Abstract
HIV-1 is one of the most studied retroviruses. The role of exosomes in HIV-1 entry and pathogenesis are beginning to be appreciated. Exosomes can incorporate host proteins that are also contained in viruses (e.g., tetraspanins).
Introduction
Human immunodeficiency virus-1 (HIV-1) is one of the most studied retroviruses; however, there is still a wealth of information to be learned about this virus’s entry, lifecycle and pathogenesis. The role of exosomes in HIV-1 entry and pathogenesis is beginning to be appreciated. Exosomes are small extracellular vehicles, which traffic nucleic acids (RNA, microRNA, and DNA), proteins, and lipids. Exosomes are secreted by most cell types into the extracellular milieu and then internalized into the recipient cells. It has been well documented that exosomal composition reflects the donor cells in which they are generated. Exosomes can incorporate host proteins that are also contained in viruses (e.g., tetraspanins) [1, 2]. Furthermore, exosomes appear to affect viral infection [3–9]. It has been previously demonstrated that blocking cluster of differentiation (CD) 9 and CD81 tetraspanins, which are commonly expressed on exosomes, resulted in significant decreases in the exosomal uptake efficiency in dendritic cells [10]. Regarding HIV-1, the role of exosomes in pathogenesis is complex. Studies have shown that exosomes may promote or inhibit HIV-1 infection [11, 12]. The overlap between HIV-1 and exosome biogenesis within an infected cell suggests that HIV-1 products, including RNA and proteins may be encased within exosomes or contaminate exosome preparations from HIV-1 infected fluids [13–15].
We are just beginning to unravel the complex nature of exosomes and HIV-1 interactions via lipids, phospholipids and proteins [16, 17]. Our group has recently shown that HIV-1 entry into human immune cells is enhanced by exosome-mediated trafficking [9]. This effect was illustrated with exosomes derived from human plasma, human breast milk, mouse neural stem cells and human lung carcinoma cells. Furthermore, our study demonstrated that HIV-1 and exosome interactions were mediated partially through binding of the T cell immunoglobulin and mucin protein 4 (TIM4) to the viral envelope [9]. In this study, we demonstrated that exosomes can enhance HIV-1 entry into human T and monocytic cell lines via exosomal tetraspanin proteins CD81 and CD9.
Materials and Methods
Cell culture
Human CD4+ lymphoblastoid T cell line (line A3R5.7) was a gift from the UAB CFAR Virology core. These cells were commercially received from the NIH AIDS Research and Reference Reagent Program and subsequently genetically modified. A3R5.7 cells were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated, exosome-free fetal bovine serum, 2 mM l-glutamine, penicillin (100 U/mL), streptomycin (100 μg/mL) (Thermo Fisher Scientific, Waltham, MA, USA), and 1 mg/mL geneticin (G418; Thermo Fisher Scientific). Human monocytic cells (line THP2574) were maintained in similar medium but without geneticin [18, 19]. All other cell lines were purchased from American Type Culture Collection.
Exosome purification
Isolation of human embryonic kidney cells (HEK 293)-derived exosomes
Cell line HEK293 was grown in DMEM-F12 complete medium containing exosome-free fetal bovine serum to ~80% confluency. In brief, cells were centrifuged at 5,000 rpm for 10 min at 4°C using a Sorvall RT600 centrifuge with a swinging bucket rotor (Thermo Fisher Scientific). The supernatant was clarified by filtration through a 0.22 μm filter and centrifuged at 13,200 rpm for 70 min at 4°C using an SW41T1 swinging rotor in a Beckman Coulter (Brea, CA, USA) Optima L-70K ultracentrifuge for exosome collection [8, 9, 20]. Exosomes were resuspended in 120–450 µL sterile phosphate-buffered saline (PBS) and then quantified by Bradford protein quantitation method.
Isolation of breast milk-derived exosomes
Breast milk samples were retrieved from remnants of breast milk samples from human donors and centrifuged twice at 3,500 rpm for 10 min at 4°C. The fat layer was aspirated and the supernatant transferred to a new tube. A third spin was performed at 5,000 rpm for 30 min at 4°C, after which the remaining fat was aspirated and the supernatant transferred to a new tube. Breast milk was then filtered with a 0.22 µm filter, transferred into an ultracentrifuge tube and then the tube volume was adjusted with PBS prior to an ultracentrifugation spin at 32,000 rpm for 70 min at 4°C. The pellet was collected and resuspended in 120–450 µL sterile PBS.
Isolation of human plasma-derived exosomes
Plasma was collected from whole blood of human donors into tubes containing ethylenediaminetetraacetic acid (EDTA) and processed as described by Konadu et al [21] with some modifications. Whole-blood samples were centrifuged at 3,500 rpm for 10 min at 4°C. If the samples contained a high lipid content after the low-speed centrifugation (evidenced by color), they were incubated for 2 h at 4°C, and the precipitated fat was removed by centrifugation at 5,000 rpm for 10 min at 4°C. The supernatants were then filtered through a 0.22 μm filter and ultracentrifuged for 30 min at 13,200 rpm in a SW41T1 swinging rotor at 4°C. The pellet was collected by centrifug-ing the samples at 27,000 rpm for 2 h at 4°C. The resulting pellet was resuspended in 1 mL PBS, loaded on to OptiPrep velocity gradients, and subjected to flotation centrifugation at 27,000 rpm for 2 h. Fractions with peak exosome content (fractions 2 and 3) were pooled, diluted with PBS, and ultra-centrifuged for 2 h at 32,000 rpm. The exosome pellet was resuspended in 120 µL PBS.
SDS-PAGE and Western blot analyses of exosome-associated proteins
To analyze proteins associated with exosomes, exosomes were mixed with loading buffer (1:1), boiled, and resolved on a 4%–12% Bis–Tris gel, followed by transfer and blocking on a polyvinylidene difluoride membrane. Blotting was per-formed with two different primary antibodies: anti-clathrin monoclonal antibodies (1:1,000; BD Biosciences, San Jose, CA, USA) and anti-CD81 (1:1,000; System Biosciences, Inc., Palo Alto, CA, USA). Incubation with secondary antibodies was performed using either horseradish peroxidase (HRP)-conjugated goat anti-mouse (1:2,000; Dako Denmark A/S, Glostrup, Denmark) or anti-rabbit (1:20,000; System Biosciences, Inc.) antibodies. Proteins were detected using an enhanced chemiluminescence kit (ELC Western Blotting Substrate Pierce/Thermo Fisher Scientific) and a Bio-Rad ChemiDoc XRS system (Bio-Rad Laboratories, Hercules, CA, USA).
HIV-1 infection of human cell lines
HIV-1 experiments were conducted using an infectious molecular clone (IMC) (NL-LucR.T2A-YU2.ecto [YU-2]) engi-neered to express Renilla luciferase (LucR) [22] and ENV from the YU-2 virus strain, which was derived from the postmor-tem brain of a patient with HIV-associated neurocognitive disorder (HAND)[23]. Throughout this manuscript, this IMC will be referred to as, virus.
A3R5.7 cells were seeded on 96-well plates at a density of 1×105 cells/well with the addition of 5 µg/mL diethylam-inoethyl-dextran [24]. Exosomes derived from NSC, A549, breast milk, and plasma were incubated, respectively, with virus at a multiplicity of infection (MOI) of 0.002 for 1 h at 37°C in 5% CO2, [23] and then HIV-1/exosome mixture was co-incubated with cells for 72 h at 37°C in 5% CO2. LucR activity was determined using the Renilla Luciferase Assay System (Promega Corporation, Fitchburg, WI, USA). Relative luminescence units (RLUs) were measured in triplicate on a Victor X light luminescence counter (PerkinElmer Inc, Waltham, MA, USA) with an exposure time of 0.1 s/well.
THP2574 cells (1×104 cells/well) were classically differentiated into macrophages with 1 ng/mL phorbol 12-myristate 13-acetate for 96 h. The virus with an MOI of 0.18 was incubated with exosomes for 1 h at 37°C in 5% CO2. The HIV-1/exosome mixture was co-incubated with the cells for 72 h at 37°C in 5% CO2, and LucR activity was measured in triplicate as described earlier.
Amounts of exosomes and viral MOIs used for each cell line were determined experimentally. The following quantities of exosomes, which would saturate all HIV-1 virions, were used for viral entry experiments: 0.035 μg breast milk-derived exosomes, 0.05 μg plasma cell-derived exosomes, and 0.1 μg HEK 293-derived exosomes.
Blocking of HIV-1 infection
A protocol similar to HIV-1 infection was performed but with addition of 0.2 µg/well anti-human CD9 or CD81 (1:1,000; BD Biosciences, San Jose, CA, USA) to the YU-2/exosome/cell incubation. Virus only was used as control.
Determining exosome size and concentration
We used nanoparticle tracking analysis (NTA) using the Nano-Sight LM10 (Malvern Instruments, Inc., Malvern, UK) and NTA v2.0 software to characterize breast milk- and plasma-derived exosomes. All data were collected using five frames and in triplicate. Samples were diluted 1:1,000 prior to tracking, which led to particle sizes of 10−7–10−9 m. Mean values were recorded and analyzed for each given reading frame.
Human study participants
The study was approved by the Institutional Review Board (IRB) for the Protection of Human Subjects in Research at the University of Alabama at Birmingham (UAB) in accordance with approved guidelines and protocol. Breast milk was collected from samples prior to being discarded at the UAB Regional Newborn Intensive Care Unit with expedited IRB approval. Blood donors provided written informed consent prior to donation.
Statistical analysis
Data represent 12 independent experiments. Significant differences between treatment groups were determined by one-way ANOVA with a post hoc Tukey’s test performed on obtained data points, and results are presented as means ± SEM, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Results
Isolation and characterization of exosomes
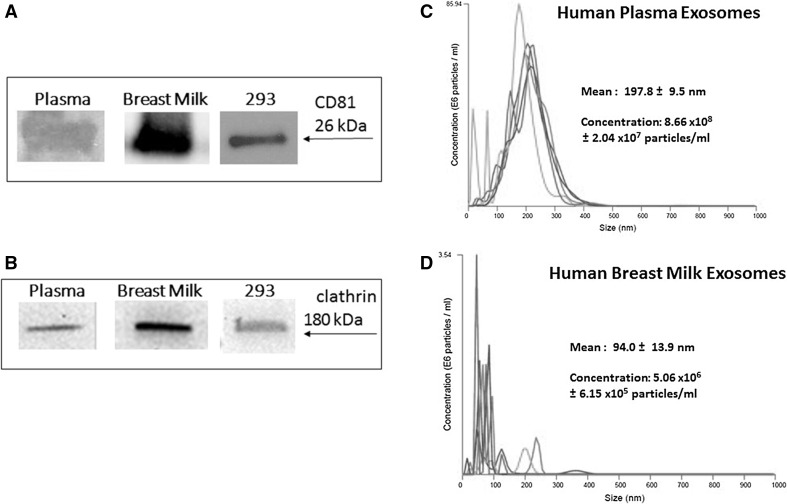
Exosomes were isolated and characterized for clathrin and exosomal markers, CD81, as described in our previous study [8]. For the current studies, we isolated exosomes from one cell line, HEK 293. We also isolated exosomes derived from human breast milk and human plasma using published methods [8, 9, 21]. To confirm successful exosome isolation, we performed a series of SDS-PAGE and western blot analyses to examine expression of well-known exosomal proteins CD81 (26 kDa) and clathrin (180 kDa) (Fig. 1A and 1B). We used nanoparticle tracking analysis (NTA) as previously performed [9] to validate our exosome preparation from human biological samples (Fig. 1C and D). Human plasma exosomes (Fig. 1C) have a mean diameter size of 197.8 ± 9.5 nm and concentration of 8.66 x108 ± 2.04 x107 particles/ml. Human breast milk-derived exosomes (Fig. 1D) have a mean size of 94.0 ± 13.9 nm.
Fig. 1.
Western blot and nanoparticle tracking analysis (NTA) validation of exosomal samples. Western blots of plasma exosomes, breast milk exosomes, and 293 exosomes. 60 μg/lane, were probed with (A) anti-CD81 or anti-clathrin (B). Arrows indicate proteins of interest. NTA-generated size and concentration plots for (C) human plasma and (D) human breast milk-derived exosomes. The different colors lines represent different pools of exosomes
Human-derived exosomes augment HIV-1 entry into cell lines and is subsequently blocked by CD81 antibodies
We previously showed that various exosomes augmented HIV-1 entry in human immune cell lines and this entry was partially blocked by TIM-4 blocking [9]. In this current study, we evaluated the impact of tetraspanins on exosome-mediated HIV-1 entry. We used IMC termed NL-LucR.T2A-YU2.ecto, which was also used in our recent work [9]. Throughout this manuscript, this IMC will be referred to as, virus. To determine the impact of exogenous exosomes (human plasma, breast milk, and HEK 293) on HIV-1 entry and subsequent gene expression, we performed a series of viral infection experiments using exosomes only, virus only, exosomes with virus, or exosomes with virus and anti-CD81 antibody. CD81 blocking experiments were based on previously published experimental dose titrations [9]. The above combinations of molecules were added to either the T lymphoblastoid cell line (A3R5.7) or the macrophage-like cell line (THP2574).
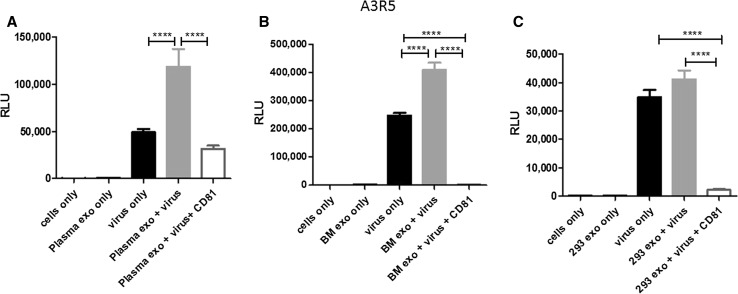
Virus alone infected A3R5.7 cells and induced viral gene expression, which was measured at approximately 49,000 relative luminescence units (RLU). Co-incubation of virus and plasma-derived exosomes enhanced infectivity significantly (p<0.0001) compared to virus only. The addition of anti-CD81 antibody significantly (p<0.0001) diminished viral gene expression, illustrating the specificity of CD81/exosome interactions (Fig. 2A). Similar results were observed when the CD81 antibody was used to block exosome-mediated virus entry of breast milk-derived exosomes (p<0.0001; Fig. 2B) and 293 derived-exosomes (p<0.0001; Fig. 2C).
Fig. 2.
Exosomes significantly enhance HIV-1 entry into a human T cell line. A3R5.7 cells were seeded at a density of 1x105 cells/well with the addition of 5 µg/ml diethylaminoethyl-dextran [9, 24]. Virus was used at a multiplicity of infection (MOI) of 0.002. Virus entry into A3R5.7 cells was evaluated in the presence or absence of (A) plasma-derived exosomes (0.05 μg) (B) breast milk-derived exosomes (0.035 μg) or (C) 293-derived exosomes (0.1 μg). Viral entry was also evaluated in the presence of exosomes and anti-CD81 antibody (0.2 µg/well). All incubations were processed for one hour. Viral gene expression in all control and treatment groups was assessed by Renilla luciferase activity at 72 h post-infection. This time point was selected because we previously observed that it allows for optimal expression in a variety of cell types, including those in the present experiments. All conditions are identical through the following experiments unless otherwise noted
In THP2574 macrophage-like cells virus alone entered the cells and yielded approximately 1.2x106 RLU (Fig. 3A). Co-incubation of virus and plasma-derived exosomes increased the number of RLU significantly p=0.0018. Administration of plasma-derived exosomes, virus, and anti-CD81 led to a significant decrease in RLU compared to the administration of plasma-derived exosomes (p<0.0001) plus virus (Fig. 3A) or virus only (p<0.0001) (Fig. 3A). Similar trends were observed with exosome-mediated virus entry using breast milk-derived exosomes (p=0.0034; Fig. 3B). Next, anti-CD81 antibody was co-incubated with exosomes and virus then subsequently added to cells. CD81 exosome-mediated blockage of virus entry and gene expression further illustrated the specificity of CD81/exosome interactions in multiple human cell types (Figures 2 and 3).
Fig. 3.
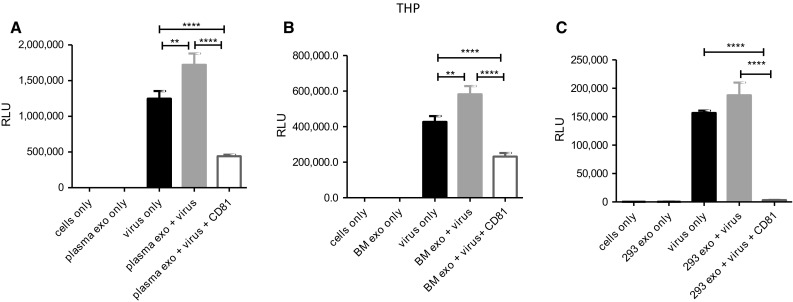
Exosomes significantly enhance HIV-1 entry into a human macrophage cell line. THP2574 cells (1x104 cells/well) were classically differentiated into macrophages with 1 ng/ml phorbol 12-myristate 13-acetate) for 96 h. Virus was used in THP2574 cells at an MOI of 0.18. Virus entry into THP2574 cells was evaluated in the presence or absence of (A) plasma-derived exosomes, (plasma exo) (B) breast milk-derived exosomes, (BM exo) or (C) 293-derived exosomes (293 exo). Viral entry was also evaluated in the presence of exosomes and anti-CD81 antibody
Breast milk-derived exosomes enhance HIV-1 entry and can be blocked using CD9 antibodies
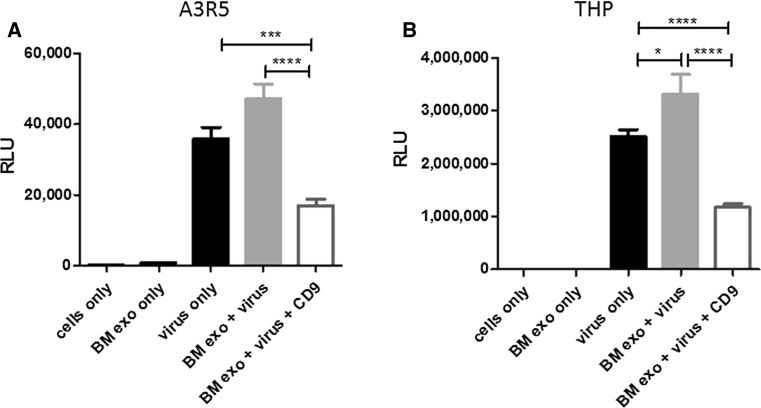
In order to evaluate the impact of CD9 on exosome-mediated HIV-1 entry, we performed several small-scale experiments identical to the virus and exosome blocking experiments shown in Figs. 2 and 3. For these studies we evaluated exosomes derived from breast milk. The addition of breast milk-derived exosomes with virus increased virus entry and subsequent gene expression in A3R5.7 cells (Fig. 4A); however, this exosome-mediated entry was significantly blocked following the addition of CD9 antibodies (p=0.0006) (Fig. 4A). When these experiments were performed in the recipient THP2574 macrophage like cell line, similar results were obtained (Fig. 4B). Breast milk-derived exosomes increased virus entry and gene expression and was subsequently blocked by the addition of CD9 antibody (p<0.0001).
Fig. 4.
Breast milk-derived exosomes significantly enhance HIV-1 entry into human immune cell lines. Virus entry into (A) A3R5.7 cells or (B) THP2574 cells was evaluated in the presence or absence of breast milk-derived exosomes (BM exo). Viral entry was also evaluated in the presence of exosomes and anti-CD9 antibody
Discussion
Several biological processes involve tetraspanins such as membrane fusion, cell motility, protein trafficking and cell adhesion, to name a few [25]. Specifically, both CD9 and CD81 are thought to bind with the cytoskeleton via ezrin-radixin-moeisin (ERM) family proteins binding to actin [26]. CD9 and CD81 also have binding partners that are critical for cell adhesion, ICAM-1 and VCAM-1 [27]. Tetraspanins, in particular CD9 and CD81, are found in extracellular vesicles such as exosomes. Thus, it is very plausible that these molecules may be involved in exosome-cellular interactions.
It has been well documented that HIV-1 exploits the exosomal machinery, as reviewed by Madison and Okeoma [28]. We now know the importance of exosomal TIM-4 on viral entry in neural stem cells [8]. We have published that HIV-1 binds to TIM-4 in exosomes and this interaction is associated with enhanced cellular entry [9]. The role of exosomes in HIV-1 pathogenesis is complex and many proteins may be involved, including CD9 and CD81. Based on the current report and our previous studies, it appears all three proteins (TIM-4, CD9 and CD81) are involved. CD9 and CD81 are critical for cell membrane interaction/fusion. CD81 associates with CD4 and is critical for HIV-1 infection [29]. These processes may still be dependent on the specific exosomes involved in exosome-cellular entry.
The role of exosomes in HIV-1 pathogenesis is complex. Näslund and group demonstrated that exosomes from breast milk inhibit HIV-1 infection of dendritic cells and subsequent viral transfer to CD4+ T cells [30]. These findings are different from our findings herein, this may be attributed to the recipient cell types examined in our experiments, T lymphoblastoid cell line or the macrophage-like cell line. In this study, we evaluated exosomes derived from HEK 293 cells to gain some preliminary knowledge related to kidney cell-derived exosomes and their role in mediating HIV-1 entry. To our knowledge, this is the first study in this context. These finding might add to the understanding and further clinical advances of HIV-1 associated nephropathy (HIVAN) which is poorly understood [31].
A role for CD81 has been identified in HIV-1 budding, cell-to-cell spread and infectivity [1, 28, 29, 32–34]. It has previously been demonstrated that blocking CD9 and CD81 tetraspanins, which are commonly expressed on exosomes, resulted in significant decreases to exosomal uptake efficiency in dendritic cells [10]. These results are similar to the results we report here. Our current study was designed to evaluate exosome-mediated entry and not virus-antibody interaction. Based on our studies with TIM-4 in exosomes, HIV-1 binds to the exosome and proteins like CD9 and CD81 may then facilitate cellular entry. Research by Luo et al., (2015) revealed the existence of two different forms of exosomes: i) AChE+/CD81 low/TSG101 low exosomes and ii) AChE-/CD81 high/TSG101 high exosomes [35]. We demonstrate that various human-derived exosomes increase the entry of virus and subsequently that this can be blocked with the addition of CD81 and CD9 antibodies, respectively. Manipulating CD81 and CD9 binding to cells may be critical for exosomal entry. Therefore, blocking exosome-mediated uptake of HIV-1 may act as a therapeutic axis to reduce HIV-1 infections, particularly in latent HIV infected targets in immune privilege sites such as the brain.
Abbreviations
- HEK 293
Human embryonic kidney cells
- TIM4
T cell immunoglobulin and mucin protein 4
- NTA
Nanoparticle tracking analysis
- HIV-1
Human Immunodeficiency virus-1
- CD
cluster of differentiation
- IMC
infectious molecular clone
- LucR
Renilla luciferase
- HAND
HIV-associated neurocognitive disorder
Author contributions
BS, AB, and QLM contributed to the design of the experiments and the writing of the manuscript. ALF, SDW and AK contributed the to the design and execution of the experiment.
Funding
This work was supported by the American Association of Immunologists Careers in Immunology Fellowship Program, Gorgas Memorial Foundation Research Grant Award. National Institutes of Health grant #5R01AI089337-04 and UAB-CCTS grant UL1TR001417, UAB CFAR, an NIH-funded program (P30 AI027767) made possible by the following institutes: NIAID, NCI, NICHD, NHLBI, NIDA, NIMH, NIA, NIDDK, NIGMS, FIC, and OAR. We are thankful to the scientific support from the UAB CFAR Virology Core, which is part of the UAB CFAR P30-AI-27767. A3R5.7 and THP2574 cell lines were a generous gift from the UAB CFAR Virology Core. We are thankful to the High-Resolution Imaging Facility Service Center who provided NTA. The High-Resolution Imaging Facility is an institutional core at UAB supported by the Office of the Vice President of Research and development and the following grants: Cancer Center Support Grant P30 CA013148 Rheumatic Disease Core Center P30 AR048311. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Compliance with ethical standards
Ethics approval and consent to participate
The study was approved by the Institutional Review Board for the Protection of Human Subjects in Research at the University of Alabama at Birmingham (UAB) in accordance with approved guidelines and protocol. Breast milk was collected from samples prior to being discarded at the UAB Regional Newborn Intensive Care Unit with expedited IRB approval. Blood donors provided written informed consent prior to donation.
Conflict interest
The authors have no conflicts of interest to disclose.
Availability of data and material
Please contact author for data requests.
Footnotes
Brian Sims and Anitra L. Farrow contributed equally to this work.
References
- 1.Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol. 2007;81(15):7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreux M, et al. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe. 2012;12(4):558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gentili M, et al. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 2015;349(6253):1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 4.da Silva LL, et al. Human immunodeficiency virus type 1 Nef protein targets CD4 to the multivesicular body pathway. J Virol. 2009;83(13):6578–6590. doi: 10.1128/JVI.00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridgeman A, et al. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 2015;349(6253):1228–1232. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baglio SR, et al. Sensing of latent EBV infection through exosomal transfer of 5’pppRNA. Proc Natl Acad Sci USA. 2016;113(5):E587–E596. doi: 10.1073/pnas.1518130113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vojtech L, et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42(11):7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sims B, et al. Neural stem cell-derived exosomes mediate viral entry. Int J Nanomed. 2014;9:4893–4897. doi: 10.2147/IJN.S70999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sims B, et al. Role of TIM-4 in exosome-dependent entry of HIV-1 into human immune cells. Int J Nanomed. 2017;12:4823–4833. doi: 10.2147/IJN.S132762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kooijmans SA, et al. Exosome mimetics: a novel class of drug delivery systems. Int J Nanomed. 2012;7:1525–1541. doi: 10.2147/IJN.S29661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mack M, et al. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med. 2000;6(7):769–775. doi: 10.1038/77498. [DOI] [PubMed] [Google Scholar]
- 12.Khatua AK, et al. Exosomes packaging APOBEC3G confer human immunodeficiency virus resistance to recipient cells. J Virol. 2009;83(2):512–521. doi: 10.1128/JVI.01658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madison MN, Jones PH, Okeoma CM. Exosomes in human semen restrict HIV-1 transmission by vaginal cells and block intravaginal replication of LP-BM5 murine AIDS virus complex. Virology. 2015;482:189–201. doi: 10.1016/j.virol.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arakelyan A, et al. Extracellular vesicles carry HIV ENV and facilitate HIV infection of human lymphoid tissue. Sci Rep. 2017;7(1):1695. doi: 10.1038/s41598-017-01739-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raymond AD, et al. HIV Type 1 Nef is released from infected cells in CD45(+) microvesicles and is present in the plasma of HIV-infected individuals. AIDS Res Hum Retroviruses. 2011;27(2):167–178. doi: 10.1089/aid.2009.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skotland T, Sandvig K, Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Nakai W, et al. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci Rep. 2016;6:33935. doi: 10.1038/srep33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLinden RJ, et al. Detection of HIV-1 neutralizing antibodies in a human CD4(+)/CXCR4(+)/CCR5(+) T-lymphoblastoid cell assay system. PLoS One. 2013;8(11):e77756. doi: 10.1371/journal.pone.0077756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun J, et al. Protein phosphatase, Mg2+/Mn2+-dependent 1A controls the innate antiviral and antibacterial response of macrophages during HIV-1 and Mycobacterium tuberculosis infection. Oncotarget. 2016;7(13):15394–15409. doi: 10.18632/oncotarget.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald MK, Capasso KE, Ajit SK. Purification and microRNA profiling of exosomes derived from blood and culture media. J Vis Exp. 2013;76:e50294. doi: 10.3791/50294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konadu KA, et al. Association of cytokines with exosomes in the plasma of HIV-1-seropositive individuals. J Infect Dis. 2015;211(11):1712–1716. doi: 10.1093/infdis/jiu676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edmonds TG, et al. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology. 2010;408(1):1–13. doi: 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, et al. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol. 1991;65(8):3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platt EJ, et al. Rapid dissociation of HIV-1 from cultured cells severely limits infectivity assays, causes the inactivation ascribed to entry inhibitors, and masks the inherently high level of infectivity of virions. J Virol. 2010;84(6):3106–3110. doi: 10.1128/JVI.01958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreu Z, Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sala-Valdes M, et al. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J Biol Chem. 2006;281(28):19665–19675. doi: 10.1074/jbc.M602116200. [DOI] [PubMed] [Google Scholar]
- 27.Barreiro O, et al. Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood. 2005;105(7):2852–2861. doi: 10.1182/blood-2004-09-3606. [DOI] [PubMed] [Google Scholar]
- 28.Madison MN, Okeoma CM. Exosomes: implications in HIV-1 pathogenesis. Viruses. 2015;7(7):4093–4118. doi: 10.3390/v7072810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon-Alonso M, et al. Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J Immunol. 2006;177(8):5129–5137. doi: 10.4049/jimmunol.177.8.5129. [DOI] [PubMed] [Google Scholar]
- 30.Naslund TI, et al. Exosomes from breast milk inhibit HIV-1 infection of dendritic cells and subsequent viral transfer to CD4+ T cells. AIDS. 2014;28(2):171–180. doi: 10.1097/QAD.0000000000000159. [DOI] [PubMed] [Google Scholar]
- 31.Khatua AK, et al. Non-productive HIV-1 infection of human glomerular and urinary podocytes. Virology. 2010;408(1):119–127. doi: 10.1016/j.virol.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grigorov B, et al. A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology. 2009;6:28. doi: 10.1186/1742-4690-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izquierdo-Useros N, et al. Capture and transfer of HIV-1 particles by mature dendritic cells converges with the exosome-dissemination pathway. Blood. 2009;113(12):2732–2741. doi: 10.1182/blood-2008-05-158642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato K, et al. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J Virol. 2008;82(2):1021–1033. doi: 10.1128/JVI.01044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo X, et al. Exosomes are unlikely involved in intercellular Nef transfer. PLoS One. 2015;10(4):e0124436. doi: 10.1371/journal.pone.0124436. [DOI] [PMC free article] [PubMed] [Google Scholar]