Summary
The RNA-guided Cpf1 (also known as Cas12a) nuclease associates with a CRISPR RNA (crRNA), and cleaves the double-stranded DNA target complementary to the crRNA guide. The two Cpf1 orthologs from Acidaminococcus sp. (AsCpf1) and Lachnospiraceae bacterium (LbCpf1) have been harnessed for eukaryotic genome editing. Cpf1 requires a specific nucleotide sequence, called a protospacer adjacent motif (PAM), for the target recognition. Besides the canonical TTTV PAM, Cpf1 recognizes suboptimal C-containing PAMs. Here, we report four crystal structures of LbCpf1 in complex with the crRNA and its target DNA, containing either TTTA, TCTA, TCCA or CCCA as the PAM. These structures revealed that, depending on the PAM sequences, LbCpf1 undergoes conformational changes to form altered interactions with the PAM-containing DNA duplexes, thereby achieving the relaxed PAM recognition. Collectively, the present structures advance our mechanistic understanding of the PAM-dependent crRNA-guided DNA cleavage by the Cpf1 family nucleases.
Introduction
The microbial CRISPR-Cas (clustered regularly interspaced short palindromic repeat-CRISPR-associated) adaptive immune system helps bacteria and archaea defend themselves against the invasion of foreign nucleic acids (Doudna and Charpentier, 2014; Marraffini, 2015; Mohanraju et al., 2016; Wright et al., 2016). In the CRISPR-Cas system, the effector nucleases associate with CRISPR RNAs (crRNAs), and degrade foreign nucleic acids complementary to the crRNA guide. Most of the CRISPR-Cas effector nucleases require a specific sequence next to the target site, called the protospacer adjacent motif (PAM), to initiate the target recognition (Garneau et al., 2010). The CRISPR-Cas system is divided into two classes, class 1 and class 2 (Makarova et al., 2015; Nishimasu and Nureki, 2017; Shmakov et al., 2017). The class 1 effectors comprise multiple Cas proteins, whereas the class 2 effectors contain a single Cas protein. The class 2 CRISPR-Cas system is divided into three types, type II, type V and type VI.
The type-II Cas9 effector nucleases, such as Streptococcus pyogenes Cas9, have been widely harnessed for genome engineering technologies, such as genome editing, epigenome editing, and transcriptional regulation (Barrangou and Doudna, 2016; Komor et al., 2016). More recently, the type-V Cpf1 (also known as Cas12a) effector nucleases from Acidaminococcus sp. BV3L6 (AsCpf1) and Lachnospiraceae bacterium ND2006 (LbCpf1) were identified and biochemically characterized (Zetsche et al., 2015), and they have been harnessed for genome editing in eukaryotic cells (Zetsche et al., 2015, 2016; Hur et al., 2016; Kim et al., 2016a, 2016b, 2016c; Kleinstiver et al., 2016; Tang et al., 2017). AsCpf1 and LbCpf1 share 34% sequence identity, while they lack similarity with Cas9 outside their RuvC domains. Cpf1 possesses several distinguishing features from Cas9. First, Cpf1 is guided by a crRNA, whereas Cas9 uses dual-guide RNAs, a crRNA and a trans-activating crRNA (Deltcheva et al., 2011). Second, Cpf1 recognizes a T-rich PAM (Zetsche et al., 2015), whereas Cas9 favors a G-rich PAM (Jinek et al., 2012; Fonfara et al., 2014; Karvelis et al., 2015; Ran et al., 2015). Third, Cpf1 generates staggered ends at its PAM-distal region (Zetsche et al., 2015), whereas Cas9 creates blunt ends at its PAM-proximal region (Garneau et al., 2010). Fourth, Cpf1 can process its own crRNA array to generate the mature crRNAs (Fonfara et al., 2016; Zetsche et al., 2016). Fifth, Cpf1 exhibits higher targeting specificity in mammalian cells, as compared with Cas9 (Kim et al., 2016b; Kleinstiver et al., 2016).
Previous structural studies provided mechanistic insights into the crRNA-guided DNA recognition and cleavage by the Cpf1 family nucleases (Dong et al., 2016; Yamano et al., 2016; Gao et al., 2016). The crystal structures of the LbCpf1-crRNA binary complex (Dong et al., 2016) and the AsCpf1-crRNA-target DNA ternary complex (Yamano et al., 2016; Gao et al., 2016) revealed the bilobed architectures of Cpf1 and the crRNA recognition mechanism. The AsCpf1-crRNA-DNA structures further revealed the crRNA-guided DNA targeting and the PAM recognition mechanisms (Yamano et al., 2016; Gao et al., 2016). In addition, these structures identified the Nuc domain next to the RuvC nuclease domain, and biochemical data demonstrated that the Nuc domain is involved in the cleavage of the target DNA strand (Yamano et al., 2016). Moreover, a structural comparison between apo-LbCpf1 and the LbCpf1-crRNA complex indicated the crRNA-induced structural rearrangements in Cpf1 (Dong et al., 2016).
Previous in vitro cleavage experiments suggested that, whereas LbCpf1 and AsCpf1 prefer the TTTV PAM, they also recognize C-containing sequences as suboptimal PAMs (Zetsche et al., 2015). Indeed, a recent study demonstrated that LbCpf1 and AsCpf1 can modify target sites with the non-canonical C-containing PAMs, such as CTTA, TCTA and TTCA, in mammalian cells, albeit with lower efficiencies than those with the canonical TTTV PAM (Kim et al., 2016c). However, the mechanism by which Cpf1 recognizes both the canonical and non-canonical PAMs has remained elusive. Moreover, the mechanism of PAM recognition by LbCpf1 also remains unknown, due to the lack of structural information about the LbCpf1-crRNA-DNA complex.
In this study, we show that AsCpf1 and LbCpf1 can recognize both the canonical and non-canonical PAMs in vitro and in vivo. We determined four crystal structures of LbCpf1 in complex with the crRNA and its target DNA, containing either TTTA, TCTA, TCCA or CCCA as the PAM, at 2.4–2.5 Å resolutions. These structures revealed that LbCpf1 undergoes conformational changes to form distinct interactions with the PAM-containing DNA duplex, depending on the PAM sequences, thereby explaining its PAM preference. A structural comparison of the LbCpf1-crRNA-DNA ternary complex with the LbCpf1-crRNA binary complex revealed the conformational rearrangements in the protein upon the crRNA-target DNA heteroduplex formation. In addition, a structural comparison between LbCpf1 and AsCpf1 revealed similarities and differences in their crRNA-guided DNA cleavage mechanisms. Furthermore, a structural comparison of Cpf1 with Cas9 highlighted the fundamental differences in the PAM recognition mechanisms of the two class 2 effector nucleases.
Results
DNA cleavage activities of LbCpf1 and AsCpf1
The in vitro DNA cleavage activities of purified LbCp1 and AsCpf1 have not been fully investigated, although their activities were compared in mammalian cells (Kim et al., 2016b; Kleinstiver et al., 2016) and plant cells (Tang et al., 2017). We thus measured the in vitro DNA cleavage activities of purified LbCpf1 and AsCpf1, using a plasmid DNA substrate with a 24-nt target sequence and the TTTA PAM (Figure 1A). LbCpf1 cleaved the plasmid target slightly more efficiently than AsCpf1 (Figure 1A). Previous studies indicated that, while LbCpf1 and AsCpf1 recognize TTTV (V is A, G or C) as the optimal PAM, they also recognize C-containing sequences, such as CTTV, TCTV and TTCV, as suboptimal PAMs (Kim et al., 2016c). To examine their preference for the fourth PAM nucleotide, we measured the cleavage activities of LbCpf1 and AsCpf1 toward four target sites, with either TTTA, TTTT, TTTG or TTTC as the potential PAM (Figure 1B). LbCpf1 and AsCpf1 efficiently cleaved the TTTA, TTTG and TTTC sites, but not the TTTT site (Figure 1B), confirming their preferences for the fourth V in the TTTV PAM. To further explore their PAM specificities, we measured their cleavage activities toward seven target sites with either TTTA, CTTA, TCTA, TTCA, CCTA, TCCA or CCCA as the potential PAM (Figure 1C). LbCpf1 and AsCpf1 cleaved the target sites with either CTTA, TCTA or TTCA as the PAM, albeit with lower efficiencies than the TTTA site (Figure 1C). In contrast, LbCpf1 and AsCpf1 were less active toward the CCTA, TCCA and CCCA sites (Figure 1C). These results confirmed that LbCpf1 and AsCpf1 recognize CTTV, TCTV and TTCV as the suboptimal non-canonical PAMs.
Figure 1. DNA cleavage activities of LbCpf1 and AsCpf1.
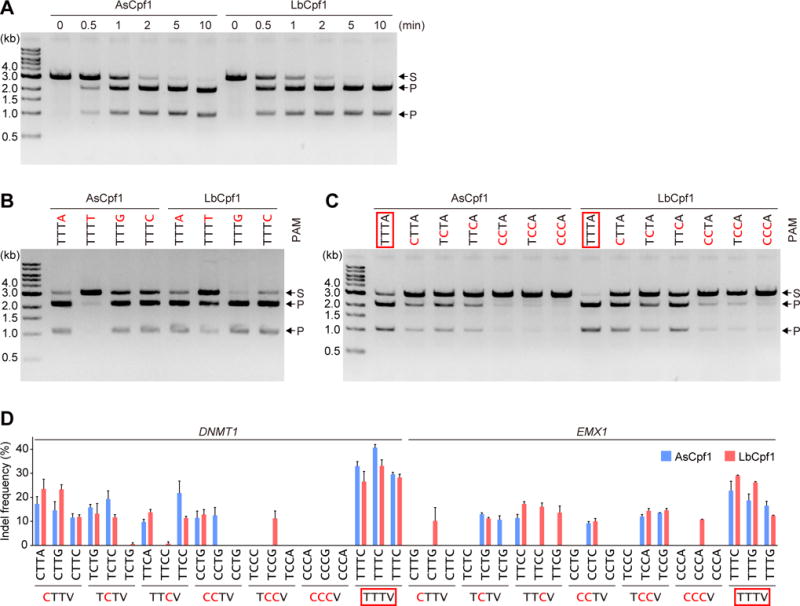
(A) In vitro DNA cleavage activities of LbCpf1 and AsCpf1. The Cpf1-crRNA complex (50 nM) was incubated at 37°C for the indicated time with a linearized plasmid target with the TTTA PAM. S, substrate; P, product.
(B and C) PAM specificities of LbCpf1 and AsCpf1. The Cpf1-crRNA complex (50 nM) was incubated at 37°C for 5 min with a linearized plasmid target with the different PAMs.
(D) In vivo cleavage activities of LbCpf1 and AsCpf1. Indel frequencies for 42 endogenous target sites with the different PAMs were measured in mammalian cells. Data are shown as mean ± s.e.m (n = 3).
In (B)–(D), the canonical PAM is boxed in red, and the substituted nucleotides are colored red.
Furthermore, we examined the activities of LbCpf1 and AsCpf1 toward 42 endogenous target sites, with either TTTV, CTTV, TCTV, TTCV, CCTV, TCCV or CCCV as the potential PAM, in the DNMT1 and EMX1 loci in HEK293 cells (Figure 1D). LbCpf1 and AsCpf1 efficiently modified all six of the target sites with the canonical TTTV PAM (Figure 1D). In contrast, LbCpf1/AsCpf1 modified the 4/3 CTTV, 3/4 TCTV, 4/3 TTCV, 2/2 CCTV, 3/2 TCCV and 1/0 CCCV sites, respectively (Figure 1D). These results revealed that LbCpf1 and AsCpf1 can edit the endogenous sites with the CTTV, TCTV or TTCV PAM, consistent with our in vitro cleavage data and a recent in vivo study (Kim et al., 2016c). Together, these results confirmed that, in addition to the canonical TTTV PAM, LbCpf1 and AsCpf1 can target CTTV, TCTV and TTCV as suboptimal non-canonical PAMs.
Overall structure of the LbCpf1-crRNA-target DNA complex
To clarify the crRNA-guided DNA cleavage mechanism of LbCpf1, we determined the crystal structure of LbCpf1 (residues 1–1,226) in complex with a 40-nt crRNA, a 29-nt target DNA strand and a 9-nt non-target DNA strand containing the TTTA PAM, at 2.5-Å resolution (Figures 2A–2D and S1 and Table 1). LbCpf1 adopts a bilobed architecture consisting of a recognition (REC) lobe and a nuclease (NUC) lobe (Figure 2C). The REC lobe includes the REC1 and REC2 domains, whereas the NUC lobe comprises the Wedge (WED), PAM-interacting (PI), RuvC and Nuc domains. A characteristic α helix (referred to as the bridge helix) between the RuvC-I and RuvC-II regions interacts with the REC2 domain. The crRNA consists of the 20-nt 5´-handle and the 20-nt guide sequence (Figure 2D). The crRNA guide sequence and the target DNA strand form the 20-bp RNA-DNA heteroduplex, which is accommodated in the central channel between the two lobes (Figures 2C, 2D and S2). Two Mg2+ ions are bound to the crRNA 5´-handle (Figure 2E). One Mg2+ is coordinated by six water molecules that interact with the phosphate groups of A(−13), U(−14) and A(−19) and the nucleobases of A(−13) and U(−18), as observed in the LbCpf1-crRNA binary complex (Dong et al., 2016). The other Mg2+ is coordinated by the main-chain carbonyl group of Thr716, the phosphate group of A(−4) and four water molecules that interact with the phosphate groups of G(−3) and U(−5) and the side chains of Asp708 and Asn718 (Figure 2E).
Figure 2. Structure of the LbCpf1-crRNA-target DNA complex.
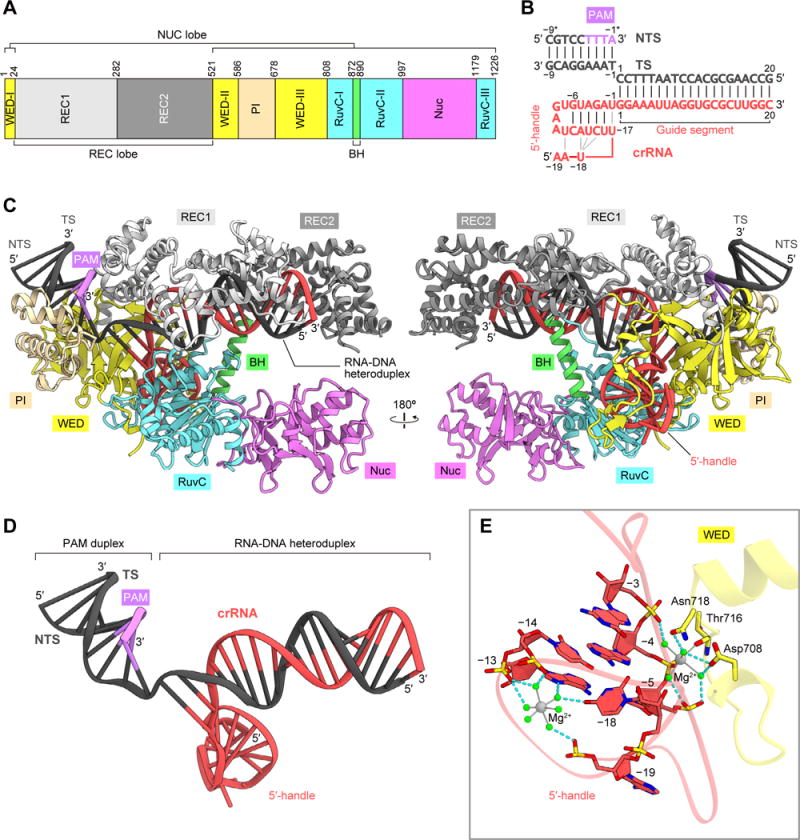
(A) Domain organization of LbCpf1. BH, bridge helix.
(B) Schematic of the crRNA and its target DNA. TS, target DNA strand; NTS, non-target DNA strand.
(C) Crystal structure of LbCpf1 in complex with the crRNA and its target DNA.
(D) Structure of the crRNA and its target DNA.
(E) Binding of Mg2+ ions to the crRNA. The bound Mg2+ ions and water molecules are indicated by gray and green spheres, respectively. Hydrogen bonds are shown as dashed lines.
See Figures S1 and S2.
Table 1.
Data collection and refinement statistics.
| TTTA | TCTA | TCCA | CCCA | |
|---|---|---|---|---|
| Data collection | ||||
| Beamline | SLS | SPring-8 | SPring-8 | SLS |
| PXI | BL41XU | BL41XU | PXI | |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Space group | P41212 | P41212 | P41212 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 103.2, 103.2, 363.9 | 102.5, 102.5, 373.9 | 102.0, 102.0, 372.5 | 102.0, 103.5, 342.7 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å)* | 49.6–2.5 (2.56–2.50) |
49.4–2.4 (2.45–2.40) |
49.2–2.5 (2.56–2.50) |
49.9–2.4 (2.44–2.40) |
| Rpim | 0.033 (0.436) | 0.014 (0.339) | 0.024 (0.518) | 0.043 (0.162) |
| I/σI | 12.7 (1.6) | 25.0 (2.1) | 15.6 (1.6) | 9.2 (2.8) |
| Completeness (%) | 99.9 (100) | 96.3 (97.8) | 99.8 (100) | 97.3 (82.8) |
| Multiplicity | 12.6 (12.7) | 8.9 (7.0) | 8.6 (9.0) | 4.0 (3.4) |
| CC(1/2) | 0.997 (0.677) | 0.999 (0.700) | 0.998 (0.543) | 0.993 (0.889) |
| Refinement | ||||
| Resolution (Å) | 49.7–2.5 (2.53–2.50) |
49.4–2.4 (2.43–2.40) |
49.2–2.5 (2.53–2.50) |
49.9–2.4 (2.43–2.40) |
| No. reflections | 69,046 (2,583) |
75,435 (2,649) |
69,014 (2,617) |
138,080 (3,632) |
| Rwork/Rfree | 0.178/0.228 (0.289/0.394) |
0.193/0.238 (0.307/0.383) |
0.193/0.244 (0.340/0.383) |
0.178/0.230 (0.215/0.277) |
| No. atoms | ||||
| Protein | 9,776 | 9,794 | 9,749 | 19,707 |
| Nucleic acid | 1,618 | 1,618 | 1,618 | 3,236 |
| Ion | 5 | 2 | 2 | 5 |
| Solvent | 143 | 53 | 27 | 471 |
| B-factors (Å2) | ||||
| Protein | 83.4 | 81.6 | 88.8 | 60.4 |
| Nucleic acid | 71.3 | 76.4 | 83.5 | 52.0 |
| Ion | 75.7 | 77.9 | 79.2 | 37.2 |
| Solvent | 66.8 | 60.1 | 68.8 | 46.2 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.007 | 0.007 | 0.007 | 0.007 |
| Bond angles (°) | 0.913 | 0.904 | 0.924 | 0.893 |
| Ramachandran plot (%) | ||||
| Favored region | 96.83 | 95.62 | 95.94 | 96.60 |
| Allowed region | 3.01 | 3.96 | 3.89 | 3.15 |
| Outlier region | 0.17 | 0.41 | 0.17 | 0.25 |
Values in parentheses are for the highest resolution shell.
Structural rearrangement upon target DNA binding
A structural comparison between the LbCpf1 binary and ternary complexes revealed that, while the individual domains are structurally similar (Figure S3A), the REC1, REC2 and PI domains undergo conformational rearrangements upon target DNA binding (Figures 3A and 3B). These structural rearrangements are consistent with a previous prediction, based on a comparison of the LbCpf1 binary complex with the AsCpf1 ternary complex (Gao et al., 2016). As compared to the binary complex, the REC1 and REC2 domains move toward and away from the NUC lobe in the ternary complex, respectively, to form the central channel that accommodates the crRNA-target DNA heteroduplex (Figures 3A, 3B and S3B). In the ternary complex, the PI domain moves toward the REC1 and WED domains to form the PAM-binding channel (Figures 3B and S3C). In addition, whereas the crRNA guide segment is disordered in the binary complex (Dong et al., 2016), the crRNA guide is ordered and forms the heteroduplex with the target DNA in the ternary complex (Figures 3B and S3D).
Figure 3. Comparison between the binary and ternary complex structures of LbCpf1.
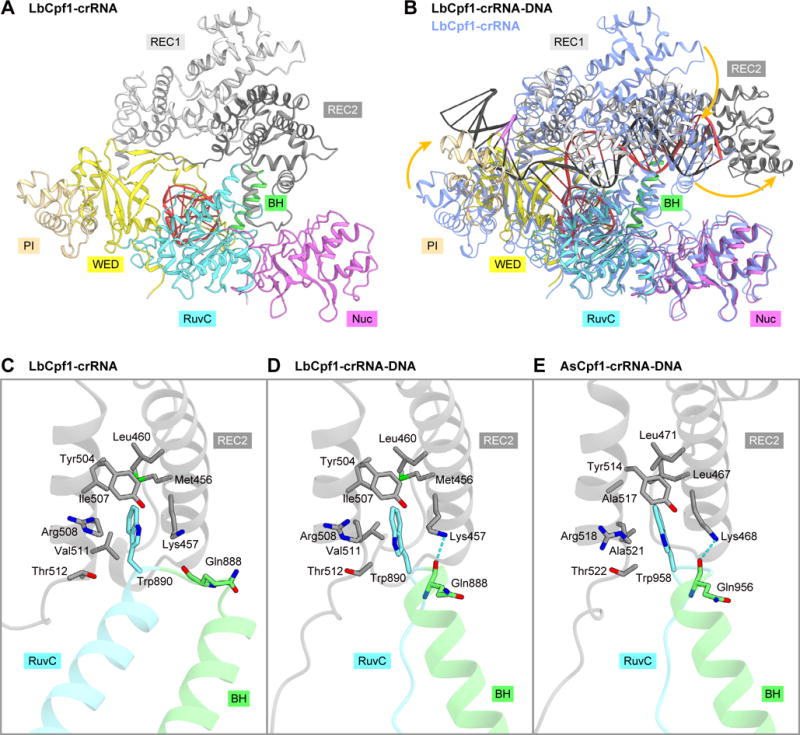
(A) Crystal structure of the LbCpf1-crRNA complex (PDB: 5ID6) (Dong et al., 2016).
(B) Superimposition of the LbCpf1-crRNA-target DNA complex (colored) and the LbCpf1-crRNA complex (blue). Structural changes are indicated by orange arrows.
(C–E) Interactions between the REC and NUC lobes in the LbCpf1 binary complex (C), the LbCpf1 ternary complex (D), and the AsCpf1 ternary complex (PDB: 5B43) (Yamano et al., 2016) (E). Hydrogen bonds are shown as dashed lines.
See Figure S3.
A structural comparison between the binary and ternary complexes also revealed a local conformational change in the bridge helix and the RuvC-II region. In the binary complex, residues 872–885/890–918 and 886–889 adopt α-helical and loop conformations, respectively (Dong et al., 2016) (Figure 3C). In contrast, in the ternary complex, residues 885–889 and 890–896 adopt α-helical and loop conformations, respectively (Figure 3D). In the binary and ternary complexes, Trp890 is inserted into a hydrophobic pocket in the REC2 domain (Figures 3C and 3D). In addition, in the ternary complex, the main-chain carbonyl group of Gln888 hydrogen bonds with the side chain of Lys457 in the REC2 domain (Figure 3D). These interdomain interactions are conserved in the AsCpf1 ternary complex (Figure 3E), and the W958A mutation reduced the DNA cleavage activity (Yamano et al., 2016). These observations highlighted the pivotal role of the conserved tryptophan residue in the RuvC domain for the structural transition from the binary to the ternary complex.
RNA-DNA heteroduplex recognition mechanism
A structural comparison between the ternary complexes of AsCpf1 and LbCpf1 revealed that they share a bilobed architecture and recognize the RNA-DNA heteroduplex in similar manners (Figures 4A and 4B). In the LbCpf1 ternary complex, the 20-bp RNA-DNA heteroduplex is recognized by the REC lobe, in which Trp355 in the REC2 domain stacks with the C20:dG20 base pair in the heteroduplex (Figure 4C). Similarly, in the AsCpf1 ternary complex, the 20-bp heteroduplex is accommodated within the REC lobe, in which Trp382, equivalent to Trp355 in LbCpf1, stacks with the C20:dG20 base pair, while AsCpf1 was crystallized with a 24-nt-guide-containing crRNA and its complementary target DNA (Yamano et al., 2016) (Figure 4D). Consistent with these structural findings, the crRNA-DNA base pairing in the PAM-distal region is dispensable for the Cpf1-mediated DNA cleavage (Zetsche et al., 2015; Kim et al., 2016b, 2016c; Kleinstiver et al., 2016). Together, these observations indicated that LbCpf1 and AsCpf1 recognize the 20-bp crRNA-target DNA heteroduplex.
Figure 4. Comparison between the ternary complex structures of LbCpf1 and AsCpf1.
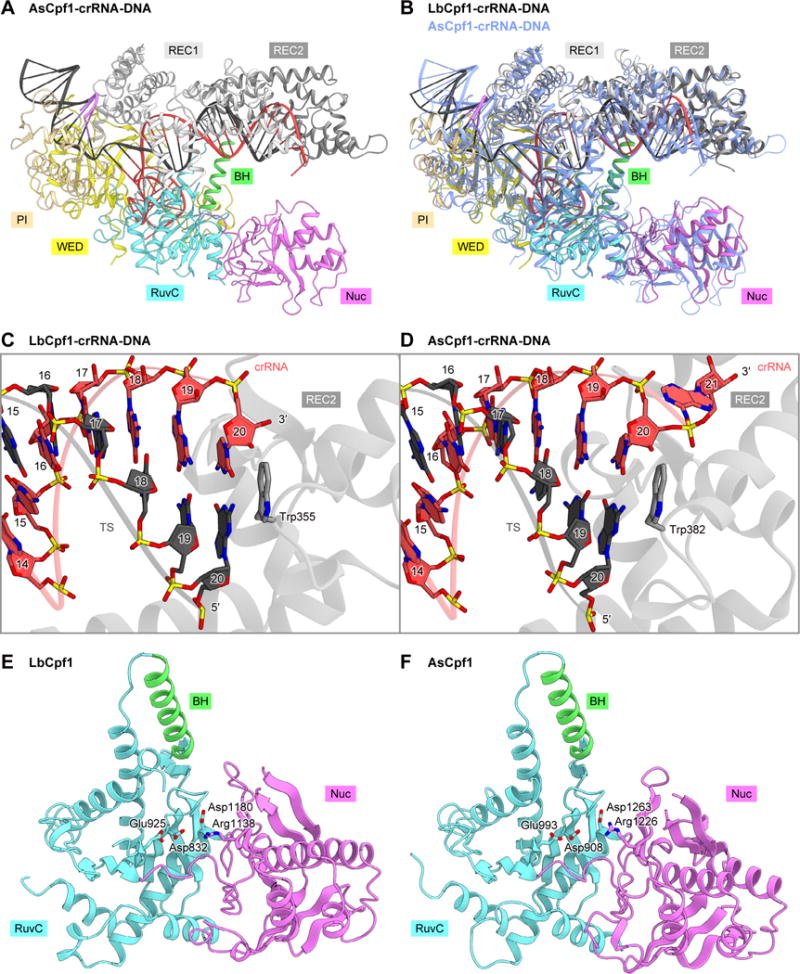
(A) Crystal structure of the LbCpf1-crRNA-DNA complex (PDB: 5B43) (Yamano et al., 2016).
(B) Superimposition of the LbCpf1-crRNA-DNA complex (colored) and the AsCpf1-crRNA-DNA complex (blue)
(C and D) RNA-DNA heteroduplex recognition by LbCpf1 (C) and AsCpf1 (D).
(E and F) RuvC and Nuc domains of LbCpf1 (E) and AsCpf1 (F).
See Figures S4, S5 and S6.
RuvC and Nuc nuclease domains
The RuvC and Nuc domains of LbCpf1 are structurally similar to those of AsCpf1 (Figures 4E and 4F). In the AsCpf1 structure, the RuvC active site is formed by the conserved acidic residues Asp908, Glu993 and Asp1263 (Yamano et al., 2016) (Figure 4F). The D908A, E993A and D1263A mutations abolished the in vitro cleavage activities, indicating that the RuvC domain is involved in the cleavage of both strands (Zetsche et al., 2015; Yamano et al., 2016). In contrast, Arg1226 in the Nuc domain participates in the target strand cleavage (Yamano et al., 2016). In the LbCpf1 structure, Asp832, Glu925, Asp1180 and Arg1138, equivalent to Asp908, Glu993, Asp1263 and Arg1226 of AsCpf1, are similarly arranged (Figures 4E and S4). In addition, our in vitro cleavage assays confirmed that the D832A, E925A and D1180A mutations abolish the DNA cleavage activity of LbCpf1, while the R1138A mutant functions as a nickase, as in the case of AsCpf1 (Zetsche et al., 2015; Yamano et al., 2016) (Figure S5). These observations indicated that LbCpf1 and AsCpf1 cleave the target DNA via similar mechanisms, in which the RuvC domain participates in the cleavage of both strands, while the Nuc domain is involved in the target strand cleavage.
Recently, the crystal structures of C2c1 (also known as Cas12b) from Alicyclobacillus acidoterrestris (AaC2c1), which was identified as a type V-B effector nuclease (Shmakov et al., 2015), elucidated its action mechanism and offered clues toward understanding the DNA cleavage mechanism of the type V-A Cpf1 nucleases (Liu et al., 2016; Yang et al., 2016). Despite their limited sequence similarity, Cpf1 and C2c1 have comparable domain architectures and share the RuvC domain, in which the catalytic residues are similarly arranged (Figures S6A–S6D). As in Cpf1, the Nuc domain is located next to the RuvC domain in C2c1, although their Nuc domains adopt distinct folds (Figures S6A and S6B). In the AaC2c1 structure, Arg911 in the Nuc domain interacts with the backbone phosphate group of the target strand, thereby guiding the target strand into the RuvC active site (Figure S6D). These structural observations suggested that C2c1 uses the RuvC active site to cleave both the target and non-target strands (Yang et al., 2016). Notably, LbCpf1 Arg1138 and AsCpf1 Arg1226 are located at positions analogous to that of AaC2c1 Arg911 (Figures S6C and S6D), and they are involved in the target strand cleavage (Yamano et al., 2016) (Figure S5). Thus, these observations suggested that the Cpf1 Nuc domain plays a role in guiding the target strand into the RuvC active site, rather than catalyzing the cleavage of the target strand. Supporting this notion, a recent study indicated that Francisella novicida Cpf1 cleaves the target and non-target DNA strands, using the same RuvC domain active site (Swarts et al., 2017).
Recognition mechanism of the canonical TTTV PAM
In the LbCpf1 ternary complex, the TTTA PAM duplex is bound to the channel formed by the REC1, WED and PI domains (Figures 5A and 5B). The PAM duplex adopts a distorted conformation with a narrow minor groove, as compared with the canonical B-form DNA (root-mean-square deviation (RMSD) is 1.4 Å for 18 equivalent phosphorus atoms), as observed in the AsCpf1 ternary complex (Yamano et al., 2016) (Figure 5C). The dT(−1):dA(−1*) base pair in the PAM duplex does not form base-specific contacts with the protein (Figure 5B). The O2 of dT(−2*) forms a hydrogen bond with Lys595, whereas the N7 of dA(−2) and the backbone phosphate group between dT(−1) and dA(−2) form hydrogen bonds with Tyr542 (Figure 5D). dA(−2) also forms van der Waals interactions with Pro587, Met592 and Lys595. The 5-methyl group of dT(−3*) is in the vicinity of the side-chain methyl group of Thr149, whereas the N3 and N7 of dA(−3) form hydrogen bonds with Lys595 and Lys538, respectively (Figure 5E). The 5-methyl group of dT(−4*) is surrounded by the side chains of Thr149 and Gln529, whereas the O4´ of dA(−4) forms a hydrogen bond with Lys595 (Figure 5F).
Figure 5. Canonical PAM recognition mechanism.
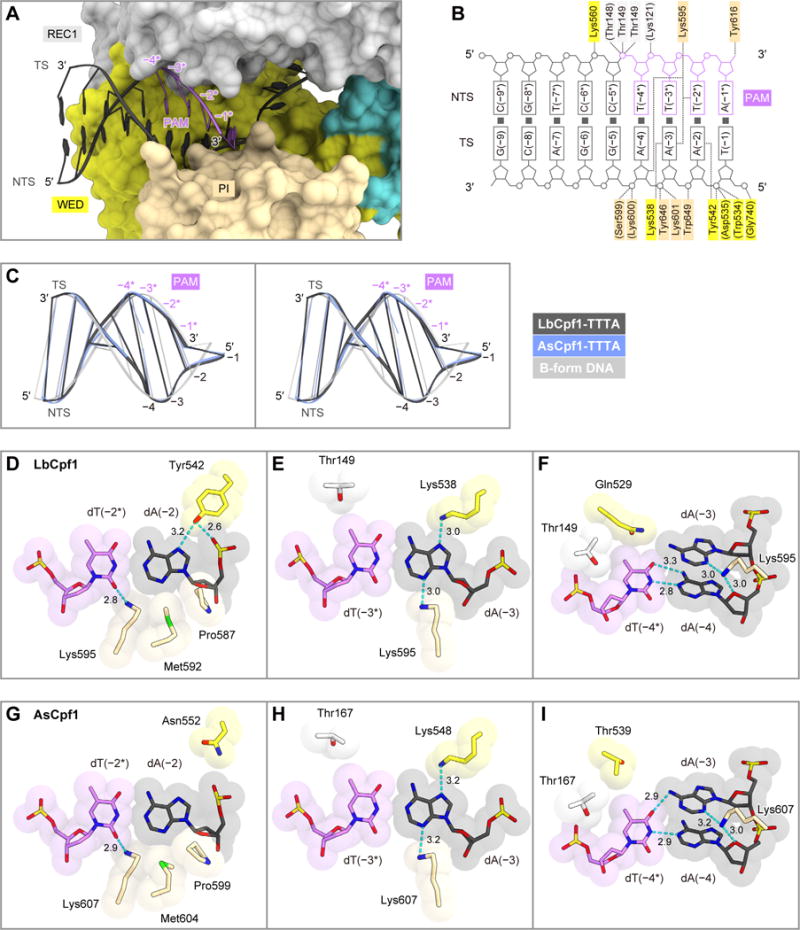
(A) PAM-duplex binding in the LbCpf1 ternary complex.
(B) Schematic of the PAM-duplex recognition by LbCpf1.
(C) Superimposition of the PAM duplexes in LbCpf1 and AsCpf1 (PDB: 5B43) (Yamano et al., 2016) onto the B-form DNA duplex (stereo view).
(D–F) Recognition of dA(−2):dT(−2*) (D), dA(−3):dT(−3*) (E), and dA(−4):dT(−4*) (F) by LbCpf1.
(G–I) Recognition of dA(−2):dT(−2*) (G), dA(−3):dT(−3*) (H), and dA(−4):dT(−4*) (I) by AsCpf1.
In (D–I), hydrogen bonds are shown as dashed lines.
A structural comparison of LbCpf1 with AsCpf1 revealed the high conservation between their PAM recognition mechanisms. In AsCpf1, Lys548 and Lys607, equivalent to Lys538 and Lys595 of LbCpf1, similarly interact with the PAM duplex (Yamano et al., 2016) (Figures 5G–5I and S4). Nonetheless, their PAM recognition mechanisms are slightly different. Whereas Tyr542 of LbCpf1 forms two hydrogen bonds with dA(−2) (Figure 5D), Asn552 of AsCpf1, equivalent to Tyr542 of LbCpf1, does not interact with the PAM duplex (Figure 5G).
Recognition mechanisms of the non-canonical PAMs
To investigate the non-canonical PAM recognition mechanism, we determined the crystal structures of the LbCpf1 ternary complex containing either TCTA, TCCA or CCCA as the PAM (Table 1). As in the TTTA complex, the TCTA and TCCA complexes crystallized in the space group P41212, with one complex molecule in the asymmetric unit. In contrast, the CCCA complex crystallized in the space group P212121, with two complex molecules in the asymmetric unit, probably due to the slightly different conformations of the bound DNA molecules. Since the two CCCA complex molecules are essentially identical (RMSD is 1.3 Å for equivalent Cα atoms), we will refer to one complex molecule for the following discussion.
In the three complexes, the PAM duplexes are recognized by the REC1, WED and PI domains, as in the TTTA complex (Figures 6A–6C), and they adopt less distorted conformations, as compared with that in the TTTA complex, probably due to the presence of the G:C base pair(s) (Figure 6D). A comparison of the four complex structures revealed that, whereas the REC1 and WED domains are similarly arranged, the PI domains undergo outward displacement in the TCTA, TCCA and CCCA complexes, as compared with that in the TTTA complex (Figures 6A–6C). The RMSD values for the equivalent Cα atoms in the PI domain between the TTTA complex and the TCTA/TCCA/CCCA complexes are 2.2, 2.4 and 2.2 Å, respectively, by superimposition based on the regions except for the PI domain. The PI domain displacement results in the opening of the PAM-binding channel, thereby allowing the binding of PAM duplexes with distinct conformations. Consistent with its conformational flexibility, the PI domain exhibits higher B-factor values than the other domains in the four complexes (Figures S7A–S7D). Furthermore, the PI domain in the TTTA complex displays lower B-factor values than those in the TCTA and TCCA complexes (Figures S7A–S7C). These structural observations suggest that LbCpf1 binds the TTTA PAM duplex more stably than the non-canonical PAM duplexes, thereby explaining the preference of LbCpf1 for the canonical TTTV PAM.
Figure 6. Comparison between the TTTA, TCTA, TCCA and CCCA complex structures.
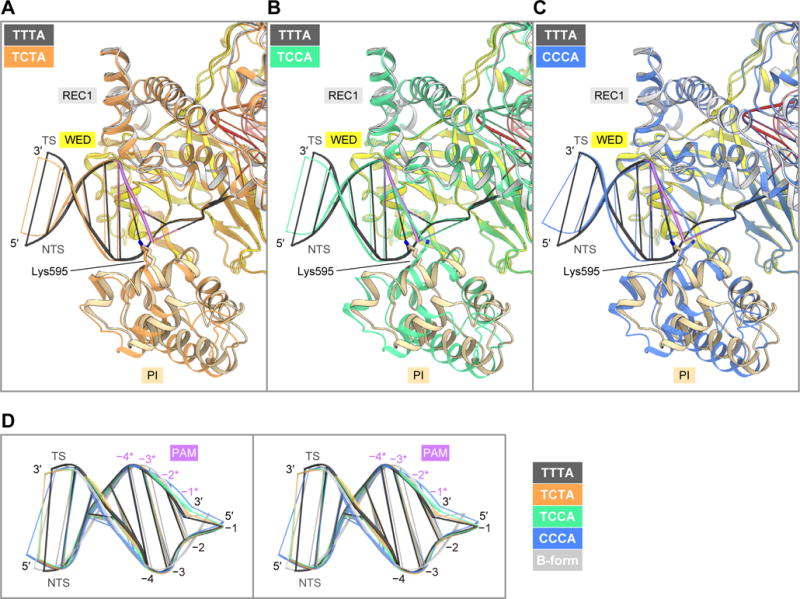
(A) Superimposition of the TCTA (orange) and TTTA (colored) complexes.
(B) Superimposition of the TCCA (green) and TTTA (colored) complexes.
(C) Superimposition of the CCCA (blue) and TTTA (colored) complexes.
(D) Superimposition of the PAM duplexes in the TTTA, TCTA, TCCA and CCCA complex structures onto the B-form DNA duplex (stereo view).
See Figure S7.
In the four complex structures, Lys538 and Tyr542 similarly interact with the second and third PAM-complementary nucleotides (Figures 7A–7H). In contrast, Lys595 interacts with the PAM nucleotides in distinct manners among the four complex structures, due to the conformational differences in the PI domain and the PAM duplex (Figures 7E–7H). In the TTTA complex, Lys595 forms hydrogen bonds with the N3 of dA(−3) and the O2 of dT(−2*) (Figure 7E). In contrast, in the TCTA complex, Lys595 does not hydrogen bond with the N3 of dG(−3) (Figure 7F). Notably, in the TCCA and CCCA complexes, Lys595 is not inserted into the minor groove of the PAM duplex (Figures 7G and 7H), probably due to steric hindrance between Lys595 and the N2 of dG(−2) (Figure S7E). These structural observations are consistent with the fact that LbCpf1 recognizes the non-canonical C-containing PAMs less efficiently than the canonical TTTV PAM. In addition, the side chain of Lys595 is less ordered in the TCTA, TCCA and CCCA complexes, as compared with that in the TTTA complex (Figures 7A–7D), consistent with the weaker interactions between Lys595 and the non-canonical PAM duplexes. Given the highly conserved PAM recognition mechanisms, AsCpf1 likely recognizes the non-canonical PAMs in similar manners. Together, these structural findings revealed the previously undescribed mechanisms of the canonical and non-canonical PAM recognition by the Cpf1 nucleases.
Figure 7. Non-canonical PAM recognition mechanism.
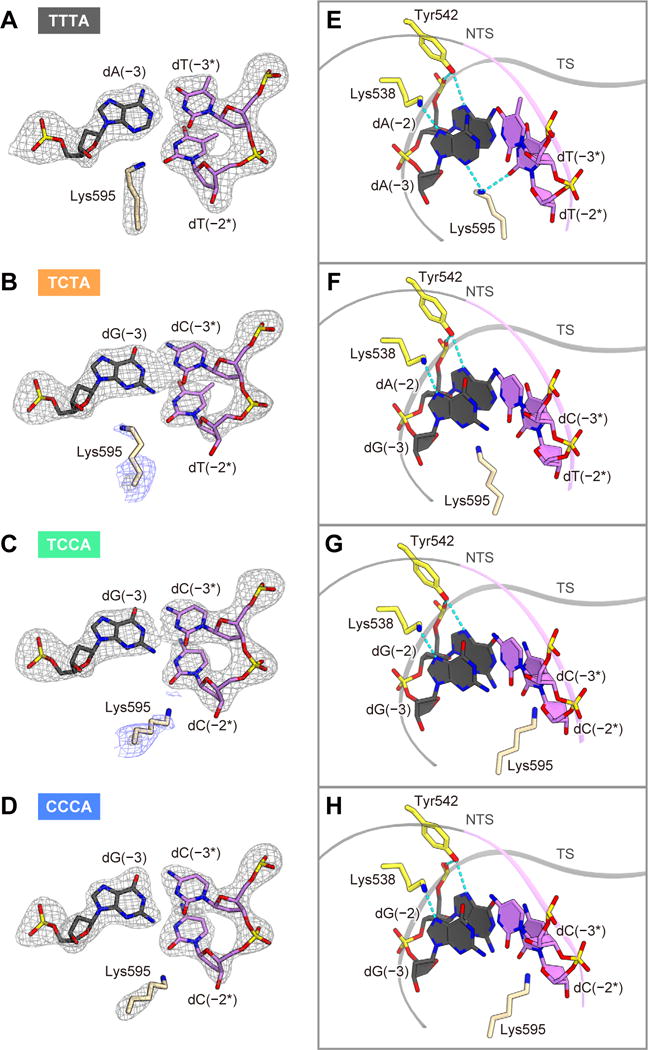
(A–D) The mFO – DFC omit electron density maps for Lys595 and the key nucleotides in the TTTA (A), TCTA (B), TCCA (C) and CCCA (D) complexes (gray, contoured at 5.0σ). In (B) and (C), the electron density map contoured at 2.0σ (blue) is also shown for Lys595.
(E–H) Recognitions of the TTTA (E), TCTA (F), TCCA (G) and CCCA (H) PAMs. Hydrogen bonds are shown as dashed lines.
See Figure S7.
Discussion
In this study, we showed that LbCpf1 and AsCpf1 recognize TTTV and CTTV/TCTV/TTCV as the canonical and non-canonical PAMs, respectively, consistent with a recent study (Kim et al., 2016c). We further determined the LbCpf1-crRNA-DNA structures with either the TTTA, TCTA, TCCA or CCCA PAM. A structural comparison of LbCpf1 with AsCpf1 highlighted the mechanistic similarities in the crRNA-guided DNA recognition and cleavage among the Cpf1 nucleases. The TTTA complex structure revealed that LbCpf1 recognizes the canonical TTTV PAM via the shape and base readout mechanism, in which Lys595 inserts into the minor groove of the PAM duplex. Lys607 of AsCpf1, equivalent to Lys595 of LbCpf1, forms similar interactions with the PAM duplex (Yamano et al., 2016), and these lysine residues are conserved among the Cpf1 family members (Zetsche et al., 2015) (Figure S3), suggesting that the Cpf1 orthologs recognize their T-rich PAMs in similar manners. Moreover, the present structures revealed that Lys595 is also important for the discrimination between the canonical and non-canonical PAMs.
The present structures also highlighted the fundamental differences in the PAM recognition mechanisms between the type-II Cas9 and type-V Cpf1 effector nucleases. In Cas9, the PAM duplex is accommodated within the PAM-binding groove in the PI domain, in which the major-groove edges of the PAM nucleotides are recognized by distinct sets of PAM-interacting residues via hydrogen-bonding interactions (i.e., base readout mechanism) (Anders et al., 2014; Nishimasu et al., 2015; Hirano et al., 2016; Yamada et al., 2017). A structural comparison between the binary and ternary complexes of Cas9 suggested that the PAM-binding groove is pre-organized for the PAM recognition (Nishimasu et al., 2014; Anders et al., 2014; Jiang et al., 2015). In contrast, in Cpf1, the PAM duplex is enveloped within the PAM-binding channel formed by the WED, REC1 and PI domains, in which both the sequence and conformation of the PAM duplex are primarily recognized by the two conserved lysine residues (LbCpf1 Lys538/Lys595 and AsCpf1 Lys548/Lys607) (i.e., base and shape readout mechanism). Importantly, the present LbCpf1 structures revealed that, in contrast to the PAM-binding groove of Cas9, the PAM-binding channel of Cpf1 has conformational flexibility, which allows the recognition of the canonical and non-canonical PAMs.
Recently, AsCpf1 has been engineered to recognize altered PAM sequences, using a structure-guided mutation screen (Gao et al., 2017), and the crystal structures of the AsCpf1 variants revealed their altered PAM recognition mechanisms (Nishimasu et al., 2017). Thus, the present findings advance our mechanistic understanding of the CRISPR-Cpf1 family nucleases, and will facilitate the engineering of Cpf1, including the development of variants with altered PAM specificities.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for further information and reagents should be directed to the Lead Contact, Osamu Nureki (nureki@bs.s.u-tokyo.ac.jp).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The plasmid DNAs were amplified in Escherichia coli Mach (Thermo Fisher Scientific), cultured in LB medium (Nacalai Tesque) at 37°C overnight. The recombinant proteins were overexpressed in E. coli Rosetta 2 (DE3) (Novagen).
Sample preparation
The gene encoding full-length LbCpf1 (residues 1–1,226) was PCR-amplified using the pcDNA3.1-LbCpf1 plasmid (Zetsche et al. 2015) as the template, and cloned between the NdeI and XhoI sites of the modified pE-SUMO vector (LifeSensors). The mutations (D832A, E925A, D1180A and R1138A) were introduced by a PCR-based method (Table S1). The LbCpf1-expressing E. coli Rosetta2 (DE3) cells were cultured at 37°C in LB medium (containing 20 mg/l kanamycin) until the OD600 reached 0.8, and protein expression was then induced by the addition of 0.1 mM isopropyl β-D-thiogalactopyranoside (Nacalai Tesque). The E. coli cells were further cultured at 20°C for 18 h, and harvested by centrifugation at 5,000 g for 10 min. The E. coli cells were resuspended in buffer A (50 mM Tris-HCl, pH 8.0, 20 mM imidazole, 300 mM NaCl and 3 mM 2-mercaptoethanol), lysed by sonication, and then centrifuged at 40,000 g for 30 min. The supernatant was mixed with 5 ml Ni-NTA Superflow (QIAGEN), and the mixture was loaded into an Econo-Column (Bio-Rad). The resin was washed with buffer A, and the protein was eluted with buffer B (50 mM Tris-HCl, pH 8.0, 300 mM imidazole, 300 mM NaCl and 3 mM 2-mercaptoethanol). The protein was loaded onto a HiTrap SP HP column (GE Healthcare) equilibrated with buffer C (20 mM Tris-HCl, pH 8.0 and 200 mM NaCl). The column was washed with buffer C, and the protein was then eluted with a linear gradient of 200–1,000 mM NaCl. To remove the His6-SUMO-tag, the eluted protein was mixed with TEV protease, and was dialyzed at 4°C for 12 h against buffer D (20 mM Tris-HCl, pH 8.0, 40 mM imidazole, 300 mM NaCl and 3 mM 2-mercaptoethanol). The protein was passed through the Ni-NTA column equilibrated with buffer D. The protein was concentrated using an Amicon Ultra 10K filter (Millipore), and was further purified by chromatography on a HiLoad Superdex 200 16/60 column (GE Healthcare) equilibrated with buffer E (10 mM Tris-HCl, pH 8.0, 150 mM NaCl and 1 mM DTT). The purified LbCpf1 proteins were stored at −80°C until use. The crRNA was purchased from Gene Design. The target and non-target DNA strands were purchased from Sigma-Aldrich. The purified LbCpf1 protein was mixed with the crRNA, the target DNA strand, and the non-target DNA strand (molar ratio, 1:1.5:2.3:2.3), and the reconstituted LbCpf1-crRNA-DNA complex was then purified by gel filtration chromatography on a Superdex 200 Increase column (GE Healthcare) equilibrated with buffer E.
Crystallography
The purified LbCpf1-crRNA-DNA complex was crystallized at 20°C, by the hanging-drop vapor diffusion method. The crystallization drops were formed by mixing 1 μl of complex solution (A260 nm = 10) and 1 μl of reservoir solution (13–18% PEG3,350 and 100 mM MIB buffer, pH 5.0), and then were incubated against 0.5 ml of reservoir solution. The crystals were improved by micro-seeding using Seed Bead (Hampton Research). The crystals of the TCTA, TCCA and CCCA complexes were cryoprotected in a solution consisting of 16–20% PEG3,350, 100 mM MIB buffer, pH 5.0, and 30% ethylene glycol. The crystals of the TTTA PAM complex were cryoprotected in the solution supplemented with 50 mM MgCl2. X-ray-diffraction data were collected at 100 K on the beamline BL41XU at SPring-8, and PXI at the Swiss Light Source. The diffraction data were processed using DIALS (Waterman et al., 2013) and AIMLESS (Evans and Murshudov, 2013). The TTTA complex structure was determined by molecular replacement with Molrep (Vagin and Teplyakov, 2010), using the coordinates of LbCpf1 (PDB: 5ID6) (Dong et al., 2016) as the search model. The TCTA, TCCA and CCCA complex structures were determined by molecular replacement, using the TTTA complex structure as the search model. The model building and structural refinement were performed using COOT (Emsley and Cowtan, 2004) and PHENIX (Adams et al., 2010), respectively. Structural figures were prepared using CueMol (http://www.cuemol.org).
In vitro cleavage assay
In vitro cleavage experiments were performed as previously described (Nishimasu et al., 2015), with minor modifications. The EcoRI-linearized pUC119 (100 ng, 4.7 nM), containing the 24-nt target sequence and the PAMs, was incubated with the Cpf1-crRNA complex (50 nM) at 37°C for 5 min, in 20 μl of reaction buffer containing 20 mM HEPES-NaOH, pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM DTT and 5% glycerol. The reaction was stopped by the addition of a solution containing EDTA (40 mM final concentration) and Proteinase K (10 μg). Reaction products were resolved on an ethidium bromide-stained 1% agarose gel, and then visualized using an Amersham Imager 600 (GE Healthcare). To examine the time course of DNA cleavage, the EcoRI-linearized pUC119 target (600 ng, 4.7 nM) was incubated with the Cpf1-crRNA complex (50 nM) at 37°C in 60 μl reaction buffer. Aliquots (10 μl) were taken at the indicated time points, and the reaction products were then analyzed as described above. To examine whether Cpf1 serves as a nickase, the circular pUC119 target (100 ng, 4.7 nM) was incubated with the Cpf1-crRNA complex (100 nM) at 37°C for 20 min in 10 μl reaction buffer, and the reaction products were then analyzed as described above.
In vivo cleavage assay
Human embryonic kidney 293T (HEK) cells were maintained in Dulbecco’s modified Eagle medium (Gibco), supplemented with 10% fetal bovine serum (FBS), at 37°C under a 5% CO2 atmosphere. HEK cells were seeded at 1.25 × 105 cells per well in 24-well plates, 24 h prior to transfection. Plasmids encoding humanized LbCpf1 (pY027) or AsCpf1 (pY026) with C-terminal nuclear localization tags and U6-driven crRNAs were transfected at 500 ng per well, using the Lipofectamine 2000 reagent (Life Technologies). Genomic DNA was extracted using 100 μl QuickExtract DNA Extraction Solution (Epicenter), 3 days post-transfection. Insertion/deletion events (indels) were analyzed by a Surveyor nuclease assay, as previously described (Ran et al., 2013). Briefly, the genomic regions flanking the target sites for DNMT1 or EMX1 were PCR-amplified (Tables S1 and S2), and the products were purified using a QIAQuick PCR purification Kit (QIAGEN) according to the manufacturer’s protocol. The purified PCR products (200 ng) were mixed with 1 μl 10 × Taq DNA Polymerase PCR buffer (Enzymatics) and ultrapure water to a final volume of 10 μl, and subjected to a re-annealing process to enable heteroduplex formation: 95°C for 10 min, 95°C to 85°C ramping at −2°C/s, 85°C to 25°C at −0.25°C/s, and 25°C hold for 1 min. After re-annealing, the products were treated with Surveyor nuclease and Surveyor enhancer S (IDT), according to the manufacturer’s recommended protocol, and analyzed on 10% Novex TBE polyacrylamide gels (Life Technologies). The gels were stained with SYBR Gold DNA stain (Life Technologies) for 10 min, and imaged with a Gel Doc gel imaging system (Bio-Rad). Quantification was based on the relative band intensities. The indel percentage was determined by the formula, 100 × (1 − (1 − (b + c)/(a + b + c))1/2), where a is the integrated intensity of the undigested PCR product, and b and c are the integrated intensities of each cleavage product.
METHOD DETAILS QUANTIFICATION AND STATISTICAL ANALYSES
In vitro cleavage experiments were performed at least three times, and representative results are shown. In vivo cleavage experiments were performed at three times, and data are shown as mean ± s.e.m (n = 3).
DATA AND SOFTWARE AVAILABILITY
The atomic coordinates of the LbCpf1 ternary complexes have been deposited in the Protein Data Bank, with the accession numbers PDB: 5XUS (the TTTA PAM), 5XUT (the TCTA PAM), 5XUU (the TCCA PAM) and 5XUV (the CCCA PAM). Data of in vitro cleavage experiments have been deposited in the Mendeley Data repository (doi:10.17632/3fbrghfbkt.1). The CueMol program is available at http://www.cuemol.org.
Supplementary Material
Acknowledgments
We thank Dr. Takanori Nakane for assistance with data processing, and Masahiro Fukuda for assistance with model building. We thank the beamline scientists at PXI at the Swiss Light Source and BL41XU at SPring-8 for assistance with data collection. H.N. is supported by JST, PRESTO (JPMJPR13L8), and JSPS KAKENHI (grants 26291010 and 15H01463). F.Z. is supported by the NIH through NIMH (5DP1-MH100706 and 1R01-MH110049) and NIDDK (5R01DK097768-03), the New York Stem Cell, Simons, Paul G. Allen Family, and Vallee Foundations, and B. Metcalfe. F.Z. is a New York Stem Cell Foundation Robertson Investigator, a founder of Editas Medicine, and a scientific advisor for Editas Medicine and Horizon Discovery. O.N. is supported by the Basic Science and Platform Technology Program for Innovative Biological Medicine from the Japan Agency for Medical Research and Development (AMED), and the Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science, and Technology.
Footnotes
Author Contributions
T.Y. solved the crystal structures and performed in vitro cleavage experiments with assistance from H.N.; B.Z. performed in vivo cleavage experiments; R.I. assisted with the structural determination; T.Y. and H.N. wrote the manuscript with help from all authors; H.N., F.Z., and O.N. supervised all of the research.
Accession Numbers
The Protein Data Bank accession numbers for the atomic coordinates of the LbCpf1-crRNA-DNA complexes reported in this paper are: 5XUS (the TTTA PAM), 5XUT (the TCTA PAM), 5XUU (the TCCA PAM), and 5XUV (the CCCA PAM).
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Ren K, Qiu X, Zheng J, Guo M, Guan X, Liu H, Li N, Zhang B, Yang D, et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532:522–526. doi: 10.1038/nature17944. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096–1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr Sect D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lécrivain AL, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- Gao P, Yang H, Rajashankar KR, Huang Z, Patel DJ. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 2016;26:901–913. doi: 10.1038/cr.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang F. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017 doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, Ishitani R, Hatada I, Zhang F, et al. Structure and engineering of Francisella novicida Cas9. Cell. 2016;164:950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Kim K, Been KW, Baek G, Ye S, Hur JW, Ryu SM, Lee YS, Kim JS. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat Biotechnol. 2016;34:807–808. doi: 10.1038/nbt.3596. [DOI] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–822. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis T, Gasiunas G, Young J, Bigelyte G, Silanskas A, Cigan M, Siksnys V. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015;16:253. doi: 10.1186/s13059-015-0818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Cheong SA, Lee JG, Lee SW, Lee MS, Baek IJ, Sung YH. Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol. 2016a;34:808–810. doi: 10.1038/nbt.3614. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur J, Been KW, Yoon S, Kim JS. Genome-wide target specificities of Cpf1 nucleases in human cells. Nat Biotechnol. 2016b;9:1–7. [Google Scholar]
- Kim HK, Song M, Lee J, Menon AV, Jung S, Kang YM, Choi JW, Woo E, Koh HC, Nam JW, et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat Methods. 2016c;14:153–159. doi: 10.1038/nmeth.4104. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK. Genome-wide specificity profiles of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2016;168:1–17. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Chen P, Wang M, Li X, Wang J, Yin M, Wang Y. C2c1-sgRNA complex structure reveals RNA-guided DNA cleavage mechanism. Mol Cell. 2016;65:1–13. doi: 10.1016/j.molcel.2016.11.040. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Nureki O. Structures and mechanisms of CRISPR RNA-guided effector nucleases. Curr Opin Struct Biol. 2017;43:68–78. doi: 10.1016/j.sbi.2016.11.013. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Yamano T, Gao L, Zhang F, Ishitani R, Nureki O. Structural basis for the altered PAM recognition by engineered CRISPR-Cpf1. Mol Cell. 2017 doi: 10.1016/j.molcel.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, van der Oost J, Jinek M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol Cell. 2017;66:221–233. doi: 10.1016/j.molcel.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3:17018. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Waterman DG, Winter G, Parkhurst JM, Fuentes-Montero L, Hattne J, Brewster A, Sauter NK, Evans G, Rosenstrom P. The DIALS framework for integration software. CCP4 Newsletter. 2013;49:16–19. [Google Scholar]
- Wright AV, Nunez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing Nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- Yamada M, Watanabe Y, Gootenberg JS, Hirano H, Ran FA, Nakane T, Ishitani R, Zhang F, Nishimasu H, Nureki O. Crystal structure of the minimal Cas9 from Campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems. Mol Cell. 2017;65:1109–1121. doi: 10.1016/j.molcel.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, Fedorova I, Nakane T, Makarova KS, Koonin EV, et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016;165:1–14. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gao P, Rajashankar KR, Patel DJ. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell. 2016;167:1814–1828. doi: 10.1016/j.cell.2016.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2016;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


