Abstract
Accumulating evidence points to a genetic contribution to explain inter-individual vulnerability to sleep deprivation. A functional polymorphism in the BDNF gene, which causes a valine (Val) to methionine (Met) amino acid substitution at Codon 66, has been associated with cognitive impairment, particularly in populations with impaired frontal functioning. We hypothesised that sleep deprivation, which affects frontal function, may lead to cognitive dysfunction in Met allele carriers. To examine this, we investigated, in different BDNF genotypes, the effects of sleep deprivation on cognitive flexibility, as measured by response inhibition using the Stroop Color Naming Task. Thirty healthy, adults of European ancestry, including 12 heterozygous Met allele carriers and 18 Val/Val homozygotes, underwent 30-h of extended wakefulness under constant routine conditions. A computerised Stroop task was administered every 2 h. Error rate and reaction times increased with time awake for all individuals. Participants with the Val/Met genotype made more errors on incongruent trials after 20 h awake. While Val/Met participants also took significantly longer to respond when inhibiting a prepotent response irrespective of time awake, this was particularly evident during the biological night. Our study shows that carriers of the BDNF Met allele are more vulnerable to the impact of prolonged wakefulness and the biological night on a critical component of executive function, as measured by response inhibition on the Stroop task.
Keywords: BDNF, Val66Met, Sleep deprivation, Cognitive flexibility, Executive function, Stroop
At the group level, sleep deprivation has a detrimental impact on cognitive function (See [1,2] for reviews). Some individuals, however, are more vulnerable than others to the adverse effects of sleep deprivation on performance [3]. Importantly, this vulnerability appears to be trait-like and likely has a genetic basis[4]. A number of candidate genes, such as PER3, COMT and ADORA2A, have been identified as contributing to individual variability in cognitive function following sleep deprivation [5–7]. More recently, a polymorphism in the Brain Derived Neurotrophic Factor (BDNF) gene has shown promise as a genetic marker of vulnerability to sleep deprivation [8]. While this gene influences cognitive function in well-rested, healthy adults, the effects of this polymorphism on vulnerability to sleep deprivation is comparatively less well-studied.
BDNF, a member of the neurotrophin family of growth factors, plays an important role in neuronal development and survival, context-dependent synaptic plasticity and long-term potentiation [9]. BDNF is expressed throughout the central nervous system, and is particularly abundant in the prefrontal cortex [9,10]. A common single nucleotide polymorphism has been identified in the BDNF gene which produces an amino acid substitution from valine (Val) to methionine (Met) at codon 66 (Val66Met) [11]. This Met substitution leads to impairments in intracellular processing, trafficking and activity dependent secretion of the BDNF protein [12], and is associated with reductions in cortical volume and thickness in prefrontal regions [10,13]. The BDNF Val66Met polymorphism has been linked to individual differences in cognitive performance, such that BDNF Met allele carriers perform more poorly on cognitive tasks, especially those relating to memory, learning and executive function [11,13–15], when well-rested. This finding is not consistent however, with other studies reporting no effects [16–18]. As Met allele impairments are more consistently found where frontal functioning is compromised, such as in schizophrenia, bipolar disorder and Obsessive Compulsive Disorder [11,19–21], we hypothesise that the Met allele may result in enhanced vulnerability to sleep deprivation where frontal function is also impaired [1,2].
To our knowledge, only one study has investigated the relationship between sleep deprivation, performance and BDNF genotype in humans. Although they reported no effect of the polymorphism on the psychomotor vigilance task, they did report that response accuracy on a working memory task was significantly poorer in Met allele carriers compared to Val/Val homozygotes during sleep deprivation [8]. To determine whether this effect may be evident across other cognitive domains, our study examined whether individuals with the BDNF Val/Met genotype, compared to Val/Val homozygotes, were more vulnerable to sleep deprivation on a task of response inhibition.
Thirty participants of self-reported European ancestry completed an 8-day inpatient study. Twelve participants carried the Val/Met genotype (4 women (33.3%); 23.17 ± 4.22 years) and 18 carried the Val/Val genotype (8 women (44.4%); 22.89 ± 5.18 years). All participants were free from medical, psychiatric and sleep disorders. Individuals with color blindness, and those who had engaged in night and/or shift work in the past 3 years, or who travelled across more than one time zone in the previous 3 months were excluded. Participants maintained a consistent, self-selected 8:16 h sleep/wake schedule for three weeks prior to the study, confirmed by time-stamped calls at sleep/wake times and actigraphy (Actiwatch-L, Minimitter, Inc, Bend, OR). The use of any medications, supplements, recreational drugs, nicotine, caffeine, and/ or alcohol were prohibited for the duration of the study, and was confirmed by urine and blood toxicology prior to study admission. The protocol was approved by the Partners Human Research Committee at the Brigham and Women’s Hospital, and participants provided full written informed consent.
Participants were continuously monitored for 8 consecutive days in a time free environment (no windows, clocks, radio, live TV, newspapers, and technicians trained to not reveal the time). Prior to commencing a 30-h constant routine (CR), participants underwent a phase advance protocol involving the 8-h advance of the sleep-wake cycle over 5 days – reported elsewhere [22,23]. During the CR, participants remained awake in dim light conditions (3 lx), in a semi-recumbent posture (head of bed at 45°), and received their equivalent daily fluid and caloric intake in hourly snacks. Participants completed a computerised version of the Stroop Color Naming Task every two hours, starting three hours post wake. The task included three trial types, where the font color and word color were the same (congruent) or different (incongruent), or simply consisted of a row of 4 colored “X”s (neutral). For all trial types, participants were asked to respond by naming the color of the text by typing the first letter of the color on a keyboard (See [24] for more details).
To determine circadian phase, Dim Light Melatonin Onset (DLMO) was defined as the time at which plasma melatonin levels rose to 25% of the nightly fitted peak. This was determined using the 3-harmonic method (See [25] for more details). DNA was extracted from whole blood and genotyped for the rs6265 (coding DNA variant Val66Met) variant of the BDNF gene using TagMan SNP genotype assays (Applied Biosystems, Assay ID: C1159275810). PCR amplification and allele specific discrimination was performed using LightCycler 480 (Roche Applied Science, USA). The reaction consisted of 10 ng DNA, 6.25 μl of 2X master mix (Applied Biosystems, catalog number: 4371357), 0.65 μl of 20X Taqman SNP genotyping assay in a PCR reaction volume of 12.5 μl. Annealing was performed at 60 °C. The LightCycler 480 software was used to detect specific SNP alleles using the end-point detection method. Genotyping was performed in duplicate to ensure reproducibility and a call rate > 95% was obtained.
Polysomnography (PSG) was recorded using a Vitaport-3 recording system (TEMEC Instruments, B.V. Kerkrade, The Netherlands). EEG (C3, C4 referenced to contralateral mastoids), EOG (upper and lower outer-canthi) and submental EMG was obtained. Electrode impedances were < 10 kΩ, and EEG signals were filtered (high-pass EEG filter 0.23 Hz; low-pass EEG filter 70.1 Hz; 24 dB/octave, sampling rate 256 Hz) and sleep staged in 30-s epochs according to the Rechtschaffen and Kales scoring system. Total sleep time (TST); sleep efficiency (% TST of time in bed) and wake after sleep onset (WASO) was calculated.
Mean reaction time (RT) and the error rate (percent incorrect responses) for congruent, incongruent and neutral trials were calculated for each Stroop session. A difference score (RT of incongruent trials - RT of neutral trials) was also calculated within each session as an index of inhibition. The inhibition score, or the cost of inhibition, reflects the additional time needed to inhibit the prepotent response for incongruent trials. Test sessions were averaged over three time periods in order optimise power for the analysis: Tertile 1: 0–10-h, Tertile 2: 11–20-h and, Tertile 3: 21–30-h. To examine the effect of sleep deprivation, PROC MIXED analyses (SAS 9.4) were performed with tertile and genotype modelled as fixed factors and participant and circadian phase modelled as random factors. Degrees of freedom were calculated using the Kenward-Rogers method and the covariance type with the lowest Schwarz Bayesian Criterion (BIC) was chosen. To control for familywise error, post hoc pairwise comparisons were conducted using a false discovery rate (FDR) comparison, reported as padj. As a secondary aim, we examined the impact of circadian time. Independent samples t-tests (SPSS Statistics 20) compared performance during the ‘biological day’ (average of three tests occurring at least one hour before DLMO) versus ‘biological night’ (average of three tests occurring at least one hour after DLMO). Only participants who had data for at least three sessions before and after DLMO were included (Val/Met: n = 11 (∼92%); Val/Val: n = 16 (∼89%)).
Data were successfully obtained from 30 participants, except for phase where we could not estimate DLMO for n = 1 (Val/Met) and sleep where there was not a polysomnography recording for n = 2 (Val/Met). Participant demographic information is shown in Table 1. The genotype frequencies observed in the current sample did not differ from the expected Hardy-Weinberg equilibrium ( , p > 0.10). There were no participants with the Met/Met genotype; however, this was expected given the low frequency of the genotype in populations of European ancestry (< 5%) [11]. To ensure the different phase advance protocols did not affect Stroop outcomes during CR, we conducted a linear mixed model to determine whether the different light or phase shift conditions during the phase advance affected any of the Stroop outcomes. There was no effect of light condition or phase advance protocol on any Stroop outcome (see Table S1 in supplemental material). Means and standard deviations for all Stroop outcomes by genotype, and the results of the linear mixed model are reported in Table 2.
Table 1.
Participant demographic information by genotype.
| Demographic | Val/Val (M ± SD) | Val/Met (M ± SD) | t (df) | p |
|---|---|---|---|---|
| n | 18 | 12 | – | – |
| Age | 22.89 ± 5.18 | 23.17 ± 4.22 | 0.16 (28) | 0.878 |
| Sex males n(%) | 10 (55.56%) | 8 (66.67%) | – | 0.709a |
| Wake time | 08.05 ± 0.86 | 8.11 ± 0.80 | 0.20 (28) | 0.847 |
| DLMO | 18.81 ± 1.92 | 20.03 ± 3.18 | 1.28 (27) | 0.212 |
| DLMO shift (h) | 2.95 ± 1.43 | 2.23 ± 2.06 | 1.11 (27) | 0.277 |
| TST (mins) | 388.22 ± 68.92 | 408.70 ± 67.35 | 0.76 (26) | 0.455 |
| SE (%) | 80.77 ± 14.46 | 85.05 ± 13.97 | 0.76 (26) | 0.455 |
| WASO (mins) | 65.08 ± 73.79 | 56.91 ± 66.62 | 0.29 (26) | 0.774 |
Dim light melatonin onset (DLMO); total sleep time (TST); sleep efficiency (SE); wake after sleep onset (WASO). Wake time refers to wake time before the 8-h phase advance and DLMO refers to DLMO assessed during the CR post phase shift.
Fisher’s exact test p-value (2-sided).
Table 2.
Performance on the Stroop task for genotype (Val/Met vs. Val/Val) and time awake (1st, 2nd and 3rd tertile) and the genotype*time awake interaction.
| Stroop Outcome | Stroop Trial Type | Time Tertile | Val/Met (M ± SD) | Val/Val (M ± SD) | Genotype F (df) | Time Awake F (df) | Genotype*Time Awake F (df) |
|---|---|---|---|---|---|---|---|
| Reaction Time (ms) | Congruent | T1 | 816.22 ± 165.67 | 780.76 ± 106.15 | 0.45 (1, 27.1) | 30.94*** (2, 53.9) | 0.32 (2, 53.9) |
| T2 | 802.30 ± 175.34 | 765.33 ± 94.56 | |||||
| T3 | 912.57 ± 186.82 | 896.33 ± 133.66 | |||||
| Neutral | T1 | 829.83 ± 149.58 | 825.17 ± 123.89 | 0.26 (1, 27.1) | 38.38*** (2, 53.9) | 0.71 (2, 53.9) | |
| T2 | 833.14 ± 169.23 | 801.43 ± 102.32 | |||||
| T3 | 951.93 ± 173.68 | 954.08 ± 134.56 | |||||
| Incongruent | T1 | 923.37 ± 206.32 | 873.57 ± 132.44 | 0.45 (1, 27.1) | 30.94*** (2, 53.9) | 0.32 (2, 53.9) | |
| T2 | 926.39 ± 229.87 | 866.65 ± 114.17 | |||||
| T3 | 1055.23 ± 217.14 | 1009.36 ± 134.98 | |||||
| Error rate | Congruent | T1 | 2.55 ± 2.27 | 2.35 ± 1.64 | 3.01 (1, 28) | 20.35*** (2, 54.5) | 1.28 (2, 54.5) |
| T2 | 3.26 ± 2.76 | 1.89 ± 1.03 | |||||
| T3 | 7.11 ± 4.22 | 4.90 ± 3.69 | |||||
| Neutral | T1 | 4.05 ± 2.52 | 2.87 ± 2.20 | 5.25* (1, 27.8) | 22.56*** (2, 54.3) | 1.30 (2, 54.3) | |
| T2 | 4.24 ± 2.86 | 3.03 ± 1.85 | |||||
| T3 | 8.99 ± 4.48 | 5.62 ± 3.89 | |||||
| Incongruent | T1 | 4.43 ± 2.55 | 4.13 ± 2.00 | 2.10 (1, 27.3) | 24.65*** (2, 53.9) | 3.65** (2, 53.9) | |
| T2 | 4.90 ± 3.18 | 4.24 ± 2.40 | |||||
| T3 | 10.15 ± 5.05 | 6.45 ± 4.41 | |||||
| Inhibition | n/a | T1 | 93.54 ± 69.85 | 48.19 ± 27.87 | 4.13* | 0.47 | 0.48 |
| T2 | 93.78 ± 68.33 | 65.89 ± 37.05 | (1, 27.1) | (2, 53.7) | (2, 53.7) | ||
| T3 | 103.31 ± 84.13 | 55.43 ± 69.03 | 0.003 | 0.03 | 0.02 |
Values for the F-statistic and degrees of freedom are shown for the main effects of time and genotype, and the time*genotype interaction. Significance is denoted by
p < 0.001;
p < 0.01;
p < 0.05.
p-values for comparison between timepoints and for post-hoc tests for the significant interaction are reported in the text.
Overall, response times were slower and the error rate of responses was greater as time awake progressed (See Table 2; all trial types p < 0.0001). Post hoc comparison revealed that reaction times were slowest and more errors were made in the third tertile of sleep deprivation (20–30 h awake), compared to both the first (0–10 h awake; congruent and incongruent: p < 0.0001) and second tertiles (10–20 h awake; all trial types: p < 0.0001). There was no difference between the first and second tertiles for either reaction time (all trial types: p > 0.3) or error rate (all trial types: p > 0.7). Post-hoc paired t-test statistics for the main effect of time are shown in supplemental Table S2.
No significant differences were found between BDNF genotypes for reaction time on any trial type, or the error rate for both congruent and incongruent trials (Table 2). For neutral trials, however, individuals with the Val/Met genotype made more errors compared to those homozygous for the Val/Val genotype (Val/Met 5.76 ± 4.03 ms; Val/Val 3.84 ± 3.02; p = 0.03). As seen in Fig. 1, a significant genotype*time interaction was found for the error rate on incongruent trials (p = 0.032). Post hoc comparison showed that Val/Mets made 3.54% [95% CI 0.9590, 6.1267] more errors, relative to Val/Val homozygotes, in Tertile 3 (t55.3 = 2.75, p = 0.008, padj = 0.017). Other interactions were not significant as described in Table 2.
Fig. 1.
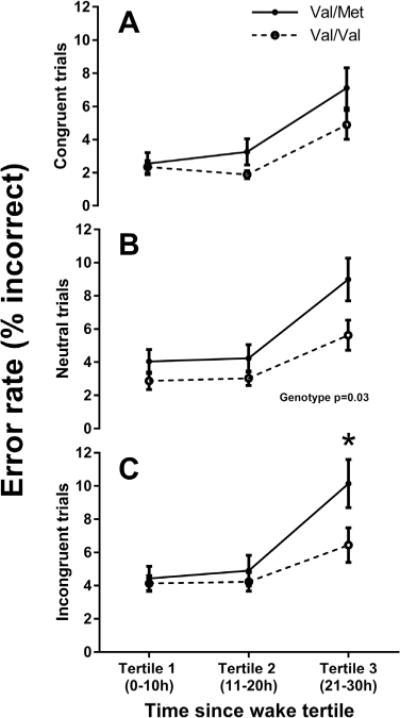
Mean and standard error of the number of errors committed for each genotype. Each trial type is plotted separately: (A) Congruent (B) Neutral, and (C) Incongruent. *p < 0.05. Untransformed data are plotted.
As seen in Fig. 2A, Val/Met heterozygotes took significantly longer to respond, relative to Val/Val homozygotes, when required to inhibit a prepotent response, such that the Val/Met homozygotes had a higher cost of inhibition (Val/Met 96.88 ± 72.43 ms; Val/Val 56.50 ± 47.66 ms; p = 0.047). There was no effect of time awake or a significant genotype*time interaction on cost of inhibition, however. As the main effect of genotype appeared to be most apparent at 0–10 h awake and 21–30 h awake (See Fig. 2A), which, due to the protocol design, corresponded with night time hours, we conducted a follow-up analysis comparing the genotypes during the biological day and the biological night. As seen in Fig. 2B, participants with the Val/Met genotype took significantly longer to respond when inhibiting the prepotent response than those with the Val/Val genotype during the biological night (Val/Met 112.46 ± 76.09 ms; Val/Val 43.54 ± 73.27 ms; t25 = 2.37, p = 0.026, d = 0.92), but not during the biological day (Val/Met 71.53 ± 59.25 ms; Val/Val 63.26 ± 44.35 ms; t25 = 0.42, p > 0.68, d = 0.16) – see Fig. 2B.
Fig. 2.

Mean and standard error of inhibition for each genotype. (A) Inhibition score for each tertile (B) Inhibition scores for the biological day and the biological night. *p < 0.05. Untransformed data are plotted.
Our data provide evidence that individuals who carry the Met allele of the BDNF genotype exhibit more impairment of cognitive flexibility following sleep deprivation, specifically in their ability to inhibit a prepotent response, as indicated by an increased error rate on the incongruent trials of the Stroop task following 20 h awake. The cost of inhibition, or the extra time required to inhibit the response, was also higher in those carrying the Met allele, but this was irrespective of sleep loss. This difference in cost of inhibition between the two groups appeared greatest at 0–10 h awake and 21–30 h which corresponded with the biological night in our study. Follow up analyses confirmed that the cost of inhibition was higher during the biological night for those with the Met allele, compared to those homozygous for the Val/Val BDNF genotype. Our results also support previous findings showing that Met allele carriers perform worse on a range of cognitive tasks even under well rested conditions [11,13,14], such that participants with the Val/Met genotype in the current study made more errors overall for neutral trials, irrespective of time awake.
The impact of BDNF genotype on vulnerability to sleep deprivation may be due to the effect of sleep loss on BDNF expression and subsequent cortical function. For instance, expression of BDNF is increased in the frontal cortex, basal forebrain and hippocampus following sleep loss in rats [26–28], which is associated with improved cognitive performance [26]. These findings suggest that BDNF may act to enhance cortical functioning in the areas where it is upregulated during sleep loss. As the BDNF Met allele is associated with impaired activity-dependent secretion of the BDNF protein, we speculate that any sleep deprivation-induced increase in cortical BDNF may be attenuated in Met allele carriers leading to poorer cognitive outcomes. In support of this interpretation, imaging studies in humans show reduced activation in the prefrontal cortex and hippocampus of BDNF Met carriers [10,11,13,29], which was attributed to impaired secretion of BDNF protein in the Met allele carriers affecting synaptic events underlying performance [29]. The Stroop task relies on activation of frontal brain regions [30], thus, the structural and functional brain differences [10,13] between the BDNF genotypes may underlie the variation in performance by affecting the way in which the brain responds to the task during sleep loss. These structural and functional abnormalities in Val/Met individuals may affect the allocation of frontal resources and alter the brain’s compensatory response to sleep loss, leading to impaired cognitive performance relative to Val/Val homozygotes. As this interpretation is speculative, future work comparing frontal activation and compensatory responses between those with and without the BDNF Met allele during sleep deprivation would provide crucial insight.
While our study outcomes are novel and provide further evidence for a genetic marker of vulnerability to sleep loss, our study has a number of limitations. Firstly, the results of this study should be interpreted with caution due to the small sample size, and our results should be replicated under similar conditions. Despite this, our sample size is similar to previous studies investigating the effects of a polymorphism on performance during sleep deprivation [5–8] and our findings replicate previous research showing impaired frontal functioning in BDNF met allele carriers in well-rested individuals [13,14,19,20] and during sleep deprivation [8]. Secondly, individuals were exposed to varying light exposure and phase shifting protocols prior to the CR as this was part of a larger study. To address whether these conditions had an impact on our outcomes, we conducted a linear mixed model (See supplementary material Table S1) which showed that there was no effect of light and phase shift condition on Stroop outcomes. There was also no difference in the average phase shift between genotypes in response to the phase advance protocol. Finally, as the main protocol induced a phase advance prior to the CR, resulting in (some) participants sleeping at a non-optimal circadian time, we again examined sleep outcomes prior to the CR as this may have exacerbated performance deficits. As we found no difference in total sleep time, wake after sleep onset, and sleep efficiency between genotypes, we argue that differences in prior sleep between the groups does not explain our observed differences in performance during sleep deprivation (CR).
In summary, our study shows that the BDNF Met allele carriers are more vulnerable to the effect of sleep deprivation and the biological night on a measure of inhibition. BDNF Met allele carriers made more inhibitory errors on the Stroop task during sleep deprivation and the cost of inhibition, the additional time taken to respond when inhibiting a response, was greater regardless of time awake, and particularly so during the biological night. These data indicate that BDNF genotype merits further inquiry as a potentially important genetic determinant for individual vulnerability to sleep loss and performance impairments at adverse circadian times. Our data both support and extend the findings presented by [8] in that the BDNF Val66Met polymorphism is associated with vulnerability to sleep deprivation, and that this vulnerability goes beyond working memory to other cognitive domains, such as cognitive flexibility, which is a critical component of executive function. Taken together, these results have important implications for individuals working extended duration shifts, particularly at night.
Supplementary Material
Acknowledgments
We thank the study participants, the staff of the Center for Clinical Investigation of the Brigham and Women’s Hospital, and the staff of the Chronobiology Core for monitoring study equipment. We also thank our recruiter Mr. David Klements.
Funding source
This study was supported by the National Space and Biomedical Research Institute (NSBRI) grant HFP01601. Inpatient studies were conducted in the CTSC and supported in part by ULI RR025758 from the National Center for Research Resources (NCRR) or the National Institutes of Health (NIH).
Ms. Grant has received scholarship funding from the Cooperative Research Centre for Alertness, Safety and Productivity. Dr Czeisler has received consulting fees from or served as a paid member of scientific advisory boards for: Bose Corporation; Boston Celtics; Columbia River Bar Pilots; Institute of Digital Media and Child Development; Klarman Family Foundation; Quest Diagnostics, Inc.; Vanda Pharmaceuticals and V-Watch/PPRS. He has also received education/research support from Cephalon Inc., Mary Ann & Stanley Snider via Combined Jewish Philanthropies, Optum, Philips Respironics, Inc., ResMed Foundation, San Francisco Bar Pilots, Schneider Inc., and Sysco. He has received lecture fees from American Academy of Sleep Medicine (AADSM), CurtCo M edia Labs LLC, Global Council on Brain Health/AARP, Hawaii Sleep Health and Wellness Foundation, National Sleep Foundation, University of Michigan, University of Washington, and Zurich Insurance Company, Ltd. The Sleep and Health Education Program of the Harvard Medical School Division of Sleep Medicine (which Dr. Czeisler directs) has received Educational Grant funding from Cephalon, Inc., Jazz Pharmaceuticals, Takeda Pharmaceuticals, Teva Pharmaceuticals Industries Ltd., Sanofi-Aventis, Inc., Sepracor, Inc. and Wake Up Narcolepsy. Dr Czeisler is the incumbent of an endowed professorship provided to Harvard University by Cephalon, Inc. and holds a number of process patents in the field of sleep/circadian rhythms (e.g., photic resetting of the human circadian pacemaker). Since 1985, he has served as an expert on various legal and technical cases related to sleep and/or circadian rhythms including those involving the following commercial entities: Bombardier, Inc.; Continental Airlines; FedEx; Greyhound; and United Parcel Service (UPS). Dr Czeisler owns or owned an equity interest in Somnus Therapeutics, Inc., and Vanda Pharmaceuticals. He received royalties from McGraw Hill, and Koninklijke Philips Electronics, N.V. for the Actiwatch-2 and Actiwatch-Spectrum devices. Dr Czeisler’s interests were reviewed and managed by Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict of interest policies. Dr. Anderson reports receiving a research award/prize from Sanofi-Aventis; contract research support from VicRoads, Rio Tinto Coal Australia, and Tontine/Pacific Brands; and lecturing fees from Brown Medical School/Rhode Island Hospital, Ausmed, HealthEd and TEVA Pharmceuticals. In addition, she has served as a consultant through her institution to the Rail, Bus and Tram Union, the Transport Accident Commission (TAC) and the National Transportation Committee (NTC). She has also served as an expert witness and/or consultant in relation to fatigue and drowsy driving. Dr Anderson is a Theme Leader, and has received research funding, in the Cooperative Research Centre for Alertness, Safety and Productivity.
Footnotes
Author contributions
All authors have contributed to this manuscript. C.A.C., A-M.C., S.C., C.A. contributed to the study concept and design. A-M.C., S.C. and C.A. contributed to the collection of data. R.S. conducted the genetic analysis. L.G., S.C. and C.A. contributed to analysis and interpretation of the data and drafting of the manuscript. All authors were involved in editing and reviewing the manuscript. The manuscript has been approved by all authors.
Disclosure statement
Drs. Cain, Chang and Saxena report no financial conflicts of interest in relation to this work.
References
- 1.Horne JA. Human sleep, sleep loss and behaviour. Implications for the prefrontal cortex and psychiatric disorder. Br J Psychiatry. 1993;162:413–419. doi: 10.1192/bjp.162.3.413. [DOI] [PubMed] [Google Scholar]
- 2.Killgore WDS. Progr Brain Res. Elsevier; 2010. Effects of sleep deprivation on cognition; pp. 105–129. [DOI] [PubMed] [Google Scholar]
- 3.Van Dongen HPA, Baynard MD, Maislin G, Dinges DF. Systematic inter-individual differences in neurobehavioral impairment from sleep loss: evidence of trait-like differential vulnerability. Sleep. 2004;27(3):423–433. [PubMed] [Google Scholar]
- 4.Kuna ST, Maislin G, Pack FM, Staley B, Hachadoorian R, Coccaro EF, Pack AI. Heritability of performance deficit accumulation during acute sleep deprivation in twins. Sleep. 2012;35(9):1223–1233. doi: 10.5665/sleep.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viola AU, Archer SN, James L, Groeger JA, Lo JCY, Skene DJ, von Schantz M, Dijk DJ. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17(7):613–618. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 6.Bodenmann S, Xu S, Luhmann UF, Arand M, Berger W, Jung HH, Landolt HP. Pharmacogenetics of modafinil after sleep loss: catechol-O-methyltransferase genotype modulates waking functions but not recovery sleep. Clin Pharmacol Ther. 2009;85(3):296–304. doi: 10.1038/clpt.2008.222. [DOI] [PubMed] [Google Scholar]
- 7.Bodenmann S, Hohoff C, Freitag C, Deckert J, Rétey JV, Bachmann V, Landolt HP. Polymorphisms of ADORA2A modulate psychomotor vigilance and the effects of caffeine on neurobehavioural performance and sleep EEG after sleep deprivation. Br J Pharmacol. 2012;165(6):1904–1913. doi: 10.1111/j.1476-5381.2011.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachmann V, Klein C, Bodenmann S, Schäfer N, Berger W, Brugger P, Landolt HP. The BDNF Val66Met polymorphism modulates sleep intensity: EEG frequency- and state-specificity. Sleep. 2012;35(3):335–344. doi: 10.5665/sleep.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murer M, Yan Q, Raisman-Vozari R. Brain-derived neurotrophic factor in the control human brain, and in Alzheimer’s disease and Parkinson’s disease. Prog Neurobiol. 2001;63(1):71–124. doi: 10.1016/s0301-0082(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 10.Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A, Weinberger DR. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24(45):10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 12.Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS. Variant brain-derived neurotrophic factor (BDNF)(Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24(18):4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schofield PR, Williams LM, Paul RH, Gatt JM, Brown K, Luty A, Cooper N, Grieve S, Dobson-Stone C, Morris C, Kuan SA, Gordon E. Disturbances in selective information processing associated with the BDNF Val66Met polymorphism: evidence from cognition, the P300 and fronto-hippocampal systems. Biol Psychol. 2009;80(2):176–188. doi: 10.1016/j.biopsycho.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Marqués-Iturria I, Garolera M, Pueyo R, Segura B, Hernan I, García-García I, Sánchez-Garre C, Vernet-Vernet M, Sender-Palacios MJ, Narberhaus A, Ariza M, Junqué C, Jurado MA. The interaction effect between BDNF val66met polymorphism and obesity on executive functions and frontal structure. Am J Med Genet Part B Neuropsychiatr Genet. 2014;165(3):245–253. doi: 10.1002/ajmg.b.32229. [DOI] [PubMed] [Google Scholar]
- 15.Miyajima F, Ollier W, Mayes A, Jackson A, Thacker N, Rabbitt P, Pendleton N, Horan M, Payton A. Brain-derived neurotrophic factor polymorphism Val66Met influences cognitive abilities in the elderly. Genes Brain Behav. 2008;7(4):411–417. doi: 10.1111/j.1601-183X.2007.00363.x. [DOI] [PubMed] [Google Scholar]
- 16.Beste C, Baune BT, Domschke K, Falkenstein M, Konrad C. Paradoxical association of the brain-derived-neurotrophic-factor val66met genotype with response inhibition. Neuroscience. 2010;166(1):178–184. doi: 10.1016/j.neuroscience.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 17.Gajewski PD, Hengstler JG, Golka K, Falkenstein M, Beste C. The Met-genotype of the BDNF Val66Met polymorphism is associated with reduced Stroop interference in elderly. Neuropsychologia. 2012;50(14):3554–3563. doi: 10.1016/j.neuropsychologia.2012.09.042. [DOI] [PubMed] [Google Scholar]
- 18.Harris S, Fox H, Wright A, Hayward C, Starr J, Whalley L, Deary I. The brain-derived neurotrophic factor Val66Met polymorphism is associated with age-related change in reasoning skills. Mol Psychiatry. 2006;11(5):505–513. doi: 10.1038/sj.mp.4001799. [DOI] [PubMed] [Google Scholar]
- 19.Rybakowski JK, Borkowska A, Czerski PM, Skibińska M, Hauser J. Polymorphism of the brain-derived neurotrophic factor gene and performance on a cognitive prefrontal test in bipolar patients. Bipolar Disord. 2003;5(6):468–472. doi: 10.1046/j.1399-5618.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 20.Rybakowski JK, Borkowska A, Skibinska M, Szczepankiewicz A, Kapelski P, Leszczynska-Rodziewicz A, Czerski PM, Hauser J. Prefrontal cognition in schizophrenia and bipolar illness in relation to Val66Met polymorphism of the brain-derived neurotrophic factor gene. Psychiatry Clin Neurosci. 2006;60(1):70–76. doi: 10.1111/j.1440-1819.2006.01462.x. [DOI] [PubMed] [Google Scholar]
- 21.Tükel R, Gürvit H, Özata B, Öztürk N, Ertekin BA, Ertekin E, Baran B, Kalem SA, Büyükgök D, Direskeneli GS. Brain-derived neurotrophic factor gene Val66Met polymorphism and cognitive function in obsessive-compulsive disorder. Am J Med Genet Part B Neuropsychiatr Genet. 2012;159 B(7):850–858. doi: 10.1002/ajmg.b.32092. [DOI] [PubMed] [Google Scholar]
- 22.Anderson C, Chang AM, Sullivan JP, Ronda JM, Czeisler CA. Assessment of drowsiness based on ocular parameters detected by infrared reflectance oculography. J Clin Sleep Med. 2013;9(9):907–920. doi: 10.5664/jcsm.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cain SW, Chang AM, Vlasac I, Tare A, Anderson C, Czeisler CA, Saxena R. Circadian rhythms in plasma brain-derived neurotrophic factor differ in men and women. J Biol Rhythms. 2017;32(1):75–82. doi: 10.1177/0748730417693124. [DOI] [PubMed] [Google Scholar]
- 24.Cain SW, Silva EJ, Chang AM, Ronda JM, Duffy JF. One night of sleep deprivation affects reaction time, but not interference or facilitation in a Stroop task. Brain Cogn. 2011;76(1):37–42. doi: 10.1016/j.bandc.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gronfier C, Wright KP, Jr, Kronauer RE, Jewett ME, Czeisler CA. Efficacy of a single sequence of intermittent bright light pulses for delaying circadian phase in humans. Am J Physiol Endocrinol Metab. 2004;287(1 50–1):E174–E181. doi: 10.1152/ajpendo.00385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng O, Li R, Zhao L, Yu L, Yang B, Wang J, Chen B, Yang J. Short-term sleep deprivation stimulates hippocampal neurogenesis in rats following global cerebral ischemia/reperfusion. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0125877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000;885(2):303–321. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- 28.Wallingford JK, Deurveilher S, Currie RW, Fawcett JP, Semba K. Increases in mature brain-derived neurotrophic factor protein in the frontal cortex and basal forebrain during chronic sleep restriction in rats: possible role in initiating allostatic adaptation. Neuroscience. 2014;277:174–183. doi: 10.1016/j.neuroscience.2014.06.067. [DOI] [PubMed] [Google Scholar]
- 29.Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23(17):6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung HC, Skudlarski P, Gatenby JC, Peterson BS, Gore JC. An event-related functional MRI study of the Stroop color word interference task. Cereb Cortex. 2000;10(6):552–560. doi: 10.1093/cercor/10.6.552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


