Abstract
Single molecule atomic force microscopy and magnetic tweezers experiments have demonstrated that titin Ig domains are capable of folding against a pulling force, generating mechanical work which exceeds that produced by a myosin motor. We hypothesize that upon muscle activation, formation of actomyosin crossbridges reduces the force on titin causing entropic recoil of the titin polymer and triggering the folding of the titin Ig domains. In the physiological force range of 4–15 pN under which titin operates in muscle, the folding contraction of a single Ig domain can generate 200% of the work of entropic recoil, and occurs at forces which exceed the maximum stalling force of single myosin motors. Thus titin operates like a mechanical battery storing elastic energy efficiently by unfolding Ig domains, and delivering the charge back by folding when the motors are activated during a contraction. We advance the hypothesis that titin folding and myosin activation act as inextricable partners during muscle contraction.
Keywords: Muscle contraction, titin, protein folding, polymer physics, single molecule, force spectroscopy
Introduction
For most of the past century, the existence of titin, the largest protein in the human body, was unknown (1). Unsurprisingly, theories of muscle contraction were based on the molecules known at the time, specifically actin and myosin. But on the eve of the discovery of the sliding filament hypothesis, there existed several competing theories that uncannily anticipated the role of a titin-like filament in the mechanism of muscle contraction. In 1952 Milton Polissar, a close collaborator of physical chemist Henry Eyring, used kinetic rate theory to predict the existence of a segmented filament composed of 100 independent contractile units that could shorten to do mechanical work under force (2–5). His ideas fell to the wayside when the sliding filament theory was developed with ample experimental support (6–9). In a fascinating twist of fate, we know today that both theories are correct and that both mechanisms contribute to muscle contraction. There is now conclusive evidence to establish that the muscle sarcomere is comprised of three main filaments that work in concert to store elastic energy and perform mechanical work: titin, actin and myosin (10). The evidence of titin as the third filament in muscle was initially taken with skepticism (11), and it took several decades before the presence of titin in the sarcomere was widely accepted (12, 13). Nearly a century elapsed between the discovery of myosin and development of the sliding filament hypothesis (14); we are undoubtedly in only the early stages of understanding the physiological functions of titin.
Titin is the largest protein in the human body, consisting of a single polypeptide chain with the largest known isoform having a molecular weight of 3.7 MDa (15). Each titin filament spans an entire half sarcomere, which is divided into four distinct regions easily recognized under the microscope (Figure 1A). The I-band of muscle is variable in length and is comprised mainly of the actin filaments, which run parallel to titin. Depending on the degree of sarcomere strain, some portion of the actin filament may run into the A-band. The A-band is demarcated by the ends of the thick filament, which is a bundle of 294 myosin molecules, and is found to be 1.65 um long in most vertebrate species (16). When cross sections of muscle are viewed with the electron microscope, one can see that the fibers are arranged in a hexagonal lattice with six actin filaments surrounding each myosin bundle (17). The thick filaments are held together by proteins in the M-line while the actin filaments are anchored to alpha-actinin in the Z-disc.
Figure 1. Magnetic tweezers force spectroscopy shows titin refolding against a pulling force.
(A) Schematic depicting a skeletal muscle fiber in rigor at a sarcomere length of ~2.6 μm. At this extension, titin is under a hypothetical force of 4 pN and all Ig domains are folded (87). Zoom into a half sarcomere: Titin is a single polypeptide that spans from the Z disc (N-terminal) to the M band (C-terminal), and is arranged as a sequence of folded domains (filled circles) and unstructured regions (grey lines). Titin consists of four regions: the N-terminal segment (green circles), is made of immunoglobulin-like (Ig) domains and anchors titin to the Z disc; the I-band segment is the elastic component and contains two types of domains, tandem Ig-domains (red circles) and intrinsically disordered structures (N2-B and PEVK); the A-band region (white circles) is comprised of Ig and fibronectin type-III (FNIII) domains responsible for titin binding tightly to the thick filament (N.B. the thick filament is truncated to magnify the I-band segment); finally, the C-terminal segment (blue circles) contains 10 Ig domains and anchors titin to the proteins of the M-band. Titin has traditionally been considered to control only the passive elasticity of muscles, while all mechanical work is performed by the actomyosin crossbridges. Note that the relative lengths of the different titin sections are not drawn to scale. (B) Typical recording of a magnetic tweezers experiment. First a high force pulse (105 pN) results in unfolding of eight Ig-domains measured as upwards steps (black arrowheads) of 25 nm (see Figure 2A). Upon reducing the force (5 pN), the molecule first experiences an instantaneous elastic contraction (1), followed by a folding contraction (2) where the individual domains refold as discrete downward steps (red arrowheads) that shorten the polypeptide. The unfolding pulse is performed at an un-physiologically high pulling force so that the eight domains unfold in a few seconds. The gray dashed trace shows a projected unfolding trajectory at a physiological force of 10 pN, requiring ~6 hours for complete unfolding. When the force is reduced to 5 pN, the magnitude of the elastic recoil is much less but the folding contraction remains the same. (Inset) Schematic depicting a single molecule magnetic tweezers experiment. The titin polyprotein is anchored to the glass surface by Halo-Tag covalent chemistry and to a superparamagnetic bead using biotin-streptavidin linkage. The force is controlled by positioning a pair of permanent magnets over the fluid chamber containing the magnetic bead.
Titin is found in each of these four regions of muscle. The N-terminus of titin contains nine immunoglobulin (Ig) domains (Figure 1A, green) that also bind to proteins in the Z-disc. Titin of the I-band region (Figure 1A, red) contains alternatively spliced exons that encode up to 100 Ig domains as well as intrinsically disordered regions, namely the N2-B sequence and PEVK sequence, which is rich in proline, glutamate, valine, and lysine residues. It is important to note that the number of Ig domains and unstructured amino acids found in the I-band depends on alternative splicing and is thought to be different for each muscle group. Although the longest skeletal isoforms may have 100 Ig domains, the predominant N2B titin isoform of cardiac tissue has only 47 Ig domains and 735 unstructured amino acid residues. The size of the titin isoform is important for understanding the forces experienced by titin at any given sarcomere length (18). The A-band region of titin (Figure 1A, gray), contains both Ig and fibronectin domains that are tightly bound to the thick filament. Finally, the M-band section of titin (Figure 1A, blue) contains the titin (pseudo)kinase (19, 20) and 10 Ig domains, and is responsible for binding to the adjacent titin and other structural proteins. This review is concerned mainly with the force generating capabilities of the elastic I-band segment of titin but there are many excellent resources discussing the titin and muscle ultrastructure (21–25).
In the sliding filament hypothesis of muscle contraction, ATP hydrolysis and inorganic phosphate release result in structural changes of the myosin neck while the myosin head is bound to actin. This motion moves the thick filament relative to actin to generate force (9). While it is beyond doubt that the chemical energy of ATP powers the cycling of myosin motors as they slide along the actin filaments during a contraction (26–29), the role of titin during shortening is poorly understood and requires the sustained focus of the muscle community to be clarified. There are several competing theories for how titin might generate force during a muscle contraction (22, 30–32), but experiments at the muscle fiber level have failed to elucidate a molecular mechanism. Over the last 20 years, technical advances in force spectroscopy combined with protein engineering have been used to study titin dynamics under force (10, 33–39). Force spectroscopy data shows that titin stores and delivers elastic energy mostly by unfolding and refolding its numerous tandem Ig domain modules. It is now our responsibility to advance the knowledge of muscle contraction by incorporating titin folding dynamics into the framework of the sliding filament hypothesis. Here we make such an attempt.
Studies of titin under force
Traditional bulk biochemistry techniques such as chemical and thermal denaturation are not practical for understanding the function of titin within muscle. Titin operates in vivo under a mechanical load up to ~15 pN, with the load applied along a vector between titin’s attachments to the Z-disc and the tip of the thick filament. Single molecule force spectroscopy techniques such as atomic force microscopy (AFM), magnetic tweezers, and optical tweezers probe the nanoscale mechanics of titin along the same reaction coordinate that is experienced in vivo, providing more physiological insight than bulk techniques. With the emerging view that force modulates a variety of cellular processes by triggering conformational changes at a protein level, these techniques are quickly becoming an indispensable tool for understanding cell mechanics.
The basic execution of a protein force spectroscopy experiment requires tethering of a single polypeptide chain between an immobile surface and microscopic probe that can be positioned to apply tension. For AFM studies, the force probe is a silicon nitride microcantilever that can be attached to the end of a protein with either covalent or noncovalent interactions (40, 41). In magnetic tweezers, the protein is tethered to a paramagnetic bead subjected to a magnetic field (10, 40, 42, 43). Optical tweezers utilize a laser trap to apply force to a polypeptide attached to a dielectric particle (44, 45). In all cases, the position of the force probe is measured with nanometer, or even Angstrom precision, using high speed laser detection or videomicroscopy. Gradual changes in polypeptide length due to stretching, or abrupt changes in length due to domain unfolding and refolding can be measured with high spatiotemporal resolution (46–49).
Early single molecule force spectroscopy experiments investigated DNA (50, 51) and polysaccharide mechanics (52, 53), receptor-ligand unbinding (54), motor protein dynamics (26, 44), and enzyme conformational changes (55). However, the very first recording of reversible protein unfolding at nanometer resolution was carried out with micrometer long titin as a substrate (37). Using the AFM, native titin purified from bovine tissue was stretched at high force and allowed to refold by approaching the cantilever tip back to the pulling surface. Titin was a natural choice for these experiments, given that it contains many nearly identical in size immunoglobulin domains that produce the “sawtooth” pattern characteristic in constant velocity single molecule pulling experiments. Similar results using optical tweezers to stretch native titin filaments at high force demonstrated yielding of the protein in a stepwise or gradual manner, indicative of single domain unfolding events (35, 39). Recombinant protein engineering and molecular cloning techniques allowed smaller, more manageable portions of the elastic titin I-band to the studied in great detail (33, 56). This revealed a mechanical hierarchy within the Ig domains, with proximal Ig domains (nearer to the N-terminal) having lower mechanical stability than the distal Ig domains of the I-band. Further engineering of a covalent HaloTag anchor to either the N- or C-terminus of the protein has permitted specific immobilization of recombinant titin proteins for study at high force and long durations (41, 49, 57).
Over the past two decades, force spectroscopy has benefitted greatly from improvements in instrumentation and technology. Reminiscent of the voltage-clamp that underpinned the first successful studies of single ion channel gating, development of the force-clamp for the AFM greatly advanced our understanding of the energetics of protein folding (34, 38, 58). Force-clamp spectroscopy unambiguously follows single polyprotein unfolding and refolding trajectories under a defined pulling force by tracking the protein end-to-end length with Angstrom resolution. This allows for the study of the kinetics and the force dependency of protein folding and other biochemical reactions (59, 60). More recently, development of protein magnetic tweezers that operate under passive, infinite bandwidth force-clamp have permitted direct observation of equilibrium folding-unfolding transitions in titin Ig domains at forces below 15 pN (10, 43). These types of experiments offer a natural scenario in which to explore titin under conditions close to the ones under which it operates in vivo. An example of such an experiment is demonstrated in Figure 1B, which summarizes much of what we know today about titin mechanics. In this experiment, a single polyprotein of eight tandem titin Ig 91 (I91) domains is held under a constant stretching force in a magnetic tweezers apparatus. It is important to note that this I91 domain is the same as the I27 domain referred to in older papers (36, 61) due to a change in the nomenclature. The protein is initially subjected to an un-physiologically high force of 105 pN to quickly unfold all Ig domains. This results first in an elastic extension followed by the unfolding and extension of the eight Ig domains in only 20 seconds. By contrast, unfolding of I91 at a physiological force of 10 pN takes several hours (Figure 1B, gray dashed line). When the force on the polypeptide is reduced to 5.0 pN, titin undergoes instantaneous elastic recoil followed by a stepwise folding reaction.
Because titin was the first protein to be unfolded via force spectroscopy, it subsequently became a model for understanding the physics of protein folding. The mechanical stability of globular folds like titin depend on electrostatic interactions, hydrogen bonding, and hydrophobic interactions that are governed by the sequence and topology of the protein. Figure 2A demonstrates a detailed picture of a titin Ig domain reversibly unfolding under a mechanical force of 6.0 pN. The majority of the force is sustained by hydrogen bonds between the first (A-A′) and last (G) beta strands (62), which eventually yield and result in complete unfolding of the domain. This is thought to be the unfolding mechanism for most Ig domains, which show high structural similarity to I91. Once unfolded, the polypeptide becomes a random coil whose mean end-to-end distance is determined by the pulling force and scales with well-known polymer physics laws (63). For example, at 6.0 pN the folded I91 domain has an end-to-end length of approximately 4 nm (inferred from atomic coordinates, see PDB 1WAA (64)) while the extended polymer has a length of ~17 nm. Thus, the steps observed during the folding and unfolding transitions will have a size of 13 nm under a constant force of 6.0 pN. Figure 1B also demonstrates this scaling at forces of 5.0, 10, and 105 pN. Scanning for folding and unfolding transitions across a broad range of forces allows the data to be fit to a freely-jointed chain model (63) with a contour length of 25 nm and a Kuhn length of 0.9 nm (Figure 2B, solid line). In contrast to the linear elasticity of a rubber band, the entropic elasticity of a polymer is highly nonlinear, reaching 50% of its maximum extension over the physiological force range. At higher forces, the step size of protein unfolding approaches the contour length asymptotically.
Figure 2. Mechanical work generated by Ig domain folding is quantifiable using force-clamp magnetic tweezers.
(A) Ig domain unfolding/refolding under a mechanical tension. Under a pulling force of ~6 pN, Ig domains unfold and refold in equilibrium, extending or shortening by ~13 nm, following scaling laws of polymer physics. (B) Magnitude of the step size measured during unfolding/refolding events as a function of the force for the I10 and I91 titin Ig domains. The dependence of the step size can be well described by a freely-jointed chain model (solid line) with Kuhn length lk = 0.9 nm and contour length Lc=25 nm. (C) Folding probability of the Ig domains, calculated from the equilibrium fraction of folded domains (blue circles), at a given pulling force. Titin Ig domains can unfold at forces as low as 4 pN and fold at forces as high as 10 pN. The data can be well represented by a sigmoid function (solid line) with midpoint of ~6 pN. (D) The expected work generated by titin Ig domain folding can be obtained as the product of the folding probability (blue line) and the work done by the folding contraction (red line), obtained from numerical integration of the freely jointed chain model used to fit the step sizes. Titin Ig domains generate a peak work of 46 zJ at 5.7 pN, comparable to the energy delivered by the myosin motors.
The rate at which each of the titin Ig domains unfolds is vastly different, so during physiological stretch, only a subset of all the titin Ig domains may be unfolded. Cloning of native segments of eight tandem Ig domains from the I-band uncovered a mechanical hierarchy within titin. Ig domains within the proximal I-band, closer to the Z-disc, unfolded at forces much lower than Ig domains from the distal I-band, closer to the thick filaments (36). Therefore, although I91 takes several hours to unfold at 10 pN (43, 65), the proximal Ig domain 10 (I10) unfolds much more rapidly. Magnetic tweezers experiments on I10 show that at a force of 6.0 pN, there are approximately 66 unfolding events during a period of twenty minutes, or ~15 seconds between each unfolding event. Therefore at 10 pN, I10 is expected to unfold in a few seconds, although this needs to be verified experimentally. Further experiments with the native segment I4–I11 at forces between 5.0 – 6.6 pN also show rapid unfolding events and therefore low mechanical stability, supporting the view that the proximal Ig domains unfold readily in the physiological force range (10).
Although there is a wealth of information about the unfolding pathways of titin domains, the observation of folding pathways has been more elusive and was the driving force behind development of technology such as the force clamp (58). The force clamp revealed three distinct transitions during the transformation of a protein from a random coil to a native fold (34), and the results paint a very different picture than what is seen using bulk techniques. First, a protein extended under force experiences elastic recoil in response to a change in force, as seen in Figure 1B. The reduction in force results in a large increase in polymer entropy that must be shed for the protein to fold. Removal of this entropy requires crossing an energetic barrier created by the pulling force known as the entropic barrier (66). This transition, known as the collapse to molten globule transition, is thought to be driven by sequestration of hydrophobic residues into the core of the protein and results in the downwards steps seen in Figure 1B. Further reduction in configurational entropy is achieved through maturation from the molten globule to the native state, likely through formation of hydrogen bonds in the mechanical clamp. This molten globule to native transition results in very small changes in the end-to-end length of the protein so the different states must be detected by the drastically different mechanical stability of the molten globule and native states (67). The classic “two-state” models assumed in bulk studies of chemical denaturation or phi-value analysis are irreconcilable with the extended, collapsed, molten globule, and native states seen in single molecule folding experiments. Single molecule force spectroscopy offers much greater detail about the folding pathways compared to these techniques.
Protein folding performs mechanical work
Before the invention of the force-clamp AFM, some very simple measurements of folding under force were possible (33). In these experiments, a protein construct containing eight tandem titin Ig domains was first unfolded by pulling the AFM cantilever at constant velocity. The tip of the cantilever was subsequently lowered to within a short distance (tens of nanometers) from the pulling surface and held there for a few seconds without touching. This procedure had the effect of leaving a residual force on the protein. Upon pulling the cantilever again, the number of domains that had folded could be counted. Those experiments showed that the refolding rate strongly depended on the degree of relaxation of the unfolded protein (33), which provided evidence that the refolding rate depended steeply on the residual force. However, in those early experiments, the force under which the protein was folding could not be measured directly. The development of the force-clamp AFM, provided the first direct observation of the full trajectory of a protein folding under a mechanical force (34). Using protein constructs consisting of tandem ubiquitin domains, Fernandez and Li demonstrated that upon “quenching” the pulling force on an unfolded protein, the protein first recoiled elastically, and then after a lag, it contracted against the pulling force doing mechanical work as it shortened towards its fully folded length. After the force quench, a subsequent high force pulse revealed the fingerprint of the folded domains, proving that these collapse trajectories were true observations of the folding process. Similar experiments using shorter quench durations revealed the presence of a mechanically weak state that served as precursor for the fully folded proteins, similar to what has been dubbed the molten globule state (67–69). This clarified the physics of folding under force, where in response to a force quench, a protein would first recoil elastically from an extended state to a collapsed state, greatly increasing its entropy. From the collapsed state the protein would contract further as it shortened in a stepwise manner to a low entropy molten globule state. In the molten globule state protein would “mature” to their mechanically stable native conformation by reforming the network of hydrogen bonds that provides mechanical stability. In this scenario, most of the folding contraction occurs as the protein moves from the collapsed to the molten globule state, whereas the reformation of the hydrogen bond network does not significantly change the end to end length of the protein (67). Using magnetic tweezers, collapse trajectories of I10 and I91 have allowed the folding and unfolding step size to be measured at forces below 30 pN (Figure 2B) and have provided more concrete evidence for the molten globule state (70). Although only these two domains have been studied in detail, they seem to undergo the folding transition in the same range of forces (4–10 pN) despite their drastically different mechanical stabilities during unfolding. Perhaps this is because each of the Ig domains, containing approximately the same number of residues, must discharge roughly the same amount of entropy during the collapse to molten globule transition. However, as it has been shown that configurational loop entropy (71) and point mutations can affect unfolding pathways (72), it is likely that the variations in the sequences of the Ig domains will also lead to a hierarchy among the folding rates around the physiological range. This must be verified experimentally, but at this point the evidence suggests that during muscle contraction, the titin Ig domains fold in a narrow force range.
The folding contractions observed with the AFM appeared continuous and only rarely showed steps (34). By contrast, the same experiments performed using magnetic tweezers, after the initial elastic recoil, show clearly resolved steps during the folding contraction (10, 43, 49). The reason for the different shapes of the folding trajectories in the AFM and magnetic tweezers is that in all single molecule force spectroscopy experiments, the dynamics of the polypeptide are tether dependent (73). The polypeptide can only move as fast as the probe that it is attached to, so the dynamics of the chain along the pulling direction have effectively the same diffusion coefficient as the probe. A triangular cantilever normally used in single molecule pulling experiments with length and width of 310 μm and 20 μm has a measured diffusion coefficient of ~1,200 nm2/s (73). Brownian dynamics simulation of folding using the diffusion coefficient of the cantilever have been able to reproduce folding trajectories similar to those seen using the force-clamp AFM (66, 74). By contrast a freely floating spherical particle with a diameter of 2.8 μm has a diffusion coefficient of ~175,000 nm2/s although the close proximity of the magnetic bead to the surface will reduce that value somewhat (75, 76). This contributes to the differences in the collapse trajectories seen in magnetic tweezers. A word of caution is warranted here. While the beautiful stepwise folding trajectories observed with magnetic tweezers allow for a better resolved picture of the folding energy landscape (77) and the folding work (10), it may not represent the true diffusional behavior of a folding titin protein in vivo. Given that titin is anchored to large molecular structures at both ends, the Z and M lines, and that it operates in a crowded environment that includes chaperones, the folding contraction in vivo may more resemble the diffusionally constrained folding behavior observed with the AFM that shows a smoother folding contraction (34, 74).
We propose that the physiological force range for the load on titin during normal muscle activity is the force range over which the Ig domains exist in equilibrium between the folded and unfolded states. This can be quantified through the folding probability, which measures the fraction of time a titin Ig domain will spend folded at any given force. The folding probability of titin Ig domains I10 and I91 measured with magnetic tweezers force spectroscopy is summarized in Figure 2C (blue circles), and can be fit with a sigmoid function that describes the midpoint and the steepness of the folding reaction (blue curve, Figure 2C and 2D). It is clear that titin Ig domains are completely folded below forces of 4 pN and do not fold at forces much higher than 10 pN. There is now evidence that extends this force range up to 15 pN. The I-band region of titin is cysteine rich (78, 79) and many of these residues form disulfide bonds that alter the polymer dynamics of the unfolded state. One consequence of this is that it allows titin Ig domains to unfold at much higher forces, up to 15 pN, without having much effect on the mechanical stability of the folded state (80).
Force-clamp spectroscopy additionally allows simple quantification of the amount of contractile work performed by a titin domain that is folding against an applied load. At any given force, the amount of contractile work done on the system or by the system is the product of the force and the step size. The work done by folding of I10 and I91 is shown by the red curve in Figure 2D, which is the integral of the freely-jointed chain curve with respect to force. Multiplying the curve representing contractile work done with the folding probability gives the expected value of energy recovery by protein folding at any given force (black curve, Figure 2D). This distribution shows that titin Ig folding generates a maximum expected amount of work of 47 zJ against a load of 5.7 pN. At higher loads, close to 10 pN, the energy recovery is maximized but the domains fold less frequently. Alternatively, at lighter loads, the energy recovery is minimal because the step size of protein folding is small and most of the energy is dissipated in the elastic recoil. Thus we are able to predict the amount of energy storage and release through titin folding if forces on single titin filaments can be determined in vivo.
How the three filaments work together
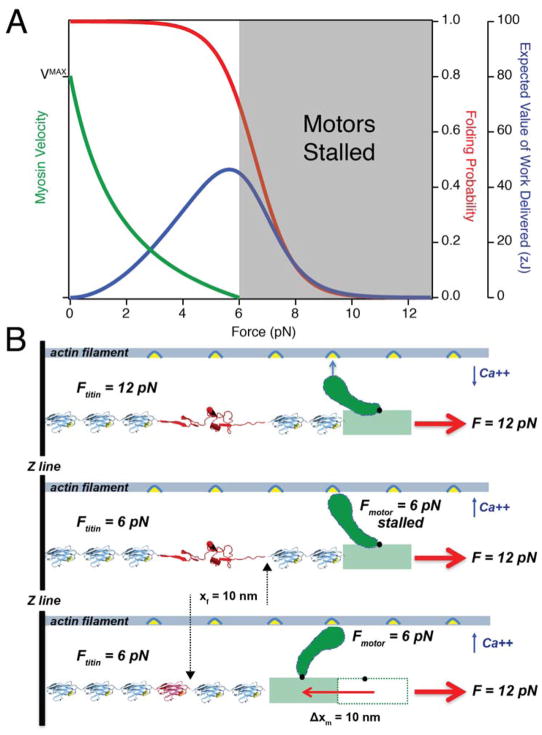
Single molecule experiments have provided a strong foundation not only for understanding the energetics of protein folding, but also for understanding the basis of force generation of single myosin motors. This provides a means for fair comparison between contractile properties of titin Ig folding versus the myosin power stroke. Single molecule force spectroscopy on single or small ensembles of myosin motors have revealed an inherent limit on the amount of useful mechanical work that ATP driven motors can perform. If a great enough load is applied to the motor, its power output will drop to zero. A cyclist pedaling uphill is keenly aware of this bit of physics. The early muscle pioneer A.V. Hill captured these effects with his famed “Hill Equation” that describes the hyperbolic relationship between contractile velocity and loading (81). In vitro assays have shown that the velocity of unloaded muscle can be very high, but applying even a small load to the muscle myosin II motors causes the sliding velocity to drop precipitously. The seminal experiments probing single skeletal myosin mechanics showed that under approximately isometric conditions, the average force produced per motor is 3.4 pN (26). Rarely, the myosin was able to generate sustained force transients in the 6–7 pN range. More recently, the same experiment was performed but with small bundles of ~8 myosin motors in an attempt to determine the single molecule force-velocity relationship (Figure 3A, green curve) (82). The myosin motors again showed net zero displacement of the actin filaments at an applied load of 6.0 pN, reaffirming that there is an upper limit to the performance of these molecular machines, which has since been reaffirmed with higher resolution optical tweezers (29). Others have shown that the stalling forces of kinesin, myosin V, and dynein are all in the 6–7 pN range (83–85). These studies used force clamp to reduce transducer compliance and maximize the force generation of the molecular motor. From these data it is logical to conclude that at forces of 6 pN, myosin II motors cannot advance along the actin filament to generate contractile work. By contrast, we showed that titin folding delivers near peak power at a load of 6 pN, where the myosin II motor is stalled (Figures 2D and 3A) (10).
Figure 3. Titin enables muscle to work at loads beyond the stalling force of myosin.
(A) Comparison of the energetics of the myosin motor and the titin Ig domain from single molecule studies: data from an experiment in which ~8 myosin heads propelled an actin filament in an optical trap show that myosin is stalled against a load of 6 pN (82). The data is fit with the A.V. Hill equation (green curve). Alternatively, titin Ig domains achieve optimal delivery of 46 zJ of work at forces of ~5.7 pN (peak of blue curve). Thus, at loads of 6 – 12 pN (gray shaded area), titin folding can generate useful contractile work, whereas the myosin power output is negative. (B) Titin folding allows the myosin motors to advance: (1) before the muscle is activated, titin bears the entire passive load of 12 pN, resulting in Ig domain unfolding. (2) Upon calcium release, actomyosin crossbridges form, but the motor can only generate a force of 6 pN so it is stalled. However, the force generated by the motor reduces the load experienced by the titin domain to 6 pN, which results in a refolding step of ~10 nm. (3) The process of protein folding shortens the titin filament, which relieves strain on the myosin motor and allows completion of the power stroke. From this mechanism, it is clear that titin cannot fold without formation of the crossbridge, but also that completion of the power stroke relies on titin folding. Thus, titin and myosin work hand-in-hand to shorten the sarcomere against high loads.
A plausible scenario in which this might occur during muscle contraction is when the sarcomere is passively loaded so that each titin filament bears 12 pN of force (86, 87). How might this load distribute between titin and myosin before and after muscle activation? If one considers the 147 double-headed myosin molecules present in the half thick filament and the 6 titin molecules per half sarcomere, then there is an absolute maximum ratio of 25 myosin molecules per titin filament. However, not all myosin heads are attaching to actin binding sites in a shortening muscle. Muscle myosin is a non-processive motor where the percentage of myosin heads attaching to actin during a contraction has been estimated at 10–20% of the maximum number of available attachment sites (88). By measuring the compliance of muscle fibers during a contraction, it was calculated that up to 84 (~28%) myosin heads/thick filament might be engaged and that the number of attached heads changes steeply both with filament overlap and with shortening velocity (89). However, titin elasticity was not considered in those experiments, and given that titin becomes stiffer as its domains fold, a fraction of the observed changes in stiffness may originate from the titin filament rather than myosin, leading to an overestimate in the number of myosin motors engaged in a contracting muscle. At physiological sarcomere extensions of 3.1 μm in rabbit psoas or 3.4 μm in human extensor digitorum, the degree of overlap between the thick and thin filaments would be reduced and further lower the number of bound myosin heads (10, 90). Hence, only 1–2 myosin heads per titin molecule are calculated to support a contraction from such a stretched length.
Returning to the hypothetical scenario, titin bears the entire 12 pN load prior to muscle contraction, which recruits proximal Ig domains to the unfolded state (Figure 3B, upper panel). This might occur during the priming of muscle for explosive shortening, for example during the windup of a baseball pitcher. In the few milliseconds after muscle activation, the maximum number of cross bridges is formed and the load is distributed through the mechanical network consisting of the three muscle filaments in the sarcomere. The myosin motors will generate a maximum of 6 pN of force, which would not be enough to shorten the sarcomere, but would reduce the load on all the titin Ig domains to 6 pN (Figure 3B, middle panel). This would result first in the extremely rapid entropic recoil of all the stretched titin molecules, followed by folding of titin at the force that recovers the maximum available work (Figure 3A, peak of blue curve). Unlike previous theories that assume titin recovers energy only via entropic elasticity, the majority of the energy recovered here actually comes from the folding contraction. At a force of 12 pN, the change in contour length achieved by going from the folded state to a random coil is 15.8 nm. Upon reduction of the force to 6.0 pN, there is 5.9 nm of entropic recoil followed by 9.9 nm of folding contraction (Figure 3B, middle panel). Protein folding comprises 63% of the energy recovered from the unfolded polypeptide while the elastic recoil contributes only 37%. The folding contraction is the dominant component of the elasticity and energy storage of titin in the physiological force range. Thus, in this scenario, the folding Ig domains relieve the stalled motors and permit the sarcomere to shorten, rather than lengthen, under the 12 pN load (Figure 3B, lower panel). The energy that is stored by stretching titin at 12 pN is returned only when the motors become engaged. This remarkable mechanism readily extends the ability of a sarcomere to deliver mechanical power at loads well beyond the stall force of its myosin II motors.
Titin folding dictates the optimal sarcomere length of a contracting muscle
The existence of the titin folding contraction may warrant the revisiting of some tenets of muscle physiology. For example, it is generally accepted that the descending limb of the active force – sarcomere length relationship is caused by the decreased overlap of the thick and thin filaments. However, the force on the ascending limb is already submaximal at sarcomere lengths of 1.7–2.6 μm in human skeletal muscles (91). The thick filament is only ~1.6 μm long, so it is unlikely that reduced force in this region of the ascending limb is caused by compression of the thick filament against the Z-discs. An alternative explanation would be that isometric force in this region is decreased because all titin Ig domains remain folded. Thus, there is no additional contractile force generated by hydrophobic collapse of the titin polypeptide. Consider a hypothetical skeletal muscle with a titin isoform of intermediate length and stiffness (Figure 4A, blue curve). This titin isoform would permit the highest performance possible from muscle because the maximum change in the titin folding probability falls squarely within the region of maximal active force generation (Figure 4A, pink shaded area). Measurement of the maximal isometric force in the gray shaded regions lacks the contribution of titin folding because titin either remains folded or unfolded at those sarcomere lengths (Figure 4A, gray shaded area). These seemingly simple experiments that support the foundation of muscle physiology are not so straightforward once considered in the context of titin folding.
Figure 4. Titin folding occurs at physiological sarcomere lengths.
(A) The active force versus sarcomere length relationship of human skeletal muscle (black curve) (91) is superimposed on the titin folding probability versus sarcomere length relationship (grey curve) for a hypothetical muscle with a titin isoform of intermediate length. Here, the folding probability quickly changes from zero to one in the range where active force is maximized, greatly enhancing the muscle contractile force. On the ascending limb of the active force curve, titin remains folded and cannot contribute to force generation at sarcomere lengths less than ~2.5 μm. On the descending limb, titin provides mainly entropic elastic restoring forces because there is very little folding at sarcomere lengths greater than ~3.1 μm. Not all muscles operate at the peak of the active force versus sarcomere length relationship, so it is likely that each muscle group has a titin isoform alternatively spliced such that the titin folding probability falls in the working range of that particular muscle group (87). (B) The segmentation of the titin I-band into many discrete Ig domains endows it with special material properties. A titin under normal physiological conditions (solid blue curve) experiences folding and unfolding of its Ig domains within the physiological force range of 4 – 15 pN (gray shaded area). The folding probability and hence the elastic properties of titin can be altered by blocking the folding reaction altogether (e.g. by S-glutathionylation, dashed blue curve). Introduction of intramolecular disulfide bonds through oxidative stress or enzymes increases the forces at which the folding reaction occurs (solid gold curve). Binding of small molecules or chaperones that increase the mechanical stability of titin would make the Ig domains more inextensible and increase the overall stiffness of titin (dashed red curve). In reality, the elasticity of titin is some mixture of all of these curves and depends on the metabolic state of the muscle.
Why protein folding?
The reversible deformation of muscle is a material property that is perhaps unmatched in other tissues. Muscle must undergo continuous cycles of stretch and relax without demonstrating material fatigue or hysteresis. For some muscle groups, the sarcomere may extend by up to 300 nm from the fully contracted to fully extended state, which is about a 10% increase in its length (90, 92, 93). The proteins that contribute to reversible muscle elasticity must fulfill two opposing roles: they must be soft enough so that the muscle can easily deform and does not hinder the contraction of the opposing muscle group, and they must be stiff enough to prevent damage by overstretching muscle and also store significant amounts of elastic energy that can be released during muscle contraction. It turns out that polypeptide filaments can be excellent material for the storage and release of elastic energy, if they are structured correctly.
Many cytoskeletal structures are rigid in the force range over which muscle operates. Microtubules have a persistence length greater than a millimeter, while f-actin has a persistence length of tens of microns (94). Intermediate filaments such as keratins and vimentins have a persistence length of hundreds of nanometers up to a micron (95). Type I collagen, which consists of a triple helix of three individual polypeptide chains, is thought to have a persistence length of tens to hundreds of nanometers (96). None of these materials are useful as an elastic storage mechanism simply because they are too stiff. Over the range of forces experienced by single protein filaments during normal muscle operation (1 – 15 pN), these proteins are already completely extended, like rigid rods, and would resist muscle elongation. Alternatively, unstructured or unfolded polypeptide chains have been characterized with a wide variety of techniques and have been shown to have a Kuhn length ranging from 0.8 nm to 1.2 nm (equates to a persistence length of 0.4 nm to 0.6 nm). Below 5 pN, a random polypeptide coil will behave approximately as a Hookean spring, but at larger forces the random coil exhibits nonlinear behavior, with the force rising more steeply as the extension increases. The recent discovery that titin is under forces of 4 to 15 pN during physiological muscle activity raises the question: is it possible to fashion a material that is able to store more elastic energy than a simple random polypeptide coil in this force range?
The structure of titin provides an extraordinary answer to this problem by containing both unstructured segments (PEVK, N2B) and many tandem Ig domains. The segmentation of the titin I-band into Ig domains greatly increases the elasticity of muscle in the physiological force range. The polypeptide stores an amount of energy W = FΔL where F is the force determined by the FJC and L is the observed length. If titin was comprised of only unstructured polypeptide, a reduction in the force from 12 pN to 6 pN would cause collapse to 63% of its extended length. For an unstructured segment containing ~90 amino acids, the length would shrink from 15.7 to 9.9 nm and the resulting elastic energy recovered would be 34.8 zJ. Now considering the same 90 amino acids in a segment that has the ability to fold, reducing the force from 12 to 6 pN would first release 34.8 zJ and then an additional 59.2 zJ upon folding. The total energy recovery is now increased to 94 zJ with nearly two thirds coming from the folding contraction. So not only does the Ig folding and unfolding permit titin to dynamically adjust to provide tension across a broad range of sarcomere lengths, it also greatly increases the energy storage and recovery within the muscle. The energy storage and recovery are only possible within a very precise force range where protein folding and unfolding occur. We propose that muscle is tuned to operate at lengths and forces that straddle the folding probability and therefore take advantage of the full elastic storage potential of the titin filaments. Furthermore, by manipulating the titin folding process through cellular signaling, the muscle can alter its ability to store elastic energy.
The titin folding reaction can be biochemically tuned
Titin is not a purely mechanical or structural protein of muscle. It is also an integrator of chemical and physical stress on the muscle that is a part of signaling pathways that ultimately affect gene expression. The elastic segment of titin has been shown to be a major target of post-translational modifications. While phosphorylation of residues within the unstructured regions of titin have been shown to cause modest changes in the elasticity of titin (97–99), far more drastic changes can be achieved by targeting the Ig domains of the I-band. In a mouse model of myocardial infarction, oxidative stress due to ischemia-reperfusion injury lead to massive S-glutathionylation of sarcomeric proteins, including some very high molecular weight proteins alleged to be titin (100). The Ig domains of the titin I-band contains a disproportionately high number of cysteine residues; however, most of these cysteine side chains are cryptic, meaning that they are buried in the hydrophobic core of the Ig fold and are solvent inaccessible. They cannot undergo S-glutathionylation unless the Ig domains become unfolded. Alegre-Cebollada and colleagues demonstrated, using single molecule force-clamp AFM, the S-glutathionylation of single titin I91 domains unfolded by force (78). A critical discovery was that S-glutathionylation created unfavorable sterics in the hydrophobic core of the I91 domain that blocked folding. It was then shown that oxidized glutathione was able to massively decrease the passive tension in single cardiomyocytes, but only after cardiomyocyte stretch to recruit Ig domains to the unfolded state. Like phosphorylation, S-glutathionylation is reversible, either through enzymatic cleavage or regeneration of reduced glutathione within the muscle tissue. However, phosphorylation only results in small changes in the stiffness, or Kuhn length, of the titin molecule while S-glutathionylation or other non-specific oxidation of the Ig fold can cause massive blockage of the folding reaction. If all titin folding was blocked, the elastic properties would behave like the dashed blue curve in Figure 4B. It is speculated that the resulting loss of passive tension can be sensed by other sarcomeric proteins and transduced into cellular signals. Another major oxidative modification of titin is introduction of intramolecular disulfide bonds. The disulfide bonds are able to greatly increase the forces at which Ig domains fold without having much affect on the mechanical stability or unfolding rate (80). The resulting shift in the folding probability will cause the folding reaction to generate higher forces during contraction (gold curve, Figure 4B). In addition to oxidative stress, acidosis a common perturbation to the homeostasis of muscle and has direct consequences on protein folding (101). Changing the pH alters the protonation of many side chains, which in turn alters the electrostatic and hydrophobic forces that drive protein folding and provide mechanical stability. Acidosis may block folding or induce misfolding events that would reduce the extent of the folding contraction and lead to changes in muscle elasticity (102). An emerging perspective is also that muscle specific chaperones can be recruited to the I-band segment of titin in response to physical stretch (103), and that there are severely altered levels of molecular chaperones in certain muscular diseases (104). Some of these chaperones, such as αB-crystallin, can bind directly to the folded Ig domains and make them more resistant to unfolding (red curve, Figure 4B) (103, 105). Single molecule force spectroscopy has recently begun to uncover the ability of molecular chaperones not only to prevent aggregation but also accelerate the folding reaction of certain proteins (Haldar et al., unpublished data). An increase in the folding kinetics in synonymous with increase power output from the titin folding reaction and would therefore have direct consequences on muscle contraction according to the three-filament model.
A rechargeable gravitational battery
Our view shifts the focus of muscle contraction from resting solely on motor activity, to the remarkable ability of mechanically extended proteins to fold and shorten while resisting a stretching force. Energy conversion from sunlight to ATP through food consumption and the metabolic machinery of animal cells is the source of the chemical energy utilized during muscle contraction. What is the source of the energy delivered by a folding protein? We know that entropic elasticity combined with short range hydrophobic collapse powers protein folding. Yet, it is clear that for a protein to do folding work, it must have been mechanically unfolded before. The segmentation of the titin polypeptide into many foldable Ig domains thus differentiates it from a simple spring with only elastic recoil. This new mechanism is compelling because in addition to chemical energy, muscle makes use of other fundamental forces of nature: gravitation and inertia to extend and unfold muscle titin during animal motions like running, and entropic and hydrophobic forces to drive the collapse and folding of extended proteins while under reduced loads (34, 106). Muscle sarcomeres are thus designed to optimally absorb and store mechanical energy from the environment in the form of unfolded titin Ig domains, and deliver this mechanical energy when triggered to do so by chemically activated myosin II motors (Figure 3B). It remains clear that at the high loads at which myosin motors are unable to advance (6 pN), titin readily folds. Thus, titin folding relieves the stalled myosin motors, setting the speed at which they advance, and permitting efficient muscle contraction.
This active role of titin in muscle contraction can be understood with the analogy of an electrical battery (see Fig. 5A). The unfolding of Ig domains in the stretching phase charges the “battery” with contractile energy when the force exceeds 10 pN. As the force lowers when the motors engage, Ig domain refolding delivers the stored energy, allowing muscle contraction together with the work done by the myosin motors. Therefore, titin and myosin are the two components of a hybrid system where the electrical battery is charged during the stretching phase (black line, Fig. 5A), and only after the engagement of the myosin heads -powered by the ATP chemical energy- this stored contractile energy is delivered (red line, Fig. 5A). Nonetheless, the rate at which the contractile energy is stored and delivered is rather different, as reflected in the different slopes in Figure 5A. Titin unfolding at 10 pN occurs at a rate of ~1.5×10−4 s (65), therefore titin stores contractile energy with low power, estimated to be ~0.02 zW (obtained as the work of an Ig domain unfolding at 10 pN (140 zJ) times the rate at which it occurs). However, refolding occurs at a much higher rate, with domain folding at 6 pN in ~7 seconds, so the power delivered by titin Ig domains refolding is almost 100 times larger, estimated to be ~6.5 zW. The power relationships will be different for the different domains, so the power delivery will be much more complex in vivo. In an oversimplification of how muscle operates in animal motion, a leg muscle such as the soleus travels from its resting state to the stretching position, transmitting a high force purely to titin (1 to 2, lower panel Fig 5A). Later, and after the engagement of molecular motors and ATP hydrolysis, the force transmitted to titin decreases, allowing the delivery of mechanical work from titin and contracting the muscle (2 to 3, lower panel Fig 5A). In terms of the energy changes upon titin domain folding/unfolding, the storage/delivery cycle proposed above can be pictured as a diffusion process along the free energy landscape of the titin molecule (Fig. 5B). During the storage phase at 12 pN (green landscape) the energy landscape is tilted towards the unfolded state (large extension) so that titin domains would unfold, sequentially hoping over the free energy barriers. Upon force reduction to 6 pN (red landscape), the molecule shortens by refolding, the domains hoping the barriers in the opposite direction, thus delivering the stored contractile energy.
Figure 5. Muscle contraction model as a hybrid mechanism.
(A) Role of titin folding/unfolding in muscle contraction. (Upper panel) Titin domains work as a “battery”, storing contractile energy by unfolding and delivering it when shortening upon folding. When a muscle stretches, the mechanical tension increases and titin domains unfold (storage phase, (1) to (2)). When muscles contract, myosin heads engage, decreasing the force on titin to 6 pN, allowing Ig domains to refold and shorten, which deliver folding contractile energy and assist muscle contraction (3). (Lower panel) Simplified model for muscle stretching-contracting during animal motion, indicating each of the three phases upon titin unfolding and refolding. (B) Free energy landscape interpretation of titin-assisted muscle contraction. During the stretching phase, the landscape is tilted by the increased force (12 pN, blue) towards the unfolded state (longer length), and titin stores contractile energy by unfolding its domains. When the myosin motors engage, they reduce the mechanical load on titin to 6 pN, so the landscape favors the folded state, and domains can fold by surmounting the free energy barriers and shorten the total length of the molecule, delivering contractile work. (C) Testing this hypothesis requires incorporation of a genetically encoded force sensor into the titin sequence. One such design is provided here, utilizing HaloTag and SNAP-tag covalent labeling technology introduced in between exons I86 and I87. Using a hypothetical green donor fluorophore and red acceptor fluorophore, the FRET ratio of the muscle changes from a maximum to a minimum throughout the stride depicted in panel (A). This provides a direct readout on the force per titin molecule in vivo.
A limitation of our current observations is the slow folding rates measured at 6 pN for our eight repeat Ig domain protein probed with magnetic tweezers (Figure 1B). Indeed, we should be cautious when extrapolating single molecule observations to intact tissue, as we still do not fully understand how these observations play out in the complex environment of the sarcomere. However, a few simple observations are warranted. In addition to unfolded domains, native titin contains large unstructured segments such as PEVK and N2B that permit titin to equilibrate to any force in the sarcomere at the speed limit of polymer diffusion, which is much faster than even the fastest muscle contractions (73). Indeed, passive sarcomere shortening from a stretched length is very rapid initially (107, 108) and from recent in vivo muscle microscopy, it also appears that the sarcomere operates at lengths at which titin is under a constitutive force of several pN, further suggesting that titin does not go slack (90, 92, 93). More significantly, in contrast to the short, engineered proteins with eight Ig repeats often used in single molecule experiments, the extensible segment of native titin contains 50–100 Ig domains depending on the isoform, greatly accelerating the initial folding rate upon a reduction in force. Over the relatively short changes in sarcomere length observed in vivo, for example ~ 50 nm per half sarcomere in the cardiac tissue, the initial rate of folding would provide a better measurement of the physiologically relevant power output of titin folding. A glimpse of these effects can be observed during in-situ measurements of titin dynamics that show titin folding events happening on a timescale of less than 100 milliseconds (10). Finally, sarcomeres are crowded environments containing titin specific chaperones that will further increase the folding rate over what we have measured (102, 103). All of these effects, absent in the experiments shown in Figure 1B, would greatly enhance the power output of titin folding inside the sarcomere.
Synchronization of titin folding with actomyosin contraction
Our hypothesis for titin enhanced muscle contraction states that titin and myosin work cooperatively because titin folding depends on a reduction in force caused by engagement of the crossbridges. This mechanism predicts that the six titin molecules attached to a thick filament might fold at the same time. Single molecule force spectroscopy studies have demonstrated the simple scaling laws of parallel polyproteins. When protein domains are unfolded in parallel, they rupture simultaneously, with twice the force and half the persistence length (109, 110). Therefore, we predict that six titin Ig domains will also fold simultaneously due to their parallel arrangement around the thick filament. When the first of the six parallel Ig domains folds stochastically, the force on the other five is instantaneously reduced. The rate of unfolding or folding depends exponentially on the force, so even a small reduction in the force would accelerate the folding of other parallel domains. Hence, the power output from titin Ig folding is highly cooperative. Furthermore, it is misleading to say that actomyosin contraction is synchronous while titin folding is not (111). It is generally accepted that myosin driven muscle shortening occurs by “cyclic asynchronous ATP-driven actin-myosin contractions” (112). The formation of a cross bridge relies on diffusional encounter of the myosin head and actin filament which is by definition a stochastic process. Additionally, because the energy released by hydrolysis of ATP is only an order of magnitude larger than kT, it is thought that subsequent structural transitions of myosin rely, at least in part, on Brownian motion (113). In this view, the cycling of myosin motors and the folding of titin Ig domains are both stochastic, but in our model occur simultaneously upon chemical activation of muscle.
Testable predictions and the techniques to examine them
Muscles are complex systems, comprising several interacting proteins arranged in hierarchical structures. Determining the particular role of each of these proteins from a molecular level is a difficult enterprise. However, it seems now clear that the tandem modular arrangement of the titin molecule is not merely devoted to a role in muscle passive tension, but indeed these domains can unfold and refold in the force range under which they operate. This entails an active function of titin on the generation of contractile work in muscle shortening. However, there remains much experimental work to provide a detailed vision on the precise role of titin in muscle contraction and how the power output of domain folding interplays with that delivered by the myosin motors. The hypothesis discussed throughout makes a key testable prediction: activation of muscle leads to the engagement of the myosin motors, which lowers the load on titin to a range that triggers folding of Ig domains previously recruited to the unfolded state (10). We must ask ourselves, how can this energetic contribution of titin be isolated during an active contraction, and are the current biophysical tools sufficient?
Force spectroscopists wish to have tools that provide them with the precision of single molecule techniques at a whole tissue level. To some extent, this is being realized with genetically encoded force sensors that have been expressed within force-bearing cytoskeletal components of cultured cells (114). These force sensors utilize two fluorescent proteins that are able to exchange energy through non-radiative transfer (FRET), separated with a short unstructured or structured peptide. These genetically encoded force sensors have been incorporated into spectrin proteins (115), vinculin and talin (116, 117), and nuclear nesprins (118), all of which are subject to tension from intracellular stress fibers. As the tension on the protein containing the force sensor increases, the FRET donor and acceptor separate and the FRET ratio decreases. These force sensors are calibrated by measuring FRET ratio under applied tension using single molecule force spectroscopy over ranges from 0–10 pN (119). The titin I-band would be an ideal place to engineer such a force sensor, as depicted in Figure 5. Unlike in cell culture experiments, where cytoskeletal tension develops after cell adhesion in an uncontrolled manner, the tension on titin can be exquisitely tuned with a muscle fiber pulling apparatus (9, 108, 120). The uniform arrangement of the titin filaments within the sarcomere ensure that the fluorophores would be highly aligned, providing an excellent signal to noise ratio of the fluorescence readout in an intact muscle. Most importantly, we predict that the force on titin can be modulated by increasing the calcium concentration of the bath, resulting in cross-bridge formation that short circuits the force away from titin. According to our three-filament hypothesis of muscle contraction, formation of the crossbridges in a muscle under physiological stretch will lower the force on each titin to the range below 10 pN where titin folding delivers peak power. Not only would the genetically encoded force sensors allow the determination of the force per titin, but also the sarcomere lengths at which the titin folding contraction is triggered. Each muscle group contains a titin isoform of a different length—as shown, e.g., for a set of 40 different rabbit muscles (87). We predict that this length is programmed so that the titin folding contraction occurs within the optimal working sarcomere lengths of that particular muscle group. Such a tool would be invaluable for both in vitro and in vivo studies of muscle performance and would help move the field towards a better understanding of the physiological forces experienced by titin and the number of crossbridges needed to reduce the load on titin. These two factors, combined with the folding velocity of titin are crucial for determining the relative power output of titin and myosin during muscle contraction.
The first steps towards development of such an in vivo tool have already been achieved. A mouse model containing a knock-in of the HaloTag enzyme in the distal portion of the I-band has served as a multi-purpose tool for the biophysical characterization of titin (121). Use of the HaloTag enzyme was pioneered by our lab as a means to covalently anchor proteins to glass coverslips for single molecule pulling experiments (41, 49) and has seen widespread adoption for fluorescent imaging (122, 123). HaloTag recognizes and covalently binds to a chloroalkane substrate that can be attached to a variety of organic fluorophores with excellent imaging properties. Unlike antibodies, the chloroalkane ligands are very small and can easily permeate the sarcomere to achieve high density labeling. By placing an orthogonal labeling site such as SNAP-Tag, CLIP-Tag, or TMP-tag (122) next to the HaloTag a genetically encoded sensor similar to those described above can be achieved (Figure 5C). This has the advantage over the previously described genetically encoded force sensors of being able to use organic dyes with properties superior to those of fluorescent proteins. The set of fluorophores used may also be adapted to the experiment at hand, be it single muscle fiber stretching experiments or in vivo imaging. In addition to the HaloTag, the current mouse model also contains a TEV (viral) protease site. Unlike previous attempts to cleave the titin molecule (107, 124, 125), this would permit highly specific cutting of either half of the titin molecules (in a mouse heterozygous for the knock-in) or all the titin molecules (in a homozygote) to better understand the role of titin in muscle elasticity and force generation.
Delving deeper into the complexity of muscle contraction will require re-evaluation of the classic muscle contraction experiments while keeping in mind that titin plays a prominent role in both the passive and active mechanics of muscle. Given the downward trend in the compliance of the myosin neck (89, 126–128), the main source of energy storage in a contracting sarcomere must be reconsidered. Perhaps a genetically encoded force sensor would demonstrate that titin is the main source of energy storage in a reimagined version of the classic quick-release experiments of Huxley and Simmons (9). And although the single molecule experiments on reconstituted actomyosin complexes are ever improving (26, 28, 29) and yielding further insight into the mechanism of force generation by myosin, there has yet to be any direct observation of the rate or stroke size of single myosin motors within an intact sarcomere. In this respect, the myosin motor field is a step behind titin. Furthermore the treatment of a muscle like a thermodynamic engine by Fenn, Wilkie, and others, who measured muscle enthalpy using a thermopile and calorimeter, and assigning the resulting heat generation solely to the myosin motors greatly oversimplifies the problem (27, 129). A simple viscoelastic polymer undergoes a large increase in entropy upon a reduction in the force and hence absorbs heat from its environment. A folding Ig domain is not a simple viscoelastic polymer. When the myosin motors are engaged and the force on titin is reduced, there will first be an increase in entropy and titin will absorb heat. However, the collapse to molten globule transition, which is the dominant part of the folding contraction, results in the formation of a highly compact and highly ordered protein fold with low entropy. Thus, protein folding can result in a net loss of entropy that results in heat loss to the surrounding bath. The results of Fenn and others must be reconsidered now that we know that the thermodynamics of titin protein folding are not as simple as that of a viscoelastic polymer.
In conclusion, our view shifts the focus of muscle contraction from resting solely on motor activity, to the ability of mechanically extended proteins to fold against a stretching force. The unique material properties of titin conferred by its foldable Ig domains differentiates it from Hookean springs, or unstructured polymers, which would only undergo elastic recoil and therefore be a passive elastomer. Single molecule data clearly indicate that above 6 pN, myosin motors stall and are unable to advance and to produce mechanical work. Accordingly, titin Ig domains readily refold in that force range, delivering contractile work that relieves stalled myosin motors, and permit efficient muscle contraction. Therefore, this new evidence denotes that, in the same way it was recognized as the third muscle filament, titin needs to be considered as an active component of the sarcomere that works alongside myosin and actin as an inextricable partner in muscle contraction.
Acknowledgments
This work was supported by NIH Grants GM116122 and HL061228 and NSF Grant DBI-1252857 to JMF and 5F30HL129662 to ECE. RTR was supported by Fundación Ramón Areces. Special thanks to Zsolt Mártonfalvi for his critical reading and feedback on the manuscript.
References
- 1.Maruyama K, Matsubara S, Natori R, Nonomura Y, Kimura S, et al. Connectin, an Elastic Protein of Muscle Characterization and Function. J Biochem (Tokyo) 1977;82(2):317–37. [PubMed] [Google Scholar]
- 2.Polissar MJ. Physical chemistry of contractile process in muscle. I. A physicochemical model of contractile mechanism. Am J Physiol. 1952;168(3):766–81. doi: 10.1152/ajplegacy.1952.168.3.766. [DOI] [PubMed] [Google Scholar]
- 3.Polissar MJ. Physical chemistry of contractile process in muscle. II. Analysis of other mechano-chemical properties of muscle. Am J Physiol. 1952;168(3):782–92. doi: 10.1152/ajplegacy.1952.168.3.782. [DOI] [PubMed] [Google Scholar]
- 4.Polissar MJ. Physical chemistry of contractile process in muscle. III. Interpretation of thermal behavior of stimulated muscle. Am J Physiol. 1952;168(3):793–804. doi: 10.1152/ajplegacy.1952.168.3.793. [DOI] [PubMed] [Google Scholar]
- 5.Polissar MJ. Physical chemistry of contractile process in muscle. IV. Estimates of size of contractile unit. Am J Physiol. 1952;168(3):805–11. doi: 10.1152/ajplegacy.1952.168.3.805. [DOI] [PubMed] [Google Scholar]
- 6.Gordon AM, Huxley AF, Julian FJ. Tension development in highly stretched vertebrate muscle fibres. J Physiol. 1966;184(1):143–69. doi: 10.1113/jphysiol.1966.sp007908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon AM, Huxley AF, Julian FJ. The variation in isometric tension with sarcomere length in vertebrate muscle fibres. J Physiol. 1966;184(1):170–92. doi: 10.1113/jphysiol.1966.sp007909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huxley AF, Niedergerke R. Structural Changes in Muscle During Contraction: Interference Microscopy of Living Muscle Fibres. Nature. 1954;173(4412):971–73. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- 9.Huxley AF, Simmons RM. Proposed Mechanism of Force Generation in Striated Muscle. Nature. 1971;233(5321):533–38. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 10.Rivas-Pardo JA, Eckels EC, Popa I, Kosuri P, Linke WA, Fernández JM. Work Done by Titin Protein Folding Assists Muscle Contraction. Cell Rep. 2016;14(6):1339–47. doi: 10.1016/j.celrep.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huxley HE. Structural Arrangements and the Contraction Mechanism in Striated Muscle. Proc R Soc Lond B Biol Sci. 1964;160(981):442–48. doi: 10.1098/rspb.1964.0054. [DOI] [PubMed] [Google Scholar]
- 12.Maruyama K, Natori R, Nonomura Y. New elastic protein from muscle. Nature. 1976;262(5563):58–60. doi: 10.1038/262058a0. [DOI] [PubMed] [Google Scholar]
- 13.Maruyama K, Kimura S, Ohashi K, Kuwano Y. Connectin, an Elastic Protein of Muscle. Identification of “Titin” with Connectin. J Biochem (Tokyo) 1981;89(3):701–9. doi: 10.1093/oxfordjournals.jbchem.a133249. [DOI] [PubMed] [Google Scholar]
- 14.Stull JT. Myosin Minireview Series. J Biol Chem. 1996;271(27):15849–15849. doi: 10.1074/jbc.271.27.15849. [DOI] [PubMed] [Google Scholar]
- 15.Bang M-L, Centner T, Fornoff F, Geach AJ, Gotthardt M, et al. The Complete Gene Sequence of Titin, Expression of an Unusual ≈700-kDa Titin Isoform, and Its Interaction With Obscurin Identify a Novel Z-Line to I-Band Linking System. Circ Res. 2001;89(11):1065–72. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- 16.Burkholder TJ, Lieber RL. Sarcomere length operating range of vertebrate muscles during movement. J Exp Biol. 2001;204(9):1529–36. doi: 10.1242/jeb.204.9.1529. [DOI] [PubMed] [Google Scholar]
- 17.Millman BM. The Filament Lattice of Striated Muscle. Physiol Rev. 1998;78(2):359–91. doi: 10.1152/physrev.1998.78.2.359. [DOI] [PubMed] [Google Scholar]
- 18.Cazorla O, Freiburg A, Helmes M, Centner T, McNabb M, et al. Differential Expression of Cardiac Titin Isoforms and Modulation of Cellular Stiffness. Circ Res. 2000;86(1):59–67. doi: 10.1161/01.res.86.1.59. [DOI] [PubMed] [Google Scholar]
- 19.Bogomolovas J, Gasch A, Simkovic F, Rigden DJ, Labeit S, Mayans O. Titin kinase is an inactive pseudokinase scaffold that supports MuRF1 recruitment to the sarcomeric M-line. Open Biol. 2014;4(5):140041. doi: 10.1098/rsob.140041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, et al. The Kinase Domain of Titin Controls Muscle Gene Expression and Protein Turnover. Science. 2005;308(5728):1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 21.Linke WA, Hamdani N. Gigantic Business Titin Properties and Function Through Thick and Thin. Circ Res. 2014;114(6):1052–68. doi: 10.1161/CIRCRESAHA.114.301286. [DOI] [PubMed] [Google Scholar]
- 22.Hessel AL, Lindstedt SL, Nishikawa KC. Physiological Mechanisms of Eccentric Contraction and Its Applications: A Role for the Giant Titin Protein. Front Physiol. 2017;8 doi: 10.3389/fphys.2017.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tskhovrebova L, Trinick J. Titin: properties and family relationships. Nat Rev Mol Cell Biol. 2003;4(9):679–89. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 24.Linke WA, Krüger M. The Giant Protein Titin as an Integrator of Myocyte Signaling Pathways. Physiology. 2010;25(3):186–98. doi: 10.1152/physiol.00005.2010. [DOI] [PubMed] [Google Scholar]
- 25.Spudich JA. Hypertrophic and Dilated Cardiomyopathy: Four Decades of Basic Research on Muscle Lead to Potential Therapeutic Approaches to These Devastating Genetic Diseases. Biophys J. 2014;106(6):1236–49. doi: 10.1016/j.bpj.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368(6467):113–19. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 27.Wilkie DR. Heat work and phosphorylcreatine break-down in muscle. J Physiol. 1968;195(1):157–83. doi: 10.1113/jphysiol.1968.sp008453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaya M, Higuchi H. Nonlinear Elasticity and an 8-nm Working Stroke of Single Myosin Molecules in Myofilaments. Science. 2010;329(5992):686–89. doi: 10.1126/science.1191484. [DOI] [PubMed] [Google Scholar]
- 29.Kaya M, Tani Y, Washio T, Hisada T, Higuchi H. Coordinated force generation of skeletal myosins in myofilaments through motor coupling. Nat Commun. 2017;8 doi: 10.1038/ncomms16036. ncomms16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herzog W, Schappacher G, DuVall M, Leonard TR, Herzog JA. Residual Force Enhancement Following Eccentric Contractions: A New Mechanism Involving Titin. Physiology. 2016;31(4):300–312. doi: 10.1152/physiol.00049.2014. [DOI] [PubMed] [Google Scholar]
- 31.Herzog W, Leonard TR. Force enhancement following stretching of skeletal muscle. J Exp Biol. 2002;205(9):1275–83. doi: 10.1242/jeb.205.9.1275. [DOI] [PubMed] [Google Scholar]
- 32.Nishikawa KC, Monroy JA, Uyeno TE, Yeo SH, Pai DK, Lindstedt SL. Is titin a “winding filament”? A new twist on muscle contraction. Proc R Soc Lond B Biol Sci. 2012;279(1730):981–90. doi: 10.1098/rspb.2011.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, et al. Mechanical and chemical unfolding of a single protein: A comparison. Proc Natl Acad Sci. 1999;96(7):3694–99. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez JM, Li H. Force-Clamp Spectroscopy Monitors the Folding Trajectory of a Single Protein. Science. 2004;303(5664):1674–78. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 35.Kellermayer MSZ, Smith SB, Granzier HL, Bustamante C. Folding-Unfolding Transitions in Single Titin Molecules Characterized with Laser Tweezers. Science. 1997;276(5315):1112–16. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Linke WA, Oberhauser AF, Carrion-Vazquez M, Kerkvliet JG, et al. Reverse engineering of the giant muscle protein titin. Nature. 2002;418(6901):998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 37.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible Unfolding of Individual Titin Immunoglobulin Domains by AFM. Science. 1997;276(5315):1109–12. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 38.Schlierf M, Li H, Fernandez JM. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc Natl Acad Sci U S A. 2004;101(19):7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 1997;387(6630):308–12. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 40.Popa I, Kosuri P, Alegre-Cebollada J, Garcia-Manyes S, Fernandez JM. Force dependency of biochemical reactions measured by single-molecule force-clamp spectroscopy. Nat Protoc. 2013;8(7):1261–76. doi: 10.1038/nprot.2013.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popa I, Berkovich R, Alegre-Cebollada J, Badilla CL, Rivas-Pardo JA, et al. Nanomechanics of HaloTag Tethers. J Am Chem Soc. 2013;135(34):12762–71. doi: 10.1021/ja4056382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H, Fu H, Zhu X, Cong P, Nakamura F, Yan J. Improved High-Force Magnetic Tweezers for Stretching and Refolding of Proteins and Short DNA. Biophys J. 2011;100(2):517–23. doi: 10.1016/j.bpj.2010.12.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H, Yuan G, Winardhi RS, Yao M, Popa I, et al. Dynamics of Equilibrium Folding and Unfolding Transitions of Titin Immunoglobulin Domain under Constant Forces. J Am Chem Soc. 2015;137(10):3540–46. doi: 10.1021/ja5119368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365(6448):721–27. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 45.Martonfalvi Z, Bianco P, Linari M, Caremani M, Nagy A, et al. Low-force transitions in single titin molecules reflect a memory of contractile history. J Cell Sci. 2014;127(4):858–70. doi: 10.1242/jcs.138461. [DOI] [PubMed] [Google Scholar]
- 46.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5(6):491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huhle A, Klaue D, Brutzer H, Daldrop P, Joo S, et al. Camera-based three-dimensional real-time particle tracking at kHz rates and Ångström accuracy. Nat Commun. 2015;6 doi: 10.1038/ncomms6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dulin D, Cui TJ, Cnossen J, Docter MW, Lipfert J, Dekker NH. High Spatiotemporal-Resolution Magnetic Tweezers: Calibration and Applications for DNA Dynamics. Biophys J. 2015;109(10):2113–25. doi: 10.1016/j.bpj.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Popa I, Rivas-Pardo JA, Eckels EC, Echelman DJ, Badilla CL, et al. A HaloTag Anchored Ruler for Week-Long Studies of Protein Dynamics. J Am Chem Soc. 2016;138(33):10546–53. doi: 10.1021/jacs.6b05429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265(5178):1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 51.Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nat Struct Mol Biol. 1999;6(4):346–49. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 52.Rief M, Oesterhelt F, Heymann B, Gaub HE. Single Molecule Force Spectroscopy on Polysaccharides by Atomic Force Microscopy. Science. 1997;275(5304):1295–97. doi: 10.1126/science.275.5304.1295. [DOI] [PubMed] [Google Scholar]
- 53.Marszalek PE, Oberhauser AF, Pang Y-P, Fernandez JM. Polysaccharide elasticity governed by chair–boat transitions of the glucopyranose ring. Nature. 1998;396(6712):661–64. doi: 10.1038/25322. [DOI] [PubMed] [Google Scholar]
- 54.Florin EL, Moy VT, Gaub HE. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264(5157):415–17. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 55.Radmacher M, Fritz M, Hansma HG, Hansma PK. Direct observation of enzyme activity with the atomic force microscope. Science. 1994;265(5178):1577–79. doi: 10.1126/science.8079171. [DOI] [PubMed] [Google Scholar]
- 56.Carrion-Vazquez M, Marszalek PE, Oberhauser AF, Fernandez JM. Atomic force microscopy captures length phenotypes in single proteins. Proc Natl Acad Sci. 1999;96(20):11288–92. doi: 10.1073/pnas.96.20.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valle-Orero J, Rivas-Pardo JA, Tapia-Rojo R, Popa I, Echelman DJ, et al. Mechanical Deformation Accelerates Protein Ageing. Angew Chem Int Ed. doi: 10.1002/anie.201703630. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oberhauser AF, Hansma PK, Carrion-Vazquez M, Fernandez JM. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc Natl Acad Sci. 2001;98(2):468–72. doi: 10.1073/pnas.021321798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiita AP, Ainavarapu SRK, Huang HH, Fernandez JM. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proc Natl Acad Sci. 2006;103(19):7222–27. doi: 10.1073/pnas.0511035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiita AP, Perez-Jimenez R, Walther KA, Gräter F, Berne BJ, et al. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450(7166):124–27. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ainavarapu SRK, Brujić J, Huang HH, Wiita AP, Lu H, et al. Contour Length and Refolding Rate of a Small Protein Controlled by Engineered Disulfide Bonds. Biophys J. 2007;92(1):225–33. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sotomayor M, Schulten K. Single-Molecule Experiments in Vitro and in Silico. Science. 2007;316(5828):1144–48. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 63.Flory PJ. Principles of Polymer Chemistry. Cornell University Press; 1953. p. 696. [Google Scholar]
- 64.Stacklies W, Vega MC, Wilmanns M, Gräter F. Mechanical Network in Titin Immunoglobulin from Force Distribution Analysis. PLOS Comput Biol. 2009;5(3):e1000306. doi: 10.1371/journal.pcbi.1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan G, Le S, Yao M, Qian H, Zhou X, et al. Elasticity of the Transition State Leading to an Unexpected Mechanical Stabilization of Titin Immunoglobulin Domains. Angew Chem Int Ed. 2017 doi: 10.1002/anie.201700411. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 66.Berkovich R, Garcia-Manyes S, Klafter J, Urbakh M, Fernández JM. Hopping around an entropic barrier created by force. Biochem Biophys Res Commun. 2010;403(1):133–37. doi: 10.1016/j.bbrc.2010.10.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia-Manyes S, Dougan L, Badilla CL, Brujić J, Fernández JM. Direct observation of an ensemble of stable collapsed states in the mechanical folding of ubiquitin. Proc Natl Acad Sci. 2009;106(26):10534–39. doi: 10.1073/pnas.0901213106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Redfield C, Smith RAG, Dobson CM. Structural characterization of a highly–ordered “molten globule” at low pH. Nat Struct Mol Biol. 1994;1(1):23–29. doi: 10.1038/nsb0194-23. [DOI] [PubMed] [Google Scholar]
- 69.Mártonfalvi Z, Bianco P, Naftz K, Ferenczy GG, Kellermayer M. Force generation by titin folding. Protein Sci. 2017 doi: 10.1002/pro.3117. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eckels EC, Rivas-Pardo JA, Valle-Orero J, Popa I, Fernandez JM. The Science of Stretching: Mechanical Anisotropy in Titin Ig Domains. Biophys J. 2016;110(3):393a. [Google Scholar]
- 71.Li H, Wang H-C, Cao Y, Sharma D, Wang M. Configurational Entropy Modulates the Mechanical Stability of Protein GB1. J Mol Biol. 2008;379(4):871–80. doi: 10.1016/j.jmb.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 72.Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Point mutations alter the mechanical stability of immunoglobulin modules. Nat Struct Mol Biol. 2000;7(12):1117–20. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 73.Berkovich R, Hermans RI, Popa I, Stirnemann G, Garcia-Manyes S, et al. Rate limit of protein elastic response is tether dependent. Proc Natl Acad Sci. 2012;109(36):14416–21. doi: 10.1073/pnas.1212167109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berkovich R, Garcia-Manyes S, Urbakh M, Klafter J, Fernandez JM. Collapse Dynamics of Single Proteins Extended by Force. Biophys J. 2010;98(11):2692–2701. doi: 10.1016/j.bpj.2010.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neupane K, Woodside MT. Quantifying Instrumental Artifacts in Folding Kinetics Measured by Single-Molecule Force Spectroscopy. Biophys J. 2016;111(2):283–86. doi: 10.1016/j.bpj.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brenner H. The slow motion of a sphere through a viscous fluid towards a plane surface. Chem Eng Sci. 1961;16(3–4):242–51. [Google Scholar]
- 77.Valle-Orero J, Eckels EC, Stirnemann G, Popa I, Berkovich R, Fernandez JM. The elastic free energy of a tandem modular protein under force. Biochem Biophys Res Commun. 2015;460(2):434–38. doi: 10.1016/j.bbrc.2015.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alegre-Cebollada J, Kosuri P, Giganti D, Eckels E, Rivas-Pardo JA, et al. S-Glutathionylation of Cryptic Cysteines Enhances Titin Elasticity by Blocking Protein Folding. Cell. 2014;156(6):1235–46. doi: 10.1016/j.cell.2014.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li H, Fernandez JM. Mechanical Design of the First Proximal Ig Domain of Human Cardiac Titin Revealed by Single Molecule Force Spectroscopy. J Mol Biol. 2003;334(1):75–86. doi: 10.1016/j.jmb.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 80.Pardo JAR, Eckels E, Valle-Orero J, Fernadez JM. Disulfide Bonding in the Contractile Work of Titin. Biophys J. 2017;112(3):456a. [Google Scholar]
- 81.Hill AV. The Heat of Shortening and the Dynamic Constants of Muscle. Proc R Soc Lond B Biol Sci. 1938;126(843):136–95. [Google Scholar]
- 82.Debold EP, Patlak JB, Warshaw DM. Slip Sliding Away: Load-Dependence of Velocity Generated by Skeletal Muscle Myosin Molecules in the Laser Trap. Biophys J. 2005;89(5):L34–36. doi: 10.1529/biophysj.105.072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Visscher K, Schnitzer MJ, Block SM. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400(6740):184–89. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 84.Clemen AE-M, Vilfan M, Jaud J, Zhang J, Bärmann M, Rief M. Force-Dependent Stepping Kinetics of Myosin-V. Biophys J. 2005;88(6):4402–10. doi: 10.1529/biophysj.104.053504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-Induced Bidirectional Stepping of Cytoplasmic Dynein. Cell. 2007;131(5):952–65. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Linke WA, Stockmeier MR, Ivemeyer M, Hosser H, Mundel P. Characterizing titin’s I-band Ig domain region as an entropic spring. J Cell Sci. 1998;111(11):1567–74. doi: 10.1242/jcs.111.11.1567. [DOI] [PubMed] [Google Scholar]
- 87.Prado LG, Makarenko I, Andresen C, Krüger M, Opitz CA, Linke WA. Isoform Diversity of Giant Proteins in Relation to Passive and Active Contractile Properties of Rabbit Skeletal Muscles. J Gen Physiol. 2005;126(5):461–80. doi: 10.1085/jgp.200509364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Campbell KS. Dynamic coupling of regulated binding sites and cycling myosin heads in striated muscle. J Gen Physiol. 2014;143(3):387–99. doi: 10.1085/jgp.201311078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piazzesi G, Reconditi M, Linari M, Lucii L, Bianco P, et al. Skeletal Muscle Performance Determined by Modulation of Number of Myosin Motors Rather Than Motor Force or Stroke Size. Cell. 2007;131(4):784–95. doi: 10.1016/j.cell.2007.09.045. [DOI] [PubMed] [Google Scholar]
- 90.Llewellyn ME, Barretto RPJ, Delp SL, Schnitzer MJ. Minimally invasive high-speed imaging of sarcomere contractile dynamics in mice and humans. Nature. 2008;454(7205):784–88. doi: 10.1038/nature07104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rassier DE, MacIntosh BR, Herzog W. Length dependence of active force production in skeletal muscle. J Appl Physiol. 1999;86(5):1445–1457. doi: 10.1152/jappl.1999.86.5.1445. [DOI] [PubMed] [Google Scholar]
- 92.Cromie MJ, Sanchez GN, Schnitzer MJ, Delp SL. Sarcomere lengths in human extensor carpi radialis brevis measured by microendoscopy. Muscle Nerve. 2013;48(2):286–92. doi: 10.1002/mus.23760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sanchez GN, Sinha S, Liske H, Chen X, Nguyen V, et al. In Vivo Imaging of Human Sarcomere Twitch Dynamics in Individual Motor Units. Neuron. 2015;88(6):1109–20. doi: 10.1016/j.neuron.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120(4):923–34. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fudge DS, Gardner KH, Forsyth VT, Riekel C, Gosline JM. The Mechanical Properties of Hydrated Intermediate Filaments: Insights from Hagfish Slime Threads. Biophys J. 2003;85(3):2015–27. doi: 10.1016/S0006-3495(03)74629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lovelady HH, Shashidhara S, Matthews WG. Solvent specific persistence length of molecular type I collagen. Biopolymers. 2014;101(4):329–35. doi: 10.1002/bip.22365. [DOI] [PubMed] [Google Scholar]
- 97.Hidalgo C, Hudson B, Bogomolovas J, Zhu Y, Anderson B, et al. PKC Phosphorylation of Titin’s PEVK Element A Novel and Conserved Pathway for Modulating Myocardial Stiffness. Circ Res. 2009;105(7):631–38. doi: 10.1161/CIRCRESAHA.109.198465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kötter S, Gout L, Frieling-Salewsky MV, Müller AE, Helling S, et al. Differential changes in titin domain phosphorylation increase myofilament stiffness in failing human hearts. Cardiovasc Res. 2013;99(4):648–56. doi: 10.1093/cvr/cvt144. [DOI] [PubMed] [Google Scholar]
- 99.Perkin J, Slater R, Del Favero G, Lanzicher T, Hidalgo C, et al. Phosphorylating Titin’s Cardiac N2B Element by ERK2 or CaMKIIδ Lowers the Single Molecule and Cardiac Muscle Force. Biophys J. 2015;109(12):2592–2601. doi: 10.1016/j.bpj.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Avner BS, Shioura KM, Scruggs SB, Grachoff M, Geenen DL, et al. Myocardial infarction in mice alters sarcomeric function via post-translational protein modification. Mol Cell Biochem. 2012;363(1–2):203–15. doi: 10.1007/s11010-011-1172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li Y, Linke WA. Mechanically Unfolded Titin Immunoglobulin Domains Refold Faster and More Accurately in Presence of Chaperone Alpha-B-Crystallin. Biophys J. 2017;112(3):42a. [Google Scholar]
- 102.Kotter S, Unger A, Hamdani N, Lang P, Vorgerd M, et al. Human myocytes are protected from titin aggregation-induced stiffening by small heat shock proteins. J Cell Biol. 2014;204(2):187–202. doi: 10.1083/jcb.201306077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bullard B, Ferguson C, Minajeva A, Leake MC, Gautel M, et al. Association of the Chaperone αB-crystallin with Titin in Heart Muscle. J Biol Chem. 2004;279(9):7917–24. doi: 10.1074/jbc.M307473200. [DOI] [PubMed] [Google Scholar]
- 104.Brinkmeier H, Ohlendieck K. Chaperoning heat shock proteins: Proteomic analysis and relevance for normal and dystrophin-deficient muscle. PROTEOMICS – Clin Appl. 2014;8(11–12):875–95. doi: 10.1002/prca.201400015. [DOI] [PubMed] [Google Scholar]
- 105.Zhu Y, Bogomolovas J, Labeit S, Granzier H. Single molecule force spectroscopy of the cardiac titin N2B element: effects of the molecular chaperone alphaB-crystallin with disease-causing mutations. J Biol Chem. 2009;284(20):13914–23. doi: 10.1074/jbc.M809743200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roach NT, Venkadesan M, Rainbow MJ, Lieberman DE. Elastic energy storage in the shoulder and the evolution of high-speed throwing in Homo. Nature. 2013;498(7455):483–86. doi: 10.1038/nature12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Minajeva A, Neagoe C, Kulke M, Linke WA. Titin-based contribution to shortening velocity of rabbit skeletal myofibrils. J Physiol. 2002;540(1):177–88. doi: 10.1113/jphysiol.2001.013154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Opitz CA, Kulke M, Leake MC, Neagoe C, Hinssen H, et al. Damped elastic recoil of the titin spring in myofibrils of human myocardium. Proc Natl Acad Sci. 2003;100(22):12688–93. doi: 10.1073/pnas.2133733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dugdale TM, Dagastine R, Chiovitti A, Mulvaney P, Wetherbee R. Single Adhesive Nanofibers from a Live Diatom Have the Signature Fingerprint of Modular Proteins. Biophys J. 2005;89(6):4252–60. doi: 10.1529/biophysj.105.062489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sarkar A, Caamano S, Fernandez JM. The Mechanical Fingerprint of a Parallel Polyprotein Dimer. Biophys J. 2007;92(4):L36–38. doi: 10.1529/biophysj.106.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bianco P, Reconditi M, Piazzesi G, Lombardi V. Is muscle powered by springs or motors? J Muscle Res Cell Motil. 2016:1–3. doi: 10.1007/s10974-016-9454-4. [DOI] [PubMed] [Google Scholar]
- 112.Caremani M, Pinzauti F, Reconditi M, Piazzesi G, Stienen GJM, et al. Size and speed of the working stroke of cardiac myosin in situ. Proc Natl Acad Sci. 2016;113(13):3675–80. doi: 10.1073/pnas.1525057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takano M, Terada TP, Sasai M. Unidirectional Brownian motion observed in an in silico single molecule experiment of an actomyosin motor. Proc Natl Acad Sci. 2010;107(17):7769–74. doi: 10.1073/pnas.0911830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Freikamp A, Cost A-L, Grashoff C. The Piconewton Force Awakens: Quantifying Mechanics in Cells. Trends Cell Biol. 2016;26(11):838–47. doi: 10.1016/j.tcb.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 115.Meng F, Sachs F. Orientation-based FRET sensor for real-time imaging of cellular forces. J Cell Sci. 2012;125(3):743–50. doi: 10.1242/jcs.093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Austen K, Ringer P, Mehlich A, Chrostek-Grashoff A, Kluger C, et al. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat Cell Biol. 2015;17(12):1597–1606. doi: 10.1038/ncb3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466(7303):263–66. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arsenovic PT, Ramachandran I, Bathula K, Zhu R, Narang JD, et al. Nesprin-2G, a Component of the Nuclear LINC Complex, Is Subject to Myosin-Dependent Tension. Biophys J. 2016;110(1):34–43. doi: 10.1016/j.bpj.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brenner MD, Zhou R, Conway DE, Lanzano L, Gratton E, et al. Spider Silk Peptide Is a Compact, Linear Nanospring Ideal for Intracellular Tension Sensing. Nano Lett. 2016;16(3):2096–2102. doi: 10.1021/acs.nanolett.6b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Prosser BL, Ward CW, Lederer WJ. X-ROS Signaling: Rapid Mechano-Chemo Transduction in Heart. Science. 2011;333(6048):1440–45. doi: 10.1126/science.1202768. [DOI] [PubMed] [Google Scholar]
- 121.Rivas-Pardo JA, Mártonfalvi Z, Manteca A, Eckels EC, Echelman DJ, et al. A Multi-Tool Mouse Model to Study the Elasticity of Native Titin. Biophys J. 2017;112(3):167a. [Google Scholar]
- 122.Kohl J, Ng J, Cachero S, Ciabatti E, Dolan M-J, et al. Ultrafast tissue staining with chemical tags. Proc Natl Acad Sci. 2014;111(36):E3805–14. doi: 10.1073/pnas.1411087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stagge F, Mitronova GY, Belov VN, Wurm CA, Jakobs S. Snap-, CLIP- and Halo-Tag Labelling of Budding Yeast Cells. PLoS ONE. 2013;8(10):e78745. doi: 10.1371/journal.pone.0078745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Horowits R, Kempner ES, Bisher ME, Podolsky RJ. A physiological role for titin and nebulin in skeletal muscle. Nature. 1986;323(6084):160–64. doi: 10.1038/323160a0. [DOI] [PubMed] [Google Scholar]
- 125.Higuchi H. Changes in Contractile Properties with Selective Digestion of Connectin (Titin) in Skinned Fibers of Frog Skeletal Muscle. J Biochem (Tokyo) 1992;111(3):291–95. doi: 10.1093/oxfordjournals.jbchem.a123752. [DOI] [PubMed] [Google Scholar]
- 126.Dobbie I, Linari M, Piazzesi G, Reconditi M, Koubassova N, et al. Elastic bending and active tilting of myosin heads during muscle contraction. Nature. 1998;396(6709):383–87. doi: 10.1038/24647. [DOI] [PubMed] [Google Scholar]
- 127.Piazzesi G, Reconditi M, Linari M, Lucii L, Sun Y-B, et al. Mechanism of force generation by myosin heads in skeletal muscle. Nature. 2002;415(6872):659–62. doi: 10.1038/415659a. [DOI] [PubMed] [Google Scholar]
- 128.Reconditi M, Linari M, Lucii L, Stewart A, Sun Y-B, et al. The myosin motor in muscle generates a smaller and slower working stroke at higher load. Nature. 2004;428(6982):578–81. doi: 10.1038/nature02380. [DOI] [PubMed] [Google Scholar]
- 129.Fenn WO. A quantitative comparison between the energy liberated and the work performed by the isolated sartorius muscle of the frog. J Physiol. 1923;58(2–3):175–203. doi: 10.1113/jphysiol.1923.sp002115. [DOI] [PMC free article] [PubMed] [Google Scholar]