Abstract
In bacteria and plants, serine acetyltransferase (CysE) and O-acetylserine sulfhydrylase-A sulfhydrylase (CysK) collaborate to synthesize L-Cys from L-Ser. CysE and CysK bind one another with high affinity to form the cysteine synthase complex (CSC). We demonstrate that bacterial CysE is activated when bound to CysK. CysE activation results from the release of substrate inhibition, with the Ki for L-Ser increasing from 4 mM for free CysE to 16 mM for the CSC. Feedback inhibition of CysE by L-Cys is also relieved in the bacterial CSC. These findings suggest that the CysE active site is allosterically altered by CysK to alleviate substrate and feedback inhibition in the context of the CSC.
Keywords: cysteine synthase, protein, protein interaction, serine acetyltransferase
Plants and bacteria share a common two-reaction pathway for the synthesis of L-cysteine (L-Cys) from L-serine (L-Ser; Fig. 1). Serine acetyltransferase (CysE) catalyzes an acyl transfer from acetyl-CoA to L-Ser using a random-order kinetic mechanism [1]. The second reaction is catalyzed by O-acetylserine sulfhydrylase-A (CysK), a pyridoxal 5′-phosphate (PLP)-dependent enzyme that displaces the acetoxy group from O-acetylserine with bisulfide to yield L-Cys [2–8]. Many bacteria also encode O-acetylserine sulfhydrylase-B (CysM) [9,10] that is thought to play an important role in L-Cys biosynthesis under stress conditions [11].
Fig. 1.
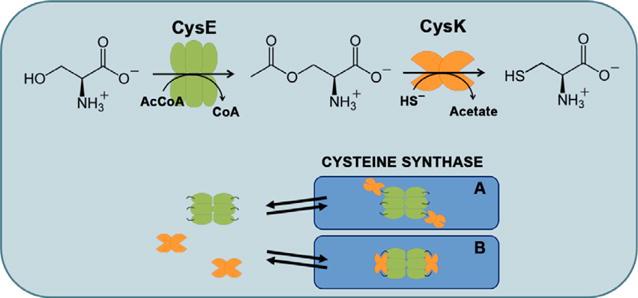
CSC and cysteine biosynthesis. The last two steps of cysteine biosynthesis are catalyzed by CysE and CysK. Bisulfide (HS) is the product of the multistep sulfate reduction pathway (not shown). CysK and CysE form the bienzymatic CSC. Because the three-dimensional structure of the complex is not known, two possible models for the complex are proposed based on previous functional studies. In model A, only one active site of each CysK dimer is occupied by the C terminus of CysE, whereas both actives sites are occupied in model B.
Kredich et al. [2,12] first discovered that CysE and CysK from Salmonella Typhimurium bind to one another with high affinity, and they called this assembly the cysteine synthase complex (CSC; Fig. 1). The CysE–CysK interaction is highly conserved across species, and the plant enzymes also form a high-affinity CSC. Although there is no experimentally solved structure available for the CSC, biochemical and spectroscopic approaches revealed that the C-terminal tail of CysE inserts into the CysK active site to anchor the interaction. CysE proteins that lack C-terminal residues are unable to bind CysK [13–15], and CSC formation is disrupted by millimolar O-acetylserine, which competes with CysE for binding to the CysK active site [12,16,17]. These findings are supported by crystal structures of CysE C-terminal peptides bound in the active site of CysK. These structures show that the C-terminal Ile residue of CysE engages in the same specific interactions with the active site as O-acetylserine substrate [18,19]. The stoichiometry of CysE to CysK has been determined to be 3:2 for CSCs from S. Typhimurium and Haemophilus influenzae. Because CysK forms homodimers and CysE exists as a dimer of trimers [20,21], the CSC is presumably composed of one CysE hexamer bound to two CysK dimers (Fig. 1).
Given the conservation of the CysE–CysK interaction from bacteria to plants, the CSC is widely thought to play important roles in L-Cys biosynthesis. Though multienzyme complexes are often exploited to transfer reaction products directly to the next enzyme in the pathway, the CSC does not mediate such substrate channeling [22]. Moreover, because the CysK active site is physically occluded by CysE, the complex actually belies its name and inefficiently produces L-Cys. Studies conducted with the plant complex suggest that the CysE–CysK interaction serves primarily to modulate substrate flux through CysE, thereby tuning the rate of L-Cys biosynthesis [23–28]. CysK binding not only promotes the activity of CysE [16,29] but also protects the enzyme from cold-inactivation and proteolytic destruction [14,29,30]. When cells are replete with sulfur, high concentrations of bisulfide stabilize the CSC likely through an allosteric anion-binding site on CysK [12,31]. Under these conditions, CysE activity is maximized and O-acetylserine can be converted into L-Cys if free CysK is available. Rising L-Cys levels exert feedback inhibition on CysE to reduce flux through the pathway. L-Cys competes with L-Ser for the CysE active site, inducing a conformational change that reduces the affinity for acetyl-CoA, thus preventing unproductive S-acetylation of L-Cys [1,20]. When sulfur is limited, O-acetylserine accumulates in the absence of bisulfide, a condition that signals sulfur starvation and quickly leads to complex dissociation. Thus, the CSC acts as a regulatory switch that allows cells to adapt L-Cys biosynthetic potential to growth conditions [26].
Despite the numerous studies of L-Cys biosynthesis in bacteria and plants, detailed kinetic analyses of the bacterial CSC have not been reported in the literature. Available data for the bacterial complex have been calculated based on inexact [32] or unspecified [12,29] kinetic mechanisms. As a consequence, the resulting kinetic parameters are either incomplete (i.e., lacking Kd for substrates in addition to KM) or not comparable to the established random-order reaction mechanism. Moreover, the activation of bacterial CysE has not been reported, and the regulatory role of the CSC in bacteria remains an open question. This study was undertaken to gain insight into the function and possible regulatory role of the CSC in bacteria. Here, we determine KM, Kd, and kcat for the L-Ser acetyltransferase reaction catalyzed by Escherichia coli CysE, both as an isolated enzyme and in complex with E. coli CysK. We find that the catalytic mechanism and kinetic parameters are unchanged between isolated CysE and the CSC. However, CysE within the CSC exhibited an apparent activation at high L-Ser concentrations. This effect is the result of a four fold increase in the L-Ser inhibition constant when CysE is in complex with CysK. In addition, feedback inhibition is relieved, with the IC50 for L-Cys increasing from 180 to 700 nM when CysE is within the complex. Together, these results suggest that CysK induces an allosteric change in CysE to regulate L-Ser acetyltransferase activity in the CSC.
Materials and methods
Reagents
Chemicals, purchased from Sigma-Aldrich (St. Louis, MO, USA), were of the best available quality and were used as received. Ninhydrin was purchased from Apollo Scientific (Stockport, UK) and acetyl-CoA from Applichem (Darmstadt, Germany). Experiments, if not otherwise indicated, were carried out in buffer A containing 20 mM sodium phosphate, 85 mM NaCl, 1 mM EDTA, and pH 7.
Protein expression and purification
CysK from E. coli [33] and CysM from S. Typhimurium [10] were expressed recombinantly in E. coli BL21(DE3) and purified by ion metal affinity chromatography (IMAC) on immobilized Co2+ ions (Talon Technology, Clontech Laboratories, Inc., Mountain View, CA, USA), following [34] with minor modifications. His-tag was removed from StCysM by incubation at 37 °C using Factor Xa in a 1:200 ratio with protein in 20 mM Hepes, 100 mM NaCl, and 4 mM CaCl2, pH 7.5. Protein purity was assessed by SDS/PAGE and shown to be higher than 95%. Protein concentration was determined by the extinction coefficient of the bound PLP, calculated by the alkali denaturation method [35]. Extinction coefficients are 9370 M−1·cm−1 at 412 nm for CysK and 6800 M−1·cm−1 at 414 nm for CysM. StCysM was used in place of EcCysM due to high sequence identity and considering that StCysK forms with EcCysE a complex which is functionally and structurally indistinguishable from the wild-type one (Fig. S1).
The CysE expression protocol was optimized to allow the preparation of a highly homogenous enzyme through a two-step chromatographic procedure. About 1% glucose was added to the induction culture to promote smooth induction conditions and hinder cysteine operon induction by OAS accumulation. The addition of 10 mM OAS to the washing buffer promotes the dissociation of CSC and the complete removal of endogenous CysK (Fig. S2). Briefly, His6-thioredoxin-tagged CysE from E. coli was expressed in BL21(DE3) Tuner™ cells (Novagen, Merck Biosciences, Billerica, MA, USA) with 1 mM IPTG induction. About 1% glucose was added to the starter culture and 1% to the induced culture. Cells were disrupted by sonication and the crude extract loaded on a FPLC column packed with Talon resin. After loading, the column was washed with a buffer containing 50 mM imidazole and 10 mM OAS. The protein was eluted with 1 M imidazole and dialyzed against 20 mM Tris-HCl, 50 mM NaCl, 1% glycerol, 1 mM DTT, 1 mM EDTA, and pH 7.5. His6-thioredoxin-tagged CysE solutions were incubated with an in-house expressed and purified His-tagged TEV protease, at 25 °C, for 4 h. Cleaved thioredoxin and TEV protease were removed with an IMAC column. The CysE preparation (80% pure) was loaded onto a FPLC column packed with Ultrogel AcA44 resin (exclusion limit 200 kDa, operating range 17–175 kDa, column volume 63 mL, and void volume 20.4 mL) and run at 0.2 mL min−1 in buffer A. CysE eluted at 28 mL, well separated from high-molecular weight contaminants, with an apparent mass of 167 200 Da, indicating the expected hexameric quaternary structure. The preparation was > 95% pure. Protein concentration was calculated using an extinction coefficient at 280 nm of 27 055 M−1·cm−1
Activity assays
CysE steady-state kinetics was measured by an adaptation of a published method [36] in buffer A at 20 °C. Briefly, L-Ser acetylation was followed in a solution containing 7 nM CysE and varying concentrations of L-Ser and acetyl-CoA by measuring the absorption at 232 nm of the thioester bond (Δε232 = 4440 M−1·cm−1). Dependence of v0 on protein concentration is linear within the 3.5–60 nM range (Fig. S3). The dependence of v0 on acetyl-CoA concentration at saturating L-Ser concentration was fitted to the Michaelis–Menten equation to calculate the apparent KM and kcat. The dependence of v0 on L-Ser concentration keeping the concentration of acetyl-CoA at 0.25 mM was fitted to Eqn (1) that takes into account substrate inhibition:
| (1) |
where Vmax is the reaction rate at saturating L-Ser concentration, KM,Ser is the apparent KM for L-Ser and Ki,Ser is the inhibition constant for L-Ser. The dependence of v0 on the concentration of both substrates was fitted to Eqn (2) for a random-order kinetic mechanism:
| (2) |
where α = KM/Kd for either L-Ser or acetyl-CoA, and Kd,Ser and Kd,AcCoA are the dissociation constants from the unligated enzyme of L-Ser and acetyl-CoA, respectively.
The IC50 for cysteine inhibition was calculated from the dependence of v0 on cysteine concentration at 0.25 mM acetyl-CoA and different concentrations of L-Ser. Data were fitted to the Eqn (3):
| (3) |
CysK steady-state kinetics were measured by a discontinuous method that exploits the quantification of cysteine following the method by Gaitonde [37] adapted to a 96-well plate format. Briefly, the sulfhydrylase reaction was initiated by the addition of 0.6 mM Na2S to a solution containing 6 nM EcCysK, 60 nM BSA, and 2 mM OAS in buffer A. Aliquots of 60 μL were taken at time intervals (about 10 time-points for each kinetics) and the reaction stopped in PCR tubes strips containing 60 μL of acetic acid. Sixty microliters of ninhydrin reagent [37] was added with a multichannel pipette and the mixture was heated at 100 °C for 10 min in a thermal cycler. The solution was cooled and 46 μL was added to the wells of a 96-well plate containing 154 μL of cold ethanol. The absorbance of the solutions at 550 nm was measured by a plate reader (Halo LED 96; Dynamica Scientific, Newport Pagnell, UK). Blanks were subtracted and kinetic data were collected at least in duplicate. The amount of L-Cys produced at each time point was calculated from a calibration curve and data were fitted to a linear equation to calculate the initial rate of cysteine production. The dependence of the initial velocity on CysE concentration was fitted to a modified Morrison’s Eqn (3) to calculate the for tight-binding inhibitors [38]:
| (4) |
where [E]T is the total enzyme concentration, [I]T is the total CysE concentration, and y0 is a vertical offset that takes into consideration the partial inhibition of CysK by CysE. For competitive inhibitors in a ping-pong reaction [38–40]:
| (5) |
Fluorescence spectroscopy
Fluorescence measurements were carried out using a FluoroMax-3 fluorometer (HORIBA, Kyoto, Japan), equipped with a thermostated cell-holder. Emission spectra of a solution containing a given concentration of CysK upon excitation at 412 nm were collected between 425 nm and 650 nm at different CysE concentrations. Fluorescence spectra were corrected for the buffer contribution. The emission intensity at 500 nm was plotted as a function of CysE concentration to calculate either the stoichiometric ratio or the dissociation constant for complex formation in the presence of 10 μM L-Cys. In the latter case, the dependence was fitted to a quadratic equation that describes tight binding:
| (6) |
where I is the fluorescence intensity at 500 nm in the presence of CysE, I0 is an horizontal offset, ΔI is the maximum fluorescence change at saturating [L], [L] is the total ligand concentration, [P] is the total protein concentration, and Kd is the dissociation constant of CSC.
Size-exclusion chromatography
The apparent molecular weights of CysK, CysE, and CSC were evaluated by size-exclusion chromatography. About 10 μL of protein (26 μM CysK, 39 μM CysE, or a mixture containing both proteins) was loaded onto a Superdex 200 increase 3.2/300 column (GE Life Sciences, Little Chalfont, UK) mounted on a Prominence HPLC system (Shimadzu, Kyoto, Japan). The column was equilibrated and developed with buffer A. The flow rate was 0.1 mL·min−1. The experiments were carried out at room temperature. The column was calibrated with gel-filtration standards carbonic anhydrase (29 kDa), ovalbumin (45 kDa), conalbumin (75 kDa), glyceraldehyde-3-phosphate dehydrogenase (144 kDa), and ferritin (440 kDa). Blue dextran was used for the determination of the void volume.
Results
CysE quaternary structure and cysteine synthase complex formation
We first used size-exclusion chromatography to monitor CSC formation. Purified CysE elutes at an elution volume corresponding to a molecular mass of 181 kDa (Fig. 2A), in good agreement with the predicted molecular mass of 174 kDa for the hexameric form. The elution profile of isolated CysK corresponds to a mass of 77 kDa (Fig. 2A), consistent with 71 kDa predicted for the homodimer. We then analyzed an equimolar mixture of CysE and CysK to monitor complex formation. The resulting complex eluted with an estimated molecular mass of 476 kDa, which is substantially greater than the expected mass of 336 kDa for the CSC. To determine the stoichiometry of the complex, we performed titrations to monitor changes in the fluorescent emission from the PLP cofactor of CysK as a function of CysE concentration [10,13] (Fig. 2B). CysK is completely saturated by CysE at a CysE:CysK ratio of approximately 1.6:1, indicating that one CysE hexamer binds to two CysK dimers, as has been previously determined for the CSC from H. influenzae [13].
Fig. 2.
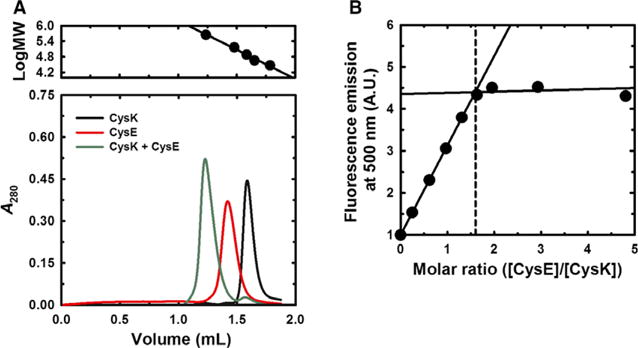
Quaternary structure of CysE and CSC stoichiometry. Panel (A) Size-exclusion chromatography of CysE, CysK, and the CSC. CysK (26 μM), CysE (39 μM), and a 1:1 molar mixture were resolved on a Superdex 200 increase 3.2/300 column. The upper panel shows the molecular mass calibration using carbonic anhydrase (29 kDa), ovalbumin (45 kDa), conalbumin (75 kDa), glyceraldehyde-3-phosphate dehydrogenase (144 kDa), and ferritin (440 kDa). Panel (B) Stoichiometry of the CSC. CysK (1 μM) was titrated with increasing concentrations of CysE and complex formation monitored by measuring the fluorescence emission of PLP at 500 nm. The dashed line indicates the intersection between the lines and corresponds to a stoichiometric ratio of 1.6.
CysE is a partial inhibitor of CysK
Because the C terminus of CysE inserts into the CysK active site, the latter enzyme is inhibited in the context of the CSC [12,32]. Thus, the dissociation constant for the CSC can be estimated from the dependence of CysK sulfhydrylase activity on CysE concentration. The initial reaction velocity (vi) decreased with increasing CysE concentrations, plateauing at about 10% of the activity for free CysK (Fig. 3). This residual activity is still present at a 100-fold molar excess of CysE over CysK, where CysK is expected to be entirely in complex with CysE. Equation (4) was used to calculate the , from which we obtained an inhibition constant of 6.2 ± 0.7 nM for the CSC using Eqn (5) [38,40]. This value is in excellent agreement with the dissociation constant calculated by measuring CysE activity (vide infra) as well as previously published data [32,41,42]. Notably, a vertical off-set was added to Morrison’s equation to account for the residual activity at 100-fold molar excess of CysE over (compare solid and dashed lines in Fig. 3).
Fig. 3.
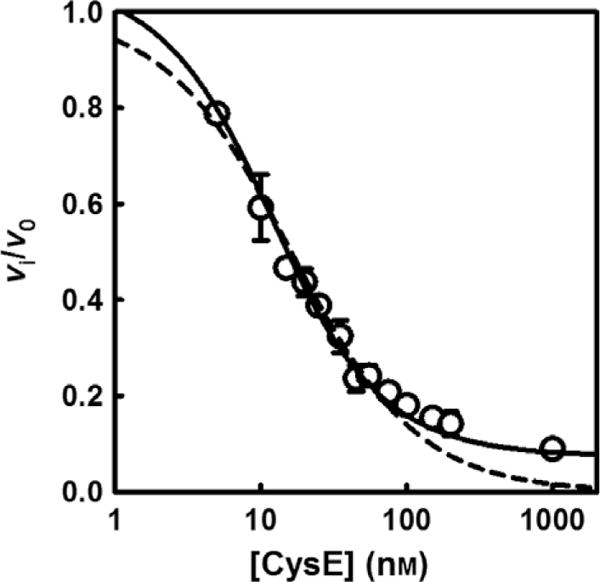
Inhibition of CysK activity by CysE. CysK (6 nM) sulfhydrylase activity was measured in buffer A containing 2 mM O-acetylserine and 0.6 mM Na2S at 20 °C. The initial reaction velocity was determined as a function of increasing CysE concentration. Fitting Eqn (4) to the dependence of vi/v0 on CysE concentration gives of 8.7 ± 1.0 nM, that is transformed with Eqn (5) to yield a Ki of 6.2 ± 0.7 nM. The standard Morrison’s equation (dashed line) fails to fit the points at high CysE concentrations as they do not reach zero relative activity.
CysK activates CysE in a concentration- and isoform-specific manner
The initial reaction velocity of CysE was measured as a function of CysK concentration, within a stoichiometric ratio range of 0.1–3.3 (Fig. 4). The concentration of L-Ser used in the assay was saturating, whereas the concentration of acetyl-CoA (0.25 mM) was very close to the physiological intracellular concentration in E. coli [43]. The initial velocity increases as a function of CysK concentration, up to fourfold at saturation. Fitting Eqn (6) to this dependence yields, albeit with some large fitting uncertainty, a dissociation constant of 4.5 nM for the CSC. Again, this value is consistent with those reported in the literature [32,41,42] and agrees with the dissociation constant obtained from the sulfhydrylase inhibition assays described above. The specificity of the effect, which results from CSC formation, is supported by the lack of CysE activation upon titration with the CysM (Fig. 4), which does not interact with CysE [14,41,44].
Fig. 4.
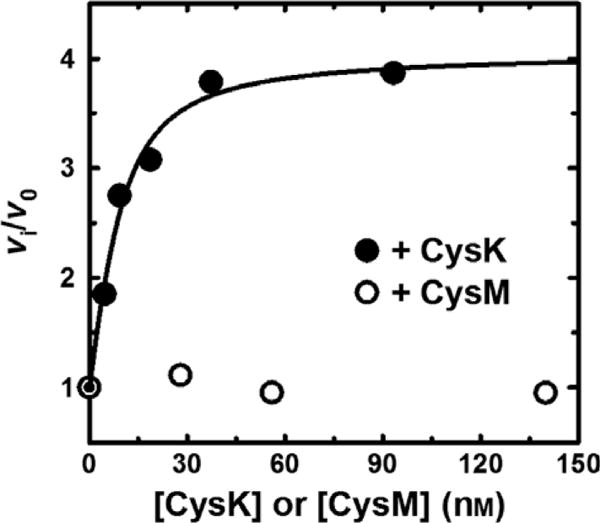
CysK binding promotes CysE activity. CysE (28 nM) L-Ser acetylation activity was determined in buffer A containing 20 mM L-Ser, 0.25 mM acetyl-CoA at 20 °C. Where indicated, reactions were supplemented with either CysK or CysM and incubated with L-Ser for 5 min prior to the addition of acetyl-CoA. The line represents the fit of Eqn (4) to the data.
Effect of CysK on CysE apparent kinetic parameters
CysE catalyzes a bisubstrate reaction described as random-order ternary complex [45]. We examined the effect of CysK (fivefold molar excess) on the kinetic parameters of CysE by varying the concentration of one substrate while keeping the concentration of the other substrate constant. When the acetyl-CoA concentration was varied, a 2.5-fold increase in the apparent catalytic efficiency (kcat/KM) was observed (Fig. 5, Table 1). On the other hand, when L-Ser concentration was varied, a smaller 1.7-fold increase in catalytic efficiency was measured, with a significant increase, from 3.7 ± 1.4 to 16 ± 5 mM, of the apparent Ki for L-Ser.
Fig. 5.
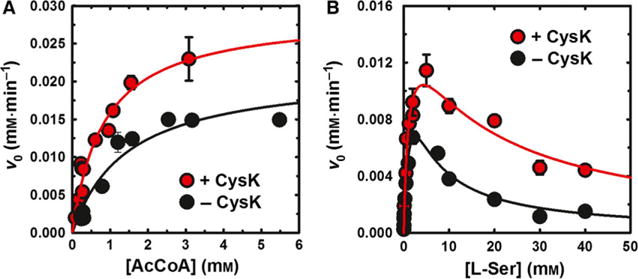
Effect of CysK binding on the apparent kinetic constants of CysE-catalyzed L-Ser acetylation. The dependence of L-Ser acetylation reaction velocity (v0) on the concentration of acetyl-CoA (Panel A) and L-Ser (Panel B) was determined in the presence and absence of fivefold molar excess CysK. L-Ser concentration was held constant at 20 mM (Panel A) and acetyl-CoA was held constant at 0.25 mM (Panel B). Lines through data points represent the Michaelis–Menten fit (Panel A), modified to account for substrate inhibition (Eqn 1, Panel B). All parameters are presented in Table 1.
Table 1.
Apparent kinetic constants for the reaction catalyzed by CysE in the absence and presence of CysK at fivefold molar excess over CysE. When acetyl-CoA was the varied substrate the concentration of L-Ser was 20 mM. When L-Ser was the varied substrate, the concentration of acetyl-CoA was 0.25 mM. The concentration of CysE for the calculation of kcat is based on the hexameric complex.
| Varied substrate | Kinetic constants | −CysK | +CysK |
|---|---|---|---|
| Acetyl-CoA | KM (mM) | 1.4 ± 0.4 | 0.8 ± 0.2 |
| kcat (s−1) | 306 ± 35 | 416 ± 38 | |
| kcat/KM (mM−1·s−1) | 212 ± 83 | 501 ± 148 | |
| L-Ser | KM (mM) | 1.7 ± 0.7 | 1.1 ± 0.4 |
| kcat (s−1) | 228 ± 60 | 228 ± 34 | |
| kcat/KM (mM−1·s−1) | 134 ± 90 | 207 ± 106 | |
| Ki,L-Ser (mM) | 3.7 ± 1.4 | 16 ± 5 |
Cysteine synthase complex formation does not affect the kinetic mechanism or the kinetic parameters of CysE
The kinetic parameters for L-Ser acetylation, mediated by free CysE and the CSC, were calculated from the dependences of v0 on the concentrations of acetyl-CoA and L-Ser (Fig. 6). In the absence of CysK, the intersection in the reciprocal plot lies to the left of y-axis and below the x-axis (Fig. S4A), which is the signature of an ordered mechanism. Global fitting of Eqn (2) to these data gives the parameters reported in Table 2, with values in agreement with those reported by Hindson and Shaw [45]. In particular, α > 2 indicates a strongly negative substrate-binding synergism, in which the affinity for the second substrate decreases after the first substrate is bound. Interestingly, in the presence of fivefold excess CysK, the kinetic parameters do not change with respect to reference conditions (Fig. 6B, Table 2), as also shown in the double reciprocal plot (Fig. S4B).
Fig. 6.
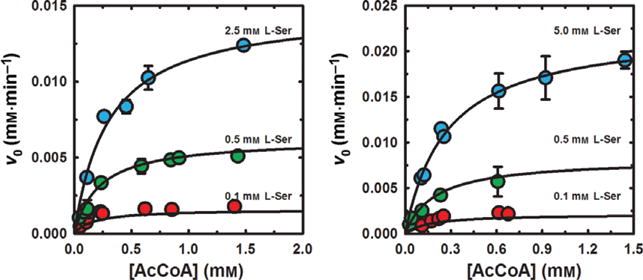
Dependence of CysE and CSC mediated L-Ser acetylation reaction velocity on substrate concentration. Panel (A) Initial L-Ser acetylation reaction velocities (v0) were determined for 7 nM CysE in the presence of varying concentrations of L-Ser. Panel (B) Initial L-Ser acetylation reaction velocities (v0) were determined for the CSC (7 nM CysE and 23 nM CysK) in the presence of varying concentrations of L-Ser. Fitting parameters obtained with Eqn (2) are presented in Table 2.
Table 2.
Kinetic parameters for the reaction catalyzed by CysE in the absence and presence of CysK at fivefold molar excess over CysE. CysE concentration for the calculation of kcat is based on the hexameric complex.
| Parameter | −CysK | +CysK |
|---|---|---|
| KM,L-Ser (mM) | 1.3 ± 0.2 | 1.2 ± 0.2 |
| Kd,L-Ser (mM) | 0.5 ± 0.2 | 0.6 ± 0.2 |
| KM,acCoA (mM) | 0.3 ± 0.1 | 0.3 ± 0.1 |
| Kd,acCoa (mM) | 0.1 ± 0.1 | 0.1 ± 0.1 |
| kcat (s−1) | 323 ± 33 | 405 ± 25 |
| α | 2.3 ± 1.5 | 2.0 ± 1.2 |
Effect of complex formation on the IC50 for cysteine
CysE activity is subject to feedback inhibition by the product L-Cys. We monitored feedback inhibition of CysE activity at constant substrate concentrations (1 mM L-Ser and 0.25 mM acetyl-CoA) in the presence and absence of a molar excess of CysK (Fig. 7). During preliminary experiments, we observed that 10 μM L-Cys destabilized the CSC, increasing the dissociation constant from 6.2 to 66 nM (Fig. S5A). Therefore, we used a larger molar excess of CysK to determine the IC50 for L-Cys. The IC50 in the presence of CysK increases by fourfold, indicating that CysE is less sensitive to inhibition by L-Cys in the context of the CSC. Similar effects were observed at L-Ser levels (0.1 mM) that mimic the physiological concentration in bacteria [46], as well as at higher concentrations (20 mM) that represent a large excess with respect to the KM (Fig. S5B).
Fig. 7.
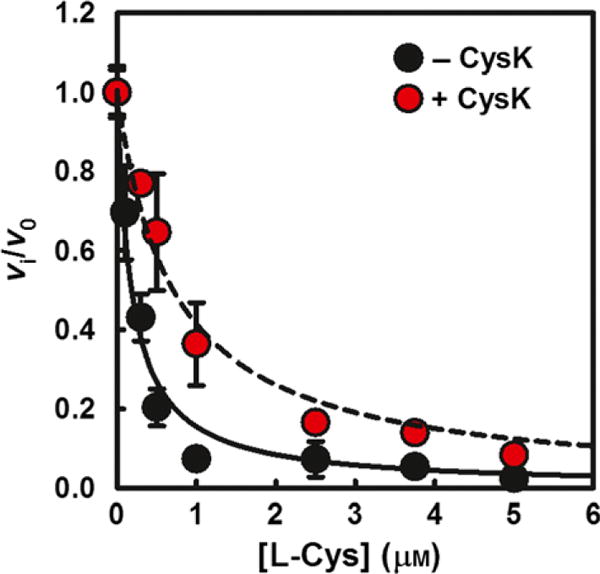
Feedback inhibition is alleviated in the CSC. Dependence of the relative rate of L-Ser acetylation by CysE on the concentration of L-Cys, in the absence and presence of a 100-fold molar excess CysK. The lines represent the fit of Eqn (3) to the data. The IC50 = 0.18 ± 0.02 μM for isolated CysE and 0.70 ± 0.07 μM for the CSC.
Discussion
Reductive sulfate assimilation and L-Cys biosynthesis have gained renewed interest as these pathways have recently been shown to play important roles in bacterial biofilm formation and virulence. E. coli cysE null mutants form biofilms more rapidly than cysE+ cells [47], and cysB mutants also show significant increases in biofilm mass compared to wild-type [48]. By contrast, mutations in the L-Cys synthetic pathway interfere with biofilm formation in Vibrio fischeri [49]. L-Cys metabolism has also been linked to antibiotic resistance in S. Typhimurium [50,51] and is actively being explored as a target for novel antimicrobial therapies and the development of antibiotic enhancers [31,52–57]. This approach is beginning to produce some encouraging results, particularly in the case of Mycobacterium tuberculosis [58,59]. Intriguingly, the CysK–CysE interaction is commonly exploited by other proteins to promote so-called ‘moonlighting’ activities of CysK [60]. For example, the contact-dependent growth inhibition (CDI) toxin from uro-pathogenic E. coli forms a high-affinity complex with CysK, and this interaction is required for the toxin’s nuclease activity [33,61]. Remarkably, the C terminus of the CDI toxin mimics CysE and inserts into the CysK active site [33,62]. Similar moonlighting interactions have been described between CymR and CysK in Bacillus subtilis [63] and between EGL-9 and the CysK paralog CYSL-1 in Caenorhabditis elegans [64]. These latter interactions are modulated to regulate transcription. The CymR–CysK complex binds DNA and represses the cys regulon under sulfur replete conditions. Under sulfur starvation, accumulating O-acetylserine dissociates the complex to derepress genes needed for sulfur assimilation [63,65]. Caenorhabditis elegans EGL-9 indirectly down-regulates the transcription of hypoxia-induced genes by hydroxylating the HIF transcription factor. This repressive effect is relieved under hypoxic conditions, which cause bisulfide to accumulate in the cell. As with the CSC, bisulfide stabilizes the EGL-9–CYSL-1 interaction, thereby inhibiting hydroxylase activity and promoting HIF-dependent transcription [64]. Thus, CysK and its paralogs are commonly co-opted to regulate other processes in both prokaryotic and eukaryotic cells [60].
The correct in vitro assembly of multiprotein complexes is a crucial requirement for the collection of meaningful functional data. In the case of CSC, the calculation of stoichiometry is even more crucial, taking into account that the complex is built up by proteins that are themselves oligomers. Structural studies show that bacterial CysE forms dimers of trimers [20,21,66,67], which is quite unusual for acyltransferases. This finding prompted Hindson et al. [67] to suggest an evolutionary transition from ancestral trimeric acyltransferases to a more stable ‘stacked trimer’ form. This transition might account for the regulatory properties of the complex on the activities of the component enzymes [67]. Because there are relatively few studies on bacterial CysE enzymes, we evaluated the quaternary structure of the E. coli enzyme to ascertain whether hexamer-to-trimer equilibria affect the functional properties. Our data indicate that E. coli CysE is primarily hexameric in solution, with an estimated molecular mass of 181 kDa as determined by size-exclusion chromatography. However, the E. coli CysE–CysK complex elutes as a sharp peak corresponding to an apparent molecular weight of around 475 kDa, which is significantly larger than the predicted mass of 336 kDa. This discrepancy could reflect the predicted elongated shape of the complex, as anticipated based on molecular docking [68,69] and binding experiments [13,17,70]. Indeed, the stoichiometry calculated from fluorimetric titrations is in excellent agreement with the predicted assembly of one CysE hexamer bound to two CysK dimers (Fig. 2). This stoichiometry is compatible with two different interaction models as previously discussed for the H. influenzae CSC [13]. In the first model, all four CysK active sites are occupied with C-terminal tails from the CysE hexamer. An alternative and more widely accepted model proposes that only one active site per CysK dimer is occupied in CSC [17,29,68] (Fig. 1). However, the unoccupied active site appears to be unavailable for further binding of CysE or CysE C-terminal peptide [13,17]. The effect that we observed on CysK activity upon complex formation is expected based on structural and functional data [13,32], i.e., a concentration-dependent inhibition that can be fitted to the equation for tight binding to calculate the dissociation constant for complex formation, about 6 nM. Interestingly, even at saturating CysE concentrations, we found that about 10% of CysK activity is retained, consistent with previous observations [32]. Because incomplete enzyme inhibition is usually associated with the binding of negative allosteric modulators, it seems likely that the residual activity is due to an unoccupied CysK active site within CSC, rather than true partial inhibition. This finding is also in agreement with elegant protein dynamic work performed on the Arabidopsis thaliana complex [69]. Based on previous work where the structure of the CSC was modeled with CysK having one active site bound to CysE and the other site unoccupied, the authors demonstrated the allosteric closure of the unoccupied CysK active site. We note that the physiological significance of this latter finding is unclear because bacterial CysK is thought to be in excess over CysE, especially under sulfur limiting conditions [71], similar to what has been observed in plants [24,72]. Nevertheless, if instances exist where CysK is present at comparable levels to CysE, this residual activity would allow for L-Cys production without assistance from the CysM isozyme. Indeed, recent data indicate that CysK and CysE transcript abundances are comparable in S. Typhimurium grown under a variety of conditions including exponential growth, bile shock, cold shock, and oxidative stress [73]. These observations suggest that the relative concentrations of CysK and CysE could be modulated under certain growth conditions.
Interestingly, CysE is a member of a relatively small group of enzymes that show a KM well above the physiological concentration of its substrate [46]. The KM for L-Ser is 1.3 mM, but the cytosolic concentration of the amino acid is about 68 μM for E. coli cells grown with glucose as a carbon source and 150 μM when glycerol is used as the carbon source [46]. The most surprising finding of this study was the observation that CysE is activated by CysK, which is in contrast to previous studies [12,32]. However, the observed activation is only apparent, as indicated by the perfect overlap of microscopic kinetic constants in the absence and presence of excess CysK. The activation results from an increase in the L-Ser substrate inhibition constant from 3.7 to 16 mM. This effect is of uncertain physiological significance, given that in vivo L-Ser concentrations are typically in the micromolar range. To the best of our knowledge, the activation parameters on plant enzymes were determined based on apparent KM and kcat values, i.e., using constant, and possibly saturating, concentrations of one substrate to calculate kinetic constants for the varying ligand [16,29,74,75]. In the absence of a full kinetic characterization of the isolated plant CysE compared to the CSC, it is possible that the observed activation for the plant complex is also apparent and due to the relief from substrate inhibition. Feedback inhibition of CysE by L-Cys plays a pivotal role in the control of sulfur assimilation both in bacteria and plants [74,76,77]. However, bacterial CysE is known to be more sensitive to L-Cys inhibition than its plant counterpart [29,74]. In addition, plant CysE has been reported to be less sensitive to L-Cys inhibition when in the CSC, with a fourfold increase in the IC50 [74] and more than 35-fold increase for Ki [29]. In contrast to what has been reported previously, we observed a fourfold increase in the L-Cys IC50 upon formation of the bacterial CSC, which is comparable to data reported for the cytosolic isoform of A. thaliana enzyme [74]. We also measured a 10-fold increase in the dissociation constant of the CSC in the presence of 10 μM L-Cys, in good agreement with previous pre-steady-state studies where a decrease in the efficiency of complex formation was measured for the H. influenzae enzymes [70]. We believe that this L-Cys-dependent effect on the complex affinity might explain the discrepancy between the present and published data with respect to the effect of complex formation on the susceptibility of CysE to L-Cys inhibition. In fact, we needed to increase the concentration of CysK in the activity assays to achieve measurable effects, due to the interference of L-Cys with complex formation. Indeed, the C terminus of CysE is engaged in both CSC formation and intrasteric inhibition in the presence of L-Cys [20], and an effect of this ligand on the affinity of the complex was expected. This result is also in line with experimental evidence that deletion of the last 10 C-terminal residues of CysE leads to a relevant decrease in the sensitivity to cysteine inhibition [74,78,79]. The engagement of the CysE C terminus with CysK is thus responsible for the allosteric modulation of the L-Cys-/L-Ser-binding site, which is functionally reflected in a less effective feedback/substrate inhibition. We anticipate that this investigation will fuel further studies aimed at the characterization of the mechanism of complex formation and its regulation within bacterial cells. A better understanding of how complex formation is regulated under different conditions will also enable the development of synthetic CysK inhibitors. Indeed, inhibition of CysK, an enzyme that is absent in mammals and could be exploited as a target for innovative antibiotics/antibiotic enhancers, could lead to complex dissociation whose final effects on cysteine biosynthesis are, at the moment, difficult to predict. For instance, the BB1 mutant strain of S. Typhimurium, characterized by an altered interaction between CysE and CysK, is a cysteine auxotroph [80,81].
Supplementary Material
Fig. S1. Heterospecific CSC formation measured by the dependence of v0 for L-Ser acetylation on CysK concentration.
Fig. S2. SDS/PAGE of > 95% pure, untagged CysE.
Fig. S3. Dependence of v0 for L-Ser acetylation on CysE concentration.
Fig. S4. Double reciprocal plots of the dependence of v0 on acCoA and L-Ser concentration in the absence (panel A) and presence (panel B) of fivefold molar excess CysK.
Fig. S5. Panel (A) Increase in fluorescence emission intensity at 500 nm upon excitation at 412 nm of a 80 nM CysK solution as a function of CysE concentration in the presence of 10 μM L-Cys. Fitting with Eqn (6) gives a Kd of 66 ± 20 nM. Panel (B) Effect of physiological (0.1 mM) and saturating (20 mM) L-Ser concentrations on the inhibition by L-Cys of CysE activity in the absence and presence of a 100-fold molar excess CysK. The concentration of L-Cys used varies in the two conditions and is around the IC50, i.e., 0.07 μM in the presence of 0.1 mM L-Ser and 0.5 μM in the presence of 20 mM L-Ser. The 1 mM L-Ser condition is reported for comparison using data of Fig. 7.
Acknowledgments
The authors gratefully acknowledge Paul F. Cook, University of Oklahoma, for the stimulating discussions that inspired this paper. The work described in this paper was partly carried out under the MSCA-ITN-2014-ETN project INTEGRATE (grant number 642620) and was partly supported by grants from the University of Parma (prot. FIL2014).
Abbreviations
- CDI
contact-dependent growth inhibition
- CSC
cysteine synthase complex
- CysE
serine acetyltransferase
- CysK
O-acetylserine sulfhydrylase-A
- CysM
O-acetylserine sulfhydrylase-B
- IMAC
ion metal affinity chromatography
- PLP
pyridoxal 5′-phosphate
Footnotes
Edited by Stuart Ferguson
Author contributions
BC, SB, and AM conceived and supervised the study; RB, ODB, GP, and NF performed experiments; CSH provided the expression vectors; BC, RB, and ODB analyzed the data; BC prepared the original draft; BC, SB, CSH, and AM reviewed and edited the manuscript.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- 1.Hindson VJ. Serine acetyltransferase of Escherichia coli: substrate specificity and feedback control by cysteine. Biochem J. 2003;375:745–752. doi: 10.1042/BJ20030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kredich NM, Tomkins GM. The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem. 1966;241:4955–4965. [PubMed] [Google Scholar]
- 3.Becker MA, Kredich NM, Tomkins GM. The purification and characterization of O-acetylserine sulfhydrylase-A from Salmonella typhimurium. J Biol Chem. 1969;244:2418–2427. [PubMed] [Google Scholar]
- 4.Cook PF, Wedding RT. A reaction mechanism from steady state kinetic studies for O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2. J Biol Chem. 1976;251:2023–2029. [PubMed] [Google Scholar]
- 5.Cook PF, Wedding RT. Overall mechanism and rate equation for O-acetylserine sulfhydrylase. J Biol Chem. 1977;252:3459. [PubMed] [Google Scholar]
- 6.Tai CH, Nalabolu SR, Jacobson TM, Minter DE, Cook PF. Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 using the natural and alternative reactants. Biochemistry. 1993;32:6433–6442. doi: 10.1021/bi00076a017. [DOI] [PubMed] [Google Scholar]
- 7.Mozzarelli A, Bettati S, Campanini B, Salsi E, Raboni S, Singh R, Spyrakis F, Kumar VP, Cook PF. The multifaceted pyridoxal 5′-phosphate-dependent O-acetylserine sulfhydrylase. Biochem Biophys Acta. 2011;1814:1497–1510. doi: 10.1016/j.bbapap.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Bettati S, Benci S, Campanini B, Raboni S, Chirico G, Beretta S, Schnackerz KD, Hazlett TL, Gratton E, Mozzarelli A. Role of pyridoxal 5′-phosphate in the structural stabilization of O-acetylserine sulfhydrylase. J Biol Chem. 2000;275:40244–40251. doi: 10.1074/jbc.M007015200. [DOI] [PubMed] [Google Scholar]
- 9.Chattopadhyay A, Meier M, Ivaninskii S, Burkhard P, Speroni F, Campanini B, Bettati S, Mozzarelli A, Rabeh WM, Li L, et al. Structure, mechanism, and conformational dynamics of O-acetylserine sulfhydrylase from Salmonella typhimurium: comparison of A and B isozymes. Biochemistry. 2007;46:8315–8330. doi: 10.1021/bi602603c. [DOI] [PubMed] [Google Scholar]
- 10.Salsi E, Guan R, Campanini B, Bettati S, Lin J, Cook PF, Mozzarelli A. Exploring O-acetylserine sulfhydrylase-B isoenzyme from Salmonella typhimurium by fluorescence spectroscopy. Arch Biochem Biophys. 2011;505:178–185. doi: 10.1016/j.abb.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Filutowicz M, Wiater A, Hulanicka D. Delayed inducibility of sulphite reductase in cysM mutants of Salmonella typhimurium under anaerobic conditions. J Gen Microbiol. 1982;128:1791–1794. doi: 10.1099/00221287-128-8-1791. [DOI] [PubMed] [Google Scholar]
- 12.Kredich NM, Becker MA, Tomkins GM. Purification and characterization of cysteine synthetase, a bifunctional protein complex, from Salmonella typhimurium. J Biol Chem. 1969;244:2428–2439. [PubMed] [Google Scholar]
- 13.Campanini B, Speroni F, Salsi E, Cook PF, Roderick SL, Huang B, Bettati S, Mozzarelli A. Interaction of serine acetyltransferase with O-acetylserine sulfhydrylase active site: evidence from fluorescence spectroscopy. Protein Sci. 2005;14:2115–2124. doi: 10.1110/ps.051492805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mino K, Hiraoka K, Imamura K, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K. Characteristics of serine acetyltransferase from Escherichia coli deleting different lengths of amino acid residues from the C-terminus. Biosci Biotechnol Biochem. 2000;64:1874–1880. doi: 10.1271/bbb.64.1874. [DOI] [PubMed] [Google Scholar]
- 15.Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K. Purification and characterization of serine acetyltransferase from Escherichia coli partially truncated at the C-terminal region. Biosci Biotechnol Biochem. 1999;63:168–179. doi: 10.1271/bbb.63.168. [DOI] [PubMed] [Google Scholar]
- 16.Droux M, Ruffet ML, Douce R, Job D. Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants–structural and kinetic properties of the free and bound enzymes. Eur J Biochem. 1998;255:235–245. doi: 10.1046/j.1432-1327.1998.2550235.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang T, Leyh TS. Three-stage assembly of the cysteine synthase complex from Escherichia coli. J Biol Chem. 2012;287:4360–4367. doi: 10.1074/jbc.M111.288423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang B, Vetting MW, Roderick SL. The active site of O-acetylserine sulfhydrylase is the anchor point for bienzyme complex formation with serine acetyltransferase. J Bacteriol. 2005;187:3201–3205. doi: 10.1128/JB.187.9.3201-3205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnell R, Oehlmann W, Singh M, Schneider G. Structural insights into catalysis and inhibition of O-acetylserine sulfhydrylase from Mycobacterium tuberculosis. Crystal structures of the enzyme alpha-aminoacrylate intermediate and an enzyme-inhibitor complex. J Biol Chem. 2007;282:23473–23481. doi: 10.1074/jbc.M703518200. [DOI] [PubMed] [Google Scholar]
- 20.Olsen LR, Huang B, Vetting MW, Roderick SL. Structure of serine acetyltransferase in complexes with CoA and its cysteine feedback inhibitor. Biochemistry. 2004;43:6013–6019. doi: 10.1021/bi0358521. [DOI] [PubMed] [Google Scholar]
- 21.Pye VE, Tingey AP, Robson RL, Moody PC. The structure and mechanism of serine acetyltransferase from Escherichia coli. J Biol Chem. 2004;279:40729–40736. doi: 10.1074/jbc.M403751200. [DOI] [PubMed] [Google Scholar]
- 22.Cook PF, Wedding RT. Initial kinetic characterization of the multienzyme complex, cysteine synthetase. Arch Biochem Biophys. 1977;178:293–302. doi: 10.1016/0003-9861(77)90194-1. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura K, Tamura G. Isolation of serine acetyltransferase complexed with cysteine synthase from Allium tuberosum. Agric Biol Chem. 1990;54:649–656. [Google Scholar]
- 24.Ruffet ML, Droux M, Douce R. Purification and kinetic properties of serine acetyltransferase free of O-acetylserine(thiol)lyase from spinach chloroplasts. Plant Physiol. 1994;104:597–604. doi: 10.1104/pp.104.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito K, Yokoyama H, Noji M, Murakoshi I. Molecular cloning and characterization of a plant serine acetyltransferase playing a regulatory role in cysteine biosynthesis from watermelon. J Biol Chem. 1995;270:16321–16326. doi: 10.1074/jbc.270.27.16321. [DOI] [PubMed] [Google Scholar]
- 26.Hell R, Hillebrand H. Plant concepts for mineral acquisition and allocation. Curr Opin Biotechnol. 2001;12:161–168. doi: 10.1016/s0958-1669(00)00193-2. [DOI] [PubMed] [Google Scholar]
- 27.Wirtz M, Droux M, Hell R. O-acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot. 2004;55:1785–1798. doi: 10.1093/jxb/erh201. [DOI] [PubMed] [Google Scholar]
- 28.Wirtz M, Hell R. Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. J Plant Physiol. 2006;163:273–286. doi: 10.1016/j.jplph.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Kumaran S, Yi H, Krishnan HB, Jez JM. Assembly of the cysteine synthase complex and the regulatory role of protein-protein interactions. J Biol Chem. 2009;284:10268–10275. doi: 10.1074/jbc.M900154200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mino K, Imamura K, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K. Increase in the stability of serine acetyltransferase from Escherichia coli against cold inactivation and proteolysis by forming a bienzyme complex. Biosci Biotechnol Biochem. 2001;65:865–874. doi: 10.1271/bbb.65.865. [DOI] [PubMed] [Google Scholar]
- 31.Campanini B, Pieroni M, Raboni S, Bettati S, Benoni R, Pecchini C, Costantino G, Mozzarelli A. Inhibitors of the sulfur assimilation pathway in bacterial pathogens as enhancers of antibiotic therapy. Curr Med Chem. 2015;22:187–213. doi: 10.2174/0929867321666141112122553. [DOI] [PubMed] [Google Scholar]
- 32.Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K. Effects of bienzyme complex formation of cysteine synthetase from Escherichia coli on some properties and kinetics. Biosci Biotechnol Biochem. 2000;64:1628–1640. doi: 10.1271/bbb.64.1628. [DOI] [PubMed] [Google Scholar]
- 33.Diner EJ, Beck CM, Webb JS, Low DA, Hayes CS. Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI) Genes Dev. 2012;26:515–525. doi: 10.1101/gad.182345.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian H, Guan R, Salsi E, Campanini B, Bettati S, Kumar VP, Karsten WE, Mozzarelli A, Cook PF. Identification of the structural determinants for the stability of substrate and aminoacrylate external Schiff bases in O-acetylserine sulfhydrylase-A. Biochemistry. 2010;49:6093–6103. doi: 10.1021/bi100473v. [DOI] [PubMed] [Google Scholar]
- 35.Peterson EA, Sober HA. Preparation of crystalline phosphorylated derivatives of vitamin B6. J Am Chem Soc. 1954;76:169–175. [Google Scholar]
- 36.Hama H, Kayahara T, Ogawa W, Tsuda M, Tsuchiya T. Enhancement of serine-sensitivity by a gene encoding rhodanese-like protein in Escherichia coli. J Biochem. 1994;115:1135–1140. doi: 10.1093/oxfordjournals.jbchem.a124469. [DOI] [PubMed] [Google Scholar]
- 37.Gaitonde MK. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem J. 1967;104:627–633. doi: 10.1042/bj1040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Copland RA. Evaluation of Enzyme Inhibitors in Drug Discovery – A Guide for Medicinal Chemists and Pharmacologists. John Wiley and Sons; Hoboken: 2005. [PubMed] [Google Scholar]
- 39.Bevans CG, Krettler C, Reinhart C, Tran H, Kossmann K, Watzka M, Oldenburg J. Determination of the warfarin inhibition constant Ki for vitamin K 2,3-epoxide reductase complex subunit-1 (VKORC1) using an in vitro DTT-driven assay. Biochim Biophys Acta. 2013;1830:4202–4210. doi: 10.1016/j.bbagen.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 40.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 41.Berkowitz O, Wirtz M, Wolf A, Kuhlmann J, Hell R. Use of biomolecular interaction analysis to elucidate the regulatory mechanism of the cysteine synthase complex from Arabidopsis thaliana. J Biol Chem. 2002;277:30629–30634. doi: 10.1074/jbc.M111632200. [DOI] [PubMed] [Google Scholar]
- 42.Zhao C, Moriga Y, Feng B, Kumada Y, Imanaka H, Imamura K, Nakanishi K. On the interaction site of serine acetyltransferase in the cysteine synthase complex from Escherichia coli. Biochem Biophys Res Commun. 2006;341:911–916. doi: 10.1016/j.bbrc.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 43.Albe KR, Butler MH, Wright BE. Cellular concentrations of enzymes and their substrates. J Theor Biol. 1990;143:163–195. doi: 10.1016/s0022-5193(05)80266-8. [DOI] [PubMed] [Google Scholar]
- 44.Zhao C, Kumada Y, Imanaka H, Imamura K, Nakanishi K. Cloning, overexpression, purification, and characterization of O-acetylserine sulfhydrylase-B from Escherichia coli. Protein Expr Purif. 2006;47:607–613. doi: 10.1016/j.pep.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Hindson VJ, Shaw WV. Random-order ternary complex reaction mechanism of serine acetyltransferase from Escherichia coli. Biochemistry. 2003;42:3113–3119. doi: 10.1021/bi0267893. [DOI] [PubMed] [Google Scholar]
- 46.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sturgill G, Toutain CM, Komperda J, O’Toole GA, Rather PN. Role of CysE in production of an extracellular signaling molecule in Providencia stuartii and Escherichia coli: loss of CysE enhances biofilm formation in Escherichia coli. J Bacteriol. 2004;186:7610–7617. doi: 10.1128/JB.186.22.7610-7617.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren D, Zuo R, Gonzalez Barrios AF, Bedzyk LA, Eldridge GR, Pasmore ME, Wood TK. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl Environ Microbiol. 2005;71:4022–4034. doi: 10.1128/AEM.71.7.4022-4034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh P, Brooks JF, II, Ray VA, Mandel MJ, Visick KL. CysK plays a role in biofilm formation and colonization by Vibrio fischeri. Appl Environ Microbiol. 2015;81:5223–5234. doi: 10.1128/AEM.00157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turnbull AL, Surette MG. L-Cysteine is required for induced antibiotic resistance in actively swarming Salmonella enterica serovar Typhimurium. Microbiology. 2008;154:3410–3419. doi: 10.1099/mic.0.2008/020347-0. [DOI] [PubMed] [Google Scholar]
- 51.Turnbull AL, Surette MG. Cysteine biosynthesis, oxidative stress and antibiotic resistance in Salmonella typhimurium. Res Microbiol. 2010;161:643–650. doi: 10.1016/j.resmic.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Pieroni M, Annunziato G, Beato C, Wouters R, Benoni R, Campanini B, Pertinhez TA, Bettati S, Mozzarelli A, Costantino G. Rational design, synthesis, and preliminary structure-activity relationships of alpha-substituted-2-phenylcyclopropane carboxylic acids as inhibitors of Salmonella typhimurium O-acetylserine sulfhydrylase. J Med Chem. 2016;59:2567–2578. doi: 10.1021/acs.jmedchem.5b01775. [DOI] [PubMed] [Google Scholar]
- 53.Salsi E, Bayden AS, Spyrakis F, Amadasi A, Campanini B, Bettati S, Dodatko T, Cozzini P, Kellogg GE, Cook PF, et al. Design of O-acetylserine sulfhydrylase inhibitors by mimicking nature. J Med Chem. 2010;53:345–356. doi: 10.1021/jm901325e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spyrakis F, Felici P, Bayden AS, Salsi E, Miggiano R, Kellogg GE, Cozzini P, Cook PF, Mozzarelli A, Campanini B. Fine tuning of the active site modulates specificity in the interaction of O-acetylserine sulfhydrylase isozymes with serine acetyltransferase. Biochim Biophys Acta. 2013;1834:169–181. doi: 10.1016/j.bbapap.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Spyrakis F, Singh R, Cozzini P, Campanini B, Salsi E, Felici P, Raboni S, Benedetti P, Cruciani G, Kellogg GE, et al. Isozyme-specific ligands for O-acetylserine sulfhydrylase, a novel antibiotic target. PLoS One. 2013;8:e77558. doi: 10.1371/journal.pone.0077558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Annunziato G, Pieroni M, Benoni R, Campanini B, Pertinhez TA, Pecchini C, Bruno A, Magalhaes J, Bettati S, Franko N, et al. Cyclopropane-1,2-dicarboxylic acids as new tools for the biophysical investigation of O-acetylserine sulfhydrylases by fluorimetric methods and saturation transfer difference (STD) NMR. J Enzyme Inhib Med Chem. 2016;31:78–87. doi: 10.1080/14756366.2016.1218486. [DOI] [PubMed] [Google Scholar]
- 57.Benoni R, Pertinhez TA, Spyrakis F, Davalli S, Pellegrino S, Paredi G, Pezzotti A, Bettati S, Campanini B, Mozzarelli A. Structural insight into the interaction of O-acetylserine sulfhydrylase with competitive, peptidic inhibitors by saturation transfer difference-NMR. FEBS Lett. 2016;590:943–953. doi: 10.1002/1873-3468.12126. [DOI] [PubMed] [Google Scholar]
- 58.Brunner K, Maric S, Reshma RS, Almqvist H, Seashore-Ludlow B, Gustavsson AL, Poyraz O, Yogeeswari P, Lundback T, Vallin M, et al. Inhibitors of the cysteine synthase CysM with antibacterial potency against dormant Mycobacterium tuberculosis. J Med Chem. 2016;59:6848–6859. doi: 10.1021/acs.jmedchem.6b00674. [DOI] [PubMed] [Google Scholar]
- 59.Palde PB, Bhaskar A, Pedró Rosa LE, Madoux F, Chase P, Gupta V, Spicer T, Scampavia L, Singh A, Carroll KS. First-in-class Inhibitors of sulfur metabolism with bactericidal activity against non-replicating M. tuberculosis. ACS Chem Biol. 2016;11:172–184. doi: 10.1021/acschembio.5b00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campanini B, Benoni R, Bettati S, Beck CM, Hayes CS, Mozzarelli A. Moonlighting O-acetylserine sulfhydrylase: new functions for an old protein. Biochim Biophys Acta. 2015;1854:1184–1193. doi: 10.1016/j.bbapap.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beck CM, Diner EJ, Kim JJ, Low DA, Hayes CS. The F pilus mediates a novel pathway of CDI toxin import. Mol Microbiol. 2014;93:276–290. doi: 10.1111/mmi.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson PM, Beck CM, Morse RP, Garza-Sanchez F, Low DA, Hayes CS, Goulding CW. Unraveling the essential role of CysK in CDI toxin activation. Proc Natl Acad Sci USA. 2016;113:9792–9797. doi: 10.1073/pnas.1607112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanous C, Soutourina O, Raynal B, Hullo MF, Mervelet P, Gilles AM, Noirot P, Danchin A, England P, Martin-Verstraete I. The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis. J Biol Chem. 2008;283:35551–35560. doi: 10.1074/jbc.M805951200. [DOI] [PubMed] [Google Scholar]
- 64.Ma DK, Vozdek R, Bhatla N, Horvitz HR. CYSL-1 interacts with the O2-sensing hydroxylase EGL-9 to promote H2S-modulated hypoxia-induced behavioral plasticity in C. elegans. Neuron. 2012;73:925–940. doi: 10.1016/j.neuron.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Even S, Burguiere P, Auger S, Soutourina O, Danchin A, Martin-Verstraete I. Global control of cysteine metabolism by CymR in Bacillus subtilis. J Bacteriol. 2006;188:2184–2197. doi: 10.1128/JB.188.6.2184-2197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gorman J, Shapiro L. Structure of serine acetyltransferase from Haemophilus influenzae Rd. Acta Crystallogr D Biol Crystallogr. 2004;60:1600–1605. doi: 10.1107/S0907444904015240. [DOI] [PubMed] [Google Scholar]
- 67.Hindson VJ, Moody PC, Rowe AJ, Shaw WV. Serine acetyltransferase from Escherichia coli is a dimer of trimers. J Biol Chem. 2000;275:461–466. doi: 10.1074/jbc.275.1.461. [DOI] [PubMed] [Google Scholar]
- 68.Feldman-Salit A, Wirtz M, Hell R, Wade RC. A mechanistic model of the cysteine synthase complex. J Mol Biol. 2009;386:37–59. doi: 10.1016/j.jmb.2008.08.075. [DOI] [PubMed] [Google Scholar]
- 69.Feldman-Salit A, Wirtz M, Lenherr ED, Throm C, Hothorn M, Scheffzek K, Hell R, Wade RC. Allosterically gated enzyme dynamics in the cysteine synthase complex regulate cysteine biosynthesis in Arabidopsis thaliana. Structure. 2012;20:292–302. doi: 10.1016/j.str.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 70.Salsi E, Campanini B, Bettati S, Raboni S, Roderick SL, Cook PF, Mozzarelli A. A two-step process controls the formation of the bienzyme cysteine synthase complex. J Biol Chem. 2010;285:12813–12822. doi: 10.1074/jbc.M109.075762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kredich NM, Tomkins GM. The biosynthesis of L-cysteine in Escherichia coli and Salmonella typhimurium by a multifunctional enzyme complex. In: Vogel HJ, Lampen JO, Brysonk V, editors. Organizational Biosynthesis. Academic Press; New York, NY: 1967. pp. 189–198. [Google Scholar]
- 72.Droux M, Martin J, Sajus P, Douce R. Purification and characterization of O-acetylserine (thiol) lyase from spinach chloroplasts. Arch Biochem Biophys. 1992;295:379–390. doi: 10.1016/0003-9861(92)90531-z. [DOI] [PubMed] [Google Scholar]
- 73.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, et al. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 74.Wirtz M, Birke H, Heeg C, Muller C, Hosp F, Throm C, Konig S, Feldman-Salit A, Rippe K, Petersen G, et al. Structure and function of the heterooligomeric cysteine synthase complex in plants. J Biol Chem. 2010;285:32810–32817. doi: 10.1074/jbc.M110.157446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yi H, Dey S, Kumaran S, Lee SG, Krishnan HB, Jez JM. Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone. J Biol Chem. 2013;288:36463–36472. doi: 10.1074/jbc.M113.527143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Noji M, Inoue K, Kimura N, Gouda A, Saito K. Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J Biol Chem. 1998;273:32739–32745. doi: 10.1074/jbc.273.49.32739. [DOI] [PubMed] [Google Scholar]
- 77.Johnson CM, Huang B, Roderick SL, Cook PF. Kinetic mechanism of the serine acetyltransferase from Haemophilus influenzae. Arch Biochem Biophys. 2004;429:115–122. doi: 10.1016/j.abb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 78.Denk D, Bock A. L-cysteine biosynthesis in Escherichia coli: nucleotide sequence and expression of the serine acetyltransferase (cysE) gene from the wild-type and a cysteine-excreting mutant. J Gen Microbiol. 1987;133:515–525. doi: 10.1099/00221287-133-3-515. [DOI] [PubMed] [Google Scholar]
- 79.Nakamori S, Kobayashi SI, Kobayashi C, Takagi H. Overproduction of L-cysteine and L-cystine by Escherichia coli strains with a genetically altered serine acetyltransferase. Appl Environ Microbiol. 1998;64:1607–1611. doi: 10.1128/aem.64.5.1607-1611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Becker MA, Tomkins GM. Pleiotrophy in a cysteine-requiring mutant of Samonella typhimurium resulting from altered protein-protein interaction. J Biol Chem. 1969;244:6023–6030. [PubMed] [Google Scholar]
- 81.Wirtz M, Berkowitz O, Droux M, Hell R. The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction. Eur J Biochem. 2001;268:686–693. doi: 10.1046/j.1432-1327.2001.01920.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Heterospecific CSC formation measured by the dependence of v0 for L-Ser acetylation on CysK concentration.
Fig. S2. SDS/PAGE of > 95% pure, untagged CysE.
Fig. S3. Dependence of v0 for L-Ser acetylation on CysE concentration.
Fig. S4. Double reciprocal plots of the dependence of v0 on acCoA and L-Ser concentration in the absence (panel A) and presence (panel B) of fivefold molar excess CysK.
Fig. S5. Panel (A) Increase in fluorescence emission intensity at 500 nm upon excitation at 412 nm of a 80 nM CysK solution as a function of CysE concentration in the presence of 10 μM L-Cys. Fitting with Eqn (6) gives a Kd of 66 ± 20 nM. Panel (B) Effect of physiological (0.1 mM) and saturating (20 mM) L-Ser concentrations on the inhibition by L-Cys of CysE activity in the absence and presence of a 100-fold molar excess CysK. The concentration of L-Cys used varies in the two conditions and is around the IC50, i.e., 0.07 μM in the presence of 0.1 mM L-Ser and 0.5 μM in the presence of 20 mM L-Ser. The 1 mM L-Ser condition is reported for comparison using data of Fig. 7.


