Abstract
Huanglongbing (HLB, citrus greening) is a devastating citrus disease affecting citrus production worldwide. It is associated with the bacterium “Candidatus Liberibacter asiaticus” (CLas) and is vectored by the Asian citrus psyllid (ACP). Currently, diagnosis of CLas in regulatory samples is based on real-time quantitative polymerase chain reaction (qPCR) using 16S rRNA gene specific primers/probe. The detection of CLas using qPCR is challenging due to low pathogen titer and uneven distribution in infected plants and exacerbated by sampling issues and presence of inhibitors. This study evaluated a duplex droplet digital polymerase chain reaction (ddPCR) using multi-copy gene targets, 16S and RNR, to simultaneously detect CLas DNA targets in the same sample for unambiguous detection of the HLB pathogen in DNA extracts from citrus leaves and ACP. Standard curve analyses on tenfold dilution series with plasmid, citrus leaf and ACP DNA showed that both ddPCR and qPCR exhibited good linearity and efficiency in the duplex assay. CLas-infected low titer samples were used to validate the duplex ddPCR and qPCR performance and demonstrated that detection rate is higher when both 16S and RNR primers were used in duplex assay. However, the receiver operating characteristic analysis indicated that area under the curve for RNR primer was significantly broader, compared to 16S primers for CLas detection at low target titer. The absolute quantification of CLas at variable titers was reproducible and repeatable for both primer sets and the ddPCR showed higher resilience to PCR inhibitors with citrus leaf and ACP extracts. Hence, the resultant duplex ddPCR assay resulted in a significantly improved detection platform for diagnosis of CLas in samples with low pathogen titer.
Introduction
“Candidatus Liberibacter asiaticus” (CLas) is a Gram-negative, α –proteobacterium [1] associated with a devastating citrus disease known as Huanglongbing (HLB) (aka citrus greening) which is present worldwide except in the Australia, New Zealand and European Union countries [2]. HLB is characterized by blotchy mottling of young leaves with yellow shoots, malformed fruit, twig dieback and tree decline. This fastidious bacterium resides in phloem sieve cells of host plants. The bacterium causes multiple pockets of necrotic phloem leading to blockage of translocation and accumulation of starch in plastids, aberrations in cambial activity and excessive phloem formations observed in the infected leaf tissues under light microscopy [3]. CLas is vectored by the Asian citrus psyllid (ACP) (Diaphorina citri).
The detection of CLas is challenging due to low titer with uneven distribution in planta. HLB symptoms can be confused with nutritional deficiencies or other pathogens such as Spiroplasma citri, causal agent of citrus stubborn disease in California and other arid citrus-growing regions. Following CLas infection, there is a latency period of several months to multiple years before HLB symptoms appear and trees become unproductive or can die. Currently, no sustainable remedies are available for effective management of HLB. In CLas quarantine areas, infected trees are removed at the earliest stage possible to prevent further acquisition and spread of CLas by ACP. A robust and accurate detection method plays an important role in reduction of inoculum and limiting disease spread.
The available nucleic acid-based detection methods such as electron microscopy [4], fluorescence imaging techniques [5], and loop mediated isothermal amplification [6, 7] results are not sensitive at low titers. The chromosome of CLas has three copies of the 16S rRNA gene [8] and five copies of nrdB, encoding the β-subunit of ribonucleotide reductase (RNR), a critical enzyme involving bacterial proliferation [9]. Polymerase chain reaction (PCR), real-time PCR (qPCR) and ddPCR have been developed for 16S rDNA [10, 11, 12]. CLas detection using the RNR gene by PCR [13] and qPCR [9] was developed.
Droplet digital PCR (ddPCR) has been widely used in medical and clinical research in recent years for absolute quantification of nucleic acids without dependence on external standard curves [14]. The PCR reaction containing DNA templates, primers and a fluorescently labeled hydrolysis probe or a nucleic acid intercalating dye (Eva-Green) is partitioned into thousands of nanoliter-sized water-in-oil droplets. A droplet with target DNA amplification shows fluorescence and is defined as positive; whereas, a non-fluorescent droplet with no target DNA is defined as negative [15, 16]. The total number of target DNA molecules detected by the ddPCR reaction of a sample is then calculated from the fraction of positive droplets by Poisson statistics [15]. Zhong et al [12] recently reported on a ddPCR assay to detect CLas using the 16S rRNA target using plasmid DNA and citrus leaf samples.
The aim of this study was to develop duplex ddPCR assays using primers for 16S rRNA and RNR genes for more accurate, robust, and unambiguous detection of CLas in low titer infected citrus leaf and ACP DNA samples. The linearity, dynamic range, sensitivity and low titer detection performance of qPCR and ddPCR duplex assays were compared. The repeatability, reproducibility and tolerance to residual matrix inhibitors for ddPCR assays were performed with CLas-infected citrus leaf tissue and ACP extracts.
Materials and methods
CLas-infected DNA samples
Experimental citrus leaf petiole and ACP samples infected with CLas were obtained from the Contained Research Facility, University of California, Davis, California and the U.S. Horticultural Research Laboratory, USDA-ARS, Fort Pierce, Florida. DNA was extracted from citrus leaf and ACP by the cetyl trimethyl ammonium bromide (CTAB) method [17]. Nucleic acid quality and quantity was measured using Qubit 3.0 (Thermo Fisher Scientific, USA).
Primers and probes
Primers and probes used in qPCR and ddPCR assays are shown in Table 1. The TaqMan probes were synthesized by labeling the 5' -terminal nucleotide with the 6-carboxy-fluorescein (FAM) and VIC reporter dyes for the 16S and RNR genes, respectively. The 3’ -terminal nucleotide was labelled with minor groove binder/non-fluorescent quencher (Thermo Fisher Scientific, USA).
Table 1. Primer and probe sequences used for the qPCR and ddPCR assay for detection of “Candidatus Liberibacter asiaticus”.
| Target Gene | Primer/Probe name | Sequence (5'-3') | Amplicon length | Reference |
|---|---|---|---|---|
| 16S rRNA gene | 16S F | TCGAGCGCGTATGCAATACG | 76 bp | [11] |
| 16S R | GCGTTATCCCGTAGAAAAAGGTAG | |||
| 16S P | 6FAM/AGACGGGTGAGTAACGCG/ MGB/NFQ | |||
| nrdB, β-subunit of ribonucleotide reductase | RNR F | CATGCTCCATGAAGCTACCC | 80 bp | [9] |
| RNR R | GGAGCATTTAACCCCACGAA | |||
| RNR P | VIC/CCTCGAAATCGCCTATGCAC/MGB/NFQ |
Preparation of cloned plasmid and standard curves
The 16S rRNA (76 bp) and RNR (80 bp) genes were amplified using DNA extracted from HLB-symptomatic citrus leaves using 16S F/R and RNR F/R primers, respectively (Table 1). The amplicons were ligated into pGEM-T Easy vector (Promega) and transformed in JM-109 (Promega) separately. Plasmids isolated from white colonies were linearized using SpeI restriction enzyme (New England Biolabs, UK) and the concentrations were measured using Qubit dsDNA BR Assay Kit in Qubit 3.0 fluorometer (Thermofisher). Ten-fold serial dilutions were made using linearized 16S and RNR plasmids to generate the standard curves to assess analytical sensitivity, linearity and dynamic range of the ddPCR and qPCR assays.
CLas-infected citrus leaf and ACP DNA were extracted by the CTAB method and used for 10-fold serial dilution ranging from 2 ng to 200 fg per reaction and 1 ng to 100 fg per reaction, respectively. Three replicates of each concentration were tested simultaneously in the same run. The linear relationship was produced by plotting the log DNA concentration against the cycle quantitation (Cq).
Duplex quantitative qPCR assay
qPCR was performed in a CFX96 Real-Time System (Biorad). Duplex qPCR for ten-fold serial dilutions of linearized 16S and RNR plasmids was optimized using different concentrations of primer:probe (50:100 nm, 100:200 nm, 150:300 nm) in a final volume of 20 μl reaction which contained 10 μl of SsoAdvanced™ Universal Probes Supermix (Biorad), 1 μl of plasmid DNA and final volume made up with double distilled water. The thermocycling conditions consisted of initial denaturation at 95°C for 5 min, then 40 cycles of denaturation at 95°C for 10 s, annealing at 58°C for 40 s. Each run included log10 dilutions of linearized 16S and RNR plasmids, negative control and no template control (NTC).
Thermal gradient optimization and duplex ddPCR
To determine optimal annealing temperatures for 16S and RNR gene targets, the thermal gradient ranging from 54°C to 64°C was performed in the S1000 thermal cycler (BioRad) using the same amount of linearized plasmid DNA and primers/probes concentrations (900 nM/ 250 nM) in singleplex and duplex assay.
The duplex ddPCR reaction mixture (20 μl) contained 2x ddPCR Supermix for probes (no dUTP) (Biorad), 900 nM of each forward and reverse primer, 250 nM of probe and 1 μl of CLas DNA. The reaction mixture was transferred in individual wells of disposable eight channel DG8 cartridge and the wells were filled with 70 μl of droplet generation oil. The prepared cartridge was then placed into a cartridge holder and loaded in to the QX 200 droplet generator. The prepared droplet emulsions were further loaded in a semi-skirted, PCR-clean 96-well plate (Eppendorf) using a multichannel pipet (Mettler-Toledo Rainin LLC, CA, USA), by aspirating 40 μl from the DG8 cartridge. The plate was then heat sealed with pierceable foil using a PX1 PCR plate sealer (Biorad) and PCR amplification was carried out in a S1000 thermal cycler (Bio-Rad). The thermal cycling conditions consisted of 10 min. initial denaturation at 95°C, followed by 40 cycles of denaturation at 94°C for 30 sec. and annealing/elongation at 58°C for 1 min. with a ramp of 2°C/sec. and a final 10 min. incubation at 98°C for enzyme deactivation. After thermal cycling, the plate containing the droplets was placed in a QX 200 droplet reader (Bio-Rad, CA, USA) for analyzing each individual droplet by a detector.
Precision of CLas detection at low titer
To estimate the diagnostic performance of 16S and RNR primers in duplex assay at low target concentrations, CLas-positive leaf and ACP DNA (Cq 32) were diluted in healthy citrus DNA extracts at different ratios (1:5; 1:10; 1:15; 1:20; 1:25; 1:30; 1:35 and 1:40). The duplex ddPCR and qPCR assays were performed in ten replicates of each dilution.
Assessing inter-assay and intra-assay variability of duplex ddPCR
To assess the reproducibility (inter-assay variation) and repeatability (intra-assay variation) of duplex ddPCR assays, triplicate experiments were performed with CLas-infected leaf and ACP samples using 16S and RNR primers and probes. The reproducibility was determined by measuring the samples in triplicate within the same experiment (to assess the intra-assay variation) and between three different assays (inter-assay variation). The coefficient of variation (CV) was calculated by standard deviation/mean.
Estimation of tolerance to inhibitors
The influence of inhibitors with CLas citrus leaf and ACP DNA during sample preparation was estimated for the duplex ddPCR assay. The reactions were spiked with the equal amount of CLas plasmid DNA (16S and RNR) in different quantities (1 μl to 5 μl) of citrus leaf and ACP extract. The influence of leaf and ACP extracts were assessed relative to the mean measured signals in each sample with no inhibitors (no inhibition control administered by adding double-distilled water to the CLas plasmid DNA). To obtain citrus leaf extracts, 0.5 g healthy citrus leaves were excised and homogenized in 10 ml TE buffer (pH 7.4) using a Homex 6 homogenizer (BioReb AG, Switzerland) and centrifuged at 10,000x g for 10 min at 4°C. The healthy ACP was crushed in 0.5 ml of TE buffer and centrifuged at 10,000x g for 10 min at 4°C. The above supernatants were collected and used to test the tolerance of duplex ddPCR assays.
Data analysis
The standard curves and Cq values for qPCR were generated by Bio-Rad CFX Manager Software version 3.1. Linear regression of the qPCR standard curves were recalculated with Microsoft Excel software (Microsoft, USA). The Cq values were regressed against the logarithmically transformed copy number and DNA concentration. The qPCR amplification efficiency was estimated from the slopes of the standard curves using the equation E = 10−1/slope– 1. The ddPCR data were analyzed with QuantaSoft analysis software version 1.7 (Bio-Rad). The positive droplets containing amplified products were discriminated from negative droplets by applying a threshold above the negative droplets. Reactions with more than 10,000 accepted droplets per well were used for analysis. The copy number concentration of each sample was reported automatically by ddPCR software. The linear regression and P-value of the ddPCR assay were determined by plotting the measured copies of ddPCR and comparing them with expected values of serial dilution of plasmid DNA, citrus leaf and ACP DNA in Excel. The Poisson error and total error were calculated by QuantaSoft software. To be a true instrument technical replicate, total error bars are always greater than or equal to the Poisson error bars. Receiver operating characteristic (ROC) curves were constructed to evaluate the detection precision of the 16S and RNR primers in duplex ddPCR assay. The t-test was performed to compare the differences in measurement between inhibitors and no inhibitor control by ddPCR assay. Statistical analyses were performed with IBM SPSS Statistics version 24.
Results
Singleplex and duplex qPCR assay
The calibration curves at different concentrations of primer:probe (50:100 nm, 100:200, 150:300nm) showed that the duplex qPCR assays had the better linearity and efficiency at a 100:200 ratio with 16S and RNR plasmid DNA (R2 = 0.9973 and 0.9961, respectively) over the dynamic range tested in both the plasmid DNA from 1.48E+05 to 1.48E+00 copies/μl and 1.470E+05 to 1.47E+00 copies/μl, respectively. The slopes were -3.3371 and -3.3468, equivalent to qPCR efficiency of 99.37% and 98.97% for 16S and RNR, respectively. The singleplex qPCR efficiencies at 100:200 ratio with 16S and RNR plasmid DNA (R2 = 0.9933 and 0.9949, respectively) were 98.34% and 95.21%, respectively (Fig 1A and 1B). The standard curve showed the sensitivities of the duplex qPCR assays were 1.48 copies/20 μl and 1.47 copies/20 μl for both 16S and RNR respectively (S1 and S2 Tables).
Fig 1. Calibration curve of qPCR singleplex and duplex assays with tenfold serially diluted 16S and RNR plasmid DNA (1.48E+05 to 1.48E+00 copies/μl and 1.47E+05 to 1.47E+00 copies/μl, respectively) using 16S (broken line) and RNR (unbroken line) primers at 100:200 nm primer:probe ratio.
(a) The singleplex qPCR assay efficiency for 16S and RNR Plasmid DNA standard curve is 98.34%, 95.21%, respectively, and (b) for duplex qPCR assay is 99.37%, 98.97%, respectively.
The qPCR efficiencies for primer:probe 50:100 nm were 96.82% (R2 = 0.9914) and 89.47% (R2 = 0.992), for 16S and RNR plasmid DNA, respectively, in singleplex assays and 92.64% (R2 = 0.9973) and 92.42% (R2 = 0.995), respectively, in duplex assay (S1 Fig). The qPCR efficiency at primer:probe 150:300 nm were 98.12% (R2 = 0.9918) and 96.05% (R2 = 0.9953), for 16S and RNR plasmid DNA, respectively, in singleplex assay and 94.34% (R2 = 0.9839) and 92.42% (R2 = 0.9912), respectively, in duplex assay (S2 Fig). Based on these results, a prime:probe ratio of 100:200 nm was further used in this study for quantification of CLas in citrus leaf and ACP in duplex qPCR and ddPCR.
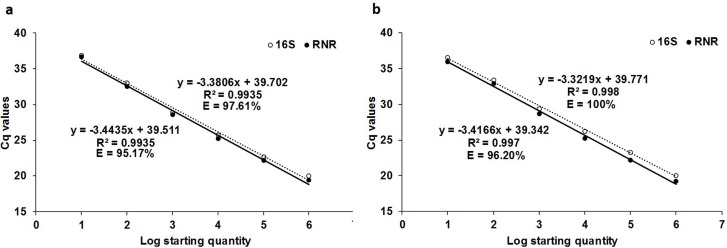
The CLas-infected citrus leaf DNA standard curve showed the efficiencies of 97.61% (R2 = 0.9935, slope = -3.3806) and 95.17% (R2 = 0.9935, slope = -3.4435) for 16S and RNR, respectively, in singleplex assay (Fig 2A). The duplex qPCR efficiencies were 100% (R2 = 0.998, slope = -3.3219) and 96.20% (R2 = 0.997, slope = -3.4166), respectively, for 16S and RNR (Fig 2B). The detection limits were 0.0002 ng of leaf DNA for 16S and RNR in duplex assay (S4 and S5 Tables).
Fig 2. Calibration curve of qPCR singleplex and duplex assays with tenfold serially diluted CLas-infected citrus leaf DNA (20 ng to 0.0002 ng) using 16S (unbroken line) and RNR (broken line) primers.
(a) The singleplex qPCR assay efficiency for 16S and RNR DNA standard curve is 97.61% and 95.17%, respectively, and (b) for duplex qPCR assay is 100% and 96.20%, respectively.
The CLas-infected ACP DNA standard curve showed the efficiencies of 100.42% (R2 = 0.9731, slope = -3.3118) and 91.33% (R2 = 0.9929, slope = -3.5489), for 16S and RNR, respectively, in singleplex assay (Fig 3A). The duplex qPCR efficiencies were 100.87% (R2 = 0.9913, slope = -3.3012) and 97.56% (R2 = 0.9954, slope = -3.3819), respectively, for 16S and RNR (Fig 3B). The detection limits were 0.0001 ng ACP DNA for 16S and RNR in duplex assay (S6 and S7 Tables).
Fig 3. Calibration curve of qPCR singleplex and duplex assays with tenfold serially diluted CLas-infected ACP DNA (10 ng to 0.0001 ng) using 16S (unbroken line) and RNR (broken line) primers.
(a) The singleplex qPCR assay efficiency for 16S and RNR DNA standard curve is 100.42% and 91.33%, respectively. (b) The duplex qPCR assay is 100.87% and 97.56%, respectively.
Singleplex and duplex ddPCR assay
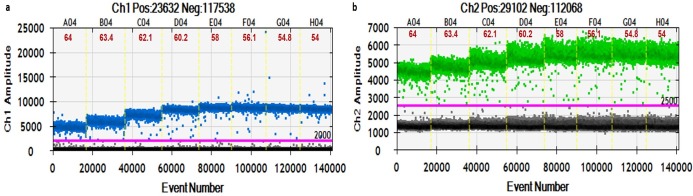
The optimum annealing temperature is the one that results in the largest fluorescence amplitude difference between the positives and negatives and that avoids nonspecific amplification. An annealing temperature of 58°C was chosen for the subsequent ddPCR experiments (Fig 4A and 4B).
Fig 4.
Thermal gradient duplex ddPCR for optimization of annealing temperature with (a) 16S (b) RNR primers. Eight ddPCR reactions are divided by vertical dotted yellow lines with an annealing temperature gradient ranged from 64°C to 54°C. The pink line is the threshold, above which are positive droplets (blue and green) and below that are negative droplets (gray) without any target DNA.
The linear regression curve was made by plotting Log10 transformed copy number concentrations measured by ddPCR against Log10-transformed predicted values of serially diluted plasmid DNA in singleplex and duplex ddPCR assays. The 16S and RNR Plasmid DNA showed R2 = 0.9998 and 0.9994, respectively, in singleplex assay and R2 = 0.9996 and 0.9995, respectively, in duplex assay (Fig 5). The sensitivity of duplex ddPCR assay for 16S and RNR plasmid DNA was 2.2 copy/20μl reaction and 3 copies/20μl, respectively (S3 Table).
Fig 5. Linear regression of the singleplex and duplex ddPCR assays for Plasmid DNA.
The Pearson correlation coefficient of singleplex 16S plasmid DNA regression curve (y = 0.7702x+22.319) is 0.9998 and RNR plasmid DNA (0.7963x-77.668) is 0.9994, respectively. Pearson correlation coefficient of duplex 16S plasmid DNA regression curve (y = 0.6994x+30.482) is 0.9996 and RNR plasmid DNA (0.8275x-76.539) is 0.9995, respectively. The inner error bars indicate the Poisson 95% confidence interval (CI) and the outer error bars show the total 95% CI of replicates. (P<0.0001).
The CLas-infected citrus leaf DNA showed good linearity for16S and RNR gene with R2 = 0.9999 and 0.9996, respectively, in singleplex assay and R2 = 0.9999 and 0.9995, respectively, in duplex assay (Fig 6). The sensitivity of ddPCR assay for 16S and RNR was 0.0002 ng (S4 and S5 Tables).
Fig 6. Linear regression of the singleplex and duplex ddPCR assays for CLas-infected leaf DNA.
Pearson correlation coefficient of singleplex 16S leaf DNA regression curve is 0.9999 and RNR leaf DNA is 0.9996, respectively. Pearson correlation coefficient of duplex 16S leaf DNA regression curve is 0.9999 and RNR leaf DNA is 0.9995, respectively. The inner error bars indicate the Poisson 95% CI and the outer error bars show the total 95% CI of replicates (P <0.0001).
CLas-infected ACP DNA showed good linearity with R2 = 0.9999 and 1.0 in singleplex assay for 16S and RNR gene, respectively, and R2 = 1.0 for both the genes in duplex assay (Fig 7). The sensitivity of ddPCR assay for 16S and RNR was 0.0001 ng (S6 and S7 Tables).
Fig 7. Linear regression of the singleplex and duplex ddPCR assays for ACP DNA.
Pearson correlation coefficient of singleplex and duplex assay for 16S and RNR with citrus leaf and ACP DNA regression curve is 0.9999. The inner error bars indicate the Poisson 95% confidence interval (CI) and the outer error bars show the total 95% CI of replicates. (P<0.0001).
Singleplex and duplex ddPCR assays showed five orders of magnitude between the target input amounts and ddPCR measured values. Droplets were positively saturated at target concentrations >106 copies/μl, making the Poisson algorithm invalid and resulting in a relative narrower dynamic range compared to qPCR. There were no significant difference (P<0.0001) in DNA copy number between singleplex and duplex ddPCR assays with plasmids, citrus leaf and ACP DNA.
Precision of CLas detection at low titer
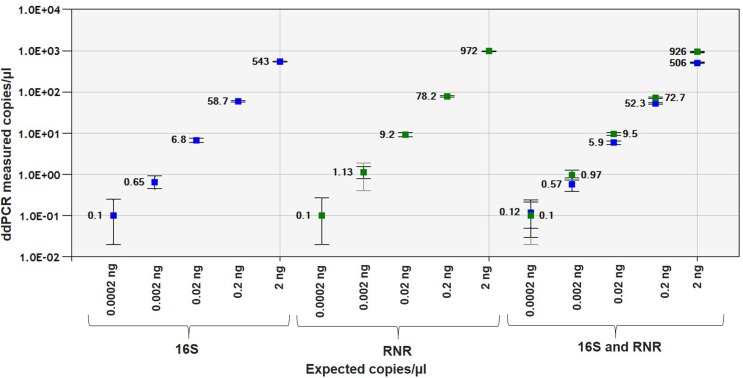
The 16S primers showed 100 percent detection up to a 1:5 ratio, whereas RNR primers showed 100 percent detection up to a 1:10 ratio with leaf and ACP DNA in both qPCR and ddPCR assays. The percent detection of RNR was greater when compared to 16S in all the dilutions with leaf and ACP DNA in both qPCR and ddPCR assays (Fig 8). The duplex ddPCR and qPCR assays showed that the overall positive detection rates were higher with both 16S and RNR primers. The robustness of ddPCR and qPCR assays increased when both 16S and RNR primers were used in duplex assays for detection of CLas.
Fig 8.
Precision of detection of CLas with citrus leaf and ACP DNA in duplex (a and b) qPCR and (c and d) ddPCR assay. Equal quantities of CLas DNA spiked in healthy extract at different dilution. Error bars indicate standard error of quantification between ten replicates of each dilution. Line shows the overall positive detection rate at different dilutions.
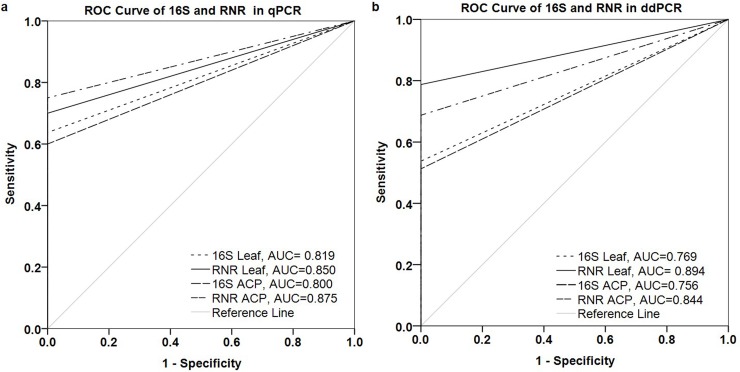
In the qPCR assay, the ROC analysis to compare the accuracy of detection between 16S and RNR primers showed that the area under curve (AUC) for 16S was 0.819 (standard error (SE) 0.050, 95% CI 0.720–0.918) and 0.800 (SE 0.054, 95% CI 0.694–0.906) for citrus leaf DNA and ACP, respectively. The AUC for the RNR primer was 0.850 (SE 0.044, 95% CI 0.763–0.937) and 0.875 (SE 0.039, 95% CI 0.798–0.952) for citrus leaf DNA and ACP, respectively (Fig 9A).
Fig 9. Diagnostic performance of 16S and RNR primers in qPCR and ddPCR assays for CLas-infected citrus leaf and ACP DNA.
Receiver operating characteristic (ROC) curve indicates better diagnostic performance with RNR primer compared to 16S primer for differentiating between healthy and CLas infected leaf and ACP DNA with significantly (P <0.05) broader AUC in (a) qPCR and (b) ddPCR assays.
In ddPCR assay, ROC analysis between 16S and RNR primers showed that the AUC for 16S was 0.769 (SE 0.060, 95% CI 0.651–0.886) and 0.756 (SE 0.062, 95% CI 0.634–0.878) for citrus leaf DNA and ACP, respectively. The AUC for RNR primer was 0.894 (SE 0.036, 95% CI 0.824–0.963) and 0.844 (SE 0.046, 95% CI 0.754–0.933) for citrus leaf DNA and ACP, respectively. The AUC of RNR primer was significantly (P <0.05) broader compared to the 16S primer in qPCR and ddPCR assays, indicating that RNR primer was better than 16S for CLas detection in leaf and ACP DNA (Fig 9B).
Reproducibility and repeatability of duplex ddPCR assay
The absolute quantification of CLas for 16S and RNR genes by the ddPCR assay revealed better reproducibility between runs (inter-assay) using RNR primers, especially for low target samples of citrus leaf and ACP. The lower coefficient of variance for 16S primer in intra-assay showed better repeatability within runs compared to RNR primers (Table 2).
Table 2. Repeatability (Intra-assay variation) and reproducibility (Inter-assay variation) of duplex ddPCR assay for detection of “Candidatus Liberibacter asiaticus".
| Assay | Inter-assay variability | Intra-assay variability | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Leaf/ACP DNAa | Assay 1b | Assay 2b | Assay 3b | CV%c | Replicate 1b | Replicate 2b | Replicate 3b | CV%c |
| 16S | Sample 1 | 257 | 283 | 252 | 6.3 | 262 | 266 | 257 | 0.6 |
| Sample 2 | 24.4 | 24.4 | 25.7 | 3.0 | 22.5 | 22.8 | 24.4 | 1.5 | |
| Sample 3 | 0.9 | 0.9 | 1.3 | 22.3 | 0.9 | 1.1 | 0.9 | 4.0 | |
| Sample 4 | 7.3 | 7.5 | 6.5 | 7.5 | 6.9 | 8 | 7.3 | 2.5 | |
| Sample 5 | 0.08 | 0.14 | 0.09 | 31.1 | 0.16 | 0.9 | 0.08 | 16 | |
| RNR | Sample 1 | 405 | 421 | 406 | 2.2 | 400 | 416 | 405 | 2 |
| Sample 2 | 37.2 | 39.4 | 37.5 | 3.1 | 42.1 | 39.4 | 37.2 | 6.2 | |
| Sample 3 | 1.8 | 1.3 | 2 | 21.2 | 1.8 | 2.3 | 1.8 | 14.7 | |
| Sample 4 | 12.3 | 11.4 | 13.6 | 8.9 | 12.2 | 11.4 | 12.3 | 4.1 | |
| Sample 5 | 0.4 | 0.4 | 0.36 | 6.0 | 0.32 | 0.15 | 0.4 | 44 | |
a Citrus leaf DNA (sample 1–3), Asian citrus psyllid DNA (sample 4, 5)
b Values reflect copies/μl of 20 μl ddPCR reaction
c CV means coefficient of variation
Influence of inhibitors on duplex ddPCR assay
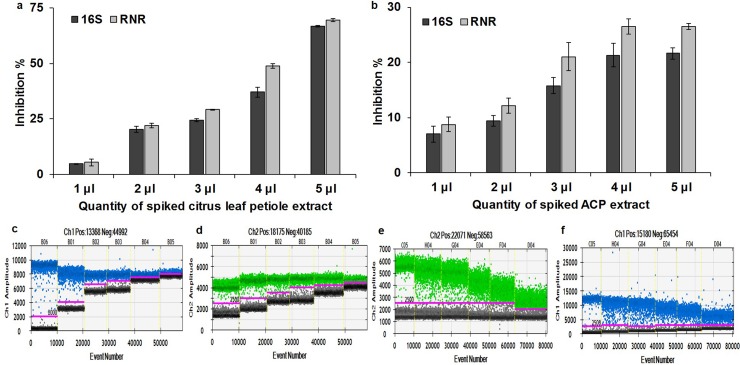
The ddPCR assays showed more tolerance to inhibition with ACP extract compared to citrus leaf petiole extract using 16S and RNR primers (Fig 10A & 10B). In contrast, the key parameters affected by the presence of the residual matrices for the ddPCR were CLas titer and fluorescent signal levels for both negative and positive droplets. Fluorescent signals of negative droplets were increased with increasing amounts of spiked citrus leaf petiole extracts and remain similar with ACP extract. Fluorescent signals of positive droplets remained similar and decreased with increasing amounts of citrus leaf petiole and ACP extracts, respectively. The t-test results showed significant (P <0.05) difference between different measurements, compare to no inhibition control (Fig 10C, 10D, 10E & 10F).
Fig 10. Influence of citrus leaf petiole extract and ACP extract on quantification of CLas by ddPCR assays for 16S and RNR genes and 1-D plot of ddPCR reactions.
Samples spiked with different quantity of (a) citrus leaf petiole, (b) ACP extract and equal amount of CLas plasmid DNA. Error bars denote standard error of inhibition between three replicates of each reaction. The 1-D plot shows only one of three replications for 16S and RNR with citrus leaf petiole (c & d, respectively) and ACP extract (e & f, respectively).
Discussion
Currently, the detection of CLas is based on 16S rRNA-specific primers for diagnostic and quarantine purposes using citrus leaf and ACP material in the United States [18]. When the pathogen titers are at low levels and unevenly distributed in the infected plant, qPCR based detection of CLas is challenging and unreliable due to sampling issues and the presence of inhibitors [19, 20]. The use of a single gene component in qPCR assays for detection of the pathogen with low and variable titers results in high Cq values that are generally considered inconclusive and the pathogen remains unverifiable. The use of two different genomic regions with multi-copy gene characteristics can be beneficial in discriminating between true and false positives [21, 22]. In this study, we developed a duplex assay using primers specific to the 16S rRNA gene (three copies) and the RNR gene (five copies) in qPCR and ddPCR assay that significantly improved the accuracy of CLas detection in citrus leaf and ACP.
The absolute quantification of CLas using ddPCR, a direct measurement of target titer, compared to qPCR, a relative measurement of titer, showed the ddPCR assay was more precise and reliable confirming results of Zhong et al. [12]. The reliability of titer data generated by qPCR is based on the accuracy of the standard curve obtained from a series of dilutions of known concentrations. This step adds expense, labor, and time for assay completion [23]. The ddPCR has several advantages over qPCR such as absolute quantification without the need of standard curve, improved accuracy, reliability and reproducibility between inter and intra assays [24]. The large-scale partitioning in ddPCR increases the precision of quantification and lessens interference due to PCR inhibitors [25]. The ddPCR technology produces more precise, reproducible and statistically significant results at low concentration of target nucleic acid [26].
In this study, the linearity, dynamic range and sensitivity of ddPCR was compared with qPCR in singleplex and duplex assays. The quantitative detection between the singleplex and duplex assay was not significantly different (P<0.0001). Both ddPCR and qPCR assays showed good linearity with plasmid, citrus leaf and ACP DNA. The ddPCR showed saturation of positive droplets at higher concentrations of template, which resulted in lower dynamic range compared to qPCR. The ratio between RNR and 16S copy number was ~1.7 in ddPCR and in qPCR a difference of ~0.7 Cq was observed. RNR primers showed better detection of CLas at low titer up to the lowest dilution (1:40) with both infected leaf and ACP. ROC analyses showed that AUC was greater for RNR primers compared to 16S primers for healthy and low titer citrus leaf and ACP samples infected with CLas. The higher AUC indicates greater sensitivity and more reliable diagnostic performance. Duplex ddPCR and qPCR assay offered more robust, accurate and sensitive quantification of CLas than the singleplex assays based on 16S rRNA or RNR detection. The duplex ddPCR assay exhibited repeatable and reproducible quantitative results with citrus leaf and ACP samples measured at both at high and low CLas titer. Therefore, the duplex ddPCR technology utilizing two different multicopy DNA targets in the CLas chromosome provided a more robust method for quantitative and unambiguous detection of CLas for diagnostic purposes with more precise and reproducible results than qPCR without the need of standard curves.
Conclusions
To the best of our knowledge, this work is the first to demonstrate absolute quantification of a single pathogen using two multi-copy genes in a duplex ddPCR assay. Our results demonstrated the applicability of simultaneous use of 16S and RNR primers for detecting CLas in a duplex ddPCR and qPCR assay. The data on the linearity, dynamic range, repeatability, reproducibility, tolerance to residual matrix inhibitors and the diagnostic performance supports this conclusion. This work suggests that when CLas titer is very low, the use of both 16S and RNR targets in duplex ddPCR assay is more reliable than the use of the 16S or RNR target in a singleplex for HLB diagnosis. The duplex assay provided greater reliability, sensitivity and cross reference capability for early pathogen detection in asymptomatic citrus samples.
Supporting information
(a) singleplex and (b) duplex assays with tenfold serially diluted 16S and RNR Plasmid DNA (1.48E+05 to 1.48E+00 copies/μl and 1.47E+05 to 1.47E+00 copies/μl, respectively) using 16S (broken line) and RNR (unbroken line) primers at 50:100 nm primer:probe ratio.
(PDF)
(a) singleplex and (b) duplex assays with tenfold serially diluted 16S and RNR Plasmid DNA (1.48E+05 to 1.48E+00 copies/μl and 1.47E+05 to 1.47E+00 copies/μl, respectively) using 16S (broken line) and RNR (unbroken line) primers at 150:300 nm primer:probe ratio.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Dr. Kris Godfrey for providing CLas infected leaf and ACP samples from Contained Research Facility, University of California, Davis, California. We also thank Robert DeBorde and Casey Crockett of the United States Department of Agriculture-Agricultural Research Service, San Joaquin Valley Agricultural Sciences Center, Parlier, CA for technical assistance. Mention of trade names or commercial products in this publication is solely for providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the California Citrus Research Board YOK-17 (ARS 58-2034-7-018) (http://www.ccnb.info) to RKY and Citrus Research Board 5300-17AA (ARS17-5300-191) (http://citrusresearch.org/) to RKY. Funders provided priority areas of research in their Request for Funds (RFP) and awarded the grant on a competitive basis. The funders had no role in the design, data collection or preparation of the manuscript.
References
- 1.Jagoueix S, Bove JM, Garnier M, The Phloem-limited bacterium of greening disease of citrus is a member of the alpha subdivision of the Proteobacteria. Int. J. Syst. Bacteriol. 1994; 44:379–386. doi: 10.1099/00207713-44-3-379 [DOI] [PubMed] [Google Scholar]
- 2.Hossein ANK, Susan EH, Susan PW, Ariena HCB. Global climate suitability of citrus huanglongbing and its vector, the Asian citrus psyllid, using two correlative species distribution modeling approaches, with emphasis on the USA. Eur. J. Plant Pathol. 2016; 144:655–670. [Google Scholar]
- 3.Schneider H. Anatomy of greening-disease sweet orange shots. Phytopathology. 1968; 58:1155–1160. [Google Scholar]
- 4.Bove JM. Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Pathol. 2006; 88:7–37. [Google Scholar]
- 5.Cen H, Weng H, Yao J, He M, Lv J, Hua S, et al. Chlorophyll fluorescence imaging uncovers photosynthetic fingerprint of Citrus Huanglongbing. Frontiers in plant science. 2017; 8:1509 doi: 10.3389/fpls.2017.01509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okuda M, Matsumoto M, Tanaka Y, Subandiyah S, Iwanami T. Characterization of the tufB-secE-nusG-rplKSJL-ropB gene cluster of the citrus greening organism and detection by loop-mediated isothermal amplification. Plant Dis. 2005; 89:705–711. [DOI] [PubMed] [Google Scholar]
- 7.Keremane ML, Ramadugu C, Rodriguez E, Kubota R, Shibata S, Hall D G, et al. A rapid field detection system for citrus huanglongbing associated “Candidatus Liberibacter asiaticus” from the psyllid vector, Diaphorina citri Kuwayama and its implications in disease management. Crop Protection. 2015; 68:41–48. [Google Scholar]
- 8.Duan Y. et al. Complete genome sequence of citrus Huanglongbing bacterium, “Candidatus Liberibacter asiaticus” obtained through metagenomics. Mol Plant-Microbe Interact. 2009; 22:1011–1020. doi: 10.1094/MPMI-22-8-1011 [DOI] [PubMed] [Google Scholar]
- 9.Zheng Z, Xu M, Bao M, Wu F, Chen J, Deng X. Unusual five copies and dual forms of nrdB in “Candidatus Liberibacter asiaticus”: Biological implications and PCR detection application. Scientific Reports. 2016; 6:39020 doi: 10.1038/srep39020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jagoueix S, Bove JM, Garnier M. PCR detection of the two “Candidatus” Liberobacter species associated with greening disease of citrus. Mol Cell Probes. 1996; 10:43–50. doi: 10.1006/mcpr.1996.0006 [DOI] [PubMed] [Google Scholar]
- 11.Li W. B., Hartung J. S. & Levy L. Quantitative real-time PCR for detection and identification of “Candidatus Liberibacter” species associated with citrus Huanglongbing. J. Microbiol. Meth. 2006; 66:104–115. [DOI] [PubMed] [Google Scholar]
- 12.Zhong X, Liu X, Lou B, Zhou C, Wang X. Development of a sensitive and reliable droplet digital PCR assay for the detection of “Candidatus Liberibacter asiaticus”. J. of Integ. Agri. 2018; 17:60345–7. [Google Scholar]
- 13.Razi MF, Keremane ML, Ramadugu C, Roose M, Khan IA, Lee RF. Detection of citrus huanglongbing-associated ‘Candidatus Liberibacter asiaticus’ in citrus and Diaphorina citri in Pakistan, seasonal variability, and implications for disease management. Phytopathology. 2014; 104:257–268. doi: 10.1094/PHYTO-08-13-0224-R [DOI] [PubMed] [Google Scholar]
- 14.Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for the polymerase chain reaction by use of limiting dilution. Biotechniques. 1992; 13:444–449. [PubMed] [Google Scholar]
- 15.Hindson B, Ness K. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011; 83:8604–8610. doi: 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012; 83:1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyle JJ. DNA protocols for plants In: Molecular Techniques in Taxonomy. Hewitt G, Johnson AWB, Young JPW. (eds.). NATO ASI Ser. H, Cell Biol. 57. Springer-Verlag, Berlin: 1991; 283–293. [Google Scholar]
- 18.United States Department of Agriculture, Animal and Plant Health Inspection Service Plant Protection and Quarantine. New Pest Response Guidelines-Citrus Greening Disease. 02 February 2012. Appendix E. Available from: https://www.aphis.usda.gov/plant_health/plant_pest_info/citrus_greening/downloads/pdf_files/cg-nprg.pdf
- 19.Tatineni S, Sagaram US, Gowda S, Robertson CJ, Dawson WO, Iwanami T, et al. In planta distribution of “Candidatus Liberibacter asiaticus” as revealed by polymerase chain reaction (PCR) and real-time PCR. Phytopathology. 2008; 98:592–9. doi: 10.1094/PHYTO-98-5-0592 [DOI] [PubMed] [Google Scholar]
- 20.Li W, Levy L, Hartung JS. Quantitative distribution of “Candidatus Liberibacter asiaticus” in citrus plants with citrus huanglongbing. Phytopathology. 2009; 99:139–44. doi: 10.1094/PHYTO-99-2-0139 [DOI] [PubMed] [Google Scholar]
- 21.Morgan JK, Zhou L, Li W, Shatters RG, Keremane M, Duan YP. Improved real-time PCR detection of ‘Candidatus Liberibacter asiaticus’ from citrus and psyllid hosts by targeting the intragenic tandem-repeats of its prophage genes. Mol. Cell. probes. 2012. 26:90–98. doi: 10.1016/j.mcp.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 22.Maheshwari Y, Selvaraj V, Hajeri S, Yokomi R. Application of droplet digital PCR for quantitative detection of Spiroplasma citri in comparison with real time PCR. PLoS ONE. 2017; 12(9):e0184751 doi: 10.1371/journal.pone.0184751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim TG, Yi T, Cho KS. Use of artificial DNA with multiple probe sites as reference DNA templates for quantitative real-time PCR to examine methanogen communities. J. Environ. Sci. Health A. 2013; 48:417–21. [DOI] [PubMed] [Google Scholar]
- 24.Dingle TC, Sedlak RH, Cook L, Jerome KR. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin. Chem. 2013; 59:1670–1672. doi: 10.1373/clinchem.2013.211045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods. 2013; 10:1003–1005. doi: 10.1038/nmeth.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor SC, Laperriere G, Germain H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Scientific Reports. 2017; 7:2409 doi: 10.1038/s41598-017-02217-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) singleplex and (b) duplex assays with tenfold serially diluted 16S and RNR Plasmid DNA (1.48E+05 to 1.48E+00 copies/μl and 1.47E+05 to 1.47E+00 copies/μl, respectively) using 16S (broken line) and RNR (unbroken line) primers at 50:100 nm primer:probe ratio.
(PDF)
(a) singleplex and (b) duplex assays with tenfold serially diluted 16S and RNR Plasmid DNA (1.48E+05 to 1.48E+00 copies/μl and 1.47E+05 to 1.47E+00 copies/μl, respectively) using 16S (broken line) and RNR (unbroken line) primers at 150:300 nm primer:probe ratio.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.