Abstract
BACKGROUND
Laboratory technologies have highlighted the progressive accumulation of the so-called “storage lesion,” a wide series of alterations to stored red blood cells (RBCs) that may affect the safety and effectiveness of the transfusion therapy. New improvements in the field are awaited to ameliorate this lesion, such as the introduction of washing technologies in the cell processing pipeline. Laboratory studies that have tested such technologies so far rely on observational qualitative or semiquantitative techniques.
STUDY DESIGN AND METHODS
A state-of-the-art quantitative proteomics approach utilizing quantitative concatamers (QconCAT) was used to simultaneously monitor fluctuations in the abundance of 114 proteins in AS-3 RBC supernatants (n =5; 11 time points, including before and after leukoreduction, at 3 hours, on Days 1 and 2, and weekly sampling from Day 7 through Day 42).
RESULTS
Leukoreduction-dependent depletion of plasma proteins was observed at the earliest time points. A subset of proteins showed very high linear correlation (r2 >0.9) not only with storage time, but also with absolute levels of hemoglobin α1 and β, a proxy for RBC hemolysis and vesiculation. Linear regression was performed to describe the temporal relationship between these proteins. Our findings suggest a role for supernatant glyceraldehyde-3-phosphate dehydrogenase; peroxiredoxin-1, -2, and -6; carbonic anhydrase-1 and -2; selenium binding protein-1; biliverdin reductase; aminolevulinate dehydratase; and catalase as potential biomarkers of RBC quality during storage.
CONCLUSION
A targeted proteomics technology revealed novel biomarkers of the RBC storage lesion and promises to become a key analytical readout for the development and testing of alternative cell processing strategies.
Transfusion of red blood cell (RBC) concentrates is a lifesaving intervention for approximately 4.5 million Americans every year.1 The latest World Health Organization data suggest a trend toward increase of volunteer unpaid blood donations (+8.6 million units) and total donations (from approx. 80 to 108 million) between 2004 and 2012.1 Despite reassuring evidence from randomized clinical trials,2 in 2014 the American Red Cross reported that the use of blood has significantly declined by almost 30% during the past 5 years.
Despite a century of improvements in the field of transfusion medicine,3 stored RBCs accumulate a wide series of biochemical, morphologic, and functional alterations, collectively referred to as the “storage lesion.”4,5 In the past few years, omics technologies, especially proteomics and metabolomics, allowed such lesions to be extensively documented. Even though an encyclopedic description of the storage lesion is now available, the clinical relevance of such lesions has yet to be determined.6 Nonetheless, omics technologies may turn out to be play a key role in the advancement of the field of transfusion medicine.7
Proteomics studies on transfusion medicine–relevant issues performed to date have primarily been observational in nature. Indeed, even though pioneering studies from Anniss and colleagues8 have paved the way for in-depth investigations of the storage lesion to the proteomes of RBC membranes9 and supernatants,10 RBC proteomics studies9,11,12 performed so far have primarily relied on qualitative or relative quantitation technologies. Fostered by critical advances in the field of mass spectrometry (MS)-based RBC proteomics,13–15 light has been shed on the unanticipated complexity of the RBC proteome in the past decade and a half. Such data have paved the way for the bioinformatics elaboration of RBC protein and metabolic interaction pathways,16,17 a key reference database for hypothesis-driven research in the field.
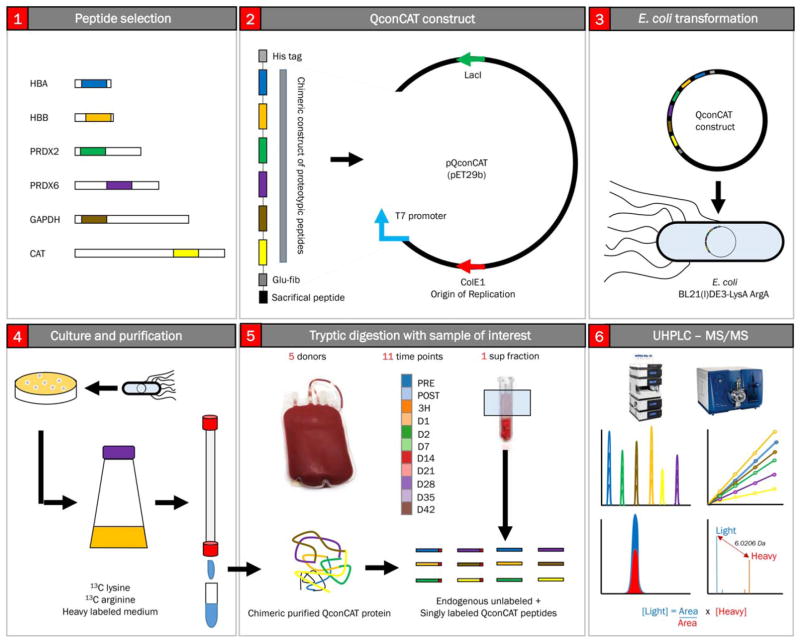
While early generations of proteomics approaches were designed to deliver qualitative or relative quantitation information at best,18–20 the focus has recently shifted toward the ideation of analytical workflows that sacrifice the broad scope of the analysis that characterizes discovery mode approaches for the absolute quantification of a subset of the proteome. Targeted approaches such as multiple reaction monitoring coupled with the adoption of heavy labeled spiked internal standard peptides have been designed and optimized by several groups.18–20 Quantitative concatamer (QconCATs; Fig. 1)21–23 approaches are based on the selection of proteotypic peptides, peptides that are experimentally observable and are composed of a sequence that uniquely identifies a given protein isoform. Such peptides are selected on the basis of existing experimental evidence from our own work and supplemented from continuously growing data repositories (e.g., PeptideAtlas and SRMAtlas).24,25 Once selected, gene sequences coding for up to approximately 40 proteotypic peptides are merged in a chimeric gene that can be produced synthetically. The gene is cloned into an expression vector and then transformed and expressed in a bacterial auxotroph where arginine and lysine 13C6 isotopologues have been incorporated at an efficiency greater than 99.99%. Expressed chimeric proteins are purified and quantified for use as internal standards. These standards control for trypsin cleavage efficiency and analytical variability at the liquid chromatography (LC)-MS step and perform absolute quantitation by determining ratios against light peptide peak areas. Although time-consuming in its development (peptide selection, chimeric protein expression, and purification), the QconCAT approach enables fast absolute quantitation of hundreds of proteins in a single analytical run with high precision.
Fig. 1.
Overview of the QconCAT workflow. (A) Proteotypic peptides are selected for proteins of interest (peptides selected in the present study are enlisted in Table S1). Proteotypic peptides are selected on the basis of strict rules, including the presence of one C-term arginine or lysine in the sequence (to enable controlled tryptic digestion), the absence of residues that can be potentially artificially affected by sample handling or digestion (e.g., oxidation of methionine), presence of amino acid residues with good ionizability, and so forth.21–23 A gene construct coding for a chimeric protein including all the proteotypic peptides is loaded onto an expression vector (B), which is used to transform a specific auxotrophic E. coli strain (C). Bacteria are grown in medium containing only heavy isotopologues of lysine and arginine at all carbon atoms (D). Expressed recombinant protein is thus purified and quantified (D) and used as internal standard for protein digestion (E), retention time reproducibility, and absolute quantification through direct ratios of peak areas for the light endogenous peptide versus the known amounts of the spiked in proteotypic peptide (labeled at C-term arginine or lysine; F). A color version of the figure can be found in the online version of the paper.
Availability of absolute quantitative data is of relevance to clinical and research laboratories by allowing interlaboratory, interprotein, and heterogeneous sample comparisons that are not achieved through qualitative or relative quantitation methods. Applications include, but are not limited to, the determination of compositional specificities and patient to patient variation in the extracellular matrix proteome of ex vivo decellularized or recellularized organs for tissue engineering and transplantation purposes or delineating the metabolic proteome in model systems.26–28 In the field of transfusion medicine, we recently adopted this approach to address the sex-dependent compositional specificities of apheresis platelet (PLT) supernatants during routine storage in the blood bank.29
Recently, we demonstrated the progressive accumulation of hydrophilic30 and lipophilic31 metabolites in the supernatants of AS-3 RBCs during routine storage, as well as the accumulation of oxidized hemoglobin (Hb) at functional amino acid residues, including proximal histidine 93 and cysteine 94 of Hbβ.32 Here we performed a methodologic study to show the potential relevance of quantitative proteomics technologies in the field of transfusion medicine. A QconCAT strategy was adopted to perform absolute quantification of 114 proteins in RBC units from five donors at 11 time points during routine storage in the blood bank in AS-3. Collection of early time points allowed us to monitor the efficiency of approximately 3.5-log and approximately 2-log white blood cell (WBC) and PLT filtration strategies by assaying RBC supernatants before, immediately after, and 3 hours after leukoreduction. Determination of absolute protein quantities enabled the calculation of time-dependent linear regression coefficients and accumulation kinetics of a subset of proteins during storage duration. A subset of proteins were identified showing strong correlation (r >0.95, Pearson) with storage duration and accumulation of Hbα1, β, and δ (a bona fide proxy and more direct measure for hemolysis or vesiculation than classic spectrophotometric absorbance). These proteins are candidate biomarkers of the RBC storage lesion and include the previously suggested marker peroxiredoxin-2 (PRDX2),33,34 as well as other redox enzymes PRDX1; PRDX6; catalase (CAT); selenium-binding protein; peptidyl-prolyl isomerase A; and the enzymes aminolevulinate dehydratase (ALAD), carbonic anhydrase 1 and 2 (CA1 and CA2), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
This study reveals the potential to use these, or other yet to be appreciated proteins, as predictive markers of the RBC storage lesion. In addition, it provides a proof-of-principle demonstration of the usefulness of MS-based quantitative proteomics in monitoring RBC storage quality under currently approved Food and Drug Administration (FDA) and European Council guidelines. Additionally, this workflow can be used to monitor donor variability or streamline the optimization and testing of alternative cell processing strategies.35,36
MATERIALS AND METHODS
Materials
Reagents were purchased from Sigma Aldrich (St Louis, MO) unless otherwise noted.
Sample collection
One unit of whole blood (500 ±50 mL) was collected from five healthy donors per AABB and FDA guidelines, using CP2D in a collection bag system (Nutricel, Pall Medical, Westborough, MA). Plasma was separated from RBCs by centrifugation and resulting supernatant was removed. AS-3 (Nutricel) was added to a final hematocrit of approximately 60%. The estimated amount of residual plasma was 5 to 10 mL/unit. RBC units were leukoreduced before storage via filtration using an in-line leukoreduction filter (RC2D, Haemonetics, Braintree, MA) and stored at 1 to 6°C. Samples (0.5 mL) were obtained through sterile couplers before or after leukoreduction; at 3 hours after leukoreduction; or on Days 1, 2, 7, 14, 21, 28, 35, and 42. The supernatant was separated from RBCs via centrifugation (1500 × g for 10 minutes at 4°C) followed by a second centrifugation at 12,500 × g for 6 minutes at 4°C to sediment residual cellular material and contaminating PLTs.
QconCAT-based absolute quantitation of 114 proteins in stored RBC supernatants
Four QconCAT constructs (Table S1, available as supporting information in the online version of this paper) were designed to quantify 114 proteins in RBC supernatants at each time point. Proteotypic peptides were selected from previous experimental data (nontargeted proteomics on RBCs, PLTs, and patient serum samples) and publicly accessible databases, including PeptideAtlas,24 SRM Atlas,25 and Global Proteome Database. QconCAT DNA constructs were synthesized de novo by Genscript (Piscat-away, NJ) and cloned into pET21b. Escherichia coli auxotrophic strain BL21(l)DE3-LysA ArgA was transformed with the plasmid and cultured in minimal medium (1×M9 salts, 1 mmol/L MgSO4, 0.1 mmol/L CaCl2, 0.00005% [wt/vol] thiamine, 0.2% [wt/vol] glucose) supplemented with 13C6 arginine and 13C6 lysine at 0.1 mg/mL (Sigma Aldrich). The cells were grown to mid log phase (A600 0.6–0.8), at which point expression was induced by adding 1 mmol/L isopropyl-D-1-thiogalactopyranoside. After 4 hours of growth at 37° C the cells were harvested by centrifugation and processed as previously described with minor modifications.26
Briefly, the cells were lysed with protein extraction reagent (BugBuster, EMD Millipore, Darmstadt, Germany). Inclusion bodies were suspended in 20 mmol/L phosphate buffer, 6 mol/L guanidinium chloride, 0.5 mol/L NaCl, 20 mmol/L imidazole (pH 7.4). QconCAT proteins were purified by affinity chromatography using a nickel-based resin. The purified QconCAT was desalted by three rounds of dialysis against 100 volumes of 10 mmol/L ammonium bicarbonate (ABC; pH 8.5).
Depletion of albumin and IgG from RBC supernatants
The two most abundant plasma proteins, albumin and IgG, were removed from the RBC supernatants using serum protein immunodepletion resins (Proteome Purify 2, R&D Systems, Inc., Minneapolis, MN) according to manufacturer’s protocol. Briefly, 600 μg of RBC supernatants was mixed with 1 mL of suspended immunodepletion resin and incubated on rotary shaker at room temperature for 30 minutes. After the incubation period, the immunodepletion resin was transferred into the upper chamber of a 0.22-μm filter unit (Corning Costar Spin-X, Sigma Aldrich) and centrifuged for 2 minutes at 2000 × g. The protein concentrations were determined by a Brad-ford protein assay before and after removal of the top two plasma proteins.
Proteomics analysis
Recombinant isotopically labeled QconCAT proteins26,27 (Table S1) were mixed with the depleted RBC supernatants at 200 or 100 fmol per injection. The samples were digested according to the FASP protocol.37 In brief, samples were mixed in a 10-kDa cutoff filter unit with 8 mol/L urea in 0.1 mol/L ABC (pH 8.5) and centrifuged at 14,000 × g for 15 minutes. Proteins were reduced by addition of 100 μL of 10 mmol/L dithiothreitol in 8 mol/L urea and 0.1 mol/L ABC (pH 8.5) and incubation for 30 minutes at room temperature, and the device was centrifuged. Subsequently, 100 μL of 55 mmol/L iodoacetamide in 8 mol/L urea in 0.1 mol/L ABC (pH 8.5) was added to the samples, and the samples were incubated for 30 minutes at room temperature in dark followed by centrifugation. Three washing steps with 100 μL of 8 mol/L urea in 0.1 mol/L ABC (pH 8.5) solution were performed, followed by three washing steps with 100 μL of 0.1 mol/L ABC buffer. Protein digestion was carried out with the presence 0.02% of surfactant tryptin enhancer (ProteaseMax, Promega, Madison, WI) surfactant at 37°C overnight. Peptides were recovered by transferring the filter unit to a new collection tube and centrifuging at 14,000 × g for 10 minutes. To complete peptide recovery, we rinsed filters twice with 50 μL of 0.2% formic acid in 10 mmol/L ABC that was collected by centrifugation. Samples were then concentrated to approximately 2 μL and reconstituted to 50 μL with 0.1% formic acid. The resultant peptide mixture was analyzed by LC–single reaction monitoring (SRM). Briefly, a targeted SRM approach was performed using the an LC-MS/MS system interfaced with a an UPLC system (QTRAP 5500 and Ultimate 3000, respectively, Thermo Fisher Scientific, San Jose, CA). Five micrograms of proteins was separated on an UPLC BEH C18 1.7 μm 1.0 × 150-mm column (ACQUITY, Waters, Milford, MA) kept at constant 50°C. The mobile phases consisted of 0.1% formic acid in double-distilled (18 mΩ) water (A) and 0.1% formic acid in 80% acetonitrile with (B), respectively. Samples were eluted at a flow rate of 150 μL/min using a gradient of 5% to 32% B for 32 minutes followed by a wash step of 5 minutes at 100% B ending with a reequilibration step of 7 minutes at 5%. The mass spectrometer was run in positive ionization mode with the following settings: a source temperature of 200°C, spray voltage of 5300 V, curtain gas of 20 psi, and a source gas of 35 psi (nitrogen gas). Multiple SRM transitions were monitored using unit resolution in both Q1 and Q3 quadrupoles to maximize specificity. SRM assay optimization was performed with the aid of computer software (Skyline, Version 3.1, UW, Seattle, WA). Collision energies and declustering potential were optimized for each transition. Method building and acquisition were performed using the instrument-supplied software (Analyst, Version 1.5.2, AB Sciex, Framingham, MA). Raw SRM data files were imported to Skyline Version 3.1 software for data processing. Quantification was based on the ratio of corresponding light and heavy peak areas. Data were further normalized for technical variability on the basis of an internal heavy peptide, encoded in each one of the four Qcon-CATs, specific to the yeast protein alcohol dehydrogenase (highlighted in red in Table S1).26,27 The method is linear (across 4- to 5-log range of concentrations) and sensitive (fmol/μg injected proteins), specific (three to four transitions were monitored for each peptide), and robust.26,27 The peak integration was performed automatically using Savitzky-Golay smoothing, and all the data were manually inspected to confirm correct peak detection. Quantitative results from Skyline were exported into Excel files. Each sample was run in duplicate. Coefficients of variations were determined (standard deviation divided by the mean of technical replicate runs). Values were normalized to the total amount (fmol/mg) of protein after depletion, as to allow for direct comparison of the preleukoreduction time points. Statistical analyses and data representations were prepared with computer software (GraphPad Prism 5.0, GraphPad Software Inc., La Jolla, CA; Excel, Microsoft, Redmond, CA; GENE-E, Broad Institute, Cambridge, MA; and Photoshop CS6, Adobe, Mountain View, CA).
RESULTS
The QconCAT approach was used to monitor 114 proteins in AS-3 RBC supernatants before and immediately after approximately 3.5-log WBC and approximately 2-log PLT reduction. Time points were then assayed at 3 hours post-reduction and sequentially on Days 1, 2, and 7 and on a weekly basis until the end of the storage period (Fig. 1; Table S1). Results are detailed in the extended in Table S2 (available as supporting information in the online version of this paper) including extended and abbreviated UniProt names, a representative chromatogram (Fig. S1, available as supporting information in the online version of this paper), absolute concentrations per each time point (fmol/μg total protein), sparkline representation of relative trends, and coefficients of variation for technical variability.
A subset of proteins, primarily those involved in coagulation and complement cascades were removed by leukoreduction (Fig. 2A, left panel), including coagulation factor (F)V, FXI, and FXIII; fibrinogen α, β, and γ; immunoglobulin components; and serum amyloid protein A-4. Of note, residual albumin and immunoglobulin were still detectable even after preliminary depletion of the top 2 abundant plasma proteins.
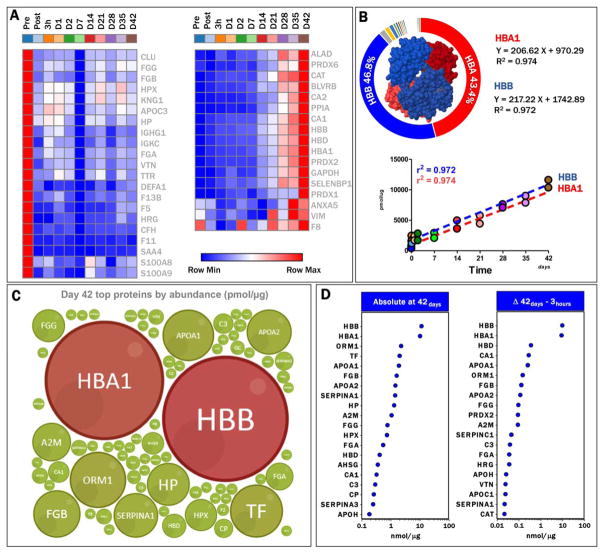
Fig. 2.
Quantitation of proteins in RBC supernatants. (A) Heat maps show protein removed by leukoreduction (red to blue, left panel) or increasing during storage in AS-3 RBC supernatants (blue to red, right panel). (B) Hbα1 and β (HBA1 and HBB) represent the 43.4 and 46.8% of the total proteins in RBC supernatants upon depletion of top two plasma proteins during sample preparation. Linearity of HBA1 and HBB increases in RBC supernatants during storage durations are indicated through quadratic correlation coefficients and first-degree equations. (C) Bubble plot representation of the relative abundance of Hb with relation to the top 50 proteins, as determined through this targeted analysis. (D) Absolute quantitative values (nmol/μg protein) are shown for the top 20 proteins in RBC supernatants at the end of the storage period (left panel). Delta increases, as determined by subtracting Day 1 absolute abundances from Day 42 values for each protein, are shown for the top 20 proteins increasing their levels during storage duration (right panel). Further details for all the 114 proteins are shown in Table S2. A color version of the figure can be found in the online version of the paper.
In parallel, a cluster of proteins progressively accumulating in RBC supernatants during storage was observed (Fig. 2B), of which, Hbα1 and β (HBA and HBB) were the most abundant, representing approximately 43.3 and 46.8% of the supernatant proteome by Storage Day 42 (Figs. 2B and 2C). As expected, molar ratios for HBA and HBB were 2:2, with linear correlations between HBA1 and HBB and storage duration of more than 0.985 (Fig. 2C), allowing us to determine the relationship between quantitative results for both proteins in a temporal fashion. At the end of the storage period, the proteome of the supernatants was predominantly composed of 20 proteins of the 114 monitored here using the QconCATs (Figs. 2D and 2E). A simple calculation of the changes in abundance between Storage Day 42 and Day 1 (Δ increases) highlighted the progressive accumulation in levels of CA1, apolipoproteins A1 and H (APOA1 and APOH), PRDX2, CAT, and coagulation components (fibrinogens FGA, FGB, FGG, complement component 3-C3 serine protease inhibitors A1 and C1 SERPINA1, and C1). These results are suggestive of a minor yet appreciable effect on the composition of RBC supernatants by residual contaminating cells, as some of these components are not reported to be from RBCs (Fig. 2D, left panel).
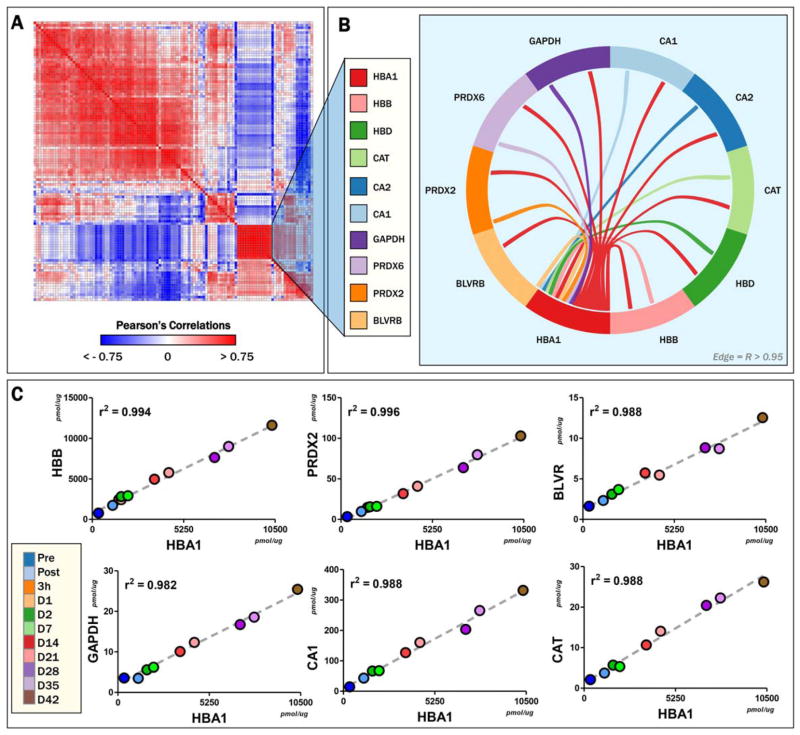
Correlative analyses of absolute levels at each time point for all the proteins (Pearson’s linear correlations; Table S3, available as supporting information in the online version of this paper) were plotted as hierarchically clustered (1 – Pearson) heat maps, to reveal protein groups following similar trends (Fig. 3A, extended vectorial version in Fig. S2, available as supporting information in the online version of this paper). Of note, a group of proteins was found to cluster with HBA1 and HBB, including HBD; ALAD; BLVRB; CA1 and 2; CAT; GAPDH; and PRDX1, 2, and 6 (Fig. 3B). These proteins had positive linear correlations with HBA1 of more than 0.9 (Figs. 3B and 3C).
Fig. 3.
Correlative analysis. (A) Protein levels at all time points were correlated among each other to show high (|r| > 0.75) positive (red) or negative linear correlations. A core of proteins strongly (r > 0.95) correlated among each other was found, including proteins listed in (B). Central hub of this group of protein included Hbα1 (B), β, and δ. Linear correlations are plotted for the other proteins versus HbA1, the latter representing a bona fide proxy for RBC hemolysis or vesiculation in RBC supernatants, since approximately 50% of Hb in RBC supernatants is either from hemolyzed RBCs or in RBC vesicles.8 A color version of the figure can be found in the online version of the paper.
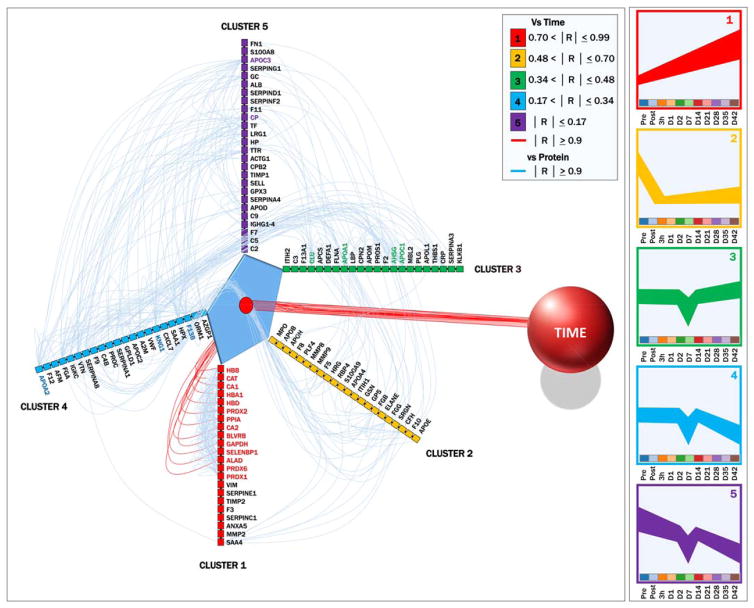
Sorting on the basis of linear correlation ranges with storage duration, five main trends (eigenvectors) were identified (Fig. 4). Proteins in each cluster were assigned to different arbitrary axes and connected to time (red line [in color in the online version of this article]) or other proteins (light blue lines [in color in the online version of this article]) if linear Pearson’s correlations were higher than 0.9 (Fig. 4). This hive plot representation allowed us to identify hub proteins in each one of the clusters showing a high degree of connectivity, including apolipoproteins A1, A2, C1, C3, and CLU and coagulation inflammatory components F13B, AHSG, and CP (Fig. 4).
Fig. 4.
Hive plot. Of the 114 proteins monitored in supernatants of AS-3 RBCs during routine storage, we could identify five main eigenvectors (clusters of trends), as color-coded in the rights-hand panel. The results of these five clusters ranged from having very high (Cluster 1) to very poor correlation with storage progression (as detailed in the legend on the top right corner). The five clusters were thus plotted as separate axes and connected with time (here represented as a sphere to the right) if showing Pearson’s linear correlations with storage progression higher than 0.9. Values from correlation matrices (proteins vs. proteins; Table S3) were plotted as light blue connecting lines if two proteins had modules of Pearson’s linear correlations or more than 0.9. The figure reveals that in Cluster 1, the top 14 proteins have more than 0.9 correlation with storage duration and were highly correlated among each other. However, other proteins were found to show extremely high correlations among each other in different clusters (bold fonts, color-coded consistently with each cluster). These hub proteins include previously recognized potential markers39 of RBC lesions through vesiculation, such as apolipoproteins A1, A2, A3, and C3 and apolipoprotein J/clusterin (CLU). A color version of the figure can be found in the online version of the paper.
DISCUSSION
Early investigations on the proteome of RBC supernatants8 revealed that omics technologies could provide precious insights on the quality of blood-derived therapeutics during routine storage in the blood bank. Recently, we applied semiquantitative discovery mode proteomics to reveal alterations to the proteome of RBC supernatants with and without leukoreduction, as a strategy to determine the biologic and, indirectly, the clinical relevance of filtration strategies.10 Discovery mode proteomics is a powerful tool to investigate fluctuations in the proteomes of RBCs and supernatants during routine storage. This is especially evident when monitoring substoichiometric protein modifications, such as oxidation of reactive thiol groups in key protein targets of redox lesions to stored RBCs, such as Hb, GAPDH and PRDX2.11,32,36
However, strides in the field of quantitative proteomics18–20,38 now enable absolute determination of protein levels in clinically relevant matrices, such as supernatants of RBCs. Here, we describe a methodologic workflow exploiting the QconCAT technology to showcase the relevance of absolute quantitative proteomics approaches in the field of transfusion medicine. As a proof of principle, quantitative changes of 114 proteins were accurately monitored in filtered RBC supernatants (approx. 3.5-log and approx. 2-log WBCs and PLTs, respectively10), early upon filtration and during storage duration on a weekly basis. These proteins were selected based on their importance in stored RBC biology,8,9,33,39 coagulation, and complement cascades. Preliminary immunoaffinity depletion of albumin and IgG, accounting for approximately 60% of the plasma proteome, was performed to enrich for lower abundant proteins, making them amenable to detection. While this process is extremely specific and offers obvious advantages, it also holds some pitfalls as some of the low-abundance proteins may be removed due to interaction with albumin (“sponge effect”). Additionally, while the proteins monitored here were selected based on their abundance and functional relevance, they are just a subset of the proteome and do not represent the full complexity of the RBC supernatant proteome. Despite these considerations, we show the effectiveness of the filtration strategy in removing coagulation, adaptive immunity, and complement components. We also observe increased levels of some of these components at the end of the storage period, possibly due to residual contaminating WBCs and PLTs. Of note, leukofiltration of whole blood has been shown to reduce the microparticle content and the pro-coagulant potential (FVIII, FXII, and fibrinogen content) of fresh-frozen plasma.40 The technology described in this study should 1) provide a platform for assessing effects of leukoreduction or pretransfusion washing strategies and 2) provide a platform for assessing effects of pathogen inactivation technologies on the stability of the proteomes of RBCs and supernatants and 3) be used to integrate with other biologically relevant information gathered from other omics platforms, such as metabolomics.36
By obtaining accurate quantitative information, the present approach has allowed us to identify candidate markers (with >95% confidence) of protein abundances at the end of the storage period based on sampling during early storage time points. This targeted approach utilizes less than 1 hour of instrument time per sample whereas an untargeted proteomics approach based on electrophoretic gel separation, excision of gel bands, trypsin digestion, and MS analysis would take approximately 10 times longer to observe all of the proteins represented here. In addition, the stochastic data sampling of traditional proteomic approaches would result in missing data points across multiple samples.
Of note, markers of hemolysis (though the relative contribution of vesiculation cannot be excluded) were identified other than Hbs HBA1, HBB and HBD, including metabolic enzymes (ALAD, CA1, CA2, GAPDH, selenium-binding protein), antioxidant enzymes (CAT and PRDX1, 2, and 6), or stress-related chaperone proteins (peptidyl-prolyl isomerase A). While PRDX2 had already been proposed as a potential marker of the oxidative lesion to stored RBCs,33 the additional proteins represent candidates for future translational application, either in the blood bank or in the pipeline leading to the design and development of new RBC storage additives or processing strategies. These proteins had significant correlations with storage duration and Hb levels, a bona fide proxy for RBC hemolysis and vesiculation and thus a key predictor of transfusion outcomes. It can be safely stated that RBCs that lyse will certainly not circulate, nor function, and thus be associated with potential impaired effectiveness of the transfusion therapy or to transfusion-related complications.
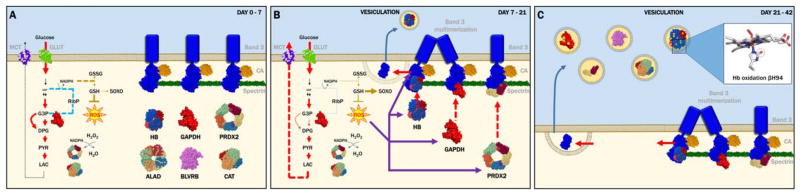
Appreciation of the progressive increase in the levels of these proteins in RBC supernatants can be reconciled with existing literature in the field of RBC storage lesions (Fig. 5).4 For example, GAPDH accumulation in RBC supernatants is consistent with the observed progressive loss of metabolic modulation in stored RBCs, resulting in a compromised capacity to fuel the pentose phosphate pathway to generate reducing equivalents (NADPH).4,30 In turn, NADPH is an essential cofactor for antioxidant enzymes such as CAT and PRDXs, in addition to enzymes involved in membrane migration and vesiculation.4,9,33,41,42 Here, these enzymes were shown to be increasing in the supernatant with a first-degree linear relationship. Of note, recycling of oxidized PRDX2 dimers has recently been shown to depend on the redox status of cysteine 94 of Hbβ (counting initiator methionine),34 a mechanism that is impaired during routine storage due to the irreversible modification to sulfinic/sulfonic acid or desulfurization to dehydroalanine of the functional thiol of this key residue.32 Even though RBCs are naturally equipped to cope with irreversibly oxidized proteins, some of these antioxidant mechanisms are impaired during storage. For example, damaged protein removal through proteasome-mediated degradation is compromised by the progressive release of proteasome components in the supernatants.4 Vesiculation of oxidized components has been indirectly suggested by colorimetric evidence (dinitrophenylhydrazine-reactive species as a marker of protein carbonylation)43,44 and, more recently, MS-based approaches.32
Fig. 5.
An overview of the RBC storage lesion, merging the hereby presented findings and existing literature.4 Three main stages are proposed in a time-dependent fashion, from (A) to (C). A color version of the figure can be found in the online version of the paper.
Vesiculation is a tightly regulated phenomenon in RBCs, involving structural proteins at the interface between lipid homeostasis and membrane integrity such as apolipoproteins.39 Here we observed a subgroup of proteins showing poor correlations with storage duration, probably owing to the initial depletion by leukoreduction and delayed increase only after the second week of storage, when the rates of vesiculation events are known to increase.4,41,42 This cluster included proteins with a high degree of connectivity with other proteins, including several apolipoproteins. It is worth noting the presence of apolipoprotein J/CLU in this group, a previously appreciated marker of RBC storage lesions and vesiculation of damaged proteins.39 These high correlations likely reflect the accuracy of the assay, highlighting the stability of molar ratios and potential multimerization45 of these key proteins in RBC supernatants during storage progression.
Limitations of the presented technology include 1) its supervised hypothesis-driven nature (only preselected proteins can be monitored); 2) the high level of expertise required in the fields of molecular biology, MS, and proteomics data analysis to perform such experiments; and 3) the time-consuming preanalytical steps to select proteotypic peptides, generate chimeric vectors, transform and culture bacterial strains, and purify and test chromatographic and MS responses of the peptides of interest. Once these burdens are overcome, the heavy isotope standards are available for basic science and clinical translational application for many years from the development of the technique. This makes it a promising strategy for quantitative proteomics applications for the foreseeable future until the advent of transformative technologies (e.g., nanochips, top-down proteomics) that supersede those of today.
In this methodologic study, we describe a recent advancement in the field of quantitative proteomics, the QconCAT technology, and discuss the relevance of its application in the field of transfusion medicine. Here, we exploit the QconCAT reagents and a targeted proteomics workflow to assay the supernatants of leukoreduced AS-3 RBCs during storage in the blood bank. Results mirror the effectiveness of the leukoreduction strategies adopted here, confirming and expanding on previous reports.40 However, coagulation and complement components accumulated in supernatants after the third week of storage, probably released by residual contaminating cells. Such components may contribute to some of the adverse effects related to transfusion.46 Twelve novel candidate protein biomarkers of RBC lysis or vesiculation were identified, to estimate late storage levels from as early as Day 1 assays of RBC supernatants. These findings require large-scale validation studies under a range of conditions encountered in the blood bank and clinic to determine their accuracy and general utility. In the meantime, these results represent a reference point for follow-up studies on alternative RBC storage additives or processing strategies (e.g., pretransfusion washing filters, pathogen reduction technologies). The described technology might have utility in evaluating not just the quality of blood-derived therapeutics during routine storage, but also prestorage variables such as processing strategies, interdonor variability, and donor health.
Supplementary Material
Fig. S1. A representative chromatogram of an LC-SRM run.
Fig. S2. Heat map of linear correlations among proteins in stored RBC supernatants, following hierarchical clustering. The figure is a vectorial extended version of Fig. 3A.
TABLE S1. QconCAT constructs.
TABLE S2. QconCAT report.
TABLE S3. Correlation table.
Acknowledgments
The authors are grateful to Dr Ron Hay and Ellis Jaffray of the University of Dundee for providing the auxotroph used in this study and Drs Marguerite Kelher and Christopher C. Silliman (Bonfils Blood Center) for providing the samples analyzed in this study.
ABBREVIATIONS
- ABC
ammonium bicarbonate
- ALAD
aminolevulinate dehydratase
- CA1 (or 2)
carbonic anhydrase-1 (or -2)
- CAT
catalase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PRDX2
peroxiredoxin-2
- QconCAT(s)
quantitative concatamer(s)
- SRM
single reaction monitoring
Footnotes
CONFLICT OF INTEREST
The authors have disclosed no conflicts of interest.
Additional Supporting Information may be found in the online version of this article at the publisher’s Web site
References
- 1.Blood safety and availability [Internet] Geneva: World Health Organization; 2015. [cited 2015 Oct 16]. Available from: http://www.who.int/entity/mediacentre/factsheets/fs279/en/index.html. [Google Scholar]
- 2.Holst LB, Petersen MW, Haase N, et al. Restrictive versus liberal transfusion strategy for red blood cell transfusion: systematic review of randomised trials with meta-analysis and trial sequential analysis. BMJ. 2015;350:h1354. doi: 10.1136/bmj.h1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hess JR. An update on solutions for red cell storage. Vox Sang. 2006;91:13–9. doi: 10.1111/j.1423-0410.2006.00778.x. [DOI] [PubMed] [Google Scholar]
- 4.D’Alessandro A, Kriebardis AG, Rinalducci S, et al. An update on red blood cell storage lesions, as gleaned through biochemistry and omics technologies. Transfusion. 2015;55:205–19. doi: 10.1111/trf.12804. [DOI] [PubMed] [Google Scholar]
- 5.Zolla L, D’Alessandro A, Rinalducci S, et al. Classic and alternative red blood cell storage strategies: seven years of “omics” investigations. Blood Transfus. 2015;13:21–31. doi: 10.2450/2014.0053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimring JC. Established and theoretical factors to consider in assessing the red cell storage lesion. Blood. 2015;125:2185–90. doi: 10.1182/blood-2014-11-567750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spitalnik SL, Triulzi D, Devine DV, et al. 2015 proceedings of the National Heart, Lung, and Blood Institute’s State of the Science in Transfusion Medicine symposium. Transfusion. 2015;55:2282–90. doi: 10.1111/trf.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anniss AM, Glenister KM, Killian JJ, et al. Proteomic analysis of supernatants of stored red blood cell products. Transfusion. 2005;45:1426–33. doi: 10.1111/j.1537-2995.2005.00547.x. [DOI] [PubMed] [Google Scholar]
- 9.D’Alessandro A, D’Amici GM, Vaglio S, et al. Time-course investigation of SAGM-stored leukocyte-filtered red blood cell concentrates: from metabolism to proteomics. Haematologica. 2012;97:107–15. doi: 10.3324/haematol.2011.051789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dzieciatkowska M, Silliman CC, Moore EE, et al. Proteomic analysis of the supernatant of red blood cell units: the effects of storage and leucoreduction. Vox Sang. 2013;105:210–8. doi: 10.1111/vox.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rinalducci S, Marrocco C, Zolla L. Thiol-based regulation of glyceraldehyde-3-phosphate dehydrogenase in blood bank-stored red blood cells: a strategy to counteract oxidative stress. Transfusion. 2015;55:499–506. doi: 10.1111/trf.12855. [DOI] [PubMed] [Google Scholar]
- 12.Rinalducci S, Longo V, Ceci LR, et al. Targeted quantitative phosphoproteomic analysis of erythrocyte membranes during blood bank storage. J Mass Spectrom. 2015;50:326–35. doi: 10.1002/jms.3531. [DOI] [PubMed] [Google Scholar]
- 13.Pasini EM, Kirkegaard M, Mortensen P, et al. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108:791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- 14.Roux-Dalvai F, Gonzalez de Peredo A, Simó C, et al. Extensive analysis of the cytoplasmic proteome of human erythrocytes using the peptide ligand library technology and advanced mass spectrometry. Mol Cell Proteomics. 2008;7:2254–69. doi: 10.1074/mcp.M800037-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Zaccaria A, Roux-Dalvai F, Bouamrani A, et al. Accessing to the minor proteome of red blood cells through the influence of the nanoparticle surface properties on the corona composition. Int J Nanomedicine. 2015;10:1869–83. doi: 10.2147/IJN.S70503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Alessandro A, Righetti PG, Zolla L. The red blood cell proteome and interactome: an update. J Proteome Res. 2010;9:144–63. doi: 10.1021/pr900831f. [DOI] [PubMed] [Google Scholar]
- 17.Bordbar A, Jamshidi N, Palsson BO. iAB-RBC-283: a proteomically derived knowledge-base of erythrocyte metabolism that can be used to simulate its physiological and pathophysiological states. BMC Syst Biol. 2011;5:110. doi: 10.1186/1752-0509-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aebersold R. Quantitative proteome analysis: methods and applications. J Infect Dis. 2003;187(Suppl 2):S315–20. doi: 10.1086/374756. [DOI] [PubMed] [Google Scholar]
- 19.Bantscheff M, Schirle M, Sweetman G, et al. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–31. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 20.Meissner F, Mann M. Quantitative shotgun proteomics: considerations for a high-quality workflow in immunology. Nat Immunol. 2014;15:112–7. doi: 10.1038/ni.2781. [DOI] [PubMed] [Google Scholar]
- 21.Bislev SL, Kusebauch U, Codrea MC, et al. Quantotypic properties of QconCAT peptides targeting bovine host response to Streptococcus uberis. J Proteome Res. 2012;11:1832–43. doi: 10.1021/pr201064g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brownridge PJ, Harman VM, Simpson DM, et al. Absolute multiplexed protein quantification using QconCAT technology. Methods Mol Biol. 2012;893:267–93. doi: 10.1007/978-1-61779-885-6_18. [DOI] [PubMed] [Google Scholar]
- 23.Pratt JM, Simpson DM, Doherty MK, et al. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat Protoc. 2006;1:1029–43. doi: 10.1038/nprot.2006.129. [DOI] [PubMed] [Google Scholar]
- 24.Desiere F, Deutsch EW, King NL, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–8. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picotti P, Lam H, Campbell D, et al. A database of mass spectrometric assays for the yeast protein. Nat Methods. 2008;5:913–4. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill RC, Calle EA, Dzieciatkowska M, et al. Quantification of extracellular matrix proteins from a rat lung scaffold to provide a molecular readout for tissue engineering. Mol Cell Proteomics. 2015;14:961–73. doi: 10.1074/mcp.M114.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson TD, Hill RC, Dzieciatkowska M, et al. Quantification of decellularized human myocardial matrix: a comparison of six patients. Proteomics Clin Appl. 2015;10:75–83. doi: 10.1002/prca.201500048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batth TS, Singh P, Ramakrishnan VR, et al. A targeted proteomics toolkit for high-throughput absolute quantification of Escherichia coli proteins. Metab Eng. 2014;26:48–56. doi: 10.1016/j.ymben.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Dzieciatkowska M, D’Alessandro A, Hill RC, et al. Plasma Qcon-CATs reveal a gender-specific proteomic signature in apheresis platelet plasma supernatants. J Proteomics. 2015;120:1–6. doi: 10.1016/j.jprot.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Alessandro A, Nemkov T, Kelher M, et al. Routine storage of red blood cell (RBC) units in additive solution-3: a comprehensive investigation of the RBC metabolome. Transfusion. 2015;55:1155–68. doi: 10.1111/trf.12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Alessandro A, Nemkov T, Hansen KC, et al. Red blood cell storage in additive solution-7 preserves energy and redox metabolism: a metabolomics approach. Transfusion. 2015 doi: 10.1111/trf.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wither M, Dzieciatkowska M, Nemkov T, et al. Hemoglobin oxidation at functional amino acid residues during routine storage of red blood cells. Transfusion. 2015 doi: 10.1111/trf.13363. [DOI] [PubMed] [Google Scholar]
- 33.Rinalducci S, D’Amici GM, Blasi B, et al. Peroxiredoxin-2 as a candidate biomarker to test oxidative stress levels of stored red blood cells under blood bank conditions. Transfusion. 2011;51:1439–49. doi: 10.1111/j.1537-2995.2010.03032.x. [DOI] [PubMed] [Google Scholar]
- 34.Harper VM, Oh JY, Stapley R, et al. Peroxiredoxin-2 recycling is inhibited during erythrocyte storage. Antioxid Redox Signal. 2015;22:294–307. doi: 10.1089/ars.2014.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silliman CC, Kelher MR, Khan SY, et al. Experimental prestorage filtration removes antibodies and decreases lipids in RBC supernatants mitigating TRALI in vivo. Blood. 2014;123:3488–95. doi: 10.1182/blood-2013-10-532424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel RM, Roback JD, Uppal K, et al. Metabolomics profile comparisons of irradiated and nonirradiated stored donor red blood cells. Transfusion. 2015;55:544–52. doi: 10.1111/trf.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiśniewski JR, Zougman A, Nagaraj N, et al. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 38.Marx V. Targeted proteomics. Nat Methods. 2013;10:19–22. doi: 10.1038/nmeth.2285. [DOI] [PubMed] [Google Scholar]
- 39.Antonelou MH, Kriebardis AG, Stamoulis KE, et al. Apolipoprotein J/clusterin in human erythrocytes is involved in the molecular process of defected material disposal during vesiculation. PLoS One. 2011;6:e26033. doi: 10.1371/journal.pone.0026033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan KS, Sparrow RL. Microparticle profile and procoagulant activity of fresh-frozen plasma is affected by whole blood leukoreduction rather than 24-hour room temperature hold. Transfusion. 2014;54:1935–44. doi: 10.1111/trf.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubin O, Crettaz D, Tissot JD, et al. Microparticles in stored red blood cells: submicron clotting bombs? Blood Transfus. 2010;8(Suppl 3):s31–8. doi: 10.2450/2010.006S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kriebardis AG, Antonelou MH, Stamoulis KE, et al. RBC-derived vesicles during storage: ultrastructure, protein composition, oxidation, and signaling components. Transfusion. 2008;48:1943–53. doi: 10.1111/j.1537-2995.2008.01794.x. [DOI] [PubMed] [Google Scholar]
- 43.Delobel J, Prudent M, Rubin O, et al. Subcellular fractionation of stored red blood cells reveals a compartment-based protein carbonylation evolution. J Proteomics. 2012;76(Spec No):181–93. doi: 10.1016/j.jprot.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Kriebardis AG, Antonelou MH, Stamoulis KE, et al. Progressive oxidation of cytoskeletal proteins and accumulation of denatured hemoglobin in stored red cells. J Cell Mol Med. 2007;11:148–55. doi: 10.1111/j.1582-4934.2007.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pallotta V, D’Alessandro A, Rinalducci S, et al. Native protein complexes in the cytoplasm of red blood cells. J Proteome Res. 2013;12:3529–46. doi: 10.1021/pr400431b. [DOI] [PubMed] [Google Scholar]
- 46.Flegel WA, Natanson C, Klein HG. Does prolonged storage of red blood cells cause harm? Br J Haematol. 2014;165:3–16. doi: 10.1111/bjh.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. A representative chromatogram of an LC-SRM run.
Fig. S2. Heat map of linear correlations among proteins in stored RBC supernatants, following hierarchical clustering. The figure is a vectorial extended version of Fig. 3A.
TABLE S1. QconCAT constructs.
TABLE S2. QconCAT report.
TABLE S3. Correlation table.