Summary
In the brain, many types of interneurons make functionally diverse inhibitory synapses onto principal neurons. While numerous molecules have been identified to function in inhibitory synapse development, it remains unknown whether there is a unifying mechanism for development of diverse inhibitory synapses. Here we report a general molecular mechanism underlying hippocampal inhibitory synapse development. In developing neurons, the establishment of GABAergic transmission depends on Neuroligin2 (NL2), a synaptic cell adhesion molecule (CAM). During maturation, inhibitory synapse development requires both NL2 and Slitrk3 (ST3), another CAM. Importantly, NL2 and ST3 interact with nanomolar affinity through their extracellular domains to synergistically promote synapse development. Selective perturbation of the NL2-ST3 interaction impairs inhibitory synapse development with consequent disruptions in hippocampal network activity and increased seizure susceptibility. Our findings reveal how unique postsynaptic CAMs work in concert to control synaptogenesis and establish a general framework for GABAergic synapse development.
Introduction
Synapse development is a multi-step process orchestrated by numerous molecules acting in a highly spatially and temporally controlled manner (McAllister, 2007; Sanes and Yamagata, 2009; Shen and Scheiffele, 2010; Siddiqui and Craig, 2011; Waites et al., 2005). Among these molecules, synaptic cell adhesion molecules (CAMs) are a class of membrane proteins essential for the establishment and maturation of synaptic connections. Recent studies have identified a growing number of synaptic CAMs, including: 1) Neuroligins (NLs) and leucine-rich repeat transmembrane proteins (LRRTMs) that bind to presynaptic neurexins; 2) Slit and NTRK-like family proteins (Slitrks) that bind to presynaptic protein tyrosine phosphatases (PTPs); 3) immunoglobulin superfamily proteins (IgSFs) that mediate trans-synaptic homophilic or heterophilic adhesion interactions; 4) cadherin family proteins; and 5) transmembrane tyrosine kinase receptors (Craig and Kang, 2007; Dalva et al., 2007; de Wit and Ghosh, 2016; Huang and Scheiffele, 2008; Krueger-Burg et al., 2017; Lu et al., 2016; Missler et al., 2012; Sudhof, 2008). While these molecules are involved in various stages of synapse development, how diverse synaptic CAMs work in concert to control synapse formation remains largely unclear.
GABAergic synapses are the dominant source of inhibition throughout the mammalian brain, providing critical inhibitory balance to excitatory glutamatergic drive for control of neuronal/network excitability. As GABAergic inhibition is important in almost every aspect of brain physiology and the dysregulation of GABAergic synapse development has been implicated in many neurological and neuropsychiatric disorders (Ko et al., 2015; Ramamoorthi and Lin, 2011), it is critical to understand the molecular determinants of GABAergic synapse formation. Interestingly, GABAergic synapses exhibit remarkable diversity (Cherubini and Conti, 2001; Sassoe-Pognetto et al., 2011) with over twenty different types of interneurons providing domain-specific, functionally distinct GABAergic input onto principal neurons and each other (Klausberger and Somogyi, 2008). Although many molecules and signaling pathways have been identified to regulate hippocampal GABAergic synapse development (Huang and Scheiffele, 2008; Ko et al., 2015; Krueger-Burg et al., 2017; Kuzirian and Paradis, 2011; Lu et al., 2016), a general framework for development of diverse GABAergic synapses has not been identified.
Recent studies have implicated Neuroligin2 (NL2) and Slitrk3 (ST3) as two synaptogenic molecules critical for GABAergic synapse development in hippocampus (Chih et al., 2005; Graf et al., 2004; Poulopoulos et al., 2009; Takahashi et al., 2012; Varoqueaux et al., 2004; Yim et al., 2013). In co-cultures of neurons with non-neuronal cells NL2 and ST3 are both able to induce inhibitory presynaptic differentiation (Graf et al., 2004; Scheiffele et al., 2000; Takahashi et al., 2012; Yim et al., 2013). Structurally, NL2 and ST3 are single-pass transmembrane CAMs that are highly enriched at GABAergic synapses (Takahashi et al., 2012; Varoqueaux et al., 2004). Functionally, over-expression of either NL2 or ST3 promotes GABAergic synapse formation in hippocampal primary neuronal cultures (Chih et al., 2005; Chubykin et al., 2007; Takahashi et al., 2012; Yim et al., 2013). Conversely, RNAi-mediated knockdown of NL2 or ST3 reduces GABAergic synapse density and inhibitory transmission (Chih et al., 2005; Futai et al., 2013; Graf et al., 2004; Nguyen et al., 2016; Takahashi et al., 2012; Yim et al., 2013). Furthermore, hippocampal GABAergic transmission is reduced to ~ half of control levels in NL2 or ST3 knockout (KO) mice (Poulopoulos et al., 2009; Takahashi et al., 2012). Collectively, these studies implicate NL2 and ST3 as critical CAMs for GABAergic synapse development in hippocampus. However, how NL2 and ST3 work together to control GABAergic synaptogenesis remains unknown.
Here we report a hierarchical process orchestrated by NL2 and ST3 for GABAergic synapse development. In developing hippocampal neurons, NL2, but not ST3, essentially accounts for initial GABAergic synapse development. As neurons mature, both NL2 and ST3 are required for development of GABAergic synapses. Importantly, an interaction between NL2 and ST3 in the nanomolar range facilitates ST3 trafficking to the plasma membrane and potently promotes GABAergic synapse development. Selective disruption of this NL2-ST3 interaction impairs development of GABAergic transmission, decreases hippocampal network activities associated with inhibitory pacing, and increases seizure susceptibility.
Results
ST3 is up-regulated in hippocampal neurons in NL2 KO mice
To determine the role of NL2 in GABAergic synapse development, we performed in utero electroporation to express a NL2 shRNA (small hairpin RNA) (Chih et al., 2005) along with GFP in hippocampal progenitor cells at embryonic day 14.5 (E14.5) and probed GABAergic inhibition in hippocampal slices at P8–10 (Figure 1A, Figure S1A). Measurement of miniature inhibitory postsynaptic currents (mIPSCs) showed that GABAergic transmission was barely detectable in shRNA-expressing CA1 pyramidal neurons (Figure 1B). In contrast, NL2 shRNA expression in hippocampal neurons from NL2 KOs (NL2−/−) did not significantly reduce GABAergic transmission (Figure S1C), confirming the specificity of the construct. In addition, while more mature, P19–21, CA1 neurons expressing NL2 shRNAs continued to exhibit reduced GABAergic transmission (~70% reduction of mIPSC frequency), the deficit was less severe than at P8–10 (Figure S2A). These data suggest that NL2 is essential for establishing GABAergic transmission in developing hippocampal neurons, and that as neurons mature, GABAergic transmission in NL2 knockdown neurons, although strongly reduced, can develop.
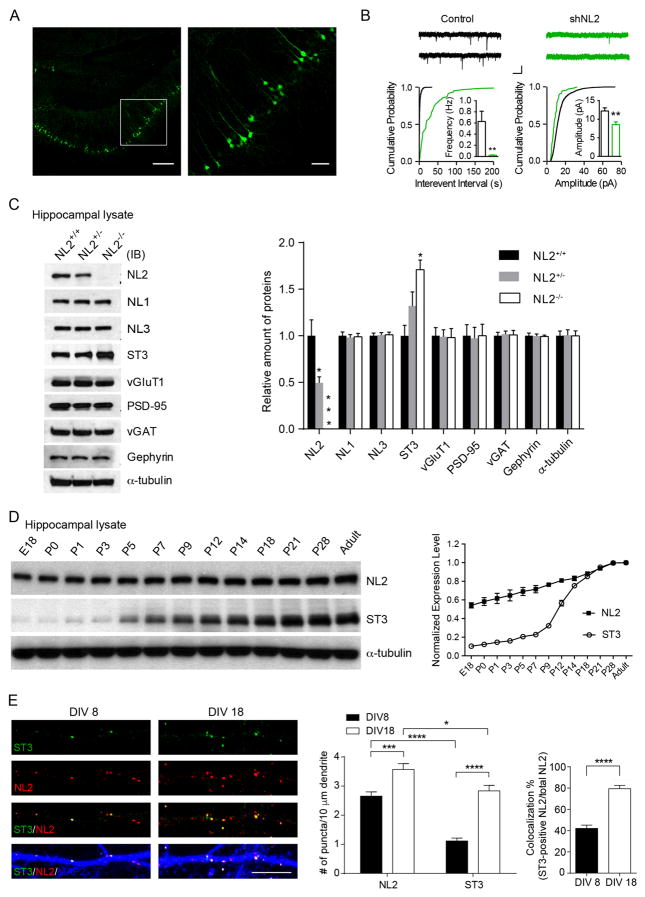
Figure 1. ST3 is up-regulated in hippocampus in NL2−/−.
(A) Representative images (left, low magnification; right, high magnification of the boxed area in left) showed mosaic expression of GFP in the CA1 neurons in an acute hippocampal slice. Scale bar, left, 100 μm; right, 50 μm.
(B) There were significant reductions of both mIPSC frequency and amplitude in NL2 knockdown CA1 neurons in acute hippocampal slices (Control, n = 10; shNL2, n = 10, p < 0.01 for both frequency and amplitude, t-test; Kolmogorov–Smirnov (K-S) test was used for cumulative distributions, p < 0.0001 for both conditions). Scale bar,10 pA and 1 s.
(C) Representative immunoblots and summary graph of protein levels in the hippocampal lysates from ~4 weeks old mice (n = 3 mice per genotype, one-way ANOVA, p < 0.05 for ST3 expression levels).
(D) Representative immunoblots and summary graph showed the expression profiles (normalized to the highest expression level) of NL2 and ST3 proteins in hippocampus.
(E) Representative images of ST3 (green), NL2 (red) and MAP2 (blue) at DIV8 and DIV18 in hippocampal cultures. t-test, p < 0.05*, 0.001***, 0.0001****. Scale bar, 10 μm.
See also Figure S1–2
In contrast to our single-cell NL2 knockdown data in vivo, previous work in adult NL2−/− observed only a modest reduction of GABAergic transmission in CA1 pyramidal neurons (~50%, (Poulopoulos et al., 2009). Similarly, we found that the reduction in GABAergic transmission in developing CA1 neurons in P8–9 NL2−/− (~70%) was less severe than the deficit induced by NL2 shRNA (Figure S2B). We thus probed in NL2−/− hippocampal lysates for other synaptic molecules that might be up-regulated. While there was no change of total expression levels of other NL family members including NL1, NL3 and several other synaptic proteins, ST3 expression was significantly increased in NL2−/− (Figure 1C). Thus, selective up-regulation of ST3 might partially compensate for the loss of NL2 in promoting GABAergic synapse development in NL2−/−.
We next examined the temporal expression profiles of NL2 and ST3 in mouse hippocampus. While NL2 was abundantly expressed in perinatal hippocampi, ST3 expression was relatively low until over one week after birth (Figure 1D). Similarly, although NL2 puncta were readily detected in immature hippocampal cultures at 8 days in vitro (DIV8), ST3 puncta were much less abundant, so that the majority of NL2 puncta did not contain ST3 at the early developmental stage (Figure 1E). However, as neurons matured ST3 puncta strongly increased and most ST3 immunolabeling co-localized with NL2 in DIV18 cultures (Figure 1E). These data show distinct expression profiles of NL2 and ST3 during development, suggesting that these two molecules play differential roles in GABAergic synapse development.
NL2, but not ST3, is essential for GABAergic synapse development in immature neurons
To further interrogate the functions of NL2 and ST3 in GABAergic synapse development, we performed single-cell shRNA-mediated knockdown of NL2 or ST3 at DIV2 hippocampal cultures and conducted electrophysiological and cell biological analyses of GABAergic synapses at DIV8. In agreement with the data collected in vivo (Figure 1A) and consistent with the expression profile of NL2 and ST3 (Figure 1D), we found that NL2 knockdown in DIV2 neurons caused an over ~95% decrease of mIPSC frequency and a strong reduction of mIPSC amplitude (Figure 2A). This functional deficit was accompanied by severe reductions in immunolabeling for vGAT (vesicular GABA transporter) and gephyrin in NL2 shRNA transfected neurons (Figure 2B), consistent with a strong reduction in GABAergic synapse density. In contrast, ST3 knockdown, induced by a ST3 shRNA (Takahashi et al., 2012), in neurons at the same developmental stage did not affect either mIPSCs or density of vGAT/gephyrin puncta density (Figure 2A–B, Figure S1B). In addition, knockdown of both NL2 and ST3 in DIV2 neurons yielded similar deficits of GABAergic synapses as NL2 knockdown alone (Figure 2A–B). These data indicate that NL2 is the dominant synaptic CAM responsible for the establishment of GABAergic transmission in developing hippocampal neurons. ST3, conversely, is expressed at relatively low levels early in development, and thus, minimally contributes to the initial formation of GABAergic synapses.
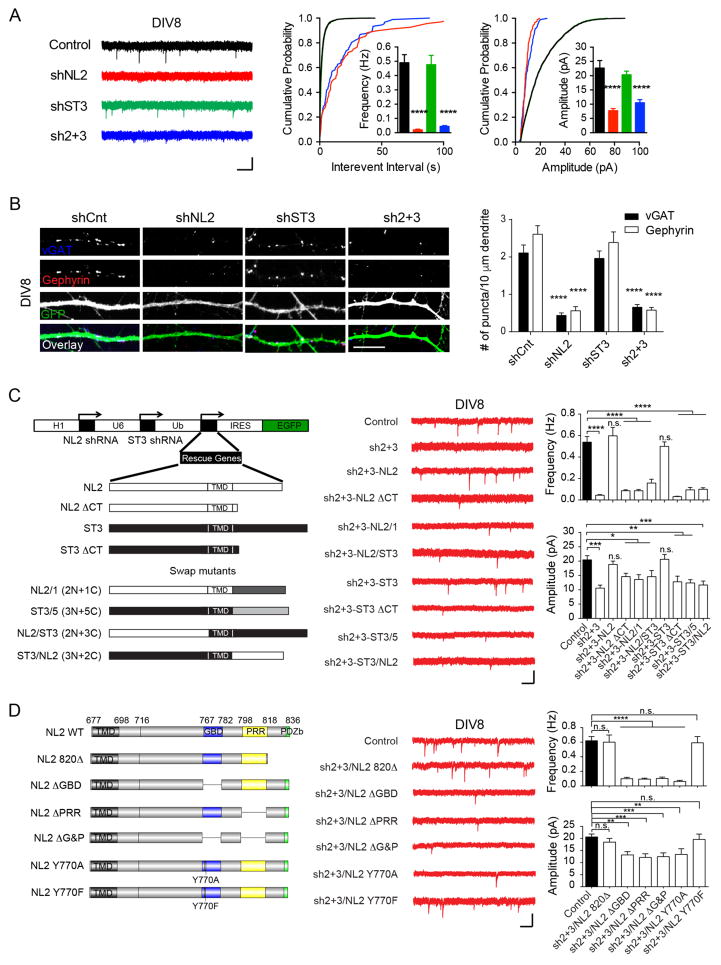
Figure 2. NL2, but not ST3, is essential for GABAergic synapse development in immature neurons.
(A) Knockdown of NL2, but not ST3, strongly reduced GABAergic transmission in DIV8 neurons (Control, n = 14; shNL2, n = 14; shST3, n = 10, sh2+3, n = 11, one-way ANOVA, p < 0.0001 for both frequency and amplitude; K-S test, p < 0.0001 between Control and shNL2 or sh2+3). Sh2+3, double knockdown of NL2 and ST3. Scale bar, 20 pA and 1 s.
(B) Knockdown of NL2, but not ST3, strongly reduced vGAT and gephyrin puncta in DIV8 neurons (one-way ANOVA, p < 0.0001). Scale bar, 10 μm.
(C) NL2 C-tail was important for establishing GABAergic transmission in developing neurons. Left, schematic of shRNA vectors. Right, sample traces and summary graphs (Control, n = 12; sh2+3, n = 11; sh2+3-NL2, n = 9; sh2+3-NL2 ΔCT, n = 16; sh2+3-NL2/1, n = 11; sh2+3-NL2/ST3, n = 13; sh2+3-ST3, n = 14; sh2+3-ST3 ΔCT, n = 10; sh2+3-ST3/5, n = 12; sh2+3-ST3/NL2, n = 13, one-way ANOVA, p < 0.05*, 0.01**, 0.001***, 0.0001****, n.s., not significant). NL2/1, NL2 C-tail was swapped with the same domain from NL1; ST3/5, ST3 C-tail was swapped with the same domain from ST5; TMD, transmembrane domain; Scale bar, 20 pA and 1 s.
(D) NL2 GBD and PRR domains were critical for GABAergic synapse development in DIV8 neurons. Left, schematic of NL2 C-tail and its variants, GBD, gephyrin binding domain; PRR, proline-rich repeat; PDZb, PDZ domain binding motif. Right, sample traces and summary graphs (Control, n = 24; sh2+3/NL2 820Δ, n = 10; sh2+3/NL2 ΔGBD, n = 14; sh2+3/NL2 ΔPRR, n = 14; sh2+3/NL2 ΔG&P, n = 14; sh2+3/NL2 Y770A, n = 11; sh2+3/NL2 Y770F, n = 6, one-way ANOVA, p < 0.01**, 0.001***, 0.0001****). Scale bar, 20 pA and 1 s.
See also Figure S1, 3
Functional dissection of the NL2 carboxyl terminus in GABAergic synapse development in immature neurons
To further understand the role of NL2 in GABAergic synapse development in immature neurons, we employed a molecular replacement approach. In this approach, the NL2 and ST3 double knockdown construct simultaneously expressed shRNA-resistant NL2, ST3 or their mutants (Figure 2C–D and Figure S3A–E). We found that while knockdown of both NL2 and ST3 at DIV2 essentially eliminated mIPSCs measured at DIV8, co-expression of shRNA-resistant NL2 with the double knockdown plasmid fully rescued mIPSC deficits (Figure 2A, 2C). In contrast, very little rescue occurred with NL2 ΔCT, a mutant lacking the carboxyl terminus (C-tail), suggesting a critical role for the NL2 C-tail (Figure 2C). In addition, an NL2 mutant (NL2/1 (2N + 1C)) in which NL2 C-tail was replaced by the same domain from NL1 that is enriched at excitatory synapses was not able to rescue mIPSC deficits (Figure 2C). Interestingly, although ST3 knockdown on its own did not affect mIPSCs in DIV8 neurons, co-expression of shRNA-resistant ST3 with the double knockdown vector could rescue mIPSC deficits (Figure 2C), suggesting that expressed ST3 is sufficient to drive GABAergic synapse development, even in the absence of NL2. The rescue of mIPSC deficits by ST3 also depended on its C-tail, as neither ST3 ΔCT that lacked the entire C-tail nor ST3/5 (3N + 5C) in which ST3 C-tail was swapped with the same domain from Slitrk5 (ST5) that regulates excitatory synapse development (Yim et al., 2013) could rescue mIPSC deficits in double knockdown neurons (Figure 2C). Finally, we made two mutants in which C-tails of NL2 and ST3 were swapped with each other. Co-expression of either of these two mutants with double knockdown vector did not rescue mIPSC deficits (Figure 2C).
Further investigation revealed that an NL2 mutant (NL2 820Δ) lacking the last 17 residues after the proline rich region (PRR), including the PDZ domain binding motif, was sufficient to restore mIPSC deficits in double knockdown neurons (Figure 2D), indicating that this region is not critical for NL2 function in GABAergic synapse development. In contrast, an NL2 C-tail mutant lacking the gephyrin binding domain (GBD) (Poulopoulos et al., 2009) failed to rescue mIPSC deficits (Figure 2D). Tyrosine 770 (Y770) within the GBD domain is critical for the NL2-gephyrin interaction (Giannone et al., 2013; Poulopoulos et al., 2009). Specifically, NL2 Y770A, but not the Y770F mutation, disrupted the binding to gephyrin. Consistently, Y770F, but not Y770A, rescued mIPSC deficits (Figure 2D), further confirming that the NL2 GBD is critical for GABAergic synapse development in immature neurons. Recently, the PRR in the NL2 C-tail was shown to regulate the function of collybistin, a guanine nucleotide exchange factor (GEF) important for GABAergic synapse formation (Kins et al., 2000; Papadopoulos et al., 2008; Papadopoulos et al., 2007; Poulopoulos et al., 2009; Soykan et al., 2014). We found that NL2 mutants lacking the PRR or both the GBD and PRR could not rescue mIPSC deficits (Figure 2D). Together, these data identify two regions in the NL2 C-tail critical for the development of GABAergic synapses in immature neurons and corroborate the important roles of gephyrin and collybistin in GABAergic synapse formation (Kins et al., 2000; Papadopoulos et al., 2008; Papadopoulos et al., 2007; Poulopoulos et al., 2009; Soykan et al., 2014; Tretter et al., 2012; Tyagarajan and Fritschy, 2014).
Both NL2 and ST3 are required for later stages of GABAergic synapse development
We next examined the function of NL2 and ST3 in GABAergic synapse development in more mature neurons when both NL2 and ST3 are highly expressed (Figure 1D–E). We transfected neurons with shRNAs constructs against NL2, ST3 or NL2 plus ST3 at DIV10, and examined GABAergic synapses at DIV18. Knockdown of NL2 induced a strong reduction of mIPSC frequency and a significant decrease of mIPSC amplitude (Figure 3A). In addition, NL2 knockdown strongly reduced vGAT and gephyrin puncta density (Figure 3B). These data together with findings in Figure 2 indicate that NL2 plays a critical role in the regulation of GABAergic synapse development in both developing and more mature neurons.
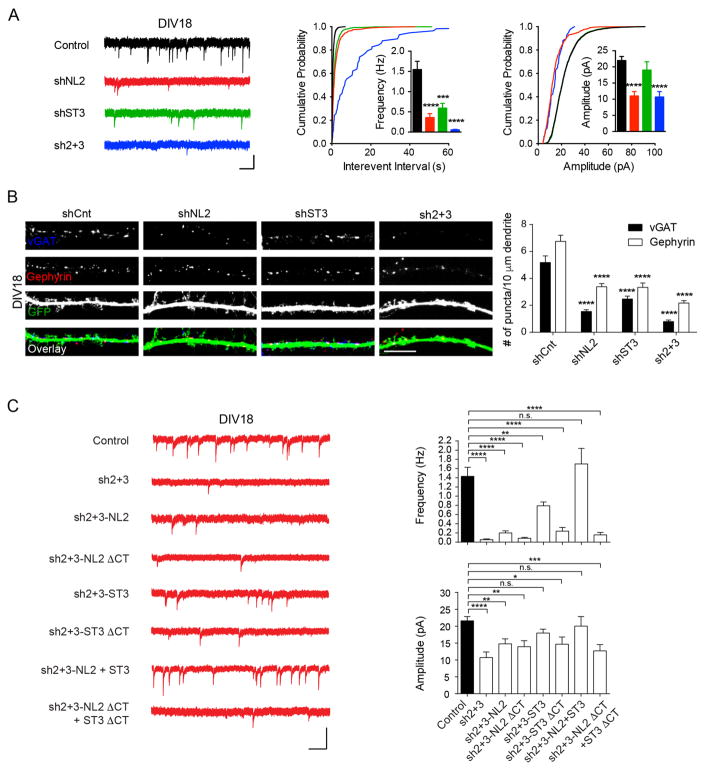
Figure 3. Both NL2 and ST3 are crucial for GABAergic synapse development in more mature neurons.
(A) Knockdown of NL2 and/or ST3 strongly impaired mIPSCs in DIV18 neurons (Control, n = 17; shNL2, n = 11; shST3, n = 9, sh2+3, n = 11, one-way ANOVA, p < 0.0001 for both frequency and amplitude; K-S test, p < 0.0001 between Control and others for Interevent interval, and p < 0.0001 between Control and shNL2 or sh2+3 for amplitude). Scale bar, 20 pA and 1 s.
(B) Knockdown of NL2, ST3 or both NL2 and ST3 dramatically reduced vGAT and gephyrin puncta in DIV18 neurons (one-way ANOVA, p < 0.0001). Scale bar, 10 μm.
(C) Rescue experiments in DIV18 NL2/ST3 double knockdown neurons (Control, n = 19; sh2+3, n = 11; sh2+3-NL2, n = 17; sh2+3-NL2 ΔCT, n = 10; sh2+3-ST3, n = 14; sh2+3-ST3 ΔCT, n = 7; sh2+3-NL2+ST3, n = 8; sh2+3-NL2 ΔCT+ST3 ΔCT, n = 11, one-way ANOVA, p < 0.05*, 0.01**, 0.001***, 0.0001****). Scale bar, 20 pA and 1 s.
Interestingly, ST3 knockdown at DIV10 also caused a strong reduction of mIPSC frequency, although it had no effect on mIPSC amplitude (Figure 3A). Similarly, both vGAT and gephyrin immunolabelling were significantly decreased in neurons expressing the ST3 shRNA from DIV10 (Figure 3B). These effects of ST3 knockdown were additive with NL2 as knockdown of both NL2 and ST3 at DIV10 eliminated nearly all GABAergic synaptic transmission and further reduced GABAergic synapse marker density in DIV18 neurons (Figure 3A–B). Thus, both NL2 and ST3 are important for GABAergic synapse development in more mature neurons, and together account for most, if not all, GABAergic synapse development in hippocampal neurons.
mIPSC deficits in double knockdown DIV18 neurons could be rescued by co-expression of shRNA-resistant NL2 and ST3, but not by NL2 on its own (Figure 3C), suggesting that NL2 and ST3 may synergistically regulate GABAergic synapse development at this stage. Interestingly, ST3 partially rescued mIPSC deficits in the double knockdown neurons at DIV18, indicating that ST3 might be sufficient to drive the development of a portion of GABAergic synapses at this age even in the absence of NL2 (Figure 3C). Importantly, NL2 ΔCT and ST3 ΔCT mutant constructs alone or in combination failed to rescue mIPSCs in neurons expressing double knockdown vector, confirming the importance of these C-tail domains (Figure 3C). Taken together, these data indicate that both NL2 and ST3 are required for GABAergic synapse development in more mature neurons.
NL2 and ST3 interact with each other in vitro and in vivo
Based on the co-localization of NL2 with ST3 in mature neurons (Figure 1E) and their co-requirement for GABAergic synapse development at this stage (Figure 3), we reasoned that NL2 and ST3 might co-operatively regulate GABAergic synapse development. To explore this possibility, we first performed a co-immunoprecipitation (co-IP) assay in detergent-solubilized mouse hippocampal lysates to test if NL2 and ST3 were present in the same protein complex. We found that ST3 was detected in the NL2 immunoprecipitates (IPs) (Figure 4A), demonstrating that native ST3 associates with NL2. In addition, in agreement with a previous report, gephyrin was also present in the IPs by the NL2 antibody (Figure 4A) (Poulopoulos et al., 2009). In contrast, two excitatory postsynaptic components, PSD-95 and GluA1, a major AMPA receptor subunit in hippocampus (Lu et al., 2009), were not co-IPed with NL2 (Figure 4A), showing the specificity of the NL2-ST3 association. Importantly, none of these proteins were detected in IPs by a control IgG (Figure 4A).
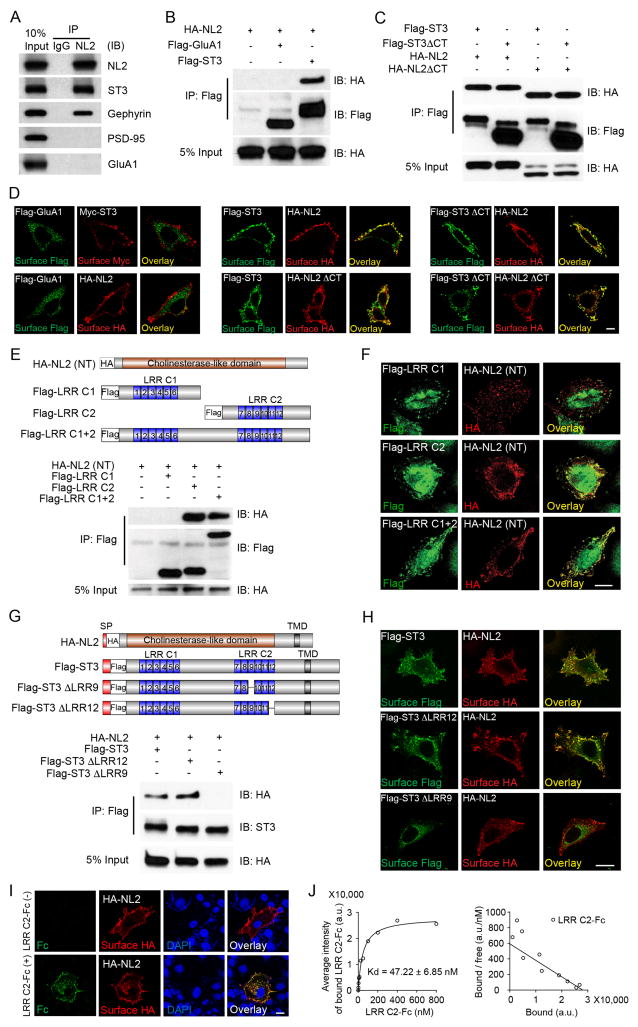
Figure 4. NL2 interacts with the LRR9 domain of ST3.
(A) ST3 associated with NL2 in mouse hippocampal lysates.
(B) NL2 interacted with ST3 in HEK293T cells. HA-NL2 was expressed alone, or together with Flag-ST3 or Flag-GluA1. Cell lysates were subjected to IP and immunoblotting (IB) assays.
(C) NL2 or ST3 mutants lacking the C-terminal domains could interact with each other. Lysates from HEK293T cells transfected with indicated plasmids were subjected to IP and IB assays.
(D) Immunostaining of HeLa cells expressing indicated plasmids. Left panel showed that cell surface GluA1 did not co-localize with surface NL2 or ST3, middle panel showed that surface ST3 co-localized with surface NL2 or NL2 ΔCT, right panel showed that surface ST3 ΔCT co-localized with surface NL2 or NL2 ΔCT. Scale bar, 10μm.
(E) The second LRR cluster (LRR C2) in the ST3 extracellular domain interacted with the NL2 extracellular domain. Top, schematic of NL2 or ST3 extracellular domain mutants. Bottom, a co-IP assay of lysates from cells transfected with indicated plasmids.
(F) Confocal images showed that ST3 N-terminal mutants containing the LRR C2 (middle and bottom panels), but not the one lacking the LRR C2 (top panel), co-localized with NL2(NT), in HeLa cells. Scale bar, 20μm.
(G) ST3 LRR9 domain mediated the interaction with NL2. Top, schematic of NL2 and ST3 or ST3 mutants. Bottom, A co-IP assay of lysates from cells transfected with indicated plasmids.
(H) Confocal images showed that ST3 mutants containing the LRR9 (top and middle panels), but not the one lacking LRR9 (bottom panel), co-localized with NL2 at the cell surface in HeLa cells. Scale bar, 20μm.
(I and J) Confocal images and Scatchard analysis showed that LRR C2-Fc proteins specifically bound to HA-NL2-expressing COS7 cells in a high affinity. Scale bar, 10μm in I, and Scatchard analysis, n = 15 in J.
See also Figure S4–5
We further characterized the NL2-ST3 association, using recombinant tagged proteins expressed in HEK293T cells. We found that HA-tagged NL2 (HA-NL2) was co-IPed with Flag-tagged ST3 (Flag-ST3) from cells co-transfected with both constructs, but not from non-transfected cells or those transfected with either plasmid alone (Figure S4). In addition, in a separate co-IP assay, HA-NL2 was co-IPed by Flag-ST3, but not by Flag-GluA1 (Figure 4B). Furthermore, C-tail deletion of either NL2 or ST3 did not affect the NL2-ST3 association (Figure 4C), showing that the domain(s) mediating the NL2-ST3 interaction lie outside the C-termini of these two molecules.
Our co-IP data were corroborated by immunocytochemical evidence in HeLa cells illustrating co-localization of cell surface expressed HA-NL2 and Flag-ST3 (Figure 4D). Once again, neither C-termini was required for the co-localization of Flag-ST3 and HA-NL2 (Figure 4D). As a control, surface Flag-GluA1 failed to co-localize with either Myc-ST3 or HA-NL2 (Figure 4D). Thus, NL2 and ST3 interact with each other in a fashion independent of their C-termini.
An extracellular ST3 LRR domain mediates the interaction with NL2
We next tested the possibility that NL2 might interact with ST3 through extracellular domains. ST3 amino-terminus (N-terminus) contains two clusters of leucine-rich repeats (LRR) consisting of six LRR modules in each cluster (Figure 4E). In HEK293T cells, HA-tagged NL2 N-terminus (HA-NL2(NT)) without the signal peptide co-IPed with Flag-tagged LRR cluster 1 and 2 (Flag-LRR C1+2) or Flag-tagged LRR cluster 2 (Flag-LRR C2), but not with Flag-tagged LRR cluster 1 (Flag-LRR C1) (Figure 4E). Similarly, Flag-LRR C1+2 or Flag-LRR C2, but not Flag-LRR C1, co-localized with HA-NL2(NT) in HeLa cells (Figure 4F). Thus, the LRR C2 domain of ST3 mediates the interaction with NL2 N-terminus.
To investigate which LRR module(s) in ST3 LRR C2 mediates binding to NL2, we made a series of deletion mutants in the ST3 LRR C2 region. All mutants lacking the LRR9, but not those containing LRR9, completely lost the ability to co-localize with HA-NL2 (NT) (Figure S5A). Similarly, HA-NL2 was co-IPed with full-length Flag-ST3, or as a control, an ST3 mutant lacking LRR12 (Flag-ST3 ΔLRR12), but not with the ST3 mutant lacking LRR9 (Flag-ST3 ΔLRR9) (Figure 4G). Thus, the LRR9 domain of ST3 is required for binding to the NL2 N-terminus. This conclusion was further supported by co-localization of Flag-ST3 and Flag-ST3 ΔLRR12, but not Flag-ST3 ΔLRR9, with HA-NL2 at the cell surface (Figure 4H).
To measure the affinity of the NL2-ST3 interaction, we purified ST3 LRR C2 fused to the Fc domain of human immunoglobulin (LRR C2-Fc). We found that LRR C2-Fc bound to HA-NL2 expressed on the surface of COS7 cells (Figure 4I). We then applied increasing amounts of LRR C2-Fc to cells expressing HA-NL2 and measured the fluorescence intensity of LRR C2-Fc on the cell surface. Scatchard analysis revealed a saturable interaction between ST3 LRR C2 and NL2 in the nanomolar range, indicating a physiologically relevant NL2-ST3 interaction (Figure 4J).
NL2-ST3 interactions facilitate ST3 cell surface trafficking
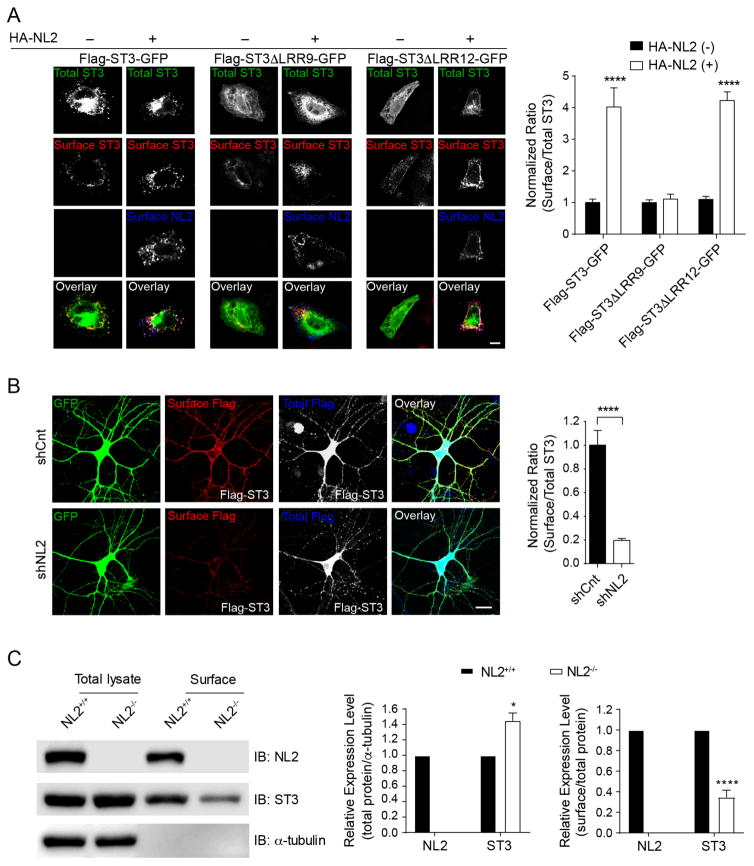
We noticed that surface levels of Flag-ST3 were higher when co-expressed with HA-NL2. To quantitate this, we expressed Flag-ST3-GFP in which Flag tag was fused to the N-terminus and GFP was fused to the C-terminus of ST3, either alone or in combination with HA-NL2 in HeLa cells, and measured the ratio of surface Flag to total ST3 expression reported by GFP fluorescence (Figure 5A). We found that co-expression of Flag-ST3-GFP with HA-NL2 strongly increased surface levels of Flag-ST3-GFP, but not HA-NL2 (Figure 5A and Figure S5B). The enhancement of Flag-ST3-GFP surface levels by HA-NL2 depended on the ST3 LRR9 domain, as NL2 did not promote surface trafficking of Flag-ST3 ΔLRR9-GFP (Figure 5A). In contrast, deletion of the ST3 LRR12 domain did not affect NL2-induced increase of surface expression (Figure 5A). In cultured hippocampal neurons surface levels of Flag-ST3 ΔLRR9 were also significantly reduced compared to surface Flag levels in the neurons transfected with either Flag-ST3 or Flag-ST3 ΔLRR12 (Figure S5C). Taken together, these data indicate that NL2 interaction with ST3 facilitates ST3 expression at the cell surface.
Figure 5. NL2 interacts with ST3 to facilitate ST3 targeting to the plasma membrane.
(A) Immunostaining (left) and summary graphs (right) showed that NL2 promoted ST3 (left, n = 12) and ST3ΔLRR12 (right, n = 12), but not ST3ΔLRR9 (middle, n = 13), trafficking to the plasma membrane in HeLa cells. Surface Flag or HA was immunolabeled under non-permeabilized condition, and total ST3 expression was indicated by GFP fluorescence. p < 0.0001, One-way ANOVA test. Scale bar, 10μm.
(B) Knockdown of NL2 significantly reduced ST3 targeting to neuronal plasma membrane. Cultured neurons expressing a control shRNA (n = 14) or NL2 shRNA (n = 16) together with Flag-ST3, respectively. p < 0.0001, t-test. Scale bar, 10μm.
(C) Surface protein biotinylation assay in hippocampal slices prepared from NL2−/− showed that surface expression of ST3 was reduced (t-test, *p < 0.05, **** p < 0.0001, n =3).
See also Figure S5
We reasoned that if the NL2-ST3 interaction facilitates ST3 surface targeting, then NL2 knockdown should impair ST3 surface expression. Indeed, while surface Flag-ST3 was readily detectable in hippocampal cultures co-transfected with a GFP-expressing empty shRNA vector, knockdown of NL2 strongly diminished surface Flag-ST3 (Figure 5B), demonstrating that endogenous NL2 is important for surface accumulation of ST3 in neurons. To further test the role of NL2 in the regulation of ST3 surface expression, we performed a cell-surface biotinylation assay in hippocampal slices prepared from NL2−/−. We found that ST3 expression at the cell surface in hippocampal slices was significantly reduced in NL2−/−, confirming that NL2 is important for surface accumulation of endogenous ST3 (Figure 5C).
The NL2-ST3 interaction promotes GABAergic presynaptic differentiation
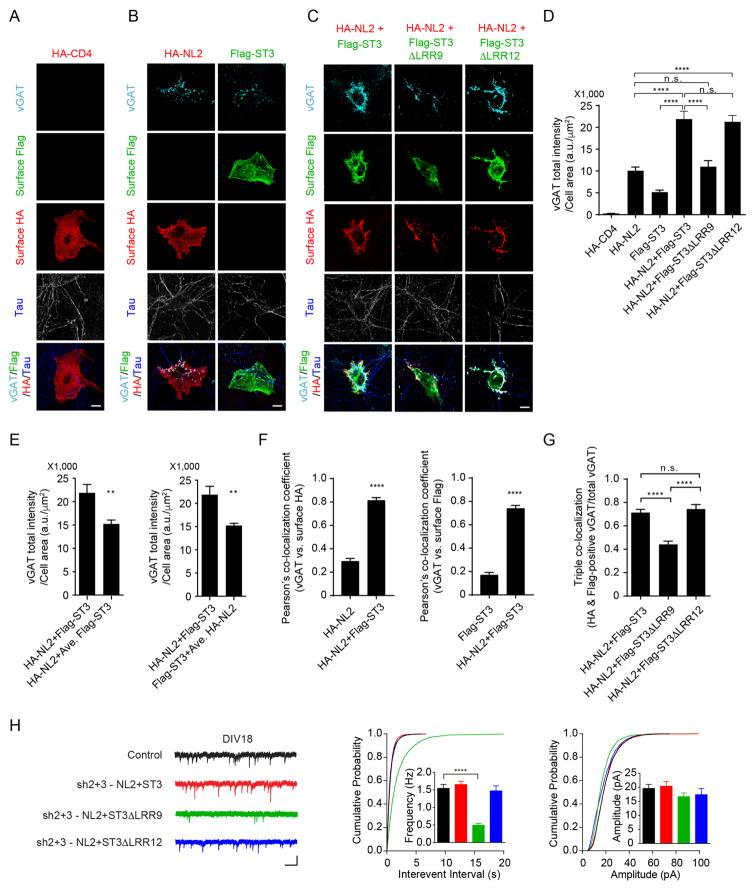
Both NL2 and ST3 are synaptogenic molecules capable of inducing inhibitory presynaptic differentiation (Graf et al., 2004; Scheiffele et al., 2000; Takahashi et al., 2012; Yim et al., 2013). To test whether the NL2-ST3 interaction participates in their synaptogenic properties, we employed a widely-used neuron-fibroblast co-culture assay (Scheiffele et al., 2000). Specifically, we quantified expression of the GABAergic presynaptic marker vGAT on axons contacting COS7 cells expressing HA-NL2, Flag-ST3 or both HA-NL2 and Flag-ST3. In agreement with previous reports (Graf et al., 2004; Scheiffele et al., 2000; Takahashi et al., 2012; Yim et al., 2013), expression of NL2 or ST3 alone both induced robust clustering of vGAT in co-cultures compared to the membrane protein HA-tagged CD4 (HA-CD4) used as a negative control (Figure 6A, B, D), confirming the synaptogenic activities of both proteins. Strikingly, co-expression of HA-NL2 and Flag-ST3 yielded a supra-additive effect recruiting significantly more vGAT input onto COS7 cells than the arithmetic sum of either construct alone (Figure 6B–E). Significantly, this enhanced synaptogenic potential was not observed when HA-NL2 was co-transfected with the Flag-ST3 mutant, Flag-ST3 ΔLRR9 that did not bind to NL2 (Figure 6C–D), revealing the importance of the NL2-ST3 interaction for enhanced synaptogenic ability of cells co-transfected with HA-NL2 and Flag-ST3. In contrast, co-expression of HA-NL2 and Flag-ST3 ΔLRR12 promoted presynaptic differentiation to a similar degree as HA-NL2 and WT Flag-ST3 co-transfection (Figure 6C–D). Despite the difference in cooperativity with NL2, both Flag-ST3 ΔLRR9 and Flag-ST3 ΔLRR12 on their own induced clustering of vGAT as efficiently as Flag-ST3 (Figure S6A). In addition, similar to WT ST3, ST3 ΔLRR9 mutant could rescue mIPSC deficits in DIV 8 neurons transfected with the double knockdown vector at DIV2, suggesting that ST3 ΔLRR9 can drive synaptic development when it is expressed in early developing neurons (Figure S6B). We also noticed that in co-cultures, surface HA-NL2 and Flag-ST3 in co-transfected cells were co-localized with vGAT clusters, in contrast to singly transfected cells where vGAT immunolabelling was more diffuse and less co-localized with surface HA-NL2 or Flag-ST3 (Figure 6B, C and F). In addition, disruption of the NL2-ST3 interaction decreased the triple co-localization of HA-NL2, Flag-ST3 and vGAT (Figure 6C and G). These data suggest an ability of the NL2-ST3 complexes in aligning NL2 and ST3 with GABAergic presynaptic release sites. Of note, the NL2-ST3 interaction did not significantly change the binding affinity of NL2 or ST3 to their presynaptic partners, neurexin 1β and protein tyrosine phosphatase δ (PTPδ), respectively (Figure S7). Taken together, these data indicate that the NL2 interaction with ST3 confers a synergistic synaptogenic effect on GABAergic presynaptic differentiation.
Figure 6. The NL2-ST3 interaction promotes GABAergic presynaptic differentiation.
(A) COS7 cells expressing HA-CD4 (n = 11) did not induce clustering of vGAT along contacting axons (indicated by Tau staining in blue) in hippocampal neuron-COS7 cell co-cultures. Scale bar, 10μm.
(B) COS7 cells expressing HA-NL2 (n = 18) or Flag-ST3 (n = 14) induced clustering of vGAT along contacting axons in co-cultures. Scale bar, 10μm.
(C) COS7 cells expressing HA-NL2 together with Flag-ST3 (n = 18) or with Flag-ST3ΔLRR12 (n = 10), but not with Flag-ST3ΔLRR9 (n = 11), induced significantly more clustering of vGAT along contacting axons in co-cultures. Scale bar, 10μm.
(D) Bar graph showed the total integrated intensity of vGAT on COS7 cells expressing the indicated plasmids in (A–C) (One-way ANOVA, p < 0.0001).
(E) Bar graphs showed that co-expression of HA-NL2 and Flag-ST3 recruited significantly more vGAT on COS7 cells than individually transfected cells combined (t-test, p < 0.01). Left, comparison of vGAT on cells co-transfected with HA-NL2 and Flag-ST3 (HA-NL2+Flag-ST3) vs. vGAT on cells transfected with HA-NL2 plus average vGAT intensity on cells transfected with Flag-ST3 (HA-NL2+Ave. Flag-ST3). Right, comparison of vGAT on cells co-transfected with HA-NL2 and Flag-ST3 (HA-NL2+Flag-ST3) vs. vGAT on cells transfected with Flag-ST3 plus average vGAT intensity on cells transfected with HA-NL2 (Flag-ST3+Ave. HA-NL2) (see experimental procedure for details).
(F) Bar graphs showed Pearson’s correlation coefficients for the co-localization analysis of vGAT puncta vs surface HA-NL2 (left) or surface Flag-ST3 (right) in the indicated groups in B and C (t-test, p < 0.0001).
(G) Triple co-localization analysis showed a reduced co-localization of surface HA, surface Flag and vGAT in cells co-transfected with HA-NL2 and Flag-ST3ΔLRR9 as shown in C (One-way ANOVA, p < 0.0001).
(H) mIPSC recordings showed that NL2 together with ST3 or with ST3ΔLRR12, but not with ST3ΔLRR9, rescued the mIPSC deficits in double knockdown neurons (Control, n = 13; sh2+3-NL2+ST3, n = 10; sh2+3-NL2+ST3ΔLRR9, n = 11, sh2+3-NL2+ST3ΔLRR12, n = 10; one-way ANOVA, p < 0.0001; K-S test, p < 0.0001 between Control and sh2+3-NL2+ ST3ΔLRR9). Scale bar, 20 pA and 1 s.
See also Figure S5–7
The NL2-ST3 interaction is required for GABAergic synapse development
To examine whether the NL2-ST3 interaction is critical for normal GABAergic synapse development, we first performed a molecular replacement experiment in hippocampal cultures. As shown before, shRNA-mediated double knockdown of NL2 and ST3 essentially eliminated mIPSCs in DIV18 neurons and the mIPSC deficits could be rescued by co-expressing shRNA-resistant NL2 and ST3 (Figures 3A and 6H). Importantly, co-expression of NL2 and ST3 ΔLRR9 with the double knockdown construct did not fully rescue mIPSC deficits at DIV18 (Figure 6H). In contrast, co-expression of NL2 and ST3 ΔLRR12 fully restored mIPSC deficits in double knockdown neurons (Figure 6H). These data indicate that the NL2-ST3 interaction is critical for establishment of GABAergic transmission in neuronal cultures.
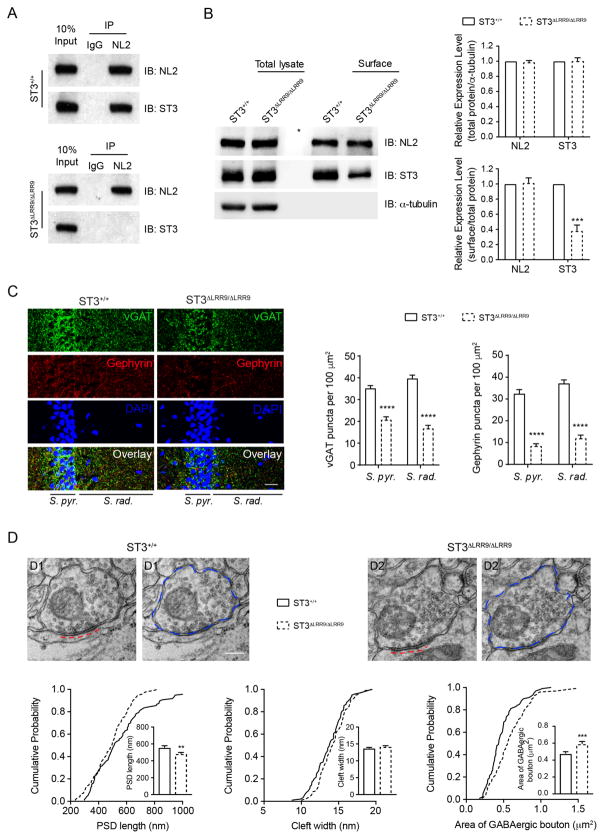
To further corroborate the role of the NL2-ST3 interaction in GABAergic synapse development, we generated a mutant mouse line, ST3ΔLRR9/ΔLRR9, in which the sequence coding for the ST3 LRR9 domain was precisely deleted by CRISPR/Cas9-mediated homologous recombination in germline (Figure S8A–C). Western blot analysis of hippocampal lysates showed that genetic deletion of the ST3 LRR9 domain in ST3ΔLRR9/ΔLRR9 did not alter the overall expression levels of ST3 (Figure 7B and Figure S8D). Importantly, while ST3 could be co-IPed with NL2 in hippocampal lysates prepared from ST3+/+, ST3 did not associate with NL2 in ST3ΔLRR9/ΔLRR9 (Figure 7A), confirming that the LRR9 domain is required for the NL2-ST3 interaction in vivo. Furthermore, a cell-surface biotinylation assay in ST3ΔLRR9/ΔLRR9 hippocampal slices demonstrated decreased surface expression of ST3, indicating that disruption of the NL2-ST3 interaction reduces surface expression of ST3 (Figure 7B).
Figure 7. GABAergic synapse development is impaired in ST3ΔLRR9/ΔLRR9 mice.
(A) ST3 did not associate with NL2 in ST3ΔLRR9/ΔLRR9 mice. co-IP assays showed that NL2 could pull down ST3 from WT (top), but not ST3ΔLRR9/ΔLRR9 (bottom), hippocampal lysates.
(B) Surface protein biotinylation assay in hippocampal slices prepared from ST3ΔLRR9/ΔLRR9 showed a significant reduction of surface expression of ST3 in ST3ΔLRR9/ΔLRR9 (t-test, p < 0.0001, n = 3).
(C) lmmunohistochemical images showed that the densities of vGAT (green) and gephyrin (red) puncta were significantly reduced in the S. pyr. and S. rad. area in the hippocampal CA1 region in ST3ΔLRR9/ΔLRR9 mice (n = 15 images from three ST3+/+ or three ST3ΔLRR9/ΔLRR9 mice; t-test, p < 0.05*, p < 0.01**, p < 0.001***). Scale bar, 20 μm.
(D) EM images showed that inhibitory PSD length, but not synaptic cleft width, was decreased, and that the GABAergic presynaptic bouton area was increased in ST3ΔLRR9/ΔLRR9 mice. Panels on the top show representative images (red dash lines indicated inhibitory PSD and blue dash circles indicated GABAergic boutons). Scale bar, 0.2 μm. Cumulative distributions and bar graphs on the bottom show quantitative analysis (PSD length: WT, n = 97; ST3ΔLRR9/ΔLRR9, n = 95, p < 0.01; cleft width: WT, n = 97, p > 0.05; ST3ΔLRR9/ΔLRR9, n = 95; GABAergic bouton area: WT, n = 100; ST3ΔLRR9/ΔLRR9, n = 98, p < 0.001; t-test; K-S test, p < 0.05. Samples were collected from two mice per genotype and there were no significant differences between mice within each genotype).
See also Figure S8
Immunohistochemistry of brain sections from ST3ΔLRR9/ΔLRR9 revealed marked reductions in the density of gephyrin puncta in both stratum pyramidale (S. pyr.) and stratum radiatum (S. rad.) compared to WT mice (Figure 7C), suggesting that postsynaptic assembly of GABAergic synapses in CA1 pyramidal neurons is impaired in ST3ΔLRR9/ΔLRR9. Significant reductions of vGAT puncta were also observed at S. pyr. and S. rad. regions (Figure 7C). These data indicate that disruption of the NL2-ST3 interaction reduces inhibitory synapse density. To provide insight into the ultrastructural properties of GABAergic synapses in ST3ΔLRR9/ΔLRR9, we examined symmetric perisomatic synapses onto CA1 pyramidal cells by transmission electron microscopy (EM). We found that the length of the postsynaptic density (PSD), but not the synaptic cleft width, was significantly reduced in ST3ΔLRR9/ΔLRR9 (Figure 7D). Furthermore, the bouton size of inhibitory presynaptic terminals was significantly increased in ST3ΔLRR9/ΔLRR9 (Figure 7D). Together, these data reveal reduced GABAergic synapse density and ultrastructural abnormalities of hipppcampal perisomatic GABAergic synapses in ST3ΔLRR9/ΔLRR9 and demonstrate that the NL2-ST3 interaction is crucial for GABAergic synapse development in vivo.
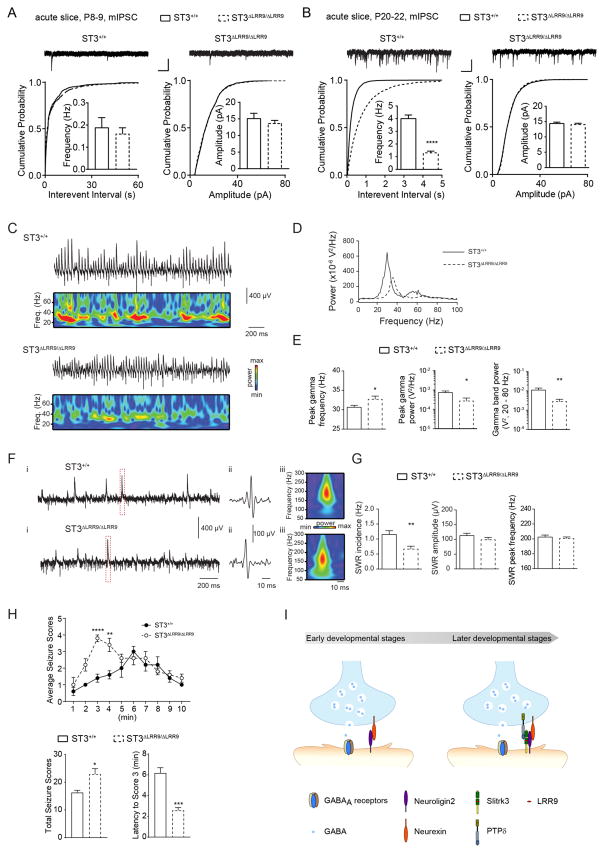
To probe for functional GABAergic transmission deficits in ST3ΔLRR9/ΔLRR9, we compared mIPSCs recorded in CA1 pyramidal neurons in acute hippocampal slices of ST3ΔLRR9/ΔLRR9 and their WT littermates. In developing neurons from P8–9 mice, we found that mIPSC frequency was slightly smaller in ST3ΔLRR9/ΔLRR9 (Figure 8A). However, at later stages of development, P20–22, ST3ΔLRR9/ΔLRR9 exhibited a strong reduction of mIPSC frequency, but not amplitude compared with WT controls (Figure 8B), demonstrating a significant deficit in GABAergic transmission of young adult ST3ΔLRR9/ΔLRR9.
Figure 8. Disruption of the NL2-ST3 interaction in vivo reduces inhibitory transmission, impairs hippocampal network activity and increases seizure susceptibility.
(A) mIPSC recordings in P8–9 slices did not show a significant reduction of GABAergic transmission in ST3ΔLRR9/ΔLRR9 (ST3+/+, n = 9; ST3ΔLRR9/ΔLRR9, n = 12; t-test, p > 0.05 for both; but K-S test used for cumulative distributions showed a significant difference for interevent interval, p < 0.05). Scale bar, 20 pA and 1 s.
(B) mIPSC recordings in P20–22 slices demonstrated a strong reduction of GABAergic transmission in ST3ΔLRR9/ΔLRR9 (ST3+/+, n = 13; ST3ΔLRR9/ΔLRR9, n = 17; t-test, p < 0.0001 for frequency; K-S test, p < 0.0001 for interevent interval). Scale bar, 20 pA and 1 s.
(C) Representative recordings illustrating gamma oscillations detected in field recordings from CA3 S. pyr. in slices from ST3+/+ (upper) or ST3ΔLRR9/ΔLRR9 (lower) after bath application of 20 μM carbachol (CCh), with corresponding Wavelet transforms shown below each trace.
(D) Power density spectra of representative recordings of CCh-evoked gamma oscillations in slices from ST3+/+ or ST3ΔLRR9/ΔLRR9 (generated from the same recordings illustrated in C but for 300 s of data).
(E) Group data plots illustrating differences in the peak gamma frequency (p < 0.05, t test), peak gamma power (p < 0.05, Mann-Whitney test) and gamma band (20–80 Hz) power (p < 0.01, Mann-Whitney test) between ST3+/+ and ST3ΔLRR9/ΔLRR9 (n = 13 slices from 5 mice in both groups).
(F) Representative unfiltered traces from ST3+/+ and ST3ΔLRR9/ΔLRR9 mice showing spontaneous SWRs (i), with the hatched area shown in (ii) filtered between 100 and 250 Hz and the corresponding wavelet transform (iii).
(G) Group data plots show that ST3ΔLRR9/ΔLRR9 mice display reduced incidence of spontaneous SWRs (ST3+/+ (n = 13 slices from 5 mice) vs ST3ΔLRR9/ΔLRR9 (n = 16 slices from 5 mice): p < 0.01, t test) with no significant differences in SWR amplitude (p = 0.22, t-test) or SWR peak frequency (p=0.57, t-test).
(H) Time course of average seizure scores induced by PTZ injection (50mg per kg) (n= 5 male mice per genotype). Behavioral responses were scored every 1 min for 10 min after the PTZ injection. Bar graphs showed total seizure scores (p < 0.05, t-test) and latency to Score 3 (p < 0.001, t-test).
(I) A model of the role of the NL2 and ST3 in GABAergic synapse development. In immature hippocampal neurons, NL2 is critical for GABAergic synapse development. In more mature neurons, both NL2 and ST3 are required for GABAergic synapse development. Importantly, at the later developmental stage, NL2 interacts with ST3 to facilitate ST3 trafficking to the neuronal surface and to synergistically promote GABAergic synapse differentiation.
Abnormal network activities and seizures in ST3ΔLRR9/ΔLRR9 mice
Hippocampal information coding requires precision in the timing, extent, and synchrony of activity within glutamatergic principal cell assemblies that is largely orchestrated by local circuits mediated GABAergic inhibition. To examine whether the reduction of GABAergic transmission in ST3ΔLRR9/ΔLRR9 promotes hippocampal network deficits, we probed carbachol (CCh) induced gamma oscillations, a network rhythm that critically relies upon entrainment by phasic inhibitory drive (Buzsaki and Wang, 2012; Fisahn et al., 1998; Mann et al., 2005). Field potential recordings in the CA3 principal cell layer of hippocampal slices revealed that gamma power was significantly impaired and that gamma frequency was modestly increased in ST3ΔLRR9/ΔLRR9 (Figure 8C–E). Moreover, during baseline recording periods prior to the induction of gamma oscillations ST3ΔLRR9/ΔLRR9 displayed a deficit in the incidence of sharp wave ripples (SWRs, Figure 8F–G). SWRs are spontaneous high frequency oscillatory events reflecting near synchronous firing of principal cell ensembles coordinated by perisomatic inhibitory input (Csicsvari et al., 1999; Ellender et al., 2010). Thus, reduced inhibitory drive in ST3ΔLRR9/ΔLRR9 precipitates deficits in the entrainment of network activities that are relevant to cognition and neuropsychiatric disorders.
A reduction of GABAergic synapses in ST3ΔLRR9/ΔLRR9 may also increase seizure susceptibility. Intraperitoneal (i.p.) administration of the convulsant pentylenetetrazole (PTZ), a GABAA receptor antagonist, can induce seizures in mice that model epilepsy. We found that PTZ administration (50 mg per kg of body weight) induced significantly more severe seizures in ST3ΔLRR9/ΔLRR9 than in WT littermates (Figure 8H). Specifically, the total seizure score for 1–10 min after PTZ administration was significantly higher, and the latency of seizure onset after PTZ administration was strongly reduced in ST3ΔLRR9/ΔLRR9 (Figure 8H). Thus, impaired GABAergic synapse development following loss of the NL2-ST3 interaction increases seizure susceptibility.
Discussion
Elucidating molecular mechanisms underlying GABAergic synapse development is essential for understanding circuit wiring and function as well as cognitive disorders. Our systematic analyses of NL2 and ST3 reveal an essential developmental framework for hippocampal GABAergic synapses (Figure 8I). These mechanistic insights into GABAergic synapse development demonstrate that although GABAergic synapses are diverse, NL2 and ST3 are required for the vast majority, if not all, GABAergic synapse development in hippocampal neurons.
NL2 and GABAergic synapse development
The synaptogenic properties of NL2 have been well established (Craig and Kang, 2007; Huang and Scheiffele, 2008; Krueger-Burg et al., 2017; Sudhof, 2008); however, its exact function in inhibitory synaptogenesis remains controversial (Chanda et al., 2017; Chih et al., 2005; Chubykin et al., 2007; Panzanelli et al., 2017; Poulopoulos et al., 2009; Varoqueaux et al., 2006; Zhang et al., 2015). Early investigations showed that NL2 is highly enriched at GABAergic synapses at both neuronal somatic and dendritic regions and in vitro co-culture assay has shown that NL2 is a potent synaptogenic CAM (Graf et al., 2004; Takahashi et al., 2012; Varoqueaux et al., 2004). Similarly, over-expression of NL2 promotes GABAergic transmission and increases inhibitory synapse density (Chih et al., 2005; Chubykin et al., 2007). In addition, NL2, through the interaction with GARLHs, is important for synaptic localization of GABAARs, indicating a role of NL2 in functional maturation of GABAergic synapses (Yamasaki et al., 2017). However, loss-of-function studies have yielded different, often contradictory, conclusions. For example, shRNA-based knockdown of NL2 in mature neuronal cultures leads to a substantial loss of GABAergic synapses (Chih et al., 2005), while germline deletion of multiple NLs, including NL2, has no effect on inhibitory synapse density in multiple brain regions despite reducing inhibitory transmission (Chubykin et al., 2007; Varoqueaux et al., 2006). A recent elegant study using conditional KO for NL2 in cerebellar Purkinje cells failed to observe significant changes of GAD65, a presynaptic marker of inhibitory synapses (Zhang et al., 2015). Nonetheless, there was a strong reduction of inhibitory transmission (Zhang et al., 2015), indicating that NL2 is important for functional maturation, but not formation, of inhibitory synapses in Purkinje cells. Similarly, global conditional deletion of NL1, 2 and 3 in hippocampal cultures does not change vGAT density, but GABAergic transmission is reduced (Chanda et al., 2017). In contrast, germline deletion of NL2 strongly reduces IPSCs in hippocampal CA1 neurons, and there is a substantial reduction of gephyrin, but not vGAT, in the CA1 region (Panzanelli et al., 2017; Poulopoulos et al., 2009), suggesting that NL2 is important for postsynaptic assembly of inhibitory synapses in hippocampus. We thus systematically analyzed NL2 regulation of GABAergic synapse development.
In NL2−/−, ST3, another synaptogenic molecule for inhibitory synapses (Takahashi et al., 2012; Yim et al., 2013), is up-regulated. Thus, compensatory up-regulation of ST3 in NL2−/− may mask the importance of NL2 in GABAergic synaptogenesis. Indeed, single-cell shRNA-based knockdown both in vitro and in vivo demonstrate that NL2 is essential for the establishment of GABAergic transmission. For example, NL2 knockdown in single developing neurons lead to an over 90% reduction of GABAergic transmission and a strong reduction of GABAergic synapse density. Similarly, NL2 knockdown in more mature neurons strongly decreases GABAergic synapses. Importantly, the effect of NL2 shRNA knockdown on GABAergic synapses is fully rescued by shRNA-resistant NL2 and NL2 shRNA expression in NL2−/− neurons does not change GABAergic transmission. These data also highlight the importance of using single-cell approaches to study the role of NL2 in synapse development. Single-cell genetic manipulation may avoid compensatory changes in network activities that may accompany broad genetic deletions. In addition, it has been reported that mosaic deletion of NL1 in cortical neurons allows transcellular synaptic competition that favors synapse development in WT neurons over neurons lacking NL1 (Kwon et al., 2012), but see (Chanda et al., 2017). Similar synaptic competition may explain the different phenotypes of NL2−/− (Poulopoulos et al., 2009) and NL2 knockdown cells.
We found that the NL2 C-tail is important for the regulation of GABAergic synapse development. Specifically, we identified both the GBD and PRR domains in the NL2 C-tail as critical for NL2’s regulation of GABAergic synapse development in immature neurons. The GBD domain interacts with gephyrin, a major inhibitory postsynaptic density protein (Tretter et al., 2012; Tyagarajan and Fritschy, 2014), and the PRR domain binds collybistin, an inhibitory synapse specific GEF (Kins et al., 2000; Soykan et al., 2014). These findings corroborate previous evidence that NL2 drives GABAergic synapse formation through gephyrin and collybistin (Poulopoulos et al., 2009). It is worth noting that in mature hippocampal neurons, the GBD and PRR domains are dispensable for NL2 function at inhibitory synapses (Nguyen et al., 2016). In addition, an NL2 mutant in which NL2 C-tail was replaced by the same domain from NL1 or NL3 functions similarly as WT NL2 at inhibitory synapses in mature hippocampal neurons (Futai et al., 2013; Nguyen et al., 2016). In contrast, a similar NL2 mutant failed to rescue mIPSC deficits in NL2 and ST3 double knockdown immature neurons (Figure 2). It is unclear what accounts for these discrepancies, although our experiments were performed in developing, immature neurons, while the previous studies examined mature neurons (Futai et al., 2013; Nguyen et al., 2016). It is also possible that overexpression of the swap mutant in WT neurons might dimerize with endogenous NLs (Poulopoulos et al., 2012), thus failing to reveal the function of NL2 C-tail. Furthermore, the functional interaction between ST3 and NL2 discovered here might have masked the role of NL2 C-tail in previous studies (Futai et al., 2013; Nguyen et al., 2016), as NL2 C-tail mutants may still promote endogenous ST3 trafficking to the cell surface in more mature neurons to regulate inhibitory transmission.
Although our data demonstrate that NL2 is important for GABAergic synapse formation in hippocampal neurons, recent studies have indicated that NL2 may play a brain region-specific role. In brainstem neurons, inhibitory transmission, but not synaptic density, is dramatically reduced, in NL2−/− (Poulopoulos et al., 2009; Varoqueaux et al., 2006), suggesting that in these neurons NL2 regulates functional maturation, but not initial formation, of inhibitory synapses. However, it remains unclear whether other inhibitory synaptogenic CAMs are up-regulated in NL2 KO brainstem neurons as shown here in hippocampus. A recent study using conditional KO of NL2 in cerebellar Purkinje cells showed that inhibitory transmission is strongly reduced, but not GAD65 staining, indicating normal inhibitory presynaptic assembly (Zhang et al., 2015). However, it is unclear whether postsynaptic assembly of inhibitory synapses is altered or not in conditional KO of NL2 in Purkinje cells. Global conditional deletion of NL1–3 in hippocampal neuronal cultures did not decrease vGAT staining (Chanda et al., 2017), but it remains to be determined whether single-cell conditional deletion of NL1–3 or NL2 on its own would change GABAergic synapse numbers in developing hippocampal neurons. In NL2 germline KOs, GABAergic transmission and postsynaptic gephyrin puncta are both reduced in hippocampal CA1 neurons (Panzanelli et al., 2017; Poulopoulos et al., 2009). Thus, further investigation into the role of NL2 in postsynaptic assembly of inhibitory synapses in different types of neurons in the brain will clarify the general function of NL2 in inhibitory synapse development.
Based on our data we propose three general conclusions about NL2. First, the function of NL2 in GABAergic synaptogenesis could partially be substituted by other synaptogenic CAMs, such as ST3. Second, NL2 is critical for GABAergic synapse development in hippocampal neurons. Third, the importance of NL2 in GABAergic synapse development depends on two domains in its C-tail that cannot be replaced by the C-tail from NL1, suggesting the importance of NL2 C-tail-mediated interactions and/or signaling in GABAergic synapse development. In addition, we recently reported that NMDA receptors play a critical role in GABAergic synaptogenesis in developing neurons (Gu et al., 2016). Thus, it will be important to determine the functional interaction of NMDA receptors and NL2 in the regulation of GABAergic synapse development.
ST3 and GABAergic synapse development
Though the GABAergic synaptogenic properties of ST3 were previously reported (Takahashi et al., 2012; Yim et al., 2013), our findings reveal a previously unappreciated temporal-specificity to the function of ST3 in regulating GABAergic synaptogenesis. In early developing neurons when GABAergic synapses are initially formed, the expression levels of ST3 are relatively low. Accordingly, knockdown of ST3 in developing neurons has little effect on GABAergic transmission and inhibitory synapse density. Interestingly, at this developmental stage, the deficits of GABAergic synapses in NL2 and ST3 double knockdown neurons can be rescued by ST3. Thus, although ST3 does not participate in GABAergic synaptogenesis in early immature neurons, it is sufficient to do so. Indeed, in NL2−/−, ST3 is up-regulated and thus might partially drive GABAergic synaptogenesis in developing neurons lacking NL2. The expression of ST3 is strongly increased during development and reaches the highest levels at ~3 weeks postnatally in mouse hippocampus. Knockdown of ST3 at this later developmental stage causes a significant reduction of GABAergic transmission and inhibitory synapse density, similar to previous reports (Takahashi et al., 2012; Yim et al., 2013). Thus, based on our data, ST3 is important for GABAergic synapse development in more mature neurons, but not in early developing neurons. Interestingly, the ST3 C-tail is critical for the establishment of GABAergic transmission during development, as the ST3 mutant lacking the C-tail or swap mutants fail to rescue mIPSC deficits in double knockdown neurons. Future studies focused on identifying protein-protein interactions and signaling mediated by the ST3 C-tail will aid our understanding of the molecular mechanisms governing GABAergic synapse development.
At the later developmental stage, mIPSC deficits in the double knockdown neurons cannot be fully rescued by either NL2 or ST3, but by co-expression of both NL2 and ST3. Thus, expression of both NL2 and ST3 in neurons is crucial for GABAergic synapse development at this stage. Knockdown of both NL2 and ST3 virtually eliminates GABAergic synaptic transmission, showing that NL2 and ST3 together account for the vast majority, if not all, GABAergic synapse development in hippocampal neurons. Thus, despite the remarkable diversity of GABAergic synapses in hippocampus, a common requirement of NL2 at the early developmental stages and of both NL2 and ST3 at the later developmental stages indicates a general framework for GABAergic synaptogenesis.
The NL2-ST3 interaction is essential for GABAergic synapse development
Many transmembrane synaptic CAMs, such as NLs, LRRTMs, neurexins, Slitrks, PTPs, and cadherins, typically use characteristic extracellular domains to mediate trans-synaptic interactions to organize and regulate synapse development and function (de Wit and Ghosh, 2016; Lu et al., 2016; Missler et al., 2012). We now report that at hippocampal GABAergic synapses, two major postsynaptic CAMs, NL2 and ST3, interact with each other in cis to control GABAergic synapse development. This interaction exhibits nanomolar affinity and is mediated by the NL2 extracellular domain with an ~20-amino acid motif, LRR9, in the ST3 extracellular domain. Functionally, the NL2-ST3 interaction is essential for hippocampal GABAergic synapse development. Indeed, disruption of the NL2-ST3 interaction strongly decreases GABAergic synaptic transmission both in vitro and in vivo. In addition, density and ultrastructural properties of GABAergic synapses are affected in hippocampal CA1 region in ST3ΔLRR9/ΔLRR9. Furthermore, hippocampal network activity is impaired and seizure susceptibility is increased in ST3ΔLRR9/ΔLRR9. These data demonstrate a critical role for the NL2-ST3 interaction in GABAergic synapse development and neural network function, and highlight a requirement of both NL2 and ST3 in the establishment of GABAergic synaptic transmission in the later developmental stages.
The in cis interaction between NL2 and ST3 does not appear to interfere with the binding of NL2 or ST3 to their presynaptic partners (Figure S7). In addition, compared to WT ST3, the ST3 mutant that does not bind to NL2 has similar synaptogenic activity (Figures S6). We also found that the NL2-ST3 interaction is important for ST3 cell surface expression, suggesting a mechanism underlying the role of the NL2-ST3 interaction in GABAergic synapse development. Although the NL2-ST3 interaction is important for ST3 expression at the cell surface, ST3 by its own can traffic, although in a less efficient manner. Furthermore, co-expression of both NL2 and ST3 in heterologous cells has a synergistic effect on inducing GABAergic presynaptic differentiation, indicating that the NL2-ST3 complex has greater synaptogenic capacity. Although the mechanism remains unclear, it is possible that the NL2-ST3 complex simultaneously binds to multiple presynaptic and postsynaptic molecules important for GABAergic synaptogenesis and facilitates the formation of a signaling complex important for GABAergic synapse development.
Conclusions and perspectives
GABAergic synapses exhibit an extraordinary diversity in terms of molecular, anatomic and physiological properties (Cherubini and Conti, 2001; Klausberger and Somogyi, 2008; Kubota et al., 2016; Sassoe-Pognetto et al., 2011). In hippocampus, principal neurons are innervated by over twenty distinct types of interneurons (Klausberger and Somogyi, 2008). While domain-specific molecular mechanisms exist in generating diverse inhibitory synapses (Ango et al., 2004; Ashrafi et al., 2014; Bloodgood et al., 2013; Cherubini and Conti, 2001; Sassoe-Pognetto et al., 2011), our findings reveal a general framework for inhibitory synaptogenesis common to hippocampal GABAergic synapses. Thus, GABAergic synapse formation is likely governed by a hierarchical but also intertwined process in which the NL2 and ST3-dependent mechanisms are essential for GABAergic synaptogenesis on one hand, and input-specific mechanisms shape highly precise synaptic contacts in a location-specific manner on the other hand. A deep understanding of GABAergic synaptogenesis requires investigations into both the general and input-specific mechanisms at the molecular level as well as how these two processes functionally interact with each other during development to sculpt inhibitory connectivity.
Recent studies have suggested a brain-region specific role of NL2 in inhibitory synapse development (Liang et al., 2015; Poulopoulos et al., 2009; Varoqueaux et al., 2006; Zhang et al., 2015). For ST3, while it is important for GABAergic synapse development in hippocampus (Takahashi et al., 2012; Yim et al., 2013), its role in other brain regions remains unknown. In addition, ST3 has restricted expression patterns in the brain with high expression levels in hippocampus and cortex, but very low expression in cerebellum and brainstem (Yim et al., 2013). Thus, NL2 and/or ST3-independent mechanisms for GABAergic synapse development may exist in different types of neurons in the brain. Future work toward a complete understanding of GABAergic synapse development in neurons across brain regions will be important to our understanding of how neural circuits in specific regions of the brain are assembled during development for distinct brain functions.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Slitrk3 | Sigma-Aldrich | Cat#: ABN356, RRID: AB_2637043 |
| Rabbit polyclonal anti-Slitrk3 | Proteintech | Cat#: 21649-1-AP, RRID: AB_10859788 |
| Rabbit polyclonal anti-Slitrk3 (K-20) | Santa Cruz | Cat#: sc-100160, RRID: AB_2190328 |
| Rabbit polyclonal anti-HA (Y-11) | Santa Cruz | Cat#: sc-805, RRID: AB_631618 |
| Rabbit polyclonal anti-vGluT1 | Synaptic Systems | Cat#: 135302, RRID: AB_887877 |
| Rabbit polyclonal anti-vGAT | Synaptic Systems | Cat#: 131002, RRID: AB_887871 |
| Rabbit polyclonal anti-Neuroligin 3 | Synaptic Systems | Cat#: 129113, RRID: AB_2619816 |
| Rabbit polyclonal anti-Flag | Sigma-Aldrich | Cat#: F7425, RRID: AB_439687 |
| Rabbit monoclonal anti-Myc-Tag (71D10) | Cell Signaling Technology | Cat#: 2278, RRID: AB_490778 |
| Mouse normal IgG | Sigma-Aldrich | Cat#: 15381, RRID: AB_1163670 |
| Mouse monoclonal anti-Flag M2, Clone M2 | Sigma-Aldrich | Cat#: F3165, RRID: AB_259529 |
| Mouse monoclonal α-tubulin | Sigma-Aldrich | Cat#: T8203, RRID: AB_1841230 |
| Mouse monoclonal anti-PSD95 | Millipore | Cat#: MAB1598, RRID: AB_94278 |
| Mouse monoclonal anti-Neuroligin 1 | Synaptic Systems | Cat#: 129111, RRID: AB_887747 |
| Mouse monoclonal anti-Neuroligin 2 | Synaptic Systems | Cat#: 129511, RRID: AB_2619813 |
| Mouse monoclonal anti-Gephyrin | Synaptic Systems | Cat#: 147021, RRID: AB_2232546 |
| Mouse monoclonal anti-Gephyrin, Clone# 807423 | R&D Systems | Cat#: MAB7519, RRID: AB_2637045 |
| Mouse monoclonal anti-HA (F-7) | Santa Cruz | Cat#: sc-7392, RRID: AB_627809 |
| Guinea pig polyclonal anti-vGAT | Synaptic Systems | Cat#: 131004, RRID: AB_887873 |
| Chicken polyclonal anti-MAP2 | Aves Labs | Cat#: MAP, RRID: AB_2313549 |
| Chicken polyclonal anti-Tau | Abcam | Cat#: ab75714, RRID: AB_1310734 |
| Alexa Fluor 488 goat anti-human IgG Fcγ fragment specific | Jackson ImmunoResearch Labs | Cat#: 109-545-008, RRID: AB_2337833 |
| Alexa Fluor 647 goat anti-chicken IgG (H+L) | Thermo Fisher Scientific | Cat#: A-21449, RRID: AB_2535866 |
| Alexa Fluor 488 goat anti-rabbit IgG (H+L) | Jackson ImmunoResearch Labs | Cat#: 111-546-003, RRID: AB_2338053 |
| Alexa Fluor 555 goat anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat#: A-21422, RRID: AB_2535844 |
| Alexa Fluor 488 goat anti-mouse IgG (H+L) | Jackson ImmunoResearch Labs | Cat#: 115-546-003, RRID: AB_2338859 |
| Alexa Fluor 555 donkey anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#: A-31572, RRID: AB_162543 |
| Alexa Fluor 405 goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Cat#: A-31556, RRID: AB_221605 |
| Alexa Fluor 405 goat anti-mouse IgG (H+L) | Thermo Fisher Scientific | Cat#: A-31553, RRID: AB_221604 |
| CF405S donkey anti-guinea pig IgG (H+L) | Sigma-Aldrich | Cat#: SAB4600230, RRID: AB_2637046 |
| Chemicals and medium | ||
| Protein A/G PLUS-agarose | Santa Cruz | Cat#: sc-2003, RRID: AB_10201400 |
| Anti-Flag M2 affinity gel | Sigma-Aldrich | Cat#: A2220, RRID: AB_10063035 |
| DNA-In Neuro transfection reagent | MTI-GlobalStem | Cat#: GST-2101 |
| protease inhibitor cocktail | Roche | Cat#: 05892791001 |
| Papain | Worthington | Cat#: LK003176 |
| Opti-MEM I | GIBCO | Cat#: 31985-070 |
| Neurobasal Medium | GIBCO | Cat#: 21103-049 |
| B27 Supplement | GIBCO | Cat#: 17504-044 |
| DMEM medium | GIBCO | Cat#: 10569-010 |
| FBS | GIBCO | Cat#: 10437-028 |
| GlutaMax | GIBCO | Cat#: 35050-061 |
| Penicillin Streptomycin | GIBCO | Cat#: 15140-122 |
| poly-D-lysine | Sigma-Aldrich | Cat#: P6407 |
| Strychnine | Abcam | Cat#: ab120416 |
| APV | Abcam | Cat#: ab120003 |
| DNQX | alomone labs | Cat#: D-131 |
| Tetrodotoxin | BIOTIUM | Cat#: 00060 |
| HiTrap Protein A HP affinity column | GE Healthcare Life Science | Cat#: 17-0402-01 |
| EZ-Link Sulfo-NHS-SS-Biotin | Thermo Fisher Scientific | Cat#: 21331 |
| Pierce Streptavidin Agarose | Thermo Fisher Scientific | Cat#: 20349 |
| Carbachol | Sigma-Aldrich | Cat#: 1092009 |
| Pentylenetetrazole (PTZ) | Sigma-Aldrich | Cat#: P6500 |
| Critical Commercial Assays | ||
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#: 23227 |
| SuperSignal West Pico Chemiluminescent Substrate | Thermo Fisher Scientific | Cat#: 34080 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | Cat#: CRL-11268 |
| HeLa | ATCC | Cat#: CCL-2 |
| COS7 | ATCC | Cat#: CRL-1651 |
| Experimental Models: Organisms/Strains | ||
| Mouse: CD1 | Charles River | Strain code: 022 |
| mouse: B6; 129-Nlgn2tm1Bros/J | The Jackson Laboratory | Stock #: 008139 |
| mouse: B6; ST3ΔLRR9/ΔLRR9 | this paper | N/A |
| Oligonucleotides | ||
| shRNA targeting sequence: mouse NL2: GGAGCAAGTTCAACAGCAA | (Chih et al., 2005) | N/A |
| shRNA targeting sequence: mouse ST3: GGCTTAGAAAGTCTGGAAT | (Takahashi et al., 2012) | N/A |
| Recombinant DNA | ||
| Plasmid: pCAGGS-HA-Neuroligin2-IRES-mCherry | (Shipman et al., 2011) | N/A |
| Plasmid: L309 | (Pang et al., 2010) | N/A |
| Plasmid: pcDNA3.1(+) | Invitrogen | Cat#: V790-20 |
| Plasmid: pEGFP-N3 | Addgene | Cat#: 6080-1 |
| plasmid: mouse slitrk3 cDNA | OriGene | Cat#: MR211375 |
| plasmid: pFUSE-hIgG1-Fc2 | InvivoGen | Cat#: pfuse-hg1fc2, Version#: 06G05-MT |
| plasmid: pcDNA3-Nrxn1βAS4(+)-Fc | Addgene | Cat#: 59314 |
| plasmid: pEB6-PTPβ-ECD-Fc | (Yoshida et al., 2011) | N/A |
| Software | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism 6.0 | GraphPad | https://www.graphpad.com |
| Igor Pro | Wavemetrics | https://www.wavemetrics.com |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Wei Lu (luw4@mail.nih.gov).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All experiments using mice were performed in accordance with animal protocols approved by the Institutional Animal Care and Use Committee at NINDS, NIH. Adult CD1 mice were purchased from Charles River, housed and bred in a conventional vivarium with standard laboratory chow and water in standard animal cages under a 12 h circadian cycle. The day of vaginal plug detection was designated as E0.5 and the day of birth as P0. Time-pregnant CD1 mice at E18 were used for dissociated hippocampal neuron cultures. Time-pregnant mice at E14.5 were used for in utero electroporation (IUE). The NL2 KO mice were obtained from Dr. Katherine Roche’s lab, NINDS, NIH (originally generated by Nils Brose’s lab, JAX Mice Database Stock Number: 008139, Strain Name: B6; 129-Nlgn2tm1Bros/J). Genotyping was performed using described methods from the Jackson Laboratory website (common forward primer: 5′-GTCTCAGTAAGCTTATTTGAGAAGCCAA-3′, wild type reverse primer: 5′-CTCTGGGCCTTCTCAGGACTGTAC-3′, mutant reverse primer: 5′-GAGCGCGCGCGGCGGAGTTGTTGAC-3′). The genotypic group NL2+/+, NL2+/− and NL2−/− indicated wild type (WT), heterozygous and homozygous KO for NL2, respectively. The Slitrk3 LRR9 domain in-frame deletion mice were generated using CRISPR/Cas9 technology with single-guide RNAs (sgRNAs) that specifically target to nucleotide sequences of LRR9 motif in the Slitrk3 genomic DNA in a C57BL/6 background (see details described below). Genotyping was performed by PCR of genomic tail DNA using the primers (forward primer: 5′-ATCCTGGTCCAAATCAACCTCCTATTGC-3′, and reverse primer: 5′-TCTCGGGGGTCCTACAGAGTACATCACC-3′) and verified by DNA sequencing. The genotypic group ST3+/+, ST3+/ΔLRR9, ST3ΔLRR9/ΔLRR9 indicated WT, heterozygous and homozygous LRR9 in-frame deletion for Slitrk3, respectively. Mice of either sex were used for the experiments unless otherwise specified.
Generation of ST3ΔLRR9/ΔLRR9 mice
Deletion of the LRR9 domain within the mouse Slitrk3 gene was achieved through homology directed recombination (HDR) directly in C57BL/6 zygotes with CRISPR/Cas9 technology in the presence of a 127 bp single-stranded donor DNA template (IDT). Briefly, a few pairs of guide RNAs (gRNAs) (for SpCas9, PAM=NGG) flanking the 66 bp LRR9 coding sequence were selected with an online gRNA selection tool (www.deskgen.com), and synthesized with T7 in vitro transcription. To select the best pair of gRNAs which can direct the intended HDR reaction efficiently, gRNAs were further tested for their in vitro cleavage activities and in cell culture target deletion efficiencies. For the in vitro cleavage assay, genomic PCR product containing the target sites of selected gRNAs was incubated with SpCas9 protein (NEB) by following the manufacturer’s suggested protocol and analyzed on 2% agarose gel stained with ethidium bromide. A pair of gRNAs that delivered the most efficient cleavage of the target DNA fragments, L72 and R58, were further tested for their target deletion efficiency in an immortalized mouse embryonic fibroblast (MEF) cell line engineered to carry a tet-inducible Cas9 expression cassette (NEI genetic engineering core, unpublished). Upon confirmation of efficient target deletion activity in MEF cells, the two gRNAs were mixed with SpCas9 protein (PNA Bio) along with a synthetic single-strand donor DNA oligo template (IDT). This single-strand donor DNA template is designed to repair the deletion gap by the gRNA pair to restore the open reading frame while having the 21 (out of the total 22) amino acid residues constituting the LRR9 domain deleted. The mixture of gRNAs, Cas9 protein and the donor oligos was microinjected into zygotes of C57BL/6 background. A small litter of 4 F0 founders were born and screened by PCR for the deletion mutation followed with sequencing confirmation. One had both chromosomes correctly modified (homozygote) and two had the correct deletion mutation on one chromosome only (heterozygotes).
Donor oligo sequence: AAGGCTGCAGGTTGGATTTCCCGGATGACGTTGAATTCGAAGTACAAGTAATGCAAGCTCTGCAGACCGGGCAGGTTGATGAAAGCCCCATCtTGGACGTAGGAAATGCGATTGTTCCCTAGGTGCA (127mer)
gRNA sequences (bold upper case) with T7 promoter sequence (lower case) 5′ upstream and part of the guide RNA constant region (lower case) downstream are shown below. The GC dinucleotide was placed upstream of the T7 promoter to enhance T7 activity (NEI genetic engineering core, unpublished).
L72: GCtaatacgactcactataggCATTTCCTACGTCCAGGATGgttttagagctagaaata
R58: GCtaatacgactcactataggAATGCAAGCTCTGCAGACCTgttttagagctagaaata
METHOD DETAILS
Plasmids
pCAGGS-HA-Neuroligin2-IRES-mCherry plasmid was a generous gift from Roger Nicoll’s lab at UCSF (Shipman et al., 2011). Mouse full-length Slitrk3 cDNA was purchased from OriGene (Cat #: MR211375), and cloned into the NheI and XhoI sites of pCAGGS-IRES-mCherry expression vector. The coding sequences of Neuroligin2 or Slitrk3 truncation and deletion mutants were generated by overlapping PCR and inserted into the pCAGGS-IRES-mCherry expression vector for neuronal expression. To construct shRNA vectors for double knockdown of NL2 and ST3, the previously described targeted sequences of mouse NL2 and ST3 (GGAGCAAGTTCAACAGCAA for NL2, GGCTTAGAAAGTCTGGAAT for ST3) were used (Chih et al., 2005; Takahashi et al., 2012), complementary oligonucleotides encoding inverted repeats were annealed and ligated into the XhoI/XbaI sites and AscI/RsrII sites of shRNA vector (the vector L309 is a gift from Dr. Zhiping Pang (Pang et al., 2010), and shRNA-resistant wild type or swap rescue sequences, which were generated by making point mutations in NL2/ST3 shRNA-targeting sites, were inserted downstream of the ubiquitin C promoter in the shRNA vector. To generate the vectors for expression experiments in HEK293T cells, HeLa cells and COS7 cells, the coding sequences of HA-tagged wild type and truncation mutants of NL2, Flag-tagged wild type and truncation mutants of ST3 were cloned into pcDNA3 vector, respectively. Flag-tagged wild type and truncation mutants of ST3 were cloned into pEGFP-N3 vector for imaging experiments. LRR C2-Fc expression vector was generated by ligation of LRR9 C2 coding sequence into the EcoRV and NcoI sites of pFUSE-hIgG1-Fc2 vector (InvivoGen, Version # 06G05-MT). pcDNA3-Nrxn1βAS4(+)-Fc was purchased from Addgene (plasmid#: 59314), and pEB6-PTPδ-ECD-Fc was generously provided by Dr. Tomoyuki Yoshida (Yoshida et al., 2011). All constructs were verified by DNA sequencing.
Cell culture and transfection
HEK293T, HeLa and COS7 cells were grown in DMEM (GIBCO) supplemented with 10% fetal bovine serum (FBS) (GIBCO), 1% Pen/Strep, 1% Glutamine, and 1% sodium pyruvate, in a humidified atmosphere in a 37°C incubator with 5% CO2. Transfection was performed in 24-well plates or 6 cm dishes with indicated cDNAs using calcium phosphate transfection or DNA-IN transfection (MTI-GlobalStem) following the manufacturer’s instructions.
Dissociated hippocampal neuronal culture
Hippocampal cultures were prepared from E18 time-pregnant mice as previously described (Gu et al., 2016). Briefly, the hippocampi were dissected out in ice-cold Hank’s balanced salt solution, and digested with papain (Worthington, LK003176) solution at 37°C for 45 min. After centrifugation for 5 min at 800 rpm, the pellet was resuspended in DNase I-containing Hank’s solution, then was mechanically dissociated into single cells by gentle trituration using a pipette. Cells were placed on top of Hank’s solution mixed with trypsin inhibitor (10 mg/ml, Sigma T9253) and BSA (10 mg/ml, Sigma A9647), and centrifuged at 800 rpm for 10 min. The pellet was resuspended in Neurobasal plating media containing 2% FBS, 2% B27 supplements and L-glutamine (2 mM). Neurons were plated at a density of ~250,000 cells/well on poly-D-lysine (Sigma P6407)-coated 12 mm glass coverslips residing in 24-well plates for electrophysiology recording, and a lower plating density (~150,000 cells/well) was adopted when neurons were used for immunocytochemistry. Culture media were changed by half volume with Neurobasal maintenance media containing 2% B27 supplements and L-glutamine (2 mM) twice a week.
Neuronal transfection
Hippocampal neurons were transfected at day 2 in vitro (DIV2) using a modified calcium phosphate transfection. Briefly, 5 μg total DNA was used to generate 200 μl total precipitates, which was added to each well at a 40 μl volume (5 coverslips/group). After 2 h incubation in a 37°C incubator, the transfected cells were incubated with 37°C pre-warmed, 10% CO2 pre-equilibrated Neurobasal medium, and placed in a 37°C, 5% CO2 incubator for 20 min to dissolve the calcium-phosphate particles. The coverslips were then transferred back to the original conditioned medium. The cells were cultured to DIV 7–8 before experiments.
At DIV9–10, neurons were transfected using DNA-IN Neuro transfection reagent following the manufacturer’s instructions (MTI-GlobalStem). Briefly, before transfection, half of neuronal culture medium was replaced. 50 μl pre-warmed Opti-MEM I was mixed with 1 μg DNA and 2 μl DNA-IN Neuro transfection reagent (per well in a 24-well plate) for 15 minutes at room temperature (RT). The mixture was then added to neuronal cultures, and the neurons were cultured to DIV17–18 before experiments.
Immunocytochemistry
Cells grown on coverslips were rinsed with PBS twice and fixed in 4% paraformaldehyde (PFA)/4% sucrose/1xPBS solution for 15 min at RT, permeabilized with 0.2% TritonX-100/1xPBS for 15 min (for surface labelling, cells were not permeabilized), and blocked with 5% normal goat serum in PBS for 1 h. Cells were labeled with primary antibodies as follows: ant-Neuroligin2 (mouse,1:1000, SYSY), anti-Slitrk3 (rabbit, 1:1000, Millipore), anti-vGAT (rabbit,1:1000, SYSY), anti-Gephyrin (mouse, 1:1000, SYSY), anti-MAP2 (chicken, 1:1000, Aves Labs), anti-Tau (chicken, 1:1000, Abcam), anti-HA (rabbit, 1:1000, Santa Cruz), anti-Flag (M2 mouse, 1:1000, Sigma) in 3%NGS/1xPBS solutions, incubated overnight at 4°C. Cells were washed three times with 1xPBS and then incubated with Alexa Fluor 405-conjugated goat anti-mouse IgG, Alexa Fluor 555-conjugated goat anti-rabbit or mouse IgG, or Alexa Fluor 647-conjugated goat anti-chicken IgG for 30 min. Coverslips were washed for three times with 1xPBS and mounted with Fluoromount-G (Southern Biotech).
Co-culture assay
Co-culture assays were conducted as described previously (Takahashi et al., 2012). Briefly, the COS7 cells were transfected with the indicated plasmid(s) using DNA-IN transfection reagent following the manufacturer’s instructions (MTI-GlobalStem). 24 h after transfection, the cells were trypsinized and plated onto hippocampal neurons at 7–8 days in vitro (DIV) at a density of 40,000/coverslip, co-cultured for 24 h in a 37°C incubator. Immunostaining was then performed as described above. Briefly, surface labeling with anti-HA/Flag antibodies was performed at RT for 2 h before cell permeabilization. After cell permeabilization with 0.2% TritonX-100/1xPBS, anti-vGAT or Tau antibodies were incubated overnight at 4°C. On the next day, cells were washed and incubated with the indicated secondary antibodies.
Fc-fusion proteins production and binding assay
Plasmids expressing LRR C2 fused to Fc (LRR C2-Fc), soluble Neurexin (Nrxn) ectodomain fused to Fc (Nrxn-Fc), or PTPδ ectodomain fused to Fc (PTPδ-Fc) were transfected into HEK293T cells for 48 h. Culture media from transfected cells were collected and Fc-fusion proteins were purified with a HiTrap Protein A HP affinity column following the manufacturer’s instructions (GE Healthcare Life Science). For testing binding affinity of LRR C2-Fc, Nrxn-Fc or PTPδ-Fc proteins, COS7 cells expressing HA-NL2, Flag-ST3 or both were incubated with purified proteins that were diluted in pre-warmed culture medium at different concentrations for 1 h at 37 °C. The cells were then washed with 1xPBS three times, fixed with 4% PFA/4% sucrose/1xPBS, and immunostained with indicated antibodies overnight at 4°C. On the next day, cells were washed and incubated with secondary antibodies together with Alexa 488 conjugated anti-human Fc IgG for 30 min at RT. After washing three times with 1xPBS, the coverslips were mounted in Fluoromount-G mounting media (Southern Biotech).
Immunohistochemistry
ST3ΔLRR9/ΔLRR9 and WT littermate mice at P21–P25 were anesthetized with isoflurane, perfused transcardially with 4% PFA + 0.1% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4 (PB). Brains were then dissected out, post-fixed in the same solution for 1 h at RT, and then cryoprotected overnight in 30% sucrose in 1× PB at 4°C. Coronal brain sections (40 μm) were collected at −20°C with a cryostat (Leica CM1050). After washing with 1xPBS, brain sections were incubated in the blocking solution (0.2 % Triton X-100 and 5 % normal goat serum in 1xPBS) for 1 h at RT under gentle agitation, and followed by incubation at 4°C with primary antibodies in 1xPBS containing 0.2 % Triton-X with 3 % normal goat serum (anti-vGAT, 1:4000, rabbit SYSY; anti-Gephyrin, 1:1000 mouse, SYSY) for 48 h. Brain sections were then washed 3 times (20 min each time) in 1xPBS, incubated with secondary antibodies (1:1000, Alexa 488, 555, Thermo Fisher Scientific) for 1 h at RT, and followed by washing for 3 times (20 min each time) in 1xPBS. Brain sections were then mounted on superfrost slides and covered with DAPI-containing Fluoromount G mounting media (Southern Biotech).
Image acquisition
Fluorescent images were acquired on a Zeiss LSM 510 or 880 laser scanning confocal microscope using a 63X oil objective (1.4 numerical aperture) lens with identical settings for laser power, gain, offset, pinhole size, and z-steps throughout the same experiments. For image acquisition in dissociated hippocampal neuronal culture and co-culture assays, serial confocal z-stack images were acquired with a resolution of 1024 × 1024, digital zoom 1.0 for low magnification images, or 512 ×128 resolution, digital zoom 4.0 for high magnification images, with a pixel dwell time of 0.79 msec and each line averaged 4 times. The maximal intensity projected images were generated by LSM browser software for analysis. For surface staining of HeLa cells, single-plane confocal images were taken with a resolution of 1024 × 1024, digital zoom 4.0. Scan speed function was set to 9 and the mean of 8 lines was collected. Pinhole was set to 1 airy unit for all experiments. Laser power, digital gain and offset settings were made identical in each experiment by using the “reuse” function in LSM software. For image acquisition of immunohistochemistry staining, single-plane confocal images were captured with a resolution of 1024 × 1024, digital zoom 2.0, using a 40X oil objective (1.4 numerical aperture). Scan speed function was set to 9 and the mean of 8 lines was collected. Pinhole was set to 1 airy unit for all experiments. Laser power, digital gain and offset settings were identical in each group. The region of interest (ROI) in brain sections was determined using DAPI images.
Co-Immunoprecipitation assay
Constructs were transfected into HEK293T cells by a calcium phosphate transient transfection method. After transfection for 48 h, cells were rinsed once with ice cold 1xPBS solution and collected in a tube. After centrifugation, the pellet was suspended with ice-cold lysis buffer containing 25 mM Tris pH 7.5, 1% Triton X-100, 150 mM NaCl, 5% glycerol, 1 mM EDTA and protease inhibitor cocktail (Roche), and underwent constant shaking for 30 min on ice. The insoluble debris was removed by centrifugation at 12,000 g for 10 min. An equal amount of the supernatant was incubated with pre-washed anti-Flag M2 antibody conjugated to affinity agarose beads for 3–4 h at 4°C while rotating. The beads were then washed 3–5 times with ice-cold lysis buffer with centrifugation for 1 min at 5,000 g after each wash. The bound proteins were eluted with 2× SDS loading buffer with β-mercaptoethanol, and denatured at 95°C for 5 min. The denatured samples were subjected to SDS-PAGE electrophoresis and Western blot as described below with indicated antibodies. For brain tissue co-immunoprecipitation, hippocampi from ~6-week old mice were dissected, and homogenized in the pre-chilled lysis buffer as mentioned above. The monoclonal anti-NL2 antibody or a control mouse IgG was used for immunoprecipitation. Hippocampal lysates were incubated with antibodies overnight at 4°C, followed by 40 μl Protein A/G beads for 2–3 h. After washing the beads with lysis buffer 3–5 times, the eluted proteins were denatured and subjected to SDS-PAGE and western blot with indicated antibodies.
Hippocampal slice biotinylation
Hippocampal slice biotinylation was performed as previously described (Gabriel et al., 2014) with minor modification. Briefly, P30–40 mice were anesthetized with isoflurane. The hippocampi were rapidly dissected out in the prechilled (4 °C) ice-cold high sucrose cutting solution, which contains (in mM) as follows: KCl 2.5, CaCl2 0.5, MgCl2 7, NaH2PO4 1.25, NaHCO3 25, glucose 7, sucrose 210 and ascorbic acid 1.3. Subsequently, hippocampi were cut into 300 μm-thick transverse slices using DTK Microslicer vibratome (Ted Pella). Then, freshly cut slices were transferred into an incubation chamber filled with 31°C carbogenated artificial cerebrospinal fluid (ACSF), containing (in mM) NaCl 119, KCl 2.5, NaHCO3 26, Na2PO4 1, glucose 11, CaCl2 2.5, MgCl2 1.3, incubated for 40 min for recovery. After recovery, slices were washed three times in ice-cold ACSF, then incubated for 45 min with EZ-Link Sulfo-NHS-SS-Biotin (1.0 mg/ml in ACSF; Thermo Fisher Scientific) on ice. After washing three times with ice-cold ACSF, the slices were incubated with Slice Quench Buffer (ACSF supplemented with 100 mM glycine) for 25 min to quench free biotin. After washing three times with ice-cold ACSF, the slices were collected by centrifuging 1000× g, 1 min, then lysed with RIPA buffer 500 μl each on ice for 30 min. Protein supernatants were collected by centrifuging 18,000× g 10 min, then bound with streptavidin agarose (Thermo Fisher Scientific) overnight at 4°C. On the next day, beads were washed three times with lysis buffer. The biotinylated proteins were eluted from agarose beads and denatured using 2× SDS loading buffer, then immunoblotted with the indicated antibodies.
Western blot
Protein concentration was quantified with the standard BCA method. An equal amount of loading samples was mixed with an equal volume of 2× SDS loading buffer and boiled for 5 min at 95°C, and then separated using pre-casted 10% SDS-PAGE gels (BioRad). The proteins were transferred onto PVDF membranes, blocked, and incubated with primary antibodies overnight a 4°C. The PVDF membranes were then washed three times with 0.1% TBST and incubated with HRP-conjugated secondary antibodies for 1 h at RT. Protein was detected with the standard enhanced chemiluminescence (ECL) method. The protein bands were quantified with Image J (NIH). For developmental profile experiments, the hippocampi were collected from mice at different developmental stages, and homogenized in lysis buffer as mentioned above. After protein quantification, the same amount of protein was loaded in each lane for Western blot analysis.
In Utero electroporation
In Utero electroporation was performed as described previously (Gu et al., 2016). Briefly, the E14.5 time-pregnant mouse was anesthetized with isoflurane. The abdominal wall was opened by a ~2 cm incision and embryos in the uterine horns were exposed and gently pulled outside the abdominal cavity. The lateral ventricles of each embryo were manually injected with approximately 1 to 2 μl of DNA solution at the concentration of 2 μg/μl mixed with 0.05% fast green (Sigma 68724). The injection glass pipettes were beveled with the BV-10 micropipette Beveller (Sutter) before injection. After each injection, voltage steps via tweezertrodes (5 mm round, platinum electrodes and BTX electroporator, BTX, ECM830) positioned on either side of the head were applied across the uterus to target hippocampal progenitor neurons. Electrical voltage pulses (45 V, 5 pulses, 50 ms duration each pulse) were used to electroporate exogenous DNA into hippocampal progenitor neurons. After electroporation, the embryos were moistened with warmed sterile 1xPBS and then returned to their original position inside the abdominal cavity. Buprenex (0.1 mg/kg) was applied and the wound was sutured. Offspring were collected at designated postnatal time points for experiments.
Electrophysiology
For mIPSCs recording in CA1 pyramidal neurons in acute slices, 300 μm transverse hippocampal slices were cut on a DTK Microslicer vibratome (Ted Pella) in ice-cold high sucrose cutting solution, which contains (in mM) as follows: KCl 2.5, CaCl2 0.5, MgCl2 7, NaH2PO4 1.25, NaHCO3 25, glucose 7, sucrose 210 and ascorbic acid 1.3. Freshly cut slices were placed in an incubating chamber containing carbogenated artificial cerebrospinal fluid (ACSF), containing (in mM) NaCl 119, KCl 2.5, NaHCO3 26, Na2PO4 1, glucose 11, CaCl2 2.5, MgCl2 1.3, and recovered at 32°C for ~30–60 min. Slices were then maintained in ACSF at RT prior to recording. After 0.5–1 h of incubation at RT, slices were transferred to the recording chamber on an upright Olympus microscope. The slices in the recording chamber were continuously perfused with 95% O2- and 5% CO2-saturated ACSF, supplemented with TTX (0.5 μM), DNQX (20 μM) and strychnine (1 μM). For mIPSC recording in hippocampal dissociated primary cultures, neurons growing on coverslips were transferred to a submersion chamber on an upright Olympus microscope, and perfused with ACSF solution supplemented with TTX (0.5 μM), DNQX (20 μM) and strychnine (1 μM). Fluorescent positive neurons in hippocampal acute slices or neuronal cultures were identified by epifluorescence microscopy. Neurons were voltage-clamped at −70 mV for detection of mIPSC events. The intracellular solution for GABAergic mIPSC recording contained (in mM) CsMeSO4 70, CsCl 70, NaCl 8, EGTA 0.3, HEPES 20, MgATP 4 and Na3GTP 0.3. Osmolality was adjusted to 285–290 mOsm and pH was buffered at 7.25–7.35. Series resistance was monitored and not compensated, and cells in which series resistance varied by 25% during a recording session were discarded. Synaptic responses were collected with a Multiclamp 700B amplifier (Axon Instruments, Foster City, CA), filtered at 2 kHz, and digitized at 10 kHz. All recordings were performed at RT. 100–300 consecutive miniature events were semiautomatically detected by offline analysis using in-house software in Igor Pro (Wavemetrics) developed in Dr. Roger Nicoll’s laboratory at UCSF, using a threshold of 6 pA. All mIPSC events were visually inspected to ensure that they were mIPSCs during analysis, and non-mIPSC traces were discarded. Experiments and analysis were conducted in a genotype-blinded manner.
Hippocampal network activities
Hippocampal slices were prepared from P30 to P45 ST3+/+ or ST3ΔLRR9/ΔLRR9 mice as indicated. Mice were anesthetized with isoflurane, and the brain was dissected in ice-cold partial sucrose substituted artificial cerebrospinal fluid (ACSF) containing (in mm): 80 NaCl, 3.5 KCl, 1.25 H2PO4, 25 NaHCO3, 4.5 MgCl2, 0.5 CaCl2, 10 glucose, and 90 sucrose equilibrated with 95% O2/5% CO2. Transverse slices (350 μm thick) were cut using a VT-1000S vibratome (Leica Microsystems, Bannockburn, IL) and then immediately transferred to an interface-style chamber (Warner Instruments, CT) containing humidified carbogen gas and perfused (1–1.5ml/min, 34°C) with aCSF containing (in mM): NaCl (126), KCl (3.5), MgCl2 (2), CaCl2 (2), NaH2PO4 (1.25), glucose (10) and NaHCO3 (26) saturated with 95% O2 and 5% CO2. Slices were incubated for at least 1h before recording. Local field potentials were recorded from CA3 stratum pyramidale using glass pipettes pulled from standard borosilicate glass (3–5 MΩ) and filled with ACSF. Signals were low-pass filtered at 2 kHz and acquired at 10 kHz using a Multiclamp 700A Axon amplifier (Molecular Devices, CA), digitized using a Digidata 1322A (Molecular Devices, CA) and captured on a computer running pClamp 9 (Molecular Devices, CA). All Data were imported into Igor Pro 6 (Wavemetrics, OR) using NeuroMatic (ThinkRandom, UK) and analyzed using custom-written procedures. Sharp wave ripple oscillation were recorded in drug-free conditions and then gamma oscillations were induced by applying 25 μM carbachol (CCh, Sigma-Aldrich, MO) as described (Fisahn et al., 1998).
For gamma oscillations, power spectra were generated from 300 s of unfiltered recordings following 30 minutes of CCh application to determine peak frequency, peak power, and gamma band power (20–80 Hz). Example traces show either the local field potential or Wavelet transforms of the unfiltered traces. SWRs were automatically detected using methods described elsewhere (Csicsvari et al., 1999; Pelkey et al., 2015), but adapted for in vitro recordings. Extracellular recordings were digitally band-pass filtered between 150 and 250 Hz to reveal SWRs, and the root mean square (RMS) of the filtered recording was used for automatic detection. Periods where the RMS crossed a threshold of 4 SDs above the background were counted as SWR events in our detection algorithm. The nearest peak to the threshold crossing was considered the center of the SWR and 100 ms on either side of the peak was extracted and the corresponding period of the unfiltered trace was transformed using a Morlet wavelet function. The boundaries of the SWRs were defined as 2 SDs above the baseline power (taken from the first 50 ms at 250 Hz) of the wavelet function. The peak frequency of the SWR was taken from the wavelet transform, and the amplitude calculated by the peak-to-trough amplitude of the band-pass filtered trace. The RMS of the filtered trace was calculated using the SWR boundaries detected from the Wavelet transform, and those events with an average RMS of less than 1.5 SDs above the background were rejected. To prevent contamination of SWR data by single unit activity, all detected events of less than 10 ms duration were automatically rejected by our detection algorithm.
Transmission electron microscopy
Mice were anesthetized and then perfused with 0.1 M phosphate buffer followed by 4% PFA plus 2% glutaraldehyde in the same buffer (Aceti et al., 2015) and brains were kept in fixative overnight. Then, brains were washed in phosphate-buffered saline and sliced at 125 μm. Sections were washed in 0.1 M cacodylate buffer and fixed in 1% osmium tetroxide, then washed and dehydrated in a series of ethanol, with 1% uranyl acetate added to the 50% ethanol. Sections were transferred to propylene oxide and embedded in epon, and heated at 64°C for 48 hours. Thin sections were cut and examined in a JEOL JEM-2100 transmission electron microscope; sections were examined blinded and micrographs of synapses were taken from the CA1 stratum pyramidale and radiatum. Inhibitory synapses with a clear pre- and post-synaptic membranes were included in the analysis for PSD length. Inhibitory synapses that had a clear synaptic cleft separating the pre- and post-synaptic membranes were measured for cleft width. The areas of presynaptic boutons with clear membrane boundary were measured for GABAergic bouton area.
PTZ-induced seizure
Five male mice of each genotype (postnatal 4–6 weeks old) were injected intraperitoneally (i.p.) with PTZ (Sigma-Aldrich, MO, USA, a GABAA receptor antagonist, dissolved in saline) at a dose of 50 mg per kg of body weight to induce seizures and monitored for 10 min after the injection. Behavioral responses were scored at every 1 min using the following scale: 0, no abnormal behavior; 1, reduced motility and prostrate position; 2, partial clonus; 3, generalized clonus including extremities; 4, tonic-clonic seizure with rigid paw extension; 5, death, as described before (Takahashi et al., 2012). Total seizure scores were calculated by summing up the minute-by-minute scores.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image analysis
For puncta density analysis, confocal images from 2–3 dendrites (35 μm in length) per neuron from at least ten neurons in each group were collected and quantified by counting the number of puncta per 10 μm dendrites or per soma with Image J software (NIH). Background was subtracted and the ROI (dendrites or soma of the neuron according to MAP2 staining) was defined and analyzed by the built-in “analyze particles” program. For fluorescent intensity analysis, the maximal intensity projected images were generated by LSM880 browser software, and the mean fluorescent intensity of ROI was measured following the subtraction for off-cell background with Image J software (NIH). For co-localization analysis, two or three secondary dendrite segments per neuron were chosen. ST3 and NL2 clusters were separately thresholded and confirmed visually to select appropriate clusters following a minimal size cut-off, which included all recognizable clusters. The ST3-positive NL2 clusters indicate the number of NL2 clusters exhibiting at least partial pixel overlapping with thresholded ST3 clusters, and co-localization percentage was quantified by the measurement of ST3-positive NL2 clusters compared to total number of thresholded NL2 clusters. For co-culture assay, the vGAT image was thresholded and the total intensity of puncta in all regions positive for surface HA or surface Flag was measured and normalized to COS7 cell area. To compare the vGAT intensity in cells co-transfected with HA-NL2 and Flag-ST3 (HA-NL2+Flag-ST3) vs combined vGAT intensity in cells transfected with HA-NL2 and cells transfected with Flag-ST3, the average vGAT intensity in cells transfected with Flag-ST3 was first calculated. The number of the average vGAT intensity was then added to vGAT intensity calculated from individual cells transfected with HA-NL2 (defined as HA-NL2 + Ave. Flag-ST3 in Figure 6E), and an unpaired t-test was used to compare the two conditions, HA-NL2+Flag-ST3 vs. HA-NL2 + Ave. Flag-ST3. Alternatively, the average vGAT intensity in cells transfected with HA-NL2 was calculated, and the number of the average vGAT intensity was added to vGAT intensity calculated from individual cells transfected with Flag-ST3 (defined as Flag-ST3 + Ave. HA-NL2 in Figure 6E), and an unpaired t-test was used to compare the two conditions, HA-NL2+Flag-ST3 vs. Flag-ST3 + Ave. HA-NL2. Pearson’s correlation coefficients were calculated with JACoP plugin in the ImageJ software to measure the co-localization of vGAT puncta versus surface HA-NL2 or surface Flag-ST3 in co-culture assays. For triple co-localization, co-localized 8-bit images of two proteins were extracted using a co-localization plugin in ImageJ software (developed by Pierre Bourdoncle, Institut Jacques Monod, Service Imagerie, Paris, France). The extracted 8-bit images were then used to quantitate triple co-localization with the third protein using a JACoP plugin as described above for double co-localization. For quantification of Fc protein binding affinity, we measured the average intensity of bound Fc protein per COS7 cell area (which was defined by HA-NL2 or Flag-ST3 immunostaining distribution), and subtracted the background intensity of the off-cell region. For analysis of puncta density of vGAT and gephyrin in the stratum pyramidale or radiatum of brain sections, quantification was carried out using Image J. vGAT and gephyrin images were converted to binary images, then threshold values were adjusted to select appropriate puncta following a minimal cut-off size. The ROI was drawn manually in a given channel. Subsequently, the total puncta number and area were automatically calculated. The puncta density was presented by puncta number per 100 μm2 ROI area.
Statistical analysis
For biochemical, cell biological, histological assays and physiological recordings, at least three independent experiments were performed (independent cultures, transfections or different mice), except in the experiments in Figure 7D and Figure S1C where two animals per group were used. Statistical analysis was performed in GraphPad Prism 6.0 software. Direct comparisons between two groups were made using two-tailed Student’s t test with Welch’s correction when the SD was significantly different. Multiple group comparisons were made with one-way ANOVA analysis with post hoc Fisher’s LSD test. The significance of the cumulative probability distributions was assessed with the Kolmogorov-Smirnov test. The statistical significance was defined as *p < 0.05, **p <0.01, ***p < 0.001 or ****p < 0.0001, respectively. All results are presented as mean ± SEM.
Supplementary Material
Acknowledgments
We are grateful to Dr. Katherine Roche for critical comments on the manuscript. We thank Dr. Mitchell Ho at NCI, NIH, and Dr. Joseph Mindell and Dr. Patricia Curran at NINDS, NIH for assistance of Fc-fusion protein purification. This work was supported by the NIH/NINDS Intramural Research Program (W.L.), NIH/NICHD Intramural Research Program (C.J.M.), NIH/NIDCD Intramural Research Program (Y.-X.W., R.S.P.), and NIH/NEI Intramural Research Program (L.J.D.).
Footnotes
Supplemental Information includes 8 figures and can be found with this article online at
AUTHOR CONTRIBUTIONS
W.L. and J.L. designed the project, and W.L. conceived the project. J.L. performed imaging and biochemical experiments, recordings in neuronal cultures and seizure test. W.Y.H., J.J.D. and X.M. performed recordings in acute hippocampal slices. K.A.P., M.T.C and C.J.M. performed and analyzed recordings of hippocampal network activities. J.L., Y.-X.W., W.Y.H., and R.S.P. performed EM experiments, and L.J.D. generated ST3ΔLRR9/ΔLRR9 mice. W.L. and J.L. wrote the manuscript, and all authors contributed to the final version of the manuscript.
References
- Aceti M, Creson TK, Vaissiere T, Rojas C, Huang WC, Wang YX, Petralia RS, Page DT, Miller CA, Rumbaugh G. Syngap1 haploinsufficiency damages a postnatal critical period of pyramidal cell structural maturation linked to cortical circuit assembly. Biol Psychiatry. 2015;77:805–815. doi: 10.1016/j.biopsych.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ango F, di Cristo G, Higashiyama H, Bennett V, Wu P, Huang ZJ. Ankyrin-based subcellular gradient of neurofascin, an immunoglobulin family protein, directs GABAergic innervation at purkinje axon initial segment. Cell. 2004;119:257–272. doi: 10.1016/j.cell.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Ashrafi S, Betley JN, Comer JD, Brenner-Morton S, Bar V, Shimoda Y, Watanabe K, Peles E, Jessell TM, Kaltschmidt JA. Neuronal Ig/Caspr recognition promotes the formation of axoaxonic synapses in mouse spinal cord. Neuron. 2014;81:120–129. doi: 10.1016/j.neuron.2013.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–125. doi: 10.1038/nature12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G, Wang XJ. Mechanisms of gamma oscillations. Annu Rev Neurosci. 2012;35:203–225. doi: 10.1146/annurev-neuro-062111-150444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S, Hale WD, Zhang B, Wernig M, Sudhof TC. Unique versus Redundant Functions of Neuroligin Genes in Shaping Excitatory and Inhibitory Synapse Properties. J Neurosci. 2017;37:6816–6836. doi: 10.1523/JNEUROSCI.0125-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherubini E, Conti F. Generating diversity at GABAergic synapses. Trends Neurosci. 2001;24:155–162. doi: 10.1016/s0166-2236(00)01724-0. [DOI] [PubMed] [Google Scholar]
- Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307:1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- Chubykin AA, Atasoy D, Etherton MR, Brose N, Kavalali ET, Gibson JR, Sudhof TC. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Kang Y. Neurexin-neuroligin signaling in synapse development. Curr Opin Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csicsvari J, Hirase H, Czurko A, Mamiya A, Buzsaki G. Oscillatory coupling of hippocampal pyramidal cells and interneurons in the behaving Rat. J Neurosci. 1999:274–287. doi: 10.1523/JNEUROSCI.19-01-00274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, Ghosh A. Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci. 2016;17:22–35. doi: 10.1038/nrn.2015.3. [DOI] [PubMed] [Google Scholar]
- Ellender TJ, Nissen W, Colgin LL, Mann EO, Paulsen O. Priming of hippocampal population bursts by individual perisomatic-targeting interneurons. J Neurosci. 2010;30:5979–5991. doi: 10.1523/JNEUROSCI.3962-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisahn A, Pike FG, Buhl EH, Paulsen O. Cholinergic induction of network oscillations at 40 Hz in the hippocampus in vitro. Nature. 1998;394:186–189. doi: 10.1038/28179. [DOI] [PubMed] [Google Scholar]
- Futai K, Doty CD, Baek B, Ryu J, Sheng M. Specific trans-synaptic interaction with inhibitory interneuronal neurexin underlies differential ability of neuroligins to induce functional inhibitory synapses. J Neurosci. 2013;33:3612–3623. doi: 10.1523/JNEUROSCI.1811-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel LR, Wu S, Melikian HE. Brain slice biotinylation: an ex vivo approach to measure region-specific plasma membrane protein trafficking in adult neurons. J Vis Exp. 2014 doi: 10.3791/51240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Mondin M, Grillo-Bosch D, Tessier B, Saint-Michel E, Czondor K, Sainlos M, Choquet D, Thoumine O. Neurexin-1beta binding to neuroligin-1 triggers the preferential recruitment of PSD-95 versus gephyrin through tyrosine phosphorylation of neuroligin-1. Cell Rep. 2013;3:1996–2007. doi: 10.1016/j.celrep.2013.05.013. [DOI] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Zhou L, Lu W. An NMDA Receptor-Dependent Mechanism Underlies Inhibitory Synapse Development. Cell Rep. 2016;14:471–478. doi: 10.1016/j.celrep.2015.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Scheiffele P. GABA and neuroligin signaling: linking synaptic activity and adhesion in inhibitory synapse development. Curr Opin Neurobiol. 2008;18:77–83. doi: 10.1016/j.conb.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kins S, Betz H, Kirsch J. Collybistin, a newly identified brain-specific GEF, induces submembrane clustering of gephyrin. Nat Neurosci. 2000;3:22–29. doi: 10.1038/71096. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Choii G, Um JW. The balancing act of GABAergic synapse organizers. Trends Mol Med. 2015;21:256–268. doi: 10.1016/j.molmed.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Krueger-Burg D, Papadopoulos T, Brose N. Organizers of inhibitory synapses come of age. Curr Opin Neurobiol. 2017;45:66–77. doi: 10.1016/j.conb.2017.04.003. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Karube F, Nomura M, Kawaguchi Y. The Diversity of Cortical Inhibitory Synapses. Front Neural Circuits. 2016;10:27. doi: 10.3389/fncir.2016.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzirian MS, Paradis S. Emerging themes in GABAergic synapse development. Prog Neurobiol. 2011;95:68–87. doi: 10.1016/j.pneurobio.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HB, Kozorovitskiy Y, Oh WJ, Peixoto RT, Akhtar N, Saulnier JL, Gu C, Sabatini BL. Neuroligin-1-dependent competition regulates cortical synaptogenesis and synapse number. Nat Neurosci. 2012;15:1667–1674. doi: 10.1038/nn.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Xu W, Hsu YT, Yee AX, Chen L, Sudhof TC. Conditional knockout of Nlgn2 in the adult medial prefrontal cortex (mPFC) induces delayed loss of inhibitory synapses. Mol Psychiatry. 2015;20:793. doi: 10.1038/mp.2015.82. [DOI] [PubMed] [Google Scholar]
- Lu W, Bromley-Coolidge S, Li J. Regulation of GABAergic synapse development by postsynaptic membrane proteins. Brain Res Bull. 2016 doi: 10.1016/j.brainresbull.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH, Nicoll RA. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EO, Suckling JM, Hajos N, Greenfield SA, Paulsen O. Perisomatic feedback inhibition underlies cholinergically induced fast network oscillations in the rat hippocampus in vitro. Neuron. 2005;45:105–117. doi: 10.1016/j.neuron.2004.12.016. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Dynamic aspects of CNS synapse formation. Annu Rev Neurosci. 2007;30:425–450. doi: 10.1146/annurev.neuro.29.051605.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Sudhof TC, Biederer T. Synaptic cell adhesion. Cold Spring Harb Perspect Biol. 2012;4:a005694. doi: 10.1101/cshperspect.a005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen QA, Horn ME, Nicoll RA. Distinct roles for extracellular and intracellular domains in neuroligin function at inhibitory synapses. Elife. 2016:5. doi: 10.7554/eLife.19236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Cao P, Xu W, Sudhof TC. Calmodulin controls synaptic strength via presynaptic activation of calmodulin kinase II. J Neurosci. 2010;30:4132–4142. doi: 10.1523/JNEUROSCI.3129-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzanelli P, Fruh S, Fritschy JM. Differential role of GABAA receptors and neuroligin 2 for perisomatic GABAergic synapse formation in the hippocampus. Brain Struct Funct. 2017 doi: 10.1007/s00429-017-1462-7. [DOI] [PubMed] [Google Scholar]
- Papadopoulos T, Eulenburg V, Reddy-Alla S, Mansuy IM, Li Y, Betz H. Collybistin is required for both the formation and maintenance of GABAergic postsynapses in the hippocampus. Mol Cell Neurosci. 2008;39:161–169. doi: 10.1016/j.mcn.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Papadopoulos T, Korte M, Eulenburg V, Kubota H, Retiounskaia M, Harvey RJ, Harvey K, O’Sullivan GA, Laube B, Hulsmann S, et al. Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. EMBO J. 2007;26:3888–3899. doi: 10.1038/sj.emboj.7601819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkey KA, Barksdale E, Craig MT, Yuan X, Sukumaran M, Vargish GA, Mitchell RM, Wyeth MS, Petralia RS, Chittajallu R, et al. Pentraxins coordinate excitatory synapse maturation and circuit integration of parvalbumin interneurons. Neuron. 2015;85:1257–1272. doi: 10.1016/j.neuron.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulopoulos A, Aramuni G, Meyer G, Soykan T, Hoon M, Papadopoulos T, Zhang M, Paarmann I, Fuchs C, Harvey K, et al. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron. 2009;63:628–642. doi: 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- Poulopoulos A, Soykan T, Tuffy LP, Hammer M, Varoqueaux F, Brose N. Homodimerization and isoform-specific heterodimerization of neuroligins. Biochem J. 2012;446:321–330. doi: 10.1042/BJ20120808. [DOI] [PubMed] [Google Scholar]
- Ramamoorthi K, Lin Y. The contribution of GABAergic dysfunction to neurodevelopmental disorders. Trends Mol Med. 2011;17:452–462. doi: 10.1016/j.molmed.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Yamagata M. Many paths to synaptic specificity. Annu Rev Cell Dev Biol. 2009;25:161–195. doi: 10.1146/annurev.cellbio.24.110707.175402. [DOI] [PubMed] [Google Scholar]
- Sassoe-Pognetto M, Frola E, Pregno G, Briatore F, Patrizi A. Understanding the molecular diversity of GABAergic synapses. Front Cell Neurosci. 2011;5:4. doi: 10.3389/fncel.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Shen K, Scheiffele P. Genetics and cell biology of building specific synaptic connectivity. Annu Rev Neurosci. 2010;33:473–507. doi: 10.1146/annurev.neuro.051508.135302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman SL, Schnell E, Hirai T, Chen BS, Roche KW, Nicoll RA. Functional dependence of neuroligin on a new non-PDZ intracellular domain. Nat Neurosci. 2011;14:718–726. doi: 10.1038/nn.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui TJ, Craig AM. Synaptic organizing complexes. Curr Opin Neurobiol. 2011;21:132–143. doi: 10.1016/j.conb.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soykan T, Schneeberger D, Tria G, Buechner C, Bader N, Svergun D, Tessmer I, Poulopoulos A, Papadopoulos T, Varoqueaux F, et al. A conformational switch in collybistin determines the differentiation of inhibitory postsynapses. EMBO J. 2014;33:2113–2133. doi: 10.15252/embj.201488143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Katayama K, Sohya K, Miyamoto H, Prasad T, Matsumoto Y, Ota M, Yasuda H, Tsumoto T, Aruga J, Craig AM. Selective control of inhibitory synapse development by Slitrk3-PTPdelta trans-synaptic interaction. Nat Neurosci. 2012;15:389–398. S381–382. doi: 10.1038/nn.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter V, Mukherjee J, Maric HM, Schindelin H, Sieghart W, Moss SJ. Gephyrin, the enigmatic organizer at GABAergic synapses. Front Cell Neurosci. 2012;6:23. doi: 10.3389/fncel.2012.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagarajan SK, Fritschy JM. Gephyrin: a master regulator of neuronal function? Nat Rev Neurosci. 2014;15:141–156. doi: 10.1038/nrn3670. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Jamain S, Brose N. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J Cell Biol. 2004;83:449–456. doi: 10.1078/0171-9335-00410. [DOI] [PubMed] [Google Scholar]
- Waites CL, Craig AM, Garner CC. Mechanisms of vertebrate synaptogenesis. Annu Rev Neurosci. 2005;28:251–274. doi: 10.1146/annurev.neuro.27.070203.144336. [DOI] [PubMed] [Google Scholar]
- Yamasaki T, Hoyos-Ramirez E, Martenson JS, Morimoto-Tomita M, Tomita S. GARLH Family Proteins Stabilize GABAA Receptors at Synapses. Neuron. 2017;93:1138–1152. e1136. doi: 10.1016/j.neuron.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim YS, Kwon Y, Nam J, Yoon HI, Lee K, Kim DG, Kim E, Kim CH, Ko J. Slitrks control excitatory and inhibitory synapse formation with LAR receptor protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 2013;110:4057–4062. doi: 10.1073/pnas.1209881110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Yasumura M, Uemura T, Lee SJ, Ra M, Taguchi R, Iwakura Y, Mishina M. IL-1 receptor accessory protein-like 1 associated with mental retardation and autism mediates synapse formation by trans-synaptic interaction with protein tyrosine phosphatase delta. J Neurosci. 2011;31:13485–13499. doi: 10.1523/JNEUROSCI.2136-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chen LY, Liu X, Maxeiner S, Lee SJ, Gokce O, Sudhof TC. Neuroligins Sculpt Cerebellar Purkinje-Cell Circuits by Differential Control of Distinct Classes of Synapses. Neuron. 2015;87:781–796. doi: 10.1016/j.neuron.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.