Abstract
Environmental temperature is one of the most important factors for the growth and survival of microorganisms. Here we describe a novel extremely halophilic archaeon (haloarchaea) designated as strain CBA1119T isolated from solar salt. Strain CBA1119T had the highest maximum and optimal growth temperatures (66 °C and 55 °C, respectively) and one of the largest genome sizes among haloarchaea (5.1 Mb). It also had the largest number of strain-specific pan-genome orthologous groups and unique pathways among members of the genus Natrinema in the class Halobacteria. A dendrogram based on the presence/absence of genes and a phylogenetic tree constructed based on OrthoANI values highlighted the particularities of strain CBA1119T as compared to other Natrinema species and other haloarchaea members. The large genome of strain CBA1119T may provide information on genes that confer tolerance to extreme environmental conditions, which may lead to the discovery of other thermophilic strains with potential applications in industrial biotechnology.
Introduction
The growth of most microorganisms is influenced by physical factors such as temperature, water activity, pH, pressure, salinity, and oxygen concentration as well as chemical factors such as availability of nutrients (e.g., carbon and nitrogen)1–4. Microorganisms are usually classified based on optimal growth temperature-i.e., as psychrophiles, mesophiles, thermophiles, and hyperthermophiles, which grow best at temperatures of ≤15 °C, 15 °C–45 °C, >45 °C, and 80 °C, respectively5. These classes also differ in terms of the amino acid composition, structure, and thermostability of proteins6. Growth temperature seems to be related to genomic features; one study showed that the average length of proteins is shorter in thermophiles (growing best at temperatures of >45 °C) as compared to their homolog in mesophiles (15 °C–45 °C), whereas the proportion of purine bases in the coding strand is higher in the former than in the latter7. Other environmental factors besides temperature affect genome size: for example, the small genomes of prokaryotes are thought to reflect adaptation to strong selective pressures in large microbial populations, while the genome size in geophytes was found to be positively correlated with early flowering and growth tendency under humid conditions8,9.
Extremely halophilic archaea (haloarchaea) belonging to the domain Archaea are usually found in hypersaline environments such as salt lakes and crystallizer ponds from artificial marine solar salterns and in salty fermented foods and salted hides10,11, as well as in avian feather12. The growth temperature of haloarchaeal type strains ranges from −1 °C to 62 °C, with few growing at temperatures >60 °C (see Supplementary information). Genus Natrinema in the family Natrialbaceae includes eight known species of haloarchaea: Natrinema altunense, Nnm. ejinorense, Nnm. gari, Nnm. pallidum, Nnm. pellirubrum, Nnm. salaciae, Nnm. soli, and Nnm. versiforme13–19. In this study we describe strain CBA1119T isolated from solar salt, which has the highest growth temperature and one of the largest genome sizes among all of the haloarchaeal members. We identified and characterized thermophilic strain CBA1119T and investigated the relationship between two strain-specific features, namely growth temperature and genome size.
Results and Discussion
Polyphasic taxonomic analysis (see Supplementary Information) revealed that strain CBA1119T belonged to the genus Natrinema and was a novel member of the genus Natrinema. Interestingly, strain CBA1119T grew at a temperature of 20 °C–66 °C; optimal growth was observed at 50 °C–55 °C. Of the four strains with an optimal growth temperature >50 °C; three belonged to the family Haloferacaceae and one was strain CBA1119T, which belongs to the family Natrialbaceae (Fig. 1a). The maximum growth temperatures of haloarchaea varied within each family (Fig. 1b). It is worth noting that there were no strains belonging to the family Halococcaceae that grew at temperatures >50 °C, and only those belonging to the family Natrialbaceae had a maximum growth temperature >60 °C, including strain CBA1119T. The maximum and optimal growth temperatures of strain CBA1119T were the highest recorded to date among haloarchaea. Environmental temperature underlies the evolution of various biological phenomena such as the density of hydrogen bonds in nucleic acid20.
Figure 1.
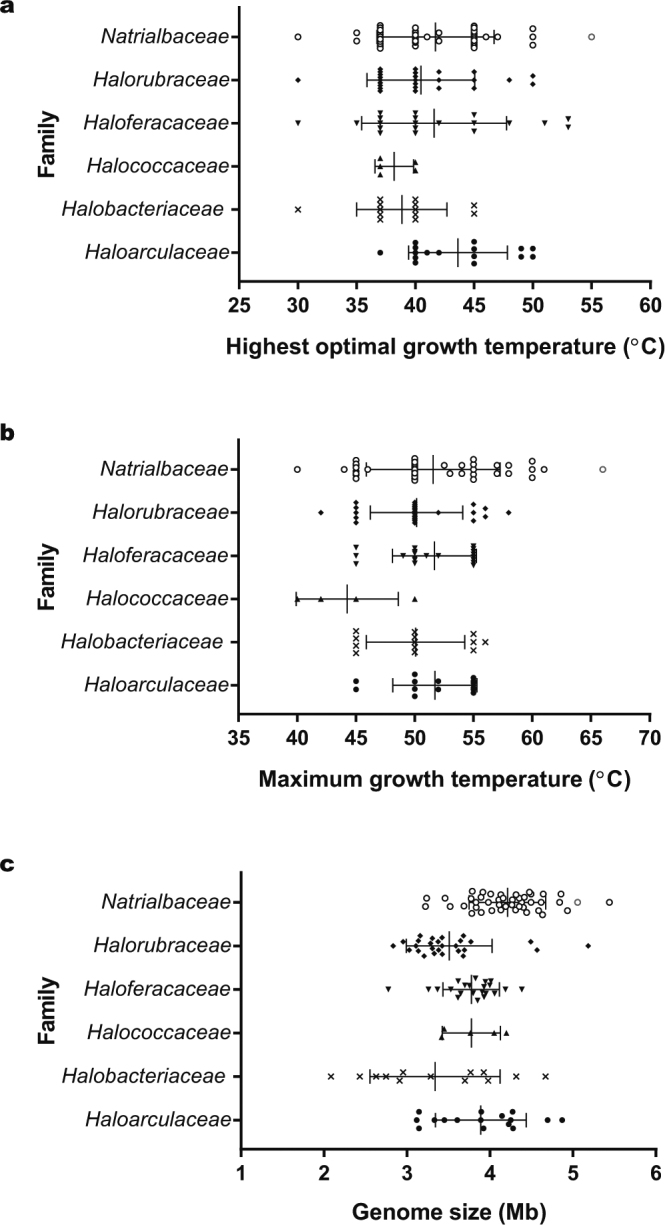
Comparison of the highest optimal (a) and maximum growth temperatures (b), and genome sizes (c) among haloarchaeal species. Strain CBA1119T has the highest optimal and maximum growth temperature, and the third largest genome size among type strains belonging to haloarchaea. Red circles indicate strain CBA1119T.
General genomic features of strain CBA1119T were described in Supplementary Information (Supplementary Tables S2 and S3; Supplementary Fig. S2b). The ways in which the microbial genome is affected by environmental factors can be understood by pan-genome comparisons21. The number of pan-genome orthologous groups (POGs) and strain-specific POGs (singletons) were compared among strain CBA1119T and seven species of the genus Natrinema (Fig. 2). The flower plot showed that strain CBA1119T had the largest number of singletons among Natrinema species. The number of singletons in strain CBA1119T was 1.4 times that in Nnm. salaciae JCM 17869T (which had the second largest number) and four times that in Nnm. altunense AJ2T (which had the smallest number). The heat map based on gene content also showed that strain CBA1119T had more exclusive POGs than other related species (Fig. 3). Additionally, each genome within the genus Natrinema had distinct KEGG pathway profiles based on POGs (Table 1). Strain CBA1119T had specific enzymes listed on the KEGG pathway named propanoate metabolism, geraniol degradation, fatty acid biosynthesis, metabolism and degradation, and valine, leucine and isoleucine degradation, with P values of zero.
Figure 2.
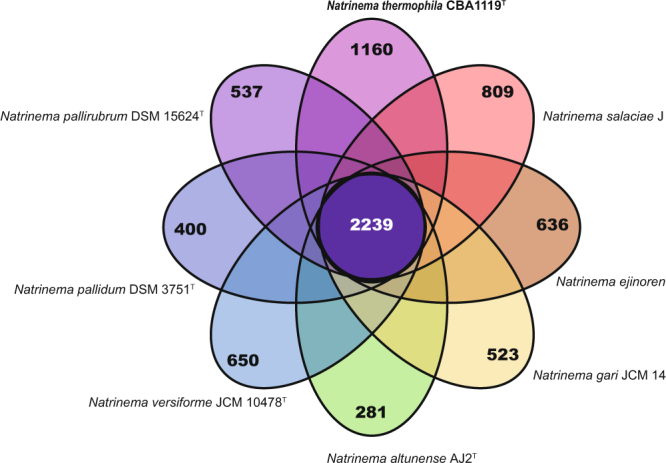
Flower plot showing strain-specific and core POGs of eight Natrinema species.
Figure 3.
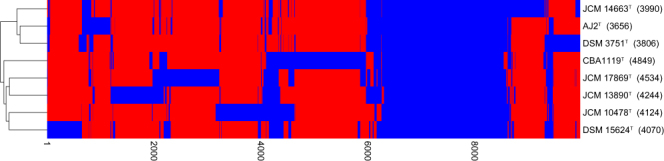
Heatmap based on gene content. A dendrogram was generated using Jaccard coefficients and unweighted pair-group method with arithmetic mean clustering. Blue and red indicate present and absent genes, respectively. Values in the brackets indicate number of POGs of each strain.
Table 1.
Strain-specific POGs listed on the KEGG pathway (P < 0.05).
| Strain | KEGG pathway ID | Pathway name | Differentially present POGs | P value |
|---|---|---|---|---|
| CBA1119T* | MAP00281 | Geraniol degradation | 13 | 0.0000 |
| MAP00640 | Propanoate metabolism | 31 | 0.0000 | |
| MAP00061 | Fatty acid biosynthesis | 16 | 0.0000 | |
| MAP01212 | Fatty acid metabolism | 36 | 0.0000 | |
| MAP00071 | Fatty acid degradation | 26 | 0.0000 | |
| MAP00280 | Valine, leucine, and isoleucine degradation | 28 | 0.0000 | |
| MAP00780 | Biotin metabolism | 12 | 0.0001 | |
| MAP00072 | Synthesis and degradation of ketone bodies | 8 | 0.0001 | |
| MAP00650 | Butanoate metabolism | 24 | 0.0002 | |
| MAP01040 | Biosynthesis of unsaturated fatty acids | 12 | 0.0011 | |
| MAP00380 | Tryptophan metabolism | 18 | 0.0026 | |
| MAP00410 | beta-Alanine metabolism | 13 | 0.0032 | |
| MAP00930 | Caprolactam degradation | 7 | 0.0064 | |
| MAP00903 | Linonene and pinene degradation | 7 | 0.0168 | |
| MAP00310 | Lysine degradation | 16 | 0.0173 | |
| MAP00620 | Pyruvate metabolism | 18 | 0.0295 | |
| MAP03022 | Basal transcription factors | 9 | 0.0408 | |
| MAP00720 | Carbon fixation pathways in prokaryotes | 21 | 0.0414 | |
| JCM 17869T | MAP00072 | Synthesis and degradation of ketone bodies | 4 | 0.0101 |
| JCM 13890T | MAP00780 | Biotin metabolism | 5 | 0.0180 |
| MAP00625 | Chloroalkane and chloroalkene degradation | 6 | 0.0298 | |
| MAP00633 | Nitrotoluene degradation | 4 | 0.0394 | |
| JCM 14663T | MAP00072 | Synthesis and degradation of ketone bodies | 4 | 0.0033 |
| JCM 10478T | MAP00983 | Drug metabolism – other enzymes | 5 | 0.0304 |
| DSM 3751T | MAP00250 | Alanine, aspartate and glutamate metabolism | 7 | 0.0356 |
| MAP00650 | Butanoate metabolism | 8 | 0.0363 | |
| DSM 15624T | MAP03022 | Basal transcription factors | 7 | 0.0067 |
*Strain CBA1119T is estimated to contain the largest number of singletons.
For genome size and growth temperature comparisons among haloarchaeal type strains, information on the strains was obtained from the GenBank database and previous studies, and is shown in Supplementary Table S4. Genome size comparison at the class level revealed that most haloarchaea (104/128 species) had a genome ranging between 3.0 and 4.5 Mb in size, with the class Natrialbaceae having the largest average genome size (Fig. 1c). Interestingly, only three strains had a genome size >5 Mb, including strain CBA1119T. Besides a high growth temperature, strain CBA1119T had an unusually large genome size. Haloarchaea species with a genome >5 Mb are uncommon; only two such type strains (and three in total) are found in the GenBank database. Strain CBA1119T had the third largest genome among haloarchaeal type strains (and the fourth among total haloarchaeal strains). Genome size was shown to be related to COG categories and pathways in bacteria; COG categories related to secondary metabolism and energy conversion were more highly represented in larger genomes, as were KEGG categories related to various cellular processes and metabolism with the exception of nucleotide metabolism22. Free-living bacteria with a genome size >6 Mb such as Bacteroides thetaiotaomicron and Streptomyces avermitili can grow in various environments and use a wide range of substrates for energy production. Thus, strain CBA1119T with its large genome size may be capable of growing under different conditions, and can potentially utilize different substrates to produce energy. Genome size increases with the level of environmental instability; that is, large genomes are also more resistant to environmental perturbations than smaller ones23. It remains to be determined whether this applies to strain CBA1119T. Clarifying the genomic and environmental factors that affect growth temperature and genome size can provide insight into environment-microbe interactions and evolutionary adaptations of various microorganisms, while additional studies on the enzymes of strain CBA1119T can reveal new tools for industrial biotechnology applications.
Materials and Methods
Isolation of archaeal strain
Strain CBA1119T was isolated from unrefined solar salt obtained from a salt field (34.587738 N, 126.105372 E) in the Republic of Korea and aerobically cultured in DBCM2 medium (JCM medium no. 574; 833 ml MDS salt water [240 g NaCl, 30 g MgCl2∙6H2O, 35 g MgSO4∙7H2O, 7 g KCl, 5 ml 1 M CaCl2 solution per liter], 1 ml FeCl2 solution [10 ml 25% HCl, 1.5 g FeCl2∙4H2O per liter], 1 ml trace element solution [70 mg ZnCl2, 100 mg MnCl2∙4H2O, 6 mg H3BO3, 190 mg CoCl2∙6H2O, 2 mg CuCl2∙2H2O, 24 mg NiCl2∙6 H2O, 36 mg Na2MoO4∙2H2O per liter], 0.25 g peptone [Oxoid, Chesire, UK], 0.05 g yeast extract [BD Biosciences, Franklin Lakes, NJ, USA], 5 ml 1 M NH4Cl, 3 ml vitamin solution [3 mg biotin, 4 mg folic acid, 50 mg pyridoxine·HCl, 33 mg thiamine·HCl, 10 mg riboflavin, 33 mg nicotinic acid, 17 mg DL-calcium pantothenate, 17 mg vitamin B12, 13 mg para-aminobenzoic acid, 10 mg lipoic acid per liter], 10 ml of 1 M sodium pyruvate solution, 2 ml potassium phosphate buffer [417 ml 1 M K2HPO4 and 83 ml 1 M KH2PO4 per liter], and 50 ml 1 M Tris-HCl, pH 7.5 per liter) at 37 °C for 4 weeks. To obtain pure culture, a single colony was transferred repeatedly to the agar medium.
Phenotypic, chemotaxonomic, and phylogenetic analyses
Phenotypic tests were performed according to the minimal standards for description of new taxa in the order Halobacteriales24. Cell morphology and size were examined by field emission transmission electron microscopy (Chuncheon Center, Korea Basic Science Institute, Korea). Gram staining was performed as previously described25. For comparative phenotypic analyses, reference strains were selected based on the relatedness of 16S rRNA gene sequences (>97%). For this purpose, Nnm. soli LMG 29247T18, Nnm. salaciae JCM 17869T17, and Nnm. ejinorense JCM 13890T14 obtained from the Japan Collection of Microorganisms (JCM) or Belgian Coordinated Collections of Microorganisms (BCCM) were cultured at 37 °C in DBCM2 medium. Growth at different temperatures (4 °C, 15 °C–60 °C at intervals of 5 °C, and 61 °C–70 °C at intervals of 1 °C), NaCl concentrations (0–30% [w/v] at intervals of 5%), pHs (5.0–11.0 at intervals of 1.0), and Mg2+ concentrations (0, 5, 10, 20, 50, 100, 200, and 500 mM) were tested using DBCM2 medium as the basal medium for 4 weeks. pH was adjusted by adding the following buffers: 10 mM 2-(N-morpholino)-ethanesulfonic acid (MES) (pH 5–6), 1,3-bis[tris(hydroxymethyl)methylamino]propane (Bis-TRIS propane) (pH 7–9), or N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) (pH 10–11). Anaerobic growth in the presence of 0.5% l-arginine, trimethylamine-N-oxide (TMAO), dimethyl sulfoxide (DMSO), or 30 mM nitrate was evaluated on DBCM2 medium at 37 °C in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI, USA) with an N2·CO2·H2 (90:5:5, v-v:v) atmosphere. Catalase and oxidase activities26 as well as the hydrolysis of starch and casein27 and of Tween 40 and Tween 8028 were evaluated according to established protocols. Antibiotic susceptibility was tested on DBCM2 medium using antibiotic discs with ampicillin (10 μg per disc), erythromycin (15 μg), gentamicin (10 μg), kanamycin (30 μg), nalidixic acid (30 μg), rifampicin (10 μg), and streptomycin (10 μg). The effectiveness of various substrates as a sole carbon and energy source and acid production were determined in HMD medium29. A total of 20 carbon sources were tested: D-fructose, D-galactose, D-mannitol, D-mannose, D-sorbitol, D-xylose, fumarate, glycerol, maltose, pyruvate, starch, succinate, sucrose, L-alanine, L-arginine, L-aspartate, L-glutamate, L-lysine, L-malate, and L-sorbose. Polar lipids from strain CBA1119T were extracted, analyzed, and compared with those of the three reference strains as previously described30. The DNA-DNA hybridization (DDH)31 was performed to determine the genetic relationship between strain CBA1119T and the three reference strains. To determine the taxonomic identity based on 16S rRNA gene sequence, chromosomal DNA was extracted using a commercial DNA extraction kit (iNtRON Biotechnology, Sungnam, Korea) and the 16S rRNA gene was amplified using PCR PreMix (iNtRON Biotechnology) with universal primers 0018 F and 1518R32. Amplified 16S rRNA PCR products were sequenced and assembled as previously described33 and 16S rRNA sequences were compared using EzTaxon-e34 or NCBI BLAST35. Phylogenetic trees were constructed based on the three 16S rRNA gene sequences of strain CBA1119T obtained from the genome sequencing data (see below) and other related species using MEGA6 software36. Phylogenetic trees were generated with neighbor-joining (NJ)37, maximum likelihood (ML)38, and maximum parsimony (MP)39 methods with 1 000 bootstrap replications based on the NJ tree.
Library preparation, sequencing, genome assembly, and annotation
To clarify the relationship between physiological characteristics (especially capacity for growth at high temperatures) and genomic features, we performed genome sequencing of strain CBA1119T and Nnm. ejinorense JCM 13890T as previously described40. In brief summary, the genomic DNA shearing and SMRTbell library preparation were carried out according to the standard PacBio 20-kb Template Preparation Using BluePippin Size-Selection System protocol by P6-C4 chemistry (Pacific Biosciences, Menlo Park, CA, USA), respectively. The strain CBA1119T genome and Nnm. ejinorense JCM 13890T genome sequences were determined using the PacBio RS II system (Pacific Biosciences). De novo genome assembly of each genome was performed using Hierarchical Genome Assembly Process v.2 software with default parameters supported by PacBio SMRT Analysis v.2.3.041. rRNA and tRNA prediction was carried out using RNAmmer v.1.242 and tRNAscan-SE v.1.2143, respectively. Genes were predicted using Glimmer3 in Rapid Annotation using Subsystem Technology server (http://rast.nmpdr.org), and functional gene annotations were performed based on the SEED, COG (http://www.ncbi.nlm.nih.gov/COG), and KEGG (http://www.genome.jp/kegg/) databases. The GenBank/EMBL/DDBJ accession numbers for the Natrinema thermophila CBA1119T and Natrinema ejinorense JCM 13890T are PDBS00000000 and NXNI00000000, respectively.
Comparative genomic analysis
For genomic comparisons, Natrinema species genomes were obtained from the NCBI genome database, except those of strains CBA1119T and JCM 13890T, which were sequenced as described above. The OrthoANI algorithm was used to analyze the genomic relatedness between strain CBA1119T and other species. OrthoANI percentages were calculated and a phylogenetic tree was constructed44. Orthologs in strain CBA1119T and the reference strains were predicted and mapped using the reciprocal best hit method in UBLAST45. Pan-genome orthologous groups (POGs) were estimated using the EzBioCloud Comparative Genomics Database (http://cg.ezbiocloud.net/)46, and their presence was calculated using the Jaccard coefficient. The unweighted pair-group method with arithmetic mean (UPGMA) clustering was then used to assess clustering between strain CBA1119T and the reference strains from a dendrogram constructed based on the presence or absence of gene content. Haloarchaea genomes for comparisons were obtained from the NCBI genome database according to the following criteria: genomes with optimal or maximum growth temperature information were selected for comparisons of optimal and maximum growth temperature, respectively; genomes of unclassified strains47 were excluded; and genomes with fewer contigs that are less incomplete were selected, when multiple genomes were available for a single strain.
Electronic supplementary material
Acknowledgements
This research was supported by a grant from the World Institute of Kimchi (KE1802-2) funded by the Ministry of Science and ICT; and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2015R1D1A1A09061039), Republic of Korea.
Author Contributions
S.W.R. conceived the project and designed the experiments. Y.B.K., H.S.S., and J.-K.R. performed the experiments. Y.B.K., J.Y.K. and S.W.R. carried out data analyses and interpreted the results. S.W.R., Y.B.K., J.Y.K., H.S.S., C.L., S.W.A., S.H.L., J.K., D.-W.H., J.-W.B., and M.Y.J. discussed the results. Y.B.K., J.Y.K., J.K., D.-W.H., J.-W.B. and S.W.R wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Yeon Bee Kim and Joon Yong Kim contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25887-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Baez A, Shiloach J. Effect of elevated oxygen concentration on bacteria, yeasts, and cells propagated for production of biological compounds. Microb Cell Fact. 2014;13:181. doi: 10.1186/s12934-014-0181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelkirk, P. G., Duben-Engelkirk, J. L. & Burton, G. R. W. Burton’s microbiology for the health sciences. Lippincott Williams & Wilkins (2011).
- 3.Forbort, J. Treatment of Organic Contaminants: Biological Treatment. In: Groundwater Treatment Technology. John Wiley & Sons, Inc. (2009).
- 4.Mota MJ, Lopes RP, Delgadillo I, Saraiva JA. Microorganisms under high pressure — Adaptation, growth and biotechnological potential. Biotechnol Adv. 2013;31:1426–1434. doi: 10.1016/j.biotechadv.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Madigan, M. T., Martinko, J. M., Bender, K. S., Buckley, D. H. & Stahl, D. A. Brock Biology of Microorganisms, 14th edn. Pearson Education Limited (2015).
- 6.Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K. H. & Stackebrandt, E. The Prokaryotes: Vol. 2: Ecophysiology and Biochemistry. Springer New York (2006).
- 7.Hickey DA, Singer GA. Genomic and proteomic adaptations to growth at high temperature. Genome Biol. 2004;5:117. doi: 10.1186/gb-2004-5-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sela I, Wolf YI, Koonin EV. Theory of prokaryotic genome evolution. Proc Natl Acad Sci USA. 2016;113:11399–11407. doi: 10.1073/pnas.1614083113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vesely P, Bures P, Smarda P, Pavlicek T. Genome size and DNA base composition of geophytes: the mirror of phenology and ecology? Ann Bot. 2012;109:65–75. doi: 10.1093/aob/mcr267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oren, A. The Order Halobacteriales. In: The Prokaryotes: Volume 3: Archaea. Bacteria: Firmicutes, Actinomycetes. Springer New York (2006).
- 11.Roh SW, et al. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J. 2009;4:1–16. doi: 10.1038/ismej.2009.83. [DOI] [PubMed] [Google Scholar]
- 12.Yim KJ, et al. Occurrence of viable, red-pigmented haloarchaea in the plumage of captive flamingoes. Sci Rep. 2015;5:16425. doi: 10.1038/srep16425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X-W, Ren P-G, Liu S-J, Wu M, Zhou P-J. Natrinema altunense sp. nov., an extremely halophilic archaeon isolated from a salt lake in Altun Mountain in Xinjiang, China. Int J Syst Evol Microbiol. 2005;55:1311–1314. doi: 10.1099/ijs.0.63622-0. [DOI] [PubMed] [Google Scholar]
- 14.Castillo AM, et al. Natrinema ejinorense sp. nov., isolated from a saline lake in Inner Mongolia, China. Int J Syst Evol Microbiol. 2006;56:2683–2687. doi: 10.1099/ijs.0.64421-0. [DOI] [PubMed] [Google Scholar]
- 15.Tapingkae W, et al. Natrinema gari sp. nov., a halophilic archaeon isolated from fish sauce in Thailand. Int J Syst Evol Microbiol. 2008;58:2378–2383. doi: 10.1099/ijs.0.65644-0. [DOI] [PubMed] [Google Scholar]
- 16.McGenity TJ, Gemmell RT, Grant WD. Proposal of a new halobacterial genus Natrinema gen. nov., with two species Natrinema pellirubrum nom. nov. and Natrinema pallidum nom. nov. Int J Syst Bacteriol. 1998;48:1187–1196. doi: 10.1099/00207713-48-4-1187. [DOI] [PubMed] [Google Scholar]
- 17.Albuquerque L, Taborda M, La Cono V, Yakimov M, da Costa MS. Natrinema salaciae sp. nov., a halophilic archaeon isolated from the deep, hypersaline anoxic Lake Medee in the Eastern Mediterranean Sea. Syst Appl Microbiol. 2012;35:368–373. doi: 10.1016/j.syapm.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Rasooli M, et al. Natrinema soli sp. nov., a novel halophilic archaeon isolated from a hypersaline wetland. Int J Syst Evol Microbiol. 2017;67:2142–2147. doi: 10.1099/ijsem.0.001909. [DOI] [PubMed] [Google Scholar]
- 19.Xin H, et al. Natrinema versiforme sp. nov., an extremely halophilic archaeon from Aibi salt lake, Xinjiang, China. Int J Syst Evol Microbiol. 2000;50:1297–1303. doi: 10.1099/00207713-50-3-1297. [DOI] [PubMed] [Google Scholar]
- 20.Jeffrey, G. A. & Saenger, W. Hydrogen Bonding in Biological Structures. Springer Berlin Heidelberg (2012).
- 21.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Konstantinidis KT, Tiedje JM. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bentkowski P, Van Oosterhout C, Mock T. A Model of Genome Size Evolution for Prokaryotes in Stable and Fluctuating Environments. Genome Biol Evol. 2015;7:2344–2351. doi: 10.1093/gbe/evv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oren A, Ventosa A, Grant WD. Proposed Minimal Standards for Description of New Taxa in the Order Halobacteriales. Int J Syst Bacteriol. 1997;47:233–238. doi: 10.1099/00207713-47-1-233. [DOI] [Google Scholar]
- 25.Dussault HP. An improved technique for staining red halophilic bacteria. J Bacteriol. 1955;70:484–485. doi: 10.1128/jb.70.4.484-485.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson, H. J. Microbiological applications: a laboratory manual in general microbiology. McGraw-Hill Higher Education Boston (2002).
- 27.Smibert, R. M. & Kreg, N. R. Phenotypic characterization. In: Methods for General and Molecular Bacteriology. Gerhardt, P., Murray, R. G. E., Wood, W. A. & Krieg, N. R. Eds American Society for Microbiology, Washington, DC, 607–654 (1994).
- 28.Gonzalez C, Gutierrez C, Ramirez C. Halobacterium vallismortis sp. nov. An amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol. 1978;24:710–715. doi: 10.1139/m78-119. [DOI] [PubMed] [Google Scholar]
- 29.Savage KN, Krumholz LR, Oren A, Elshahed MS. Haladaptatus paucihalophilus gen. nov., sp. nov., a halophilic archaeon isolated from a low-salt, sulfide-rich spring. Int J Syst Evol Microbiol. 2007;57:19–24. doi: 10.1099/ijs.0.64464-0. [DOI] [PubMed] [Google Scholar]
- 30.Minnikin, D. E. et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods, 233–241 (1984).
- 31.Ezaki T, Hashimoto H, Yabuuchi E. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol. 1989;39:224–229. doi: 10.1099/00207713-39-3-224. [DOI] [Google Scholar]
- 32.Cui HL, Zhou PJ, Oren A, Liu SJ. Intraspecific polymorphism of 16S rRNA genes in two halophilic archaeal genera, Haloarcula and Halomicrobium. Extremophiles. 2009;13:31–37. doi: 10.1007/s00792-008-0194-2. [DOI] [PubMed] [Google Scholar]
- 33.Roh SW, et al. Arthrobacter soli sp. nov., a novel bacterium isolated from wastewater reservoir sediment. J Microbiol. 2008;46:40–44. doi: 10.1007/s12275-007-0239-8. [DOI] [PubMed] [Google Scholar]
- 34.Kim OS, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 38.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 39.Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20:406–416. doi: 10.2307/2412116. [DOI] [Google Scholar]
- 40.Kim JY, et al. Genome sequence of a commensal bacterium, Enterococcus faecalis CBA7120, isolated from a Korean fecal sample. Gut Pathog. 2016;8:62. doi: 10.1186/s13099-016-0145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 42.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 45.Ward N, Moreno-Hagelsieb G. Quickly finding orthologs as reciprocal best hits with BLAT, LAST, and UBLAST: how much do we miss? PLoS One. 2014;9:e101850. doi: 10.1371/journal.pone.0101850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon SH, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.002404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta RS, Naushad S, Fabros R, Adeolu M. A phylogenomic reappraisal of family-level divisions within the class Halobacteria: proposal to divide the order Halobacteriales into the families Halobacteriaceae, Haloarculaceae fam. nov., and Halococcaceae fam. nov., and the order Haloferacales into the families, Haloferacaceae and Halorubraceae fam nov. Antonie Van Leeuwenhoek. 2016;109:565–587. doi: 10.1007/s10482-016-0660-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


