30095-7. [DOI] [PubMed] [Google Scholar]
- 51.Ruffin M, Bilodeau C, Maillé É, et al. Quorum sensing inhibition abrogates the deleterious impact of Pseudomonas aeruginosa on airway epithelial repair. FASEB J. 2016;(15):1–14. doi: 10.1096/fj.201500166R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vadekeetil A, Saini H, Chhibber S, Harjai K. Exploiting the anti-virulence efficacy of an ajoene-ciprofloxacin combination against Pseudomonas aeruginosa biofilm associated murine acute pyelonephritis. Biofouling. 2016;32(4):371–82. doi: 10.1080/08927014.2015.1137289. [DOI] [PubMed] [Google Scholar]
- 53.Jarvis C, Han Z, Kalas V, et al. Antivirulence Isoquinolone Mannosides: Optimization of the Biaryl Aglycone for FimH Lectin Binding Affinity and Efficacy in the Treatment of Chronic UTI. ChemMedChem. 2016;11(4):367–73. doi: 10.1002/cmdc.201600006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bender KO, Garland M, Ferreyra JA, et al. A small-molecule anti-virulence agent for treating Clostridium difficile infection. Sci Transl Med. 2015;7(306):306ra148. doi: 10.1126/scitranslmed.aac9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dean SN, van Hoek ML. Screen of FDA-approved drug library identifies maprotiline, an antibiofilm and antivirulence compound with QseC sensor-kinase dependent activity in Francisella novicida. Virulence. 2015;6(5):487–503. doi: 10.1080/21505594.2015.1046029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson BK, Colvin CJ, Needle DB, Mba Medie F, Champion PAD, Abramovitch RB. The Carbonic Anhydrase Inhibitor Ethoxzolamide Inhibits the Mycobacterium tuberculosis PhoPR Regulon and Esx-1 Secretion and Attenuates Virulence. Antimicrob Agents Chemother. 2015;59(8):4436–45. doi: 10.1128/AAC.00719-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gruss A, Novick RP. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol Gen Genet. 1986;202(1):58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 58.Queck SY, Jameson-Lee M, Villaruz AE, et al. RNAIII-Independent Target Gene Control by the agr Quorum-Sensing System: Insight into the Evolution of Virulence Regulation in Staphylococcus aureus. Mol Cell. 2008;32(1):150–8. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48(6):1429–49. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 60.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12(10):3967–75. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of ROT translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61(4):1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 62.Boisset S, Geissmann T, Huntzinger E, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator ROT by an antisense mechanism. Genes Dev. 2007;21(11):1353–66. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gray B, Hall P, Gresham H. Targeting agr- and agr-Like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. Sensors (Basel) 2013;13(4):5130–66. doi: 10.3390/s130405130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gong J, Li D, Yan J, Liu Y, Li D, Dong J, et al. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine intracranial abscesses model. Brazilian J Infect Dis. 2014;18(5):501–6. doi: 10.1016/j.bjid.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wardenburg JB, Patel RJ, Schneewind O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun. 2007;75(2):1040–4. doi: 10.1128/IAI.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heyer G, Saba S, Adamo R, et al. Staphylococcus aureus agr and sarA functions are required for invasive infection but not inflammatory responses in the lung. Infect Immun. 2002;70(1):127–33. doi: 10.1128/IAI.70.1.127-133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montgomery CP, Boyle-Vavra S, Daum RS. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One. 2014;5(12):e15177. doi: 10.1371/journal.pone.0015177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fowler VG, Jr, Sakoulas G, McIntyre LM, et al. Persistent Bacteremia Due to Methicillin-Resistant Staphylococcus aureus Infection Is Associated with agr Dysfunction and Low-Level In vitro Resistance to Thrombin-Induced Platelet Microbicidal Protein. J Infect Dis. 2004;190(6):1140–9. doi: 10.1086/423145. [DOI] [PubMed] [Google Scholar]
- 69.Kang CK, Cho JE, Choi YJ, et al. agr Dysfunction Affects Staphylococcal Cassette Chromosome mec Type-Dependent Clinical Outcomes in Methicillin-Resistant Staphylococcus aureus Bacteremia. Antimicrob Agents Chemother. 2015;59(6):3125–32. doi: 10.1128/AAC.04962-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shopsin B, Drlica-Wagner A, Mathema B, Adhikari RP, Kreiswirth BN, Novick RP. Prevalence of agr Dysfunction among Colonizing Staphylococcus aureus Strains. J Infect Dis. 2008;198(8):1171–4. doi: 10.1086/592051. [DOI] [PubMed] [Google Scholar]
- 71.Shopsin B, Eaton C, Wasserman GX, et al. Mutations in agr Do Not Persist in Natural Populations of Methicillin− Resistant Staphylococcus aureus. J Infect Dis. 2010:202. doi: 10.1086/656915. [DOI] [PubMed] [Google Scholar]
- 72.McDanel JS, Perencevich EN, Diekema DJ, et al. Association between microbial characteristics and poor outcomes among patients with methicillin-resistant Staphylococcus aureus pneumonia: a retrospective cohort study. Antimicrob Resist Infect Control. 2015;4:51. doi: 10.1186/s13756-015-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276(5321):2027–30. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 74.Jarraud S, Lyon GJ, Figueiredo A, et al. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol. 2000;182(22):6517–22. doi: 10.1128/jb.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lyon GJ, Mayville P, Muir TW, Novick RP. Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc Natl Acad Sci USA. 2000;97(24):13330–5. doi: 10.1073/pnas.97.24.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sully EK, Malachowa N, Elmore BO, et al. Selective Chemical Inhibition of agr Quorum Sensing in Staphylococcus aureus Promotes Host Defense with Minimal Impact on Resistance. PLoS Pathog. 2014;10(6) doi: 10.1371/journal.ppat.1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nielsen A, Manåsson M, Bojer MS, et al. Solonamide B inhibits quorum sensing and reduces Staphylococcus aureus mediated killing of human neutrophils. PLoS One. 2014;9(1):1–10. doi: 10.1371/journal.pone.0084992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baldry M, Kitir B, Frøkiær H, et al. The agr Inhibitors Solonamide B and Analogues Alter Immune Responses to Staphylococccus aureus but Do Not Exhibit Adverse Effects on Immune Cell Functions. PLoS One. 2016;11(1):e0145618. doi: 10.1371/journal.pone.0145618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Daly SM, Elmore BO, Kavanaugh JS, et al. ω-Hydroxyemodin Limits Staphylococcus Aureus Quorum Sensing-Mediated Pathogenesis and Inflammation. Antimicrob Agents Chemother. 2015;59(4):2223–35. doi: 10.1128/AAC.04564-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alksne LE, Projan SJ. Bacterial virulence as a target for antimicrobial chemotherapy. Curr Opin Biotechnol. 2000;11(6):625–36. doi: 10.1016/s0958-1669(00)00155-5. [DOI] [PubMed] [Google Scholar]
- 81.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3(9):541–8. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 82.Caballero A, Foletti D, Bierdeman M, et al. Effectiveness of Alpha-toxin Fab Monoclonal Antibody Therapy in Limiting the Pathology of Staphylococcus aureus Keratitis. Ocul Immunol Inflamm. 2014 Apr;:1–7. doi: 10.3109/09273948.2014.920035. [DOI] [PubMed] [Google Scholar]
- 83.Powers ME, Kim HK, Wang Y, Bubeck Wardenburg J. ADAM10 mediates vascular injury induced by Staphylococcus aureus α-hemolysin. J Infect Dis. 2012;206(3):352–6. doi: 10.1093/infdis/jis192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schulz B, Pruessmeyer J, Maretzky T, et al. ADAM10 regulates endothelial permeability and T-Cell transmigration by proteolysis of vascular endothelial cadherin. Circ Res. 2008;102(10):1192–201. doi: 10.1161/CIRCRESAHA.107.169805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saftig P, Reiss K. The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: Novel drug targets with therapeutic potential? Eur J Cell Biol. 2011;90(6–7):527–35. doi: 10.1016/j.ejcb.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 86.Zhu Y, Fan X, Zhang X, et al. Structure of ROT, a global regulator of virulence genes in Staphylococcus aureus. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 9):2467–76. doi: 10.1107/S1399004714015326. [DOI] [PubMed] [Google Scholar]
- 87.Jeng BH, Gritz DC, Kumar AB, et al. Epidemiology of ulcerative keratitis in Northern California. Arch Ophthalmol (Chicago, Ill 1960) 2010;128(8):1022–8. doi: 10.1001/archophthalmol.2010.144. [DOI] [PubMed] [Google Scholar]
- 88.Kerr N, Stern GA. Bacterial keratitis associated with vernal kerato-conjunctivitis. Cornea. 1992;11(4):355–9. doi: 10.1097/00003226-199207000-00015. [DOI] [PubMed] [Google Scholar]
- 89.Qiu J, Niu X, Dong J, et al. Baicalin protects mice from Staphylococcus aureus pneumonia via inhibition of the cytolytic activity of α-hemolysin. J Infect Dis. 2012;206(2):292–301. doi: 10.1093/infdis/jis336. [DOI] [PubMed] [Google Scholar]
- 90.Hua L, Hilliard JJ, Shi Y, et al. Assessment of an anti-alpha-toxin monoclonal antibody for prevention and treatment of Staphylococcus aureus-induced pneumonia. Antimicrob Agents Chemother. 2014;58(2):1108–17. doi: 10.1128/AAC.02190-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hua L, Cohen TS, Shi Y, et al. MEDI4893* promotes survival and extends the antibiotic treatment window in a Staphylococcus aureus immunocompromised pneumonia model. Antimicrob Agents Chemother. 2015;59(8):4526–32. doi: 10.1128/AAC.00510-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kennedy AD, Bubeck Wardenburg J, Gardner DJ, et al. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J Infect Dis. 2010;202(7):1050–8. doi: 10.1086/656043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Menzies BE, Kernodle DS. Passive immunization with antiserum to a nontoxic alpha-toxin mutant from Staphylococcus aureus is protective in a murine model. Infect Immun. 1996;64(5):1839–41. doi: 10.1128/iai.64.5.1839-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Menzies BE, Kernodle DS. Site-directed mutagenesis of the alpha-toxin gene of Staphylococcus aureus: Role of histidines in toxin activity in vitro and in a murine model. Infect Immun. 1994;62(5):1843–7. doi: 10.1128/iai.62.5.1843-1847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rouha H, Badarau A, Visram ZC, et al. Five birds, one stone: Neutralization of α-hemolysin and 4 bi-component leukocidins of Staphylococcus aureus with a single human monoclonal antibody. MAbs. 2015;7(1):243–54. doi: 10.4161/19420862.2014.985132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oganesyan V, Barnes A, Tkaczyk C, Ferguson A, Wu H, Dall’Acqua WF. Crystallization and preliminary X-ray diffraction analysis of the complex between a human anti-alpha toxin antibody fragment and alpha toxin. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69(Pt 3):302–5. doi: 10.1107/S1744309113002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chua KYL, Monk IR, Lin Y, et al. Hyperexpression of α-hemolysin explains enhanced virulence of sequence type 93 community-associated methicillin-resistant Staphylococcus aureus. BMC Microbiol. 2014;14(1):31. doi: 10.1186/1471-2180-14-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cheung GYC, Joo HS, Chatterjee SS, Otto M. Phenol-soluble modulins - critical determinants of staphylococcal virulence. FEMS Microbiol Rev. 2014;38(4):698–719. doi: 10.1111/1574-6976.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mehlin C, Headley CM, Klebanoff SJ. An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J Exp Med. 1999;189(6):907–18. doi: 10.1084/jem.189.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang R, Braughton KR, Kretschmer D, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13(12):1510–4. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 101.Queck SY, Khan BA, Wang R, et al. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 2009;5(7):1–12. doi: 10.1371/journal.ppat.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Surewaard BGJ, De Haas CJC, Vervoort F, et al. Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cell Microbiol. 2013 Mar;15:1427–37. doi: 10.1111/cmi.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tsompanidou E, Denham EL, Becher D, et al. Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces. Appl Environ Microbiol. 2013;79(3):886–95. doi: 10.1128/AEM.03157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang R, Khan BA, Cheung GYC, et al. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest. 2011;121(1):238–48. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Periasamy S, Joo H-S, Duong AC, et al. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci USA. 2012;109(4):1281–6. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li M, Diep BA, Villaruz AE, et al. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2009;106(14):5883–8. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Syed AK, Reed TJ, Clark KL, Boles BR, Kahlenberg JM. Staphlyococcus aureus phenol-soluble modulins stimulate the release of proinflammatory cytokines from keratinocytes and are required for induction of skin inflammation. Infect Immun. 2015;83(9):3428–37. doi: 10.1128/IAI.00401-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kaito C, Saito Y, Nagano G, et al. Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog. 2011;7(2) doi: 10.1371/journal.ppat.1001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaito C, Saito Y, Ikuo M, et al. Mobile Genetic Element SCCmec-encoded psm-mec RNA Suppresses Translation of agrA and Attenuates MRSA Virulence. PLoS Pathog. 2013;9(4) doi: 10.1371/journal.ppat.1003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gonzalez DJ, Okumura CY, Hollands A, et al. Novel phenol-soluble modulin derivatives in community-associated methicillin-resistant Staphylococcus aureus identified through imaging mass spectrometry. J Biol Chem. 2012;287(17):13889–98. doi: 10.1074/jbc.M112.349860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Surewaard BGJ, Nijland R, Spaan AN, Kruijtzer JAW, de Haas CJC, van Strijp JA. Inactivation of staphylococcal phenol soluble modulins by serum lipoprotein particles. PLoS Pathog. 2012;8(3):e1002606. doi: 10.1371/journal.ppat.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Geiger T, Francois P, Liebeke M, et al. The Stringent Response of Staphylococcus aureus and Its Impact on Survival after Phagocytosis through the Induction of Intracellular PSMs Expression. PLoS Pathog. 2012;8(11) doi: 10.1371/journal.ppat.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Adhikari RP, Ajao AO, Aman MJ, et al. Lower antibody levels to staphylococcus aureus exotoxins are associated with sepsis in hospitalized adults with invasive S. aureus infections. J Infect Dis. 2012;206(6):915–923. doi: 10.1093/infdis/jis462. [DOI] [PubMed] [Google Scholar]
- 114.Nolte FS, Kapral FA. Immunogenicity of Staphylococcus aureus Immunogenicity of Staphylococcus aureus. Delta-Toxin. 1981;31(3):1251–60. doi: 10.1128/iai.31.3.1251-1260.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chatterjee SS, Joo H-S, Duong AC, et al. Essential Staphylococcus aureus toxin export system. Nat Med. 2013;19(3):364–7. doi: 10.1038/nm.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sjöquist J, Movitz J, Johansson IB, Hjelm H. Localization of protein A in the bacteria. Eur J Biochem. 1972;30(1):190–4. doi: 10.1111/j.1432-1033.1972.tb02086.x. [DOI] [PubMed] [Google Scholar]
- 117.Guss B, Uhlén M, Nilsson B, Lindberg M, Sjöquist J, Sjödahl J. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur J Biochem. 1984;138(2):413–20. doi: 10.1111/j.1432-1033.1984.tb07931.x. [DOI] [PubMed] [Google Scholar]
- 118.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96(22):12424–9. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bjork I, Petersson B-A, Sjoquist J. Some Physicochemical Properties of Protein A from Staphylococcus aureus. Eur J Biochem. 1972;29:579–84. doi: 10.1111/j.1432-1033.1972.tb02024.x. [DOI] [PubMed] [Google Scholar]
- 120.Huntzinger E, Boisset S, Saveanu C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24(4):824–35. doi: 10.1038/sj.emboj.7600572. /articlerender.fcgi?artid=549626&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Björk I, Petersson BA, Sjöquist J. Some physiochemical properties of protein A from Staphylococcus aureus. Eur J Biochem. 1972;29(3):579–84. doi: 10.1111/j.1432-1033.1972.tb02024.x. [DOI] [PubMed] [Google Scholar]
- 122.Potter KN, Li Y, Capra JD. Staphylococcal protein A simultaneously interacts with framework region 1, complementarity-determining region 2, and framework region 3 on human VH3-encoded Igs. J Immunol. 1996;157(7):2982–8. [PubMed] [Google Scholar]
- 123.Graille M, Stura EA, Corper AL, et al. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci U SA. 2000;97(10):5399–404. doi: 10.1073/pnas.97.10.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sjödahl J. Repetitive sequences in protein A from Staphylococcus aureus. Eur J Biochem. 1977;351:343–51. doi: 10.1111/j.1432-1033.1977.tb11324.x. [DOI] [PubMed] [Google Scholar]
- 125.Moks T, Abrahmsén L, Nilsson B, Hellman U, Sjöquist J, Uhlén M. Staphylococcal protein A consists of five IgG-binding domains. Eur J Biochem. 1986;156(3):637–43. doi: 10.1111/j.1432-1033.1986.tb09625.x. [DOI] [PubMed] [Google Scholar]
- 126.Janoschek L, Freiherr von Roman M, Berensmeier S. Protein A affinity precipitation of human immunoglobulin G. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;965:72–8. doi: 10.1016/j.jchromb.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 127.Forsgren a. Significance of protein a production by staphylococci. Infect Immun. 1970;2(5):672–3. doi: 10.1128/iai.2.5.672-673.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peterson PK, Verhoef JAN, Sabath LD, Quie PG. Staphylococcal Opsonization. 1977;15(3):760–4. doi: 10.1128/iai.15.3.760-764.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Goodyear CS, Silverman GJ. Death by a B Cell Superantigen. J Exp Med. 2003;197(9):1125–39. doi: 10.1084/jem.20020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim HK, Cheng AG, Kim H-Y, Missiakas DM, Schneewind O. Nontoxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections in mice. J Exp Med. 2010;207(9):1863–70. doi: 10.1084/jem.20092514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thammavongsa V, Rauch S, Kim HK, Missiakas DM, Schneewind O. Protein A-neutralizing monoclonal antibody protects neonatal mice against Staphylococcus aureus. Vaccine. 2015;33(4):523–26. doi: 10.1016/j.vaccine.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim HK, Emolo C, DeDent AC, Falugi F, Missiakas DM, Schneewind O. Protein A-specific monoclonal antibodies and prevention of staphylococcus aureus disease in mice. Infect Immun. 2012;80(10):3460–70. doi: 10.1128/IAI.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim HK, Emolo C, Missiakas D, Schneewind O. A monoclonal antibody that recognizes the E domain of staphylococcal protein A. Vaccine. 2014 Jan 16;32(4):464–9. doi: 10.1016/j.vaccine.2013.11.054. cited 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kobayashi SD, Deleo FR. Staphylococcus aureus Protein A promotes Immune Suppression. Am Soc Microbiol. 2013;4(5):4–6. doi: 10.1128/mBio.00764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Proctor RA. Challenges for a universal Staphylococcus aureus vaccine. Clin Infect Dis. 2012;54(8):1179–86. doi: 10.1093/cid/cis033. [DOI] [PubMed] [Google Scholar]
- 136.Fowler VG, Proctor RA. Where does a Staphylococcus aureus vaccine stand? Clin Microbiol Infect. 2014;20(Suppl 5):66–75. doi: 10.1111/1469-0691.12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shokri R, Salouti M, Zanjani RS. Anti protein A antibody-gold nanorods conjugate: a targeting agent for selective killing of methicillin resistant Staphylococcus aureus using photothermal therapy method. J Microbiol. 2015;53(2):116–21. doi: 10.1007/s12275-015-4519-4. [DOI] [PubMed] [Google Scholar]
- 138.Panyala Nagender Reddy, Peña-Méndez Eladia María, Havel Josef. Gold and nano-gold in medicine: overview, toxicology and perspectives. J Appl Biomed. 2009;7:75–91. [Google Scholar]
- 139.Boyle-Vavra S, Daum RS. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab Invest. 2007;87(1):3–9. doi: 10.1038/labinvest.3700501. [DOI] [PubMed] [Google Scholar]
- 140.Van de Velde H. Etude sur le mécanisme de la virulence du staphylocoque pyogène. Cellule. 1894;10:403–460. [Google Scholar]
- 141.Panton PN, Valentine FCO. STAPHYLOCOCCAL TOXIN. Lancet. 1932;219(5662):506–8. [Google Scholar]
- 142.Panton PN, Valentine FCO, Dix V. Staphylococcal Infection and Antitoxin Treatment. Lancet. 1931;218(5648):1180–3. [Google Scholar]
- 143.Woodin aM. Purification of the two components of leucocidin from Staphylococcus aureus. Biochem J. 1960;75(1):158–65. doi: 10.1042/bj0750158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Woodin AM. Fractionation of a leucocidin from Staphylococcus aureus. Biochem J. 1959;73(1):225–37. doi: 10.1042/bj0730225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Colin DA, Mazurier I, Sire S, Finck-Barbançon V. Interaction of the two components of leukocidin from Staphylococcus aureus with human polymorphonuclear leukocyte membranes: sequential binding and subsequent activation. Infect Immun. 1994;62(8):3184–8. doi: 10.1128/iai.62.8.3184-3188.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Spaan AN, Henry T, Van Rooijen WJM, et al. The staphylococcal toxin panton-valentine leukocidin targets human C5a receptors. Cell Host Microbe. 2013;13(5):584–94. doi: 10.1016/j.chom.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 147.Jayasinghe L, Bayley H. The leukocidin pore: evidence for an octamer with four LukF subunits and four LukS subunits alternating around a central axis. Protein Sci. 2005;14:2550–61. doi: 10.1110/ps.051648505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Genestier A, Michallet M, Prévost G, et al. Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. J Clin Invest. 2005;115(11):3117–27. doi: 10.1172/JCI22684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gillet Y, Issartel B, Vanhems P, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359(9308):753–9. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 150.Crémieux AC, Saleh-Mghir A, Danel C, et al. α-hemolysin, not panton-valentine leukocidin, impacts rabbit mortality from severe sepsis with methicillin-resistant Staphylococcus aureus osteomyelitis. J Infect Dis. 2014;209(11):1773–80. doi: 10.1093/infdis/jit840. [DOI] [PubMed] [Google Scholar]
- 151.Kobayashi SD, Malachowa N, Whitney AR, et al. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect Dis. 2011;204(6):937–41. doi: 10.1093/infdis/jir441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tong A, Tong SYC, Zhang Y, et al. Panton-Valentine leukocidin is not the primary determinant of outcome for Staphylococcus aureus skin infections: evaluation from the CANVAS studies. PLoS One. 2012;7(5):e37212. doi: 10.1371/journal.pone.0037212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Löffler B, Hussain M, Grundmeier M, et al. Staphylococcus aureus panton-valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 2010;6(1):e1000715. doi: 10.1371/journal.ppat.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Diep BA, Chan L, Tattevin P, et al. Polymorphonuclear leukocytes mediate Staphylococcus aureus Panton-Valentine leukocidin-induced lung inflammation and injury. Proc Natl Acad Sci USA. 2010;107(12):5587–92. doi: 10.1073/pnas.0912403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wardenburg JB, Bae T, Otto M, DeLeo FR, Schneewind O. Poring over pores: α-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13(12):1405–6. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- 156.Tseng CW, Biancotti JC, Berg BL, et al. Increased Susceptibility of Humanized NSG Mice to Panton-Valentine Leukocidin and Staphylococcus aureus Skin Infection. PLoS Pathog. 2015;11(11):1–17. doi: 10.1371/journal.ppat.1005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Singh G, Marples RR, Kligman AM. Experimental Staphylococcus aureus infections in humans. J Invest Dermatol. 1971;57:149–62. doi: 10.1111/1523-1747.ep12261498. [DOI] [PubMed] [Google Scholar]
- 158.Elek SD. Experimental staphylococcal infections in the skin of man. Ann NY Acad Sci. 1956;65(3):85–90. doi: 10.1111/j.1749-6632.1956.tb36626.x. [DOI] [PubMed] [Google Scholar]
- 159.Otto MP, Martin E, Badiou C, et al. Effects of subinhibitory concentrations of antibiotics on virulence factor expression by community-acquired methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2013 Mar;68:1524–32. doi: 10.1093/jac/dkt073. [DOI] [PubMed] [Google Scholar]
- 160.Gullberg E, Cao S, Berg OG, et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7(7):1–9. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Andersson DI, Hughes D. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol. 2014;12(7):465–78. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 162.Sandegren L. Selection of antibiotic resistance at very low antibiotic concentrations. Ups J Med Sci. 2014 Jan;:1–5. doi: 10.3109/03009734.2014.904457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Andersson DI, Hughes D. Evolution of antibiotic resistance at non-lethal drug concentrations. Drug Resist Updat. 2012;15(3):162–72. doi: 10.1016/j.drup.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 164.Laventie B, Potrich C, Atmanène C, et al. p-Sulfonato-calix[n]arenes inhibit staphylococcal bicomponent leukotoxins by supramolecular interactions. Biochem J. 2013;450(3):559–71. doi: 10.1042/BJ20121628. [DOI] [PubMed] [Google Scholar]
- 165.Chen Y, Yeh AJ, Cheung GYC, et al. Basis of virulence in a Panton-Valentine leukocidin-negative community-associated methicillin-resistant Staphylococcus aureus strain. J Infect Dis. 2015;211(3):472–80. doi: 10.1093/infdis/jiu462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Choi YW, Kotzin B, Herron L, Callahan J, Marrack P, Kappler J. Interaction of Staphylococcus aureus toxin “superantigens” with human T cells. Proc Natl Acad Sci USA. 1989;86(22):8941–5. doi: 10.1073/pnas.86.22.8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jardetzky TS, Brown JH, Gorga JC, et al. Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen. Nature. 1994;368(6473):711–8. doi: 10.1038/368711a0. [DOI] [PubMed] [Google Scholar]
- 168.Peavy DL, Adler WH, Smith RT. The mitogenic effects of endotoxin and staphylococcal enterotoxin B on mouse spleen cells and human peripheral lymphocytes. J Immunol. 1970;105(6):1453–8. [PubMed] [Google Scholar]
- 169.Argudín MÁ, Mendoza MC, Rodicio MR. Food Poisoning and Staphylococcus aureus Enterotoxins. Toxins (Basel) 2010;2(7):1751–73. doi: 10.3390/toxins2071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13(1):16–34. doi: 10.1128/cmr.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Schantz EJ, Roessler WG, Wagman J, Spero L, Dunnary DA, Bergdoll MS. Purification of staphylococcal enterotoxin B. Biochemistry. 1965;4(6):1011–6. doi: 10.1021/bi00882a005. [DOI] [PubMed] [Google Scholar]
- 172.Krakauer T. Update on staphylococcal superantigen-induced signaling pathways and therapeutic interventions. Toxins (Basel) 2013;5(9):1629–54. doi: 10.3390/toxins5091629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lina G, Bohach GA, Nair SP, et al. Standard nomenclature for the superantigens expressed by Staphylococcus. J Infect Dis. 2004;189(12):2334–6. doi: 10.1086/420852. [DOI] [PubMed] [Google Scholar]
- 174.Krakauer T, Stiles BG. The staphylococcal enterotoxin (SE) family: SEB and siblings. Virulence. 2013;4(8):759–73. doi: 10.4161/viru.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Omoe K, Hu DL, Ono HK, Shimizu S, et al. Emetic potentials of newly identified staphylococcal enterotoxin-like toxins. Infect Immun. 2013;81(10):3627–31. doi: 10.1128/IAI.00550-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bergdoll MS, Borja CR, Avena RM. Identification of a new enterotoxin as enterotoxin C. J Bacteriol. 1965;90(5):1481–5. doi: 10.1128/jb.90.5.1481-1485.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rusnak JM, Kortepeter M, Ulrich R, Poli M, Boudreau E. Laboratory exposures to staphylococcal enterotoxin B. Emerg Infect Dis. 2004;10(9):1544–9. doi: 10.3201/eid1009.040250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cao R, Zeaki N, Wallin-Carlquist N, Skandamis PN, Schelin J, Rådström P. Elevated enterotoxin a expression and formation in Staphylococcus aureus and its association with prophage induction. Appl Environ Microbiol. 2012;78(14):4942–8. doi: 10.1128/AEM.00803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tremaine MT, Brockman DK, Betley MJ. Staphylococcal enterotoxin A gene (sea) expression is not affected by the accessory gene regulator (agr) Infect Immun. 1993;61(1):356–9. doi: 10.1128/iai.61.1.356-359.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bavari S, Ulrich RG, LeClaire RD. Cross-reactive antibodies prevent the lethal effects of Staphylococcus aureus superantigens. J Infect Dis. 1999;180(4):1365–9. doi: 10.1086/314977. [DOI] [PubMed] [Google Scholar]
- 181.Tilahun ME, Rajagopalan G, Shah-Mahoney N, et al. Potent neutralization of staphylococcal enterotoxin B by synergistic action of chimeric antibodies. Infect Immun. 2010;78(6):2801–11. doi: 10.1128/IAI.01121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Varshney AK, Wang X, Cook E, et al. Generation, Characterization, and Epitope Mapping of Neutralizing and Protective Monoclonal Antibodies against Staphylococcal Enterotoxin B-induced Lethal Shock *. 2011;286(11):9737–47. doi: 10.1074/jbc.M110.212407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Larkin EA, Stiles BG, Ulrich RG. Inhibition of toxic shock by human monoclonal antibodies against staphylococcal enterotoxin B. PLoS One. 2010;5(10):e13253. doi: 10.1371/journal.pone.0013253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Inskeep TK, Stahl C, Odle J, et al. Oral vaccine formulations stimulate mucosal and systemic antibody responses against staphylococcal enterotoxin B in a piglet model. Clin Vaccine Immunol. 2010;17(8):1163–9. doi: 10.1128/CVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Reddy PN, Paul S, Sripathy MH, Batra HV. Evaluation of recombinant SEA-TSST fusion toxoid for protection against superantigen induced toxicity in mouse model. Toxicon. 2015;103:106–13. doi: 10.1016/j.toxicon.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 186.Wang K, Wu D, Chen Z, et al. Inhibition of the superantigenic activities of Staphylococcal enterotoxin A by an aptamer antagonist. Toxicon. 2016;119:21–7. doi: 10.1016/j.toxicon.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 187.Kim T, Choi J, Lee S, Yeo KJ, Cheong HK, Kim KK. Structural Studies on the Extracellular Domain of Sensor Histidine Kinase YycG from Staphylococcus aureus and Its Functional Implications. J Mol Biol. 2016;428(15):3074–89. doi: 10.1016/j.jmb.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 188.Krell T, Lacal J, Busch A, Silva-Jiménez H, Guazzaroni M-E, Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol. 2010;64:539–59. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 189.Voyich JM, Vuong C, DeWald M, et al. The SaeR/S Gene Regulatory System Is Essential for Innate Immune Evasion by Staphylococcus aureus. J Infect Dis. 2009;199(11):1698. doi: 10.1086/598967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Flack CE, Zurek OW, Meishery DD, et al. Differential regulation of staphylococcal virulence by the sensor kinase SaeS in response to neutrophil-derived stimuli. Proc Natl Acad Sci USA. 2014;111(19):E2037–45. doi: 10.1073/pnas.1322125111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Goerke C, Fluckiger U, Steinhuber A, et al. Role of Staphylococcus aureus Global Regulators sae and B in Virulence Gene Expression during Device-Related Infection. Infect Immun. 2005;73(6):3415–21. doi: 10.1128/IAI.73.6.3415-3421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Long DR, Mead J, Hendricks JM, Hardy ME, Voyich JM. 18β-glycyrrhetinic acid inhibits methicillin-resistant Staphylococcus aureus survival and attenuates virulence gene expression. Antimicrob Agents Chemother. 2013;57(1):241–7. doi: 10.1128/AAC.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Parsons JB, Kukula M, Jackson P, et al. Perturbation of Staphylococcus aureus gene expression by the enoyl-acyl carrier protein reductase inhibitor AFN-1252. Antimicrob Agents Chemother. 2013;57(5):2182–90. doi: 10.1128/AAC.02307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhao D, Chen C, Liu H, et al. Biological evaluation of halogenated thiazolo[3,2-a]pyrimidin-3-one carboxylic acid derivatives targeting the YycG histidine kinase. Eur J Med Chem. 2014;87:500–7. doi: 10.1016/j.ejmech.2014.09.096. [DOI] [PubMed] [Google Scholar]
- 195.Okada A, Igarashi M, Okajima T, et al. Walkmycin B targets WalK (YycG), a histidine kinase essential for bacterial cell growth. J Antibiot. 2010;63(2):89–94. doi: 10.1038/ja.2009.128. [DOI] [PubMed] [Google Scholar]
- 196.Igarashi M, Watanabe T, Hashida T, et al. Waldiomycin, a novel WalK-histidine kinase inhibitor from Streptomyces sp. MK844-mF10. J Antibiot. 2013;66(8):459–64. doi: 10.1038/ja.2013.33. [DOI] [PubMed] [Google Scholar]
- 197.Velikova N, Fulle S, Manso AS, et al. Putative histidine kinase inhibitors with antibacterial effect against multi-drug resistant clinical isolates identified by in vitro and in silico screens. Sci Rep. 2016 May;6(2015):26085. doi: 10.1038/srep26085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Marshall JH, Wilmoth GJ. Pigments of Staphylococcus aureus, a series of triterpenoid carotenoids. J Bacteriol. 1981;147(3):900–13. doi: 10.1128/jb.147.3.900-913.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Liu GY, Essex A, Buchanan JT, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202(2):209–15. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Clauditz A, Resch A, Wieland KP, Peschel A, Götz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. 2006;74(8):4950–3. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Liu C-I, Liu GY, Song Y, et al. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319(5868):1391–4. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Song Y, Lin F, Yin F, et al. Phosphonosulfonates Are Potent, Selective Inhibitors of Dehydrosqualene Synthase and Staphyloxanthin Biosynthesis in Staphylococcus aureus. J Med Chem. 2009;52(4):976–88. doi: 10.1021/jm801023u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Song Y, Liu C, Lin F, et al. Inhibition of Staphyloxanthin Virulence Factor Biosynthesis in Staphylococcus aureus: In vitro, in vivo, and Crystallographic Results †. J Med Chem. 2009;52(13):3869–80. doi: 10.1021/jm9001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lin F-Y, Zhang Y, Hensler M, et al. Dual Dehydrosqualene/Squalene Synthase Inhibitors: Leads for Innate Immune System-Based Therapeutics. ChemMedChem. 2012;7(4):561–564. doi: 10.1002/cmdc.201100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chow OA, von Köckritz-Blickwede M, Bright AT, et al. Statins enhance formation of phagocyte extracellular traps. Cell Host Microbe. 2010;8(5):445–54. doi: 10.1016/j.chom.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen F, Di H, Wang Y, et al. Small-molecule targeting of a diapophytoene desaturase inhibits S. aureus virulence. Nat Chem Biol. 2016;12(3):174–9. doi: 10.1038/nchembio.2003. [DOI] [PubMed] [Google Scholar]
- 207.Fadlallah A, Chelala E, Legeais J. Corneal Infection Therapy with Topical Bacteriophage Administration. Open Ophthalmol J. 2015;9:167–8. doi: 10.2174/1874364101509010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mattila S, Ruotsalainen P, Jalasvuori M. On-demand isolation of bacteriophages against drug-resistant bacteria for personalized phage therapy. Front Microbiol. 2015;6(V):1–7. doi: 10.3389/fmicb.2015.01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pincus NB, Reckhow JD, Saleem D, Jammeh ML, Datta SK, Myles IA. Strain specific phage treatment for Staphylococcus aureus infection is influenced by host immunity and site of infection. PLoS One. 2015;10(4):1–16. doi: 10.1371/journal.pone.0124280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kaur S, Harjai K, Chhibber S. In vivo Assessment of Phage and Linezolid Based Implant Coatings for Treatment of Methicillin Resistant S. aureus (MRSA) Mediated Orthopaedic Device Related Infections. PLoS One. 2016;11(6):e0157626. doi: 10.1371/journal.pone.0157626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Walls RJ, Roche SJ, O’Rourke A, McCabe JP. Surgical site infection with methicillin-resistant Staphylococcus aureus after primary total hip replacement. J Bone Joint Surg Br. 2008;90(3):292–8. doi: 10.1302/0301-620X.90B3.20155. [DOI] [PubMed] [Google Scholar]
- 212.Trampuz A, Widmer AF. Infections associated with orthopedic implants. Curr Opin Infect Dis. 2006;19:349–56. doi: 10.1097/01.qco.0000235161.85925.e8. [DOI] [PubMed] [Google Scholar]
- 213.Schweizer ML, Chiang HY, Septimus E, et al. Association of a bundled intervention with surgical site infections among patients undergoing cardiac, hip, or knee surgery. JAMA. 2015;313:2162–71. doi: 10.1001/jama.2015.5387. [DOI] [PubMed] [Google Scholar]
- 214.Giersing BK, Dastgheyb SS, Modjarrad K, Moorthy V. Status of vaccine research and development of vaccines for Staphylococcus aureus. Vaccine. 2016;34(26):2962–6. doi: 10.1016/j.vaccine.2016.03.110. [DOI] [PubMed] [Google Scholar]
- 215.Levy J, Licini L, Haelterman E, et al. Safety and immunogenicity of an investigational 4-component Staphylococcus aureus vaccine with or without AS03B adjuvant: Results of a randomized phase I trial. Hum Vaccines Immunother. 2015;11(3):620–31. doi: 10.1080/21645515.2015.1011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Levy J, Licini L, Haelterman E, et al. component Staphylococcus aureus vaccine with or without AS03 B adjuvant: Results of a randomized phase I trial Safety and immunogenicity of an investigational 4-component Staphylococcus aureus vaccine with or without AS03 B adjuvant: Results of a random. 2016 Jan;5515 doi: 10.1080/21645515.2015.1011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Stegmiller NP, Barcelos EC, Leal JM, et al. Intranasal vaccination with adjuvant-free S. aureus antigens effectively protects mice against experimental sepsis. Vaccine. 2016;34(30):3493–9. doi: 10.1016/j.vaccine.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 218.Selle M, Hertlein T, Oesterreich B, et al. Global antibody response to Staphylococcus aureus live-cell vaccination. Sci Rep. 2016;6:24754. doi: 10.1038/srep24754. [DOI] [PMC free article] [PubMed] [Google Scholar]