Abstract
Introduction
Classical swine fever (CSF) has caused severe economic losses in pig production in many countries. Recent CSF outbreaks in China are mainly associated with sub-genotype 2.1 of CSF virus (CSFV). Although there is abundant information regarding 2.1 isolates, few data are available on whole-genome analysis.
Material and Methods
The biological and genome characteristics of three recently emerged Chinese CSFV isolates, i.e. SD2014-1, SD2014-2, and SD2014-3, were fully analysed.
Results
Sequence analysis showed that the isolates shared 83.4%–95.0% nucleotide identity with eight other CSFV isolates. In addition, the 5′ untranslated region (5′UTR) and the non-structural (NS) proteins NS3, NS4A, and NS4B were more conserved than other regions of the genome. Phylogenetic analysis based on the complete genome sequences or full-length structural protein E2 gene sequences revealed that the three isolates belonged to sub-genotype 2.1b. In addition, several unique molecular characteristics of the 5′UTR, 3′UTR, and E2 were identified.
Conclusion
The genomic variations of the three isolates will support further analysis of virulence determinants and the evolutionary trend of CSFV.
Keywords: classical swine fever virus, complete genomic analysis, sub-genotype 2.1b, genomic variations
Introduction
Classical swine fever (CSF) is a highly contagious, lethal, and widespread swine disease caused by CSF virus (CSFV) and is classified by the Office International des Epizooties (OIE) as a notifiable disease (5). CSFV is an enveloped, single-stranded, positive-sense RNA virus belonging to the Pestivirus genus within the Flaviviridae family. The family also contains bovine viral diarrhoea virus (BVDV) and border disease virus (BDV) (9, 19). The CSFV genome is approximately 12.3 kb and contains a large open reading frame (ORF) encoding a polyprotein of 3,898 amino acids, with a 5′ untranslated region (UTR) and a 3′UTR at either end (18). The polyprotein undergoes viral and cellular proteolysis to produce four structural proteins (C, E0 or Erns, E1, and E2) and eight non-structural proteins (Npro, P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (18).
Because of its utility in tracking the virus origin and developing effective control strategies against CSF, many studies focus on sequence diversity analysis of CSFV, particularly the sequence variation associated with virulence changes (8). The sequence comparability of the 5′UTR (150 nt), E2 (190 or 1119 nt), and NS5B (409 nt) have been extensively used for genetic typing and investigating CSFV variation (16, 20, 21). Currently, it has been confirmed that the full-length E2 gene is statistically reliable in phylogenetic analysis (22). At the genome level, CSFV isolates are classified into three genotypes (1, 2, and 3) and 11 sub-genotypes (1.1, 1.2, 1.3, 1.4, 2.1, 2.2, 2.3, 3.1, 3.2, 3.3, and 3.4) (21, 23). Sub-genotype 2.1 isolates have been further divided into 2.1a, 2.1b, and 2.1c (4, 11, 20). In mainland China, CSF outbreaks are largely due to the circulation of several genotypes of CSFV isolates (1.1, 2.1, 2.2, and 2.3) (26), and sub-genotype 2.1b has predominated since the 1990s (7, 22, 26). Recently, a new sub-genotype 2.1d was confirmed by our laboratory (30). Newly emerged CSFV isolates in China in 2014–2015 have been characterised (13). Here, the complete genome sequences of three field CSFV isolates, i.e., SD2014-1, SD2014-2, and SD2014-3, originating from intensive pig farms in Shandong Province, China, were described.
Material and Methods
Clinical samples and virus isolation
A total of eight clinical samples, including the lung, lymphatic nodes, and spleen, were collected from pigs suspected to have CSFV infections during 2014. The animals were from large-scale pig farms in Shandong Province, China. Tissue samples were homogenised using a TissueLyser II (Qiagen, Germany) in Dulbecco’s modified Eagle medium (Gibco, USA). The tissue homogenates or sera were centrifuged at 10,000 × g for 15 min. The supernatant was passed through a 0.22 μm filter and transferred to swine testis (ST) cells which were grown for 36–48 h, and the remaining samples were kept at –70°C until the total RNA was extracted. The cells were subsequently incubated at 37°C with 5% CO2 for 3-5 days. Viral stocks were harvested from infected cells by three cycles of freezing and thawing and stored at -70°C.
Primer design and RT-PCR
Based on the published known sequence of CSFV Zj0801 (GenBank accession no. FJ529205), six pairs of primers were designed (Table 1). Total RNA was extracted from tissue homogenates or serum samples using a viral RNA extraction kit (Qiagen). Reverse transcription (RT)-PCR was performed using the One Step RT-PCR Kit (Qiagen) according to the manufacturer’s instructions. A total of 8 μL of the RNA template was added to 42 μL of the RT-PCR master mix, including 1 μL each of the primer sets. The reaction conditions were as follows: 95°C for 5 min, 30 cycles of denaturation (95°C for 30 s), annealing at 56°C for 30 s, elongation at 72°C for 2.5 min, and final extension at 72°C for 10 min. Based on the results from the RT-PCR and viral isolation attempts, three samples from three independent pig farms were selected to determine the full-length genomic sequences of CSFV. The three CSFV isolates were designated as SD2014-1, SD2014-2, and SD2014-3.
Table 1.
Primers used for amplification of gene fragments of CSFV SD2014-1, SD2014-2, and SD2014-3
| Fragment | Primer sequence (5′-3′)a | Position in genomea | Product size (bp)a |
|---|---|---|---|
| CSFV-A | GTATACGAGGTTAGTTCATTCTCGT | 1–2,047 | 2,047 |
| GATTACCAGAGAAAGCAACAAGAAT | |||
| CSFV-B | GATAATAGGCCCCGGTAAATTTGAC | 2,023–3,313 | 1,291 |
| TTTCCTTACAGGTCCCTCGCTAGAG | |||
| CSFV-C | AAATGAGACGGGTTACAGGGTA | 3,124–4,771 | 1,648 |
| CATCCCGTAGATCTCTTCACCTCCA | |||
| CSFV-D | CATAGATGAAATAGCTGGCGGGACC | 4,564–7,171 | 2,608 |
| TAGTGCTCTGCCAGCCTCCACAGTG | |||
| CSFV-E | TCTGCTGATATCAGAGGAGCTG | 6,866–668 | 2,803 |
| GCTTACCCAGACTTAATGTTTCTAG | |||
| CSFV-F | GCCCTATGTAAGGTCGACACCGCTC | 9,572–12,296 | 2,725 |
| GGGCCGTTAGGAAATTACCTTAGTC |
The primer sequence, position in genome and product size with respect to the CSFV Zj0801 (accession no. FJ529205) genome
Genome cloning and sequencing
RT-PCR products were analysed via 1% agarose gel electrophoresis. The target fragments were excised from the gels for purification using a Gel Extraction Kit (OMEGA, USA) at a later stage. The purified PCR products were cloned into a pMD18-T vector (TaKaRa, Dalian, China). Recombinant clones were sent to Comate Bioscience (Jinlin, China) for sequencing.
Phylogenetic analysis
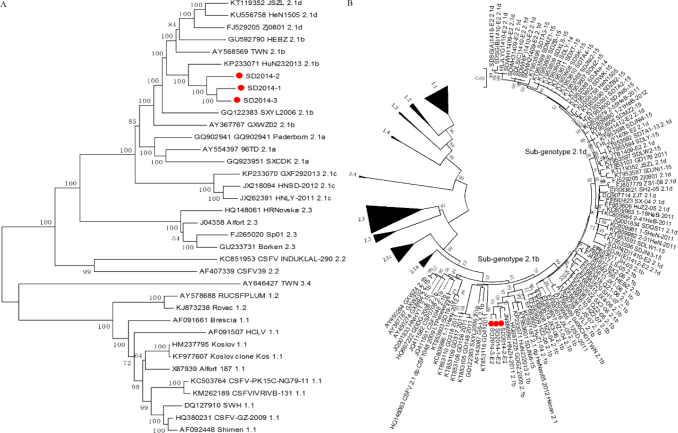
The entire genome sequences of SD2014-1, SD2014-2, and SD2014-3 were obtained after sequencing. Phylogenetic trees were constructed with MEGA (version 7) using the neighbour-joining method with 1,000 bootstraps (12). The evolutionary trees of the three CSFV isolates and other known isolates worldwide were determined based on complete genomic sequences or full-length E2 gene sequences (Fig. 1).
Fig. 1.
Phylogenetic analysis of the 3 new isolates (•) and other reference CSFV isolates based on the complete genomic sequences (A) and full-length E2 gene sequences (B)
Nucleotide and amino acid analysis
The complete genome sequences and deduced amino acid sequences of SD2014-1, SD2014-2, and SD2014-3 were analysed and their homologies with other eight representatives of CSFV strains were compared using DNAstar (DNAstar Inc., USA) (Tables 2 and 3). To explore the genetic variations of SD2014-1, SD2014-2, and SD2014-3, the nucleotide sequences of the 5′UTR and the 3′UTR and the amino acid sequences of E2 were fully analysed, together with 33 other CSFV isolates from China and other countries.
Table 2.
The genome homology of SD2014-1, SD2014-2, and SD2014-3 (%)
| Strains | SD2014-1 | SD2014-2 | SD2014-3 |
|---|---|---|---|
| SD2014-1 | – | 97.7 | 98.6 |
| SD2014-2 | 97.7 | – | 97.9 |
| SD2014-3 | 98.6 | 97.9 | – |
Table 3.
Detailed comparison of the full-length genomes of SD2014-1, SD2014-2, and SD2014-3 to other representative CSFV isolates (%)
| Nucleotides | Shimen (1.1)a | Paderborn (2.1a)a | HEBZ (2.1b)a | HNSD-2012 (2.1c)a | JSZL (2.1d)a | CSFV39 (2.2)a | Alfort/ Tuebingen (2.3)a | 94.4/IL/94/T-WN (3.4)a | |
|---|---|---|---|---|---|---|---|---|---|
| 5′UTR | 94.1∗ | 97.1∗ | 98.1∗ | 96.2∗ | 96.8∗ | 94.4∗ | 94.6∗ | 91.4∗ | |
| 92.8∗∗ | 98.1∗∗ | 96.8∗∗ | 96.5∗∗ | 97.1∗∗ | 93.0∗∗ | 95.4∗∗ | 92.8∗∗ | ||
| 93.3∗∗∗ | 97.6∗∗∗ | 97.3∗∗∗ | 96.0∗∗∗ | 97.6∗∗∗ | 93.8∗∗∗ | 94.1∗∗∗ | 92.2∗∗∗ | ||
| Npro | 87.1 | 94.2 | 95.6 | 92.9 | 95.6 | 87.5 | 87.9 | 85.3 | |
| 86.9 | 93.3 | 94.6 | 92.1 | 94.6 | 87.3 | 87.5 | 84.3 | ||
| 86.9 | 94.0 | 95.4 | 93.1 | 95.8 | 88.1 | 88.5 | 85.1 | ||
| C | 84.2 | 94.6 | 93.6 | 90.6 | 95.6 | 89.2 | 90.9 | 84.2 | |
| 83.8 | 94.9 | 93.3 | 90.9 | 95.3 | 89.6 | 91.2 | 83.8 | ||
| 84.2 | 94.3 | 92.6 | 90.2 | 94.6 | 88.9 | 90.6 | 84.5 | ||
| Erns | 84.1 | 93.0 | 94.3 | 92.7 | 95.0 | 89.3 | 90.3 | 83.1 | |
| 84.7 | 92.5 | 94.0 | 92.5 | 94.9 | 89.0 | 90.0 | 83.4 | ||
| 84.1 | 92.5 | 93.8 | 92.8 | 94.6 | 88.8 | 90.2 | 83.7 | ||
| E1 | 85.3 | 93.5 | 94.5 | 91.1 | 94.7 | 89.1 | 88.9 | 81.0 | |
| 85.3 | 93.8 | 95.2 | 91.5 | 95.4 | 90.8 | 88.9 | 81.9 | ||
| 85.1 | 94.0 | 94.7 | 91.6 | 95.2 | 89.9 | 89.4 | 81.7 | ||
| E2 | 83.3 | 93.7 | 93.9 | 90.2 | 93.7 | 87.7 | 87.8 | 81.5 | |
| 83.6 | 93.7 | 94.3 | 90.2 | 93.7 | 87.9 | 87.9 | 81.9 | ||
| 83.6 | 93.8 | 94.1 | 90.3 | 93.7 | 87.7 | 88.0 | 81.4 | ||
| P7 | 79.2 | 95.2 | 96.6 | 94.2 | 93.7 | 89.9 | 89.9 | 86.0 | |
| 81.2 | 95.2 | 96.1 | 94.2 | 94.2 | 89.9 | 89.4 | 87.0 | ||
| 80.2 | 95.2 | 96.1 | 94.2 | 93.2 | 89.9 | 89.4 | 87.0 | ||
| NS2 | 82.8 | 92.7 | 94.5 | 91.1 | 93.8 | 88.3 | 88.0 | 80.4 | |
| 82.5 | 93.1 | 94.7 | 91.2 | 93.4 | 88.0 | 88.4 | 80.1 | ||
| 82.9 | 93.0 | 94.7 | 91.5 | 93.8 | 88.4 | 88.3 | 80.2 | ||
| NS3 | 86.9 | 94.9 | 95.2 | 93.4 | 95.5 | 90.7 | 90.9 | 85.8 | |
| 86.5 | 94.7 | 94.9 | 93.2 | 95.3 | 90.6 | 90.6 | 85.2 | ||
| 97.0 | 95.1 | 95.2 | 93.3 | 95.7 | 90.8 | 91.2 | 85.4 | ||
| NS4A | 87.8 | 96.3 | 95.2 | 91.0 | 95.8 | 90.5 | 91.5 | 83.1 | |
| 87.3 | 95.2 | 94.2 | 92.1 | 94.7 | 88.4 | 88.4 | 85.2 | ||
| 89.4 | 95.8 | 94.7 | 91.5 | 95.2 | 89.9 | 91.0 | 83.6 | ||
| NS4B | 87.8 | 93.1 | 94.6 | 91.0 | 94.1 | 90.0 | 88.8 | 84.2 | |
| 88.0 | 93.7 | 95.0 | 91.9 | 95.1 | 90.6 | 89.2 | 85.0 | ||
| 88.1 | 93.8 | 95.3 | 91.6 | 94.6 | 89.9 | 89.3 | 84.6 | ||
| NS5A | 85.2 | 93.2 | 94.4 | 91.0 | 93.6 | 85.2 | 90.1 | 82.3 | |
| 85.5 | 93.4 | 94.7 | 90.9 | 93.8 | 85.4 | 90.1 | 82.4 | ||
| 85.5 | 93.4 | 94.7 | 90.8 | 94.0 | 85.3 | 90.0 | 82.5 | ||
| NS5B | 85.0 | 94.2 | 95.0 | 92.5 | 94.9 | 85.4 | 90.1 | 83.2 | |
| 85.1 | 94.7 | 95.6 | 92.7 | 95.2 | 85.6 | 90.4 | 83.6 | ||
| 85.0 | 94.8 | 95.6 | 93.2 | 95.3 | 85.4 | 90.4 | 83.4 | ||
| 3′UTR | 85.9 | 95.7 | 95.3 | 93.4 | 96.1 | 85.5 | 95.3 | 81.9 | |
| 85.0 | 94.8 | 94.4 | 93.4 | 96.1 | 84.6 | 94.4 | 81.9 | ||
| 84.1 | 93.5 | 93.1 | 93.4 | 94.8 | 83.7 | 93.1 | 81.1 | ||
| Complete | 85.5 | 94.0 | 94.9 | 92.0 | 94.7 | 88.2 | 89.8 | 83.4 | |
| 85.5 | 94.1 | 94.9 | 92.1 | 94.7 | 88.3 | 89.8 | 83.6 | ||
| 85.5 | 94.2 | 95.0 | 92.3 | 94.8 | 88.2 | 90.0 | 83.5 | ||
| Amino acid | |||||||||
| Npro | 92.9 | 97.0 | 95.2 | 95.8 | 96.4 | 93.5 | 90.5 | 91.7 | |
| 93.5 | 96.4 | 94.6 | 95.2 | 95.8 | 92.9 | 91.1 | 91.1 | ||
| 92.9 | 97.0 | 95.2 | 95.8 | 96.4 | 93.5 | 91.7 | 91.7 | ||
| C | 91.9 | 94.9 | 97.0 | 92.9 | 97.0 | 93.9 | 92.9 | 88.9 | |
| 90.9 | 94.9 | 94.9 | 92.9 | 94.9 | 93.9 | 92.9 | 88.9 | ||
| 90.9 | 94.9 | 94.9 | 92.9 | 94.9 | 93.9 | 92.9 | 88.9 | ||
| Erns | 89.9 | 97.4 | 97.8 | 98.2 | 98.2 | 95.2 | 96.0 | 91.2 | |
| 89.9 | 97.4 | 97.8 | 98.2 | 98.2 | 95.2 | 96.0 | 91.2 | ||
| 89.9 | 96.5 | 96.9 | 97.4 | 97.4 | 94.3 | 95.2 | 91.2 | ||
| El | 92.3 | 96.4 | 95.9 | 94.9 | 96.4 | 96.4 | 96.4 | 89.2 | |
| 93.8 | 97.4 | 96.9 | 95.9 | 97.4 | 97.4 | 97.4 | 90.8 | ||
| 93.8 | 97.4 | 96.9 | 95.9 | 97.4 | 97.4 | 97.4 | 90.8 | ||
| E2 | 88.5 | 95.2 | 95.7 | 94.6 | 95.7 | 90.1 | 92.2 | 88.2 | |
| 89.8 | 96.0 | 95.7 | 95.4 | 95.7 | 91.4 | 93.2 | 89.0 | ||
| 89.5 | 96.0 | 96.5 | 95.7 | 96.5 | 90.9 | 93.0 | 89.0 | ||
| P7 | 91.3 | 98.6 | 95.7 | 97.1 | 94.2 | 95.7 | 95.7 | 95.7 | |
| 92.8 | 98.6 | 95.7 | 97.1 | 94.2 | 95.7 | 95.7 | 94.2 | ||
| 91.3 | 98.6 | 95.7 | 97.1 | 94.2 | 95.7 | 95.7 | 95.7 | ||
| NS2 | 90.6 | 97.6 | 97.2 | 96.9 | 96.1 | 95.4 | 94.7 | 88.0 | |
| 90.8 | 97.8 | 97.4 | 96.7 | 95.8 | 95.4 | 95.0 | 88.2 | ||
| 90.6 | 97.4 | 97.4 | 96.7 | 95.8 | 95.4 | 95.0 | 88.2 | ||
| NS3 | 98.4 | 99.4 | 98.8 | 98.8 | 99.4 | 98.7 | 99.1 | 98.5 | |
| 98.1 | 99.1 | 98.5 | 98.5 | 99.1 | 98.4 | 98.8 | 98.2 | ||
| 98.4 | 99.4 | 98.8 | 98.8 | 99.4 | 98.7 | 99.1 | 98.5 | ||
| NS4A | 98.4 | 100 | 98.4 | 95.2 | 100 | 96.8 | 100 | 95.2 | |
| 98.4 | 100 | 98.4 | 95.2 | 100 | 96.8 | 100 | 95.2 | ||
| 98.4 | 100 | 98.4 | 95.2 | 100 | 96.8 | 100 | 95.2 | ||
| NS4B | 96.0 | 98.3 | 98.8 | 99.1 | 98.6 | 96.8 | 98.3 | 93.9 | |
| 96.0 | 98.3 | 98.8 | 99.1 | 98.6 | 96.8 | 98.3 | 93.9 | ||
| 95.7 | 98.0 | 98.6 | 98.8 | 98.3 | 96.5 | 98.0 | 93.7 | ||
| NS5A | 88.6 | 95.0 | 95.4 | 93.2 | 95.2 | 88.2 | 93.4 | 86.9 | |
| 89.2 | 95.2 | 95.6 | 93.4 | 95.2 | 88.8 | 93.2 | 87.3 | ||
| 88.8 | 95.0 | 95.4 | 93.2 | 95.6 | 88.4 | 93.0 | 86.7 | ||
| NS5B | 91.8 | 96.5 | 96.6 | 96.4 | 97.5 | 92.5 | 95.8 | 89.4 | |
| 91.9 | 97.1 | 97.6 | 97.2 | 98.5 | 92.6 | 95.9 | 89.4 | ||
| 92.6 | 97.8 | 97.9 | 97.6 | 98.7 | 93.3 | 96.6 | 90.1 |
Results
Virus isolation
The tissue homogenates or sera of suspected CSFV-infected pigs were transferred to ST cells for virus isolation. After being identified, the three strains, i.e., SD2014-1, SD2014-2, and SD2014-3, were successfully isolated.
Analysis of full-length genomic sequences
The genomic sequences of SD2014-1 (GenBank accession no. MF149061), SD2014-2 (GenBank accession no. MF149062), and SD2014-3 (GenBank accession no. MF149063) were 12,296, 12,333, and 12,296 nucleotides in length, respectively. The three strains shared 97.7%–98.6% homology between each other (Table 2).
Complete nucleotide sequences of the three isolates were further compared with eight other representative CSFV isolates, including Shimen (AF092448), Paderborn (AY072924), HEBZ (GU592790), HNSD-2012 (JX218094), JSZL (KT119352), CSFV39 (AF407339), Alfort/Tuebingen (J04358), and 94.4/IL/94/TWN (AY646427) (Table 3). The three isolates shared 94.9%–95.0% and 94.7%–94.8% homology with sub-genotype 2.1b isolate HEBZ and 2.1d isolate JSZL, respectively; these were the most closely related isolates. In addition, they shared 88.2%–94.2% homology with other genotype 2 isolates, including 2.1a Paderborn, 2.1c HNSD-2012, 2.2 CSFV39, and 2.3 Alfort/Tuebingen. They also shared only 85.5% homology with 1.1 isolate Shimen and 83.4%–83.5% homology with 3.4 isolate 94.4/IL/94/TWN. In sum, these results indicate that all three isolates belong to sub-genotype 2.1b or 2.1d.
To further examine the genomic variation in the two isolates, their genomic characteristics were analysed in detail. Compared with the eight representative isolates, the 5′UTRs of the three isolates were the most conserved regions in the genomes, which exhibited 96.0%–98.1% homology with 2.1 isolates Paderborn, HEBZ, HNSD-2012 or JSZL, 93.0%–95.4% homology with 2.2 isolate CSFV39 or 2.3 isolate Alfort/Tuebingen, and 91.4%–94.1% homology with 1.1 isolate Shimen or 3.4 isolate 94.4/IL/94/TWN. In addition, the amino acid identities of NS3, NS4A, and NS4B among the three isolates and eight other representative isolates were higher than other regions of the genome, and the homologies were 98.1%–99.4%, 95.2%–100%, and 93.7%–100%, respectively. The detailed identities of the three isolates and eight other representative strain isolates are summarised in Table 3.
Phylogenetic analysis
To understand the genetic relationships among SD2014-1, SD2014-2, SD2014-3, and other CSFV isolates, phylogenetic trees were produced using 36 CSFV entire genome sequences or 205 full-length E2 gene sequences (Fig. 1). CSFV isolates were divided into genotypes 1, 2, and 3. Genotype 2 isolates were further divided into sub-genotypes 2.1, 2.2, and 2.3, and sub-genotype 2.1 isolates could be further subdivided into 2.1a, 2.1b, 2.1c, and 2.1d. The three new isolates, i.e., SD2014-1, SD2014-2, and SD2014-3, belonged to 2.1b.
Sequence analysis of UTRs
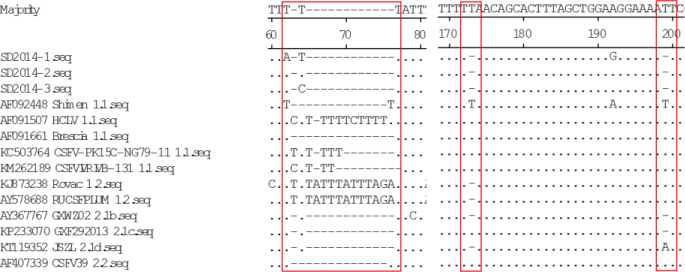
The 5′UTR and 3′UTR have been documented to be crucial regulatory elements in the CSFV genome (10, 28). Within the CSFV genome, the 5′UTR of CSFV is the most conserved region. Compared with those of other sub-genotype isolates, the 5UTRs of SD2014-2 and SD2014-3 had a nucleotide T deletion at position 44 (T44), while the 5′UTR of SD2014-1 had a nucleotide substitution at the very same position (T/C44A) (Fig. 2). In addition, different CSFV isolates showed nucleotide A, T, C, or deletions at this position, while some isolates in the same sub-genotype showed different nucleotides at position 44 (Fig. 2). Similarly to most CSFV isolates, the 5′UTRs of SD2014-1 and SD2014-2 had two continuous nucleotide A deletions at positions 357–358, while the 5′UTR of SD2014-3 had only one nucleotide A deletion at these positions (Fig. 2).
Fig. 2.
Sequence alignments of 5′UTR of the 3 new CSFV isolates and 32 reference isolates. Some mutation or deletion regions of these isolates are indicated by red boxes (□) and described in detail in the text
The 3′UTRs of all three new isolates were similar to most of the CSFV isolates. However, the HCLV vaccine strain had 12 continuous nucleotide insertions (TTTTTCTTTTTT) at positions 66–77 compared with the reference Shimen strain (Fig. 3). Similarly, the other two strains, RUCSFPLUM (AY578688) and Rovac (KJ873238), had 13 nucleotide insertions (TATTTATTTAGAT) at these positions (Fig. 3). Among the 14 analysed CSFV isolates, the CSFV-PK15C-NG79-11 (KC503764) and CSFV/IVRI/VB -131 (KM262189) strains had five and three poly T-nucleotide insertions, respectively (Fig. 3). In addition, compared with those of sub-genotype 1.1 isolates, the 3′UTRs of most sub-genotype 2.1 isolates, including the three new isolates, had two discontinuous nucleotide T deletions at positions 173 and 199, respectively (Fig. 3).
Fig. 3.
Sequence alignments of 3′UTR of the 3 new CSFV isolates and 11 reference isolates. Some mutation or insertion regions of these isolates are indicated by red boxes (□) and described in detail in the text
Amino acid analysis of E2
The E2 protein is essential for virus attachment and entry into the target cell and is also an important virulence determinant of CSFV (24). Consequently, the E2 amino acid sequences of the three new isolates and 33 reference isolates were compared and analysed (Fig. 4). There were some unique molecular characteristics for the new isolates, including the amino acid W at position 132 (W132), E228, I283, and L343 of SD2014-1 and L12 of SD2014-2. In addition, the three new isolates also had some consistent molecular characteristics, including E31, Y210 and K277. Consistently with most 2.1b and 2.1d isolates, the new isolates had an amino acid substitution at position 182 (L182 W). In contrast, at position 195, the new isolates were amino acid V, consistent with 1.1 isolates, and all other sub-genotype isolates were amino acid T. Additionally, at position 364, SD2014-1 was V, SD2014-2 was L, SD2014-3 was A, and other sub-genotype isolates were I.
Fig. 4.
Amino acid sequence alignments of E2 genes of the 3 new CSFV isolates and 33 reference isolates. The special mutation positions of these isolates are indicated by red boxes (□) and described in detail in the text
Discussion
CSF is one of the most important viral diseases of pigs in a number of countries, including China (17, 30). In recent decades, the C-strain vaccine has played a vital role in the control or elimination of CSF, and large CSF outbreaks have been rare (17, 30). However, sporadic outbreaks occur continuously in many regions of the world (30). As an RNA virus, CSFV has high error rates during replication, and genetic variability can allow the virus to easily evade the host immune response; therefore, it is important to surveil the genetic diversity of CSFV, track the virus origin and undertake effective control strategies. In the present study, the complete genome sequences of three CSFV strains isolated in 2014 were sequenced and analysed. The results will help to better understand the molecular diversity of CSFV isolates circulating in China.
Several sub-genotypes, including 1.1, 2.1, 2.2, and 2.3, exist in mainland China; especially the sub-genotype 2.1 has long been predominant (7). In the present study, the genomic sequences of the three CSFV isolates were compared against one another and with other representative CSFV isolates (Tables 2 and 3). The results indicated that the three isolates had high homology among each other. Additionally, they shared the highest nucleotide homology with sub-genotype 2.1b; they also showed high homology with the new sub-genotype 2.1d. Importantly, it was also demonstrated that the 5′UTRs of the three isolates were the most conserved regions in the genomes. NS3, NS4A, and NS4B proteins were more conserved than other regions of the CSFV genome.
Phylogenetic analysis has become a universally accepted classification criterion of CSFV isolates based on the 5′UTR (150 nt) and the E2 (190 and 1119 nt) and NS5B (409 nt) coding sequences (16, 20, 21). However, the complete genome sequence may provide a more credible basis for classification (15). In the present study, two phylogenic trees were constructed based on the complete genome sequences and the full-length E2 sequences, respectively (Fig. 1). Both trees showed that all CSFV isolates could be grouped into three clusters and several topology types. The three clusters were groups 1, 2, and 3, with topologies 1.1, 1.2, 1.3, 1.4, 2.1, 2.2, 2.3, and 3.4. The 2.1 isolates were further grouped into 2.1a, 2.1b, 2.1c, and 2.1d. All three new isolates belonged to 2.1b, which was consistent with the result of nucleotide homology alignment between the new isolates and representative CSFV isolates.
The 5′UTR of CSFV plays important roles in regulating initial translation of the pre-polyprotein and genome replication (10, 15). In addition, the studies on the 5′UTR mainly focused on the 3′ end 2/3 region and the IRES of the 5′UTR (6, 15). Sequence alignment showed that the 5′UTRs of the three new isolates had different nucleotide substitutions or deletions at positions 44 and 357–358 compared with other CSFV isolates (Fig. 2). The substitutions or deletions affect the structural characteristics of the 5′UTRs and require experimental confirmation. However, CSFV virulence may vary according to the number and shape of the pseudoknot loop in the secondary structure of 5′UTR and its positional direction in three-dimensional space (6, 15). Thus, the new isolates may have different virulence characteristics.
The 3′UTR is the region with the most variation in the CSFV genome. Previous study reported that the poly-T deletion in the 3′UTR is characteristic of CSFV virulent isolates, which suggests a direct relationship between the poly-T deletion region and viral virulence (15). Later study confirmed that the poly-T insertion in the 3′UTR was important for the attenuation of CSFV (29). However, the insertion is not believed to be a marker for virulence (1, 31). In the present study, we found that all three new isolates had no poly-T insertion region, whereas several other strains, including HCLV, CSFV-PK15C-NG79-11, CSFV/IVRI/VB-131, RUCSFPLUM, and Rovac (AF091507, KC503764, KM262189, AY578688, and KJ873238, respectively) had the insertion region (Fig. 3). Recently, a unique poly-T tract was discovered in the 3′UTR of the CSFV Pinar del Rio strain compared with other CSFV isolates (3). Whether this novel insertion affects the pathogenicity of the virus remains unknown. Additionally, many studies demonstrated that NS3, NS5A, and NS5B of CSFV can interact with 3′UTR to regulate viral RNA synthesis and replication (2, 14, 25). In addition, the 3′UTRs of the new isolates and most sub-genotype 2.1 isolates had two discontinuous nucleotide deletions compared with those of 1.1 isolates (Fig. 3). Whether the nucleotide deletions affect the interactions with other proteins requires further research.
The E2 is the most antigenic protein of CSFV which is involved in virus neutralisation (24). Four antigenic domains, A (86–176 amino acids), B (1–83 amino acids), C (1–110 amino acids), and D (86–110 amino acids), have been mapped on E2 (27). Domain A has been subdivided into A1, A2, and A3 (27). In the present study, we found several unique amino acid substitutions for the three new isolates in these domains and in other regions (Fig. 4). However, the influence of these substitutions on the structure and function of E2 requires further study. In addition, the six cysteines at positions 4, 48, 103, 129, 139, and 167, which were essential for binding by monoclonal antibodies of the four domains, showed no variation in the E2 proteins of the three isolates (27). Furthermore, the potential N-glycosylation sites in the E2 proteins of the three isolates were consistent with previous isolates.
In recent years, the reports on whole-genome analysis of CSFV have been limited. In the present study, the complete genomes of the three new CSFV isolates were fully analysed. We found that all the isolates belonged to the genetic subgroup 2.1b. Furthermore, some genomic variations were found in the UTRs and E2. These data indicate that sub-genotype 2.1b exhibits a trend for wide variation.
Footnotes
Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: The research and the article were financed with the funds of the National Natural Science Foundation of China (No. 31502097), the National Key R and D Programme (2016YFD0500100), the Scientific and Technological Project of Henan Province (182102110240), the Foundation of Nanyang Normal University (No. 15082), and the Key Programme Foundation of Higher Education of Educational Commission of Henan Province (No. 15A230026).
Animal Rights Statement: The collection of clinical samples was approved by the Animal Ethics Committee of School of Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences and performed in accordance with animal ethics guidelines and approved protocols.
References
- 1.Bjorklund H.V., Stadejek T., Vilcek S., Belak S.. Molecular characterization of the 3′ noncoding region of classical swine fever virus vaccine strains. Virus Genes. 1998;16:307–312. doi: 10.1023/a:1008095109033. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y., Xiao J., Xiao J., Sheng C., Wang J., Jia L., Zhi Y., Li G., Chen J., Xiao M.. Classical swine fever virus NS5A regulates viral RNA replication through binding to NS5B and 3′UTR. Virology. 2012;432:376–388. doi: 10.1016/j.virol.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Coronado L., Liniger M., Munoz-Gonzalez S., Postel A., Perez L.J., Perez-Simo M., Perera C.L., Frias-Lepoureau M.T., Rosell R., Grundhoff A., Indenbirken D., Alawi M., Fischer N., Becher P., Ruggli N., Ganges L.. Novel poly-uridine insertion in the 3′UTR and E2 amino acid substitutions in a low virulent classical swine fever virus. Vet Microbiol. 2017;201:103–112. doi: 10.1016/j.vetmic.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Deng M.C., Huang C.C., Huang T.S., Chang C.Y., Lin Y.J., Chien M.S., Jong M.H.. Phylogenetic analysis of classical swine fever virus isolated from Taiwan. Vet Microbiol. 2005;106:187–193. doi: 10.1016/j.vetmic.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Edwards S., Fukusho A., Lefevre P.C., Lipowski A., Pejsak Z., Roehe P., Westergaard J.. Classical swine fever: the global situation. Vet Microbiol. 2000;73:103–119. doi: 10.1016/s0378-1135(00)00138-3. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher S.P., Jackson R.J.. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5′ untranslated region important for IRES function. J Virol. 2002;76:5024–5033. doi: 10.1128/JVI.76.10.5024-5033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong W., Wu J., Lu Z., Zhang L., Qin S., Chen F., Peng Z., Wang Q., Ma L., Bai A., Guo H., Shi J., Tu C.. Genetic diversity of sub-genotype 2.1 isolates of classical swine fever virus. Infect Genet Evol. 2016;41:218–226. doi: 10.1016/j.meegid.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 8.He C.Q., Ding N.Z., Chen J.G., Li Y.L.. Evidence of natural recombination in classical swine fever virus. Virus Res. 2007;126:179–185. doi: 10.1016/j.virusres.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Horzinek M.C.. Pestiviruses--taxonomic perspectives. Arch Virol Suppl. 1991;3:1–5. [PubMed] [Google Scholar]
- 10.Hsu W.L., Chen C.L., Huang S.W., Wu C.C., Chen I.H., Nadar M., Su Y.P., Tsai C.H.. The untranslated regions of classic swine fever virus RNA trigger apoptosis. PLoS One. 2014;9:e88863. doi: 10.1371/journal.pone.0088863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang D.L., Gong W.J., Li R.C., Liu G.H., Hu Y.F., Ge M., Wang S.Q., Yu X.L., Tu C.. Phylogenetic analysis using E2 gene of classical swine fever virus reveals a new sub-genotype in China. Infect Genet Evol. 2013;17:231–238. doi: 10.1016/j.meegid.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S., Stecher G., Tamura K.. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leng C.L., Zhang H.L., Kan Y.C., Yao L.G., Li M.L., Zhai H.Y., Li Z., Liu C.X., Shi H.F., Ji J., Qiu R., Tian Z.J.. Characterisation of newly emerged isolates of classical swine fever virus in China, 2014–2015. J Vet Res. 2017;61:1–9. doi: 10.1515/jvetres-2017-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S., Feng S., Wang J.H., He W.R., Qin H.Y., Dong H., Li L.F., Yu S.X., Li Y., Qiu H.J.. eEF1A interacts with the NS5A protein and inhibits the growth of classical swine fever virus. Viruses. 2015;7:4563–4581. doi: 10.3390/v7082833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Xu Z., He Y., Yao Q., Zhang K., Jin M., Chen H., Qian P.. Genome comparison of a novel classical swine fever virus isolated in China in 2004 with other CSFV strains. Virus Genes. 2006;33:133–142. doi: 10.1007/s11262-005-0048-2. [DOI] [PubMed] [Google Scholar]
- 16.Lowings P., Ibata G., Needham J., Paton D.. Classical swine fever virus diversity and evolution. J Gen Virol. 1996;77:1311–1321. doi: 10.1099/0022-1317-77-6-1311. [DOI] [PubMed] [Google Scholar]
- 17.Luo Y., Li S., Sun Y., Qiu H.J.. Classical swine fever in China: a minireview. Vet Microbiol. 2014;172:1–6. doi: 10.1016/j.vetmic.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Meyers G., Thiel H.J.. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 19.Moennig V., Plagemann P.G.. The pestiviruses. Adv Virus Res. 1992;41:53–98. doi: 10.1016/s0065-3527(08)60035-4. [DOI] [PubMed] [Google Scholar]
- 20.Pan C.H., Jong M.H., Huang T.S., Liu H.F., Lin S.Y., Lai S.S.. Phylogenetic analysis of classical swine fever virus in Taiwan. Arch Virol. 2005;150:1101–1119. doi: 10.1007/s00705-004-0485-6. [DOI] [PubMed] [Google Scholar]
- 21.Paton D.J., McGoldrick A., Greiser-Wilke I., Parchariyanon S., Song J.Y., Liou P.P., Stadejek T., Lowings J.P., Bjorklund H., Belak S.. Genetic typing of classical swine fever virus. Vet Microbiol. 2000;73:137–157. doi: 10.1016/s0378-1135(00)00141-3. [DOI] [PubMed] [Google Scholar]
- 22.Postel A., Schmeiser S., Bernau J., Meindl-Boehmer A., Pridotkas G., Dirbakova Z., Mojzis M., Becher P.. Improved strategy for phylogenetic analysis of classical swine fever virus based on full-length E2 encoding sequences. Vet Res. 2012;43:1–15. doi: 10.1186/1297-9716-43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postel A., Schmeiser S., Perera C.L., Rodriguez L.J., Frias-Lepoureau M.T., Becher P.. Classical swine fever virus isolates from Cuba form a new sub-genotype 1.4. Vet Microbiol. 2013;161:334–338. doi: 10.1016/j.vetmic.2012.07.045. [DOI] [PubMed] [Google Scholar]
- 24.Risatti G.R., Borca M.V., Kutish G.F., Lu Z., Holinka L.G., French R.A., Tulman E.R., Rock D.L.. The E2 glycoprotein of classical swine fever virus is a virulence determinant in swine. J Virol. 2005;79:3787–3796. doi: 10.1128/JVI.79.6.3787-3796.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheng C., Chen Y., Xiao J, Xiao J., Wang J., Li G., Chen J., Xiao M.. Classical swine fever virus NS5A protein interacts with 3′-untranslated region and regulates viral RNA synthesis. Virus Res. 2012;163:636–643. doi: 10.1016/j.virusres.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Tu C., Lu Z., Li H., Yu X., Liu X., Li Y., Zhang H., Yin Z.. Phylogenetic comparison of classical swine fever virus in China. Virus Res. 2001;81:29–37. doi: 10.1016/s0168-1702(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 27.van Rijn P.A., Miedema G.K., Wensvoort G., van Gennip H.G., Moormann R.J.. Antigenic structure of envelope glycoprotein E1 of hog cholera virus. J Virol. 1994;68:3934–3942. doi: 10.1128/jvi.68.6.3934-3942.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilcek S., Belak S.. Organization and diversity of the 3′-noncoding region of classical swine fever virus genome. Virus Genes. 1997;15:181–186. doi: 10.1023/a:1007971110065. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., Wang Q., Lu X., Zhang C., Fan X., Pan Z., Xu L., Wen G., Ning Y., Tang F., Xia hY.. 12-nt insertion in 3′ untranslated region leads to attenuation of classic swine fever virus and protects host against lethal challenge. Virology. 2008;374:390–398. doi: 10.1016/j.virol.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Leng C., Feng L., Zhai H., Chen J., Liu C., Bai Y., Ye C., Peng J., An T., Kan Y., Cai X., Tian Z., Tong G.. A new sub-genotype 2.1d isolates of classical swine fever virus in China, 2014. Infect Genet Evol. 2015;34:94–105. doi: 10.1016/j.meegid.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 31.Zhou W., Gao S., Podgorska K., Stadejek T., Qiu H.J., Yin H., Drew T., Liu L.. Rovac is the possible ancestor of the Russian lapinized vaccines LK-VNIVViM and CS strains but not the Chinese strain (C-strain) vaccine against classical swine fever. Vaccine. 2014;32:6639–6642. doi: 10.1016/j.vaccine.2014.09.058. [DOI] [PubMed] [Google Scholar]