Highlights
-
•
Filoviruses can cause severe hemorrhagic fever in humans and non-human primates.
-
•
There is an urgent need for rapid diagnosis of filoviruses during outbreaks.
-
•
Filovirus diagnostics have advanced since the 2014–2016 Ebolavirus outbreak.
-
•
NATs are the gold standard for filovirus detection.
-
•
NAT-based diagnostic speed, portability and multiplexing have all improved.
Keywords: Filoviridae, Ebola, Marburg, Diagnostics, Point-of-care, NAT
Abstract
Nucleic acid testing (NAT) for pathogenic filoviruses plays a key role in surveillance and to control the spread of infection. As they share clinical features with other pathogens, the initial spread of these viruses can be misdiagnosed. Tests that can identify a pathogen in the initial stages of infection are essential to control outbreaks. Since the Ebola virus disease (EVD) outbreak in 2014–2016 several tests have been developed that are faster than previous tests and more suited for field use. Furthermore, the ability to test for a range of pathogens simultaneously has been expanded to improve clinical pathway management of febrile syndromes. This review provides an overview of these novel diagnostic tests.
1. Introduction
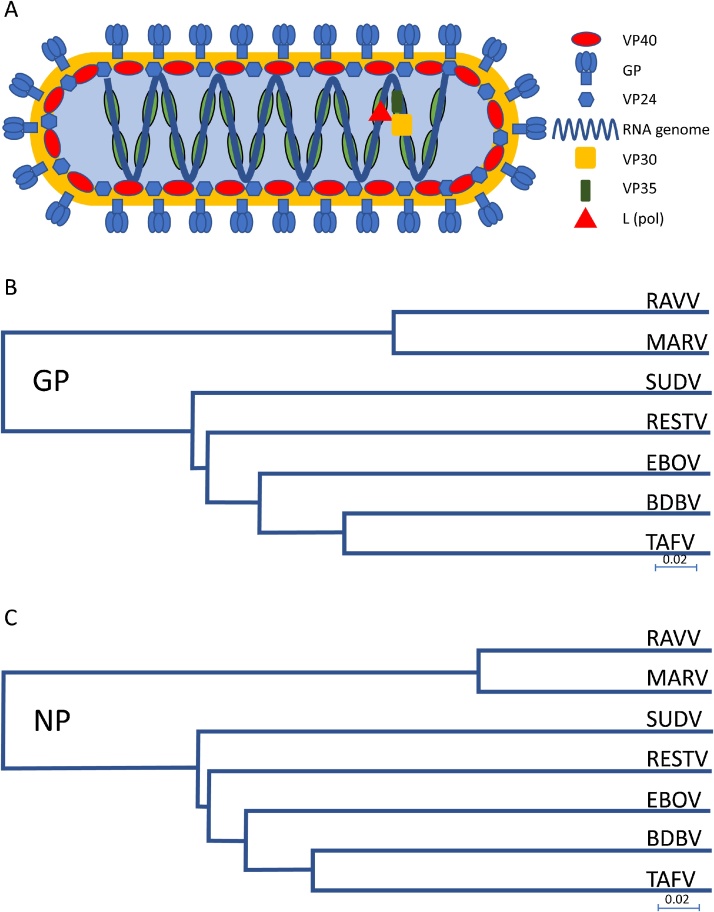
The Ebolaviruses are a group of closely related viruses in the filoviridae family. Filoviruses have negative-sense RNA genomes protected by the nucleocapsid protein (Fig. 1A). There are five distinct Ebolavirus species; Bundibugyo (BDBV), Reston (RESTV), Sudan (SUDV), Taï forest (TAFV, formerly Cote d’Ivoire Ebolavirus) and Zaire (EBOV). All five members can cause infections in humans albeit with a wide spectrum of disease severity. The Zaire and Sudan species cause Ebolavirus Disease (EVD) with a fatality rate ranging from 40 to 90% [1]. These viruses, along with other more distantly related filoviruses, Marburg virus (MARV) and Ravn virus (RAVV), are biosafety level 4 agents associated with high fatality rates and an absence of effective treatments [2].
Fig. 1.
Structure and filovirus divergence. A. Schematic of a filovirus particle. Nucleotide sequence phylogenetic trees, prepared with NCBI genomics workbench using nearest neighbour joining, showing the main targets GP (B) and NP (C) for filovirus NATs. The divergence in sequence requires specific primers for each species/subspecies. Scale bar denotes nucleotide substitutions per site.
The first documented Ebolavirus, and prototypic virus for the group, EBOV, was isolated in an outbreak of a haemorrhagic fever in Africa within the democratic Republic of Congo (formerly Zaire) in 1976 [3], nine years after MARV had been discovered in a laboratory-centred outbreak in Marburg, Germany. Both TAFV and RESTV are somewhat unusual within the Ebolavirus group as firstly; they do not appear to cause severe pathology in humans, (in non-human primates, infections are highly pathogenic) and secondly the sources were outside the central African continental region associated with filovirus outbreaks.
Filoviruses are considered to be zoonotic; there is evidence that bats are likely to be an animal reservoir for a number of viruses. Bats can harbour Ebolaviruses and Marburg virus, which replicate without causing symptoms of EVD [4]; filovirus RNA has been isolated from 3 species of bat [5,6] and more species have been shown to have antibodies against Ebolavirus [[7], [8], [9], [10], [11]]. Evidence suggests that contact between humans and fruit bats are the cause of at least one outbreak [12]. Furthermore, experiments have shown that pigs infected with EBOV can transmit virus to non-human primates kept in the same room but with no physical contact [13].
In March 2014 an outbreak of the EBOV began in western Africa. This was the largest outbreak recorded and spanned several countries in the region. Initially there were relatively few cases but they rapidly increased as transmission started to occur in densely populated areas. In 2014 the World Health Organisation (WHO) declared the epidemic as a Public Health Emergency of International Concern (PHEIC) [14]. The PHEIC was declared over, in March 2016 after the three countries, that were the main focus of the international response, completed 42 days with no newly reported cases and an additional 90 days of enhanced surveillance [15]. During the 2-year outbreak, there were more than 28,600 suspected, possible or confirmed cases of which 11,310 were fatal (∼40% case fatality rate) [16]. This epidemic highlighted the need for rapid diagnostics and epidemiology for disease tracking and containment. The unprecedented scale of the EVD outbreak spurred research into the filovirus field with the swift deployment of experimental vaccines, for phase I/II [[17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27]] and III trials [28], and development of diagnostics suited for low resource and in-field environments.
Future development of diagnostics focusing on rapid, sensitive and specific assays will be especially helpful in triage, as the symptoms of early EVD overlap with several other infections found in equatorial regions (e.g. malaria). Early isolation of EBOV infected individuals decreases the chance of perpetuating infections by breaking transmission chains. In this review, current and proposed methods and techniques for Ebolavirus diagnosis using nucleic acids (see Table 1 for advantages and disadvantages) will be described. Other technologies are not covered (e.g. ELISA, cell culture, EM).
Table 1.
Advantages and disadvantages of NATs.
| Advantages | Disadvantages |
|---|---|
| The ability to diagnose an infection prior to the adaptive immune response | Small window of diagnosis (viraemia first detectable between 3 and 10 days of initial infection) compared to antigen and antibody tests |
| A high specificity and multiplex compatibility | RNA is susceptible to rapid degradation by RNases |
| Greater sensitivity; most NATs amplify an initial sample | Cannot identify past infections, unlike antibody tests |
| Reduced operator handling (giving enhanced safety) | PCR product contamination risk due to amplification of initial sample |
| No requirement for category 4 level cell culture | Pathogen genetic drift could result in decreased sensitivity (If using DNA primers/capture probes) |
| Speed (with the exception of antigen RDTs) | |
| Definitive of virus presence; in the absence of a viral genome, there is no amplification |
2. Sample collection and storage
For most of the diagnostic methods used to determine the presence of virus (or evidence of contact with Ebolavirus and closely related filoviruses) a blood sample is required. WHO recommendations for venepuncture, in cases of suspected EBOV or MARV, state that blood should be collected into EDTA tubes with a minimum volume of 5 ml. For blood collection guidelines please refer to: http://www.who.int/csr/resources/publications/ebola/blood-collect/en/. The WHO guidelines further state that blood samples can be stored for up to 24 h at room temperature, or at 0–5° C for up to a week. For periods of longer than a week, the sample should be stored at −20 or −70° C (avoiding freeze thaw cycles) [29].
Oral swab collection has been tested in a guinea pig model of EVD [30]. The authors considered the test to be poor for samples collected ante mortem but excellent for post mortem specimens. Oral swab collection during the 2014–2016 west Africa outbreak has also been examined for post mortem surveillance [31]. The same study tested finger-stick sampling with pipette or swab collection, which in field point-of-care applications may be more feasible than venepuncture.
3. Sample preparation
As with all PCR-based assays, the purity of the input template is important for standardising tests. Due to inhibitors found in blood (reviewed in [32]) most diagnostic tests require the genome of a filovirus to be isolated from a sample; for example, to obtain a viraemia, RNA needs to be isolated from the blood plasma. One frequently used method utilises acid guanidinium thiocyanate-phenol-chloroform to separate a sample into an aqueous (containing RNA) and organic phase (containing DNA). This also has the important advantage of inactivating infectious material as proteins are denatured, although for complete inactivation of Ebolavirus, a secondary ethanol step appears to be necessary [33]. However, this method can be impracticable as it is both time consuming for an operator and uses harmful chemicals.
Commercial RNA extraction kits, many of which are based on chaotropic chemical (e.g. guanidine-isothiocyanate) extraction, use centrifugation (or vacuum) of columns containing glass fiber filters to isolate RNA, allowing batch processing. Other kits use beads, with high nucleic acid adherence, followed by magnetic separation. Most kits can concentrate nucleic acids which has the potential of improving the sensitivity of downstream assays.
Due to the sensitivity of RNA to RNases, care must be taken to minimize degradation of extracted samples prior to reverse transcription. However, there is now a move towards using all-in-one RNA isolation and reaction devices, as this minimizes the exposure of sample to RNases and of staff to potentially infectious material and reduces operator dependent variation allowing for more standardized tests.
4. Diagnostics
All members of the filoviridae have negative-sense RNA genomes and therefore require a reverse transcription step before polymerase chain reaction (PCR) can be performed. There are three main reverse transcriptase PCR techniques, as described below.
4.1. RT-PCR
There are two main methods of reverse transcriptase PCR; one step and two step. In one step RT-PCR, the sample and all required reagents are within one reaction chamber/tube and an initial reverse transcription step is directly followed by PCR cycling. cDNA libraries of total RNA in a sample can be prepared using random primers, or specific pathogen primers can be used; thereby only generating cDNA of a particular pathogen.
In two step RT-PCR, the reverse transcription and PCR are performed separately; the first to generate cDNA and the second for PCR, following the transfer of template cDNA to another chamber. With a cDNA library of total RNA, two step RT-PCR the resulting template can be used in multiple separate assays but is time consuming and sensitivity can be reduced due to splitting of the original sample.
In both one step and two step RT-PCR, the final products are assessed by agarose gel and is therefore more suited to qualitative diagnosis. The main advantage of this technique, compared to RT-qPCR, is the cost; equipment to perform PCR and electrophoresis are relatively inexpensive. However, it is not possible to multiplex within an assay; while there is some RNA sequence conservation within a species of filovirus there is far less between species (Fig. 1B and C). Published RT-PCR assays are presented in Table 2.
Table 2.
RT-PCR papers; targets, primers and sensitivity for filovirus detection.
| Paper | Target gene/sequence | Primers | Sensitivity* |
|---|---|---|---|
| Leroy [69] | Polymerase, L | Zaire F 5′- ATCGGAATTTTCTTTCTCATTGAAAGA-3′ | 100% (30/30) (95% CI 88.4–100)** |
| Zaire R 5′- ATGTGGTGGATTATAATAATCACTGACATGCAT-3′ | |||
| Towner [70] | Nucleoprotein, NP | Primary | N/A |
| SudZaiNP1(F), 5′-GAGACAACGGAAGCTAATGC-3′, | |||
| SudZaiNP1(R), 5′-AACGGAAG ATCACCATCATG-3′, | |||
| Nested | 104–105/mL (10–100/reaction) | ||
| SudZaiNP2(F), 5′-GGTCAGT TTCTATCCTTTGC-3′, | |||
| SudZaiNP2(R), 5′-CATGTGTCCAACTGATTG CC-3′ | |||
| Park [71] | NP | BDBV F GCAGAAATATGCTGAATCTCGTGAAC | 5 fg/μL |
| BDBV R ATCATCCTCGTCCTCAAGGTCAAAA | |||
| RESTV F CCAACAATATGCTGAGTCCAGAGAA | 5 fg/μL | ||
| RESTV R | |||
| CATCCTCATGATCGTCAAGATCG | |||
| SUDV F ACACGTGAGTTGGACAACCTT | 5 fg/μL | ||
| SUDV R GTCATCGTCGTCGTCCAAATTGAA | |||
| TEBOV F AATCTCGCGAGCTTGACCAT | 5 fg/μL | ||
| TEBOV R CTCGTCACCATCTTCAAGGTCAAA | |||
| EBOV F CGAACTTGACCATCTTGGACTTG | 5 fg/μL | ||
| EBOV R TCCTCGTCGTCCTCGTCTAGAT | |||
| MARV F AGGCGACATGAACATCAGGAAATT | 5 fg/μL | ||
| MARV R TCGTCCTCATTCAGCAGTGCAAAT | |||
| RAVV F GCGACATGAACACCAGGAAATTC | 500 fg/μL | ||
| RAVV R ATTTTCAAGAGTATCCTCGTCTTCG | |||
| Ogawa 2011[72] | NP | MARV FiloNP-Fm TGGCTTACYACAGGYCACATGAAAGT | 10−3 |
| MARV FiloNP-Rm GTGTGTGATTTCAGTTTTYTGGAGGTGGAA | FFU/reaction*** | ||
| L(Sanchez 1999) | EBOV FiloNP-Fe TGGCAATCAGTDGGACACATGATGGT | ||
| EBOV FiloNP-Re TGGCAATCAGTDGGACACATGATGGT | |||
| MARV FILO-A ATCGGAATTTTTCTTTCTCATT | |||
| MARV FILO-B ATGTGGTGGGTTATAATAATCACTGACATG | |||
| Bergqvist 2015[73] (NB Multiplex) | L | EV F1 Biotin CGTTTIAAIACCMIWCTSATTGC | |
| EV F2 Biotin CGATTCAACACAACTCTAATCTC | |||
| EV F3 Biotin CGATTTAATACTTTACTGATTGC | |||
| EV F4 Biotin AGGTTIAATACATCACTGATTGC | |||
| EV R Phosphor GGRTGSCCCCARTGYTTTTGVA | |||
| EBOV P C12-NH2 GCATAGACACAATCTTAAAATTG | 1500 copies/mL | ||
| SUDV P C12-NH2 GAGATTGAATATCATCTACCAGT | 150 copies/mL | ||
| TAFV P C12-NH2 GGTCAGACACTGTTTCTGGTA | 150k copies/mL | ||
| GP | MV F Biotin ACACYYYCAARHRCAACYTCAGYAC | ||
| MV R Phosphor TCAAAATCAATYKSAGYAYTTATTAACCCRTC | |||
| RAVV P C12-NH2 GCTAGTTCACGTTGTGTATCATT | |||
| MARV OZOLIN P C12—NH2 CCAACACACAAAGCATGGCCACTG | |||
| MARV MUSOKE P GATTGTGCTCTGTGTGTTGTC | |||
| MARV LEIDEN/POPP P GTGGCTGTGCTCTGTGTGTCGTA |
where available.
where antigen detection used as standard (from 26 symptomatic patients, 3 convalescent and 1 healthy).
FFU: Focus forming unit.
4.2. RT-qPCR
This technique is very similar to RT-PCR but enables the sample to be quantified against a known set of standards. It uses forward and reverse primers and internal oligo probes with fluorophores and quenchers. qPCR machines measure fluorescent signal from probe break down, which occurs relative to amplification, and can detect multiple fluorophores, allowing multiplexing within a single sample (e.g. differentiating between Ebolavirus and Marburgvirus) [34]. Photon multiplier tubes amplify fluorescence signals, potentially increasing sensitivity over RT-PCR. Furthermore, a large number of samples can also be run at the same time as the standards; depending on the format of the device, 96/384 wells. Examples of these machines include: CFX96 (Biorad), Lightcycler (Roche), ABI 7000 series (Applied Biosystems) and Rotor-Gene Q (Qiagen).
Published RT-qPCR assays for filovirus detection are presented in Table 3. The benchmark that many of the newer tests are compared against is the Trombley assay [35]. The targets described in this paper are the nucleoprotein, glycoprotein and VP40 (matrix protein). Minor groove binding (MGB) probes and standard Taqman™ probes were assessed in this paper for a number of viral pathogens; MGB probes are shorter than normal yet maintain specificity and increased primer melting temperature (relative to similar length Taqman™ probes) by adding a 3′ minor groove binding moiety that stabilises the probe-target hybrid. The Trombley assay includes primers for a human gene (ribonuclease P) as an endogenous control. Due to variation between the five subtypes of Ebolavirus, separate primers and probes are required for each. While this assay has high sensitivity for Zaire Ebolavirus (a lower limit of detection (LLOD) of 0.0001 plaque forming unit per reaction) it is expensive, time consuming and suited predominantly to well-equipped diagnostics laboratories. During the 2014–2016 Ebolavirus a number of RT-qPCR assays were authorised for emergency use (FDA EUA) [36], presented in Table 4. Two platforms used during the epidemic (Biocartis Idylla and Cepheid GeneXpert) have sample-to-result cartridges, whereas the others require sample preparation before the RT-qPCR can be performed. This is of particular interest as using a sample-to-result cartridge can reduce operator involvement, improving safety, exposure to RNases and ease-of-use. The GeneXpert system was used in Liberia during the 2014–2016 outbreak in a mobile laboratory run by Liberian laboratory technicians who had been trained and supported by the Liberian Ministry of Health, WHO and other international partners. Results from the laboratory were used for both clinical management and for determining discharge status of patients [37].
Table 3.
RT-qPCR papers, target primers/probe and sensitivity for filovirus detection.
| Paper | Target gene/sequence | Primers | Sensitivity |
|---|---|---|---|
| Towner (SEBOV)[70] | NP | Reverse transcription (and Forward primer) | One step: 103/ml (1 copy/reaction), Two step: 105/ml (100 copies/reaction) |
| F 5′-GA AAGAGCGGCTGGCCAAA-3′. | |||
| R AACGATCTCCAACCTTGATCTTT | |||
| P GACCGAAGCCATCACGACTGCAT | |||
| Trombley [35] | EBOV MGB, NP | F565 5′-TCTGACATGGATTACCACAAGATC − 3′ | 0.001 PFU/reaction |
| EBOV, GP | R640 5′-GGATGACTCTTTGCCGAACAATC − 3′ p597S 6FAM-AGGTCTGTCCGTTCAA-MGBNFQ | 0.01 (584 copies) | |
| F2000 5′ −TTTTCAATC CTCAAC CGTAAG GC − 3′ | |||
| SUDV MGB, NP | R2079 5′ − CAGTCC GGT CCCAGAATGTG − 3′ | 0.1 PFU/reaction | |
| p2058A 6FAM-CATGTGCCGCCCCATCGCTGC-TAMRA-3′ | |||
| SUDV, GP | F CAT GCA GAA CAA GGG CTC ATT C | 0.1 PFU/reaction | |
| R CTC ATC AAA CGG AAG ATC ACC ATC | |||
| P CAA CTT CCT GGC AAT | |||
| RESTV MGB, GP | F AGG ATG GAG CTT TCT TCC TCT ATG | 1.0 (34 copies) | |
| R TAC CCC CTC AGC AAA ATT GAC T | |||
| RESTV, VP40 | P CAG GCT GGC TTC AAC TGT AAT TTA CAG AGG | 1 PFU/reaction | |
| F TCA CCG CGA ACC CAA TG | |||
| R TCG CTT GTC ATG GTT GGA CTT | |||
| TEBOV MGB, GP | P ACC ATT GCC C | 1.0 (586 copies) | |
| F CTA TGG TTA TCA CCC AGG ATT GTG | |||
| R GTA ACT ATC CTG CTT GTC CAT GTG | |||
| TEBOV, GP | P TGC CAC TCT CCA GCC AGC CAT CCG | 0.1 PFU/reaction | |
| F CCC ATC TCC GCC CAC AA | |||
| R GAG TGG AAT CCT CTG AAA CCA ATT | |||
| BDBV, MGB | P CGC AGG CGA AGA C | 10−6 (RNA dilution) | |
| F TGT ACA CAA AGT CTC AGG AAC TGG | |||
| R GTC ATA CAG GAA GAA GGC TCC TTC | |||
| panMARV MGB, GP | P CCA TGC CCA GGA GGA CTC GCC TTT | 0.1 (Ravn), 1.0 (Ci67), 10 (Musoke), 1.0 (Angola) PFU/reaction | |
| panMARV, GP | F ATG GAA ACC AAG GCG AAA CTG | 0.1 (Ravn), 10 (Ci67), 1.0 Musoke), 10 (Angola) PFU/reaction | |
| R TAC TTG TGG CAT TGG CTT GTC T | |||
| P CGG GTA GCC CCC AAC | |||
| F GAT TCC CCT TTG GAA GCA TCT | |||
| F2 GAT TCC CCT TTA GAG GCA TCC | |||
| R CAA CGT TCT TGG GAG GAA CAC | |||
| P ACG ATG GGC TTT CAG | |||
| F GAT TCC CCT TTG GAA GCA TCT | |||
| F2 GAT TCC CCT TTA GAG GCA TCC | |||
| R CAA CGT TCT TGG GAG GAA CAC | |||
| P AAA CGA TGG GCC TTC AGG GCAGG | |||
| P2 AAG CGA TGG GCT TTC AGG ACAGG | |||
| Drosten /Sanchez [74,75] | L (MARV and EBOV) | Filo A ATCGGAATTTTTCTTTCTCATT | 5.3 copies/reaction (2647 copies/ml (1887 to 4964)) |
| Filo B ATGTGGTGGGTTATAATAATCACTGACATG | |||
| Gibb [76] | GP | F TGGGCTGAAAAYTGCTACAATC | LOD 8 PFU (10fg*) |
| R CTTTGTGMACATASCGGCAC | |||
| EBOV P CTACCAGCAGCGCCAGACGG | |||
| SUDV P TTACCCCCACCGCCGGATG | 3 PFU (100fg*) | ||
| Weidmann [77] | EBOV F ATGATGGAAGCTACGGCG | LOD: 10 copies/reaction Comparable to MARV and EBOV (∼10 copies/reaction) | |
| EBOV P CCAGAGTTACTCGGAAAACGGCATG | |||
| EBOV R AGGACCAAGTCATCTGGTGC | |||
| SUDV F TTGACCCGTATGATGATGAGAGTA | |||
| SUDV P CCTGACTACGAGGATTCGGCTGAAGG | |||
| SUDV R CAAATTGAAGAGATCAAGATCTCCT | |||
| MARV F CAATTCCACCTTCAGAAAACTG | LOD: 10 copies/reaction | ||
| MARV P CACACACAGTCAGACACTAGCCGTCCT | |||
| MARV R GCTAATTTTTCTCGTTTCTGGCT |
MGB: Minor groove binding. NP: Nucleoprotein. GP: Glycoprotein. L: Polymerase.
Purified RNA.
Table 4.
FDA EUA: New technologies. Targets and sensitivity (where published) for EBOV.
| Test | Target | Sensitivity | Further notes |
|---|---|---|---|
| Idylla™ Ebola Virus Triage Test[78] | GP, Human RNase P mRNA | 465 pfu/mL (1010 copies/mL) | Cartridge, Idylla™ Instrument |
| Xpert® Ebola Assay[31,[79], [80], [81]] | GP and NP (as well as a sample processing control and human DNA sample adequacy control*) | sensitivity 100%, 95% CI 84.6%–100% vs Trombley | Cartridge, GeneXpert platform |
| LightMix® Ebola Zaire rRT-PCR Test[82] | L gene (polymerase) and human housekeeping mRNA | 4781 PFU/mL | LightCycler® 480 II or cobas z 480 Analyzer |
| RealStar® Ebolavirus RT-PCR Kit 1.0[34] | L gene and heterologous target sequence | 11–67 copies/reaction | Platform dependent sensitivity |
| (CDC) Ebola Virus VP40 Real-time RT-PCR Assay[51] | VP40 | 20–60 TCID50/mL (600 TCID50/mL with whole blood) | ABI 7500 Fast Dx Real-Time PCR Instrument, BioRad CFX96 |
| (CDC) Ebola Virus NP Real-time RT-PCR Assay[51] | NP | 600–6000 TCID50/mL (6 × 103 TCID50/mL with whole blood) | ABI 7500 Fast Dx Real-Time PCR Instrument, BioRad CFX96 |
| (DoD) EZ1 Real-time RT-PCR Assay[83] | GP, Human RNase P | 5000 PFU/mL (7500 PFU/mL with whole blood) | ABI ® 7500 Fast Dx, LightCycler |
| Liferiver™ Ebola Virus (EBOV) Real Time RT-PCR Kit[84] | Not stated | 23.9/reaction 95% CI (13.4–405.9RNA/reaction) | Roche light cycler 480 |
To ensure sufficient host sample has been added.
4.3. RT-LAMP
In contrast to standard RT-PCR, reverse transcription-loop mediated isothermal amplification (RT-LAMP) is conducted at one temperature and therefore does not require high precision thermocyclers; this technique is suited to low resource settings. Within a relatively short time period, a very large pool of template can be produced (for in depth methods see [38]). A by-product of the amplification is magnesium pyrophosphate, which can even be seen by eye, and is a useful diagnostic indicator where further analysis, by agarose gel for example, is not available. While purified RNA is generally required for diagnosing filoviral infection, RT-LAMP is influenced to a lesser extent by PCR inhibitors found in the blood and can therefore be used directly with clinical samples [39,40]. As with RT-PCR, one of the main drawbacks of this technique is the inability to multiplex. Published RT-LAMP assays are presented in Table 5.
Table 5.
RT-LAMP papers, targets, primers and sensitivity for Ebolavirus detection.
| Paper | Target | Primers | Sensitivity |
|---|---|---|---|
| Kurosaki [41,42] | Trailer | EBOV F3 CAATAAACAACTATTTAAATAAC | 100% (92.5–100) compared to RT-qPCR |
| EBOV FIP GTCACACATGCTGCATTGTGTTTTCTATATTTAGCCTCTCTCCCT | |||
| EBOV BIP AACGCAACATAATAAACTCTGCATTTTATCAATAACAATATGAGCCCAG | |||
| NP | EBOV B3 CTGGCAAGATATTGATACAACA | 97.9% (88.7–100) compared to RT-qPCR | |
| EBOV LF AATTTTTTGATTATCACGC | |||
| EBOV F3 TGAAGTCAAGAAGCGTGATGG | |||
| EBOV FIP CATGGCAGCAAGTGTTCTCTTTTTAGTGAAGCGCCTTGAGGAA | |||
| EBOV BIP CAGTTTCTCTCCTTTGCAAGTCTTTTTGAACCTTCTCAAGGCAAGCC | |||
| EBOV B3 AGTCCTTGCTCTGCATGTACT | |||
| EBOV LF TGTTTTTTCCACTAGATACTGCTGG | |||
| EBOV LB TCCTTCCGAAATTGGTAGTAGGA | |||
| Xu [85] | GP | EBOV-F3 TGGTTCAAGTGCACAGTCAA | LOD: 30 copies (RNA) ≥102 TCID50/ml (Viral particle) |
| EBOV-B3 TGTCTGCTCTACGGTGATGT | |||
| EBOV-FIP(F1c + F2) GGAGGTTGAGGACTCGTGGAG GGAAGGAAAGCTGCAGTGT | |||
| EBOV-BIP(B1c + B2) CCAAAACAGGTCCGGACAACAG TCCAACTTGAGTTGCCTCAG | |||
| EBOV-LF Biotin-GCAAGGGTTGTCAGATGCG | |||
| EBOV-LB FITC-ATAATACACCCGTGTATAAACTTGAC | |||
| Benzine [44] | GP | EBOV F3 GACGGGAGTGAGTGTCTACC | LOD: 2.8 × 102 PFU/reaction (Kikwit) 1 × 103 PFU/reaction (Makona) |
| EBOV B3 AGCTTGGGGCAGTATCAGAA | |||
| EBOV FL GCACATACCGGCACC | |||
| EBOV BL CTTCCTGTATGATCGACTTGCTTC | |||
| EBOV FIP (F1c + F2) GGCACATGGTCCCGTTCCTGATTTTTTAGCGCCAGACGGGATTCG | |||
| EBOV BIP (B1c + B2) TGCCTTCCACAAAGAGGGTGCTTTTTGCGAAAGTCGTTCCTCGGT | |||
| Oloniniyi [86] | NP | EBOV F CTTAAGAATTCTCACTGATGATGTTGCAGGATTG | 256 copies/reaction |
| EBOV R CTTAAGGATCCATGGATTCTCGTCCTCAGAAAATC | |||
| EBOV R1 CTTAAGGATCCATGGATTCTCGTCCTCAGAAAGTC | |||
| SUDV F CTTAAGAATTCTCAGTCATGTTGAAGAACGGCAAG | |||
| SUDV R CTTAAGGATCCATGGATAAACGGGTGAGAGGTTC | 256 copies/reaction | ||
| BDBV F CTTAAGAATTCTCACCTGTGATGCTGGAGGA | |||
| BDBV R CTTAAGGATCCATGGATCCTCGTCCAATCAG | |||
| TAFV F1 ACTATAGGGCGAATTCATGGAGAGTCGGGCCCAC | 256 copies/reaction | ||
| TAFV F2 AAGGCTGCCCTTAGCTCGCTAGCACAACATGGAGAG | |||
| TAFV R2 CGACTCTAGAGGATCCTTACTTGTGGTGCTGAAGG | 256 copies/reaction | ||
| RESTV F CTTAAGAATTCTTACTGATGGTGCTGCAAGTTGC | |||
| RESTV R CTTAAGGATCCATGGATCGTGGGACCAGAAG | 64 copies/reaction |
RT-LAMP was assessed during the 2014–2016 EBOV outbreak in Guinea for surveillance and was directly compared with RT-qPCR [41]. In this study, buccal swabs (896) from cadavers and a small number of serum samples (21), from individuals with high-risk of EVD (based on contact tracing), were tested with both methods; none of the samples were EBOV positive [41]. The assay had been developed prior to use in Guinea [42,43] and then tested with 100 clinical samples from suspected EVD cases from Guinea. Results were assessed by measuring the turbidity (LA-200 device) of the RT-LAMP reaction or the level of fluorescence in the presence of an inter-chelating dye (Genie III device, Optigene). Compared with RT-qPCR results, the assay was nearly as sensitive (97.9% (95% CI: 88.7–100)) and positive results returned within 25 minutes [42]. Notably these assays used RNA isolated from either buccal swabs or serum. Recently a test was developed that was designed for direct whole blood use [44]. Whole blood is diluted 1:19 in lysis buffer and filtered (10 μm filter) into tubes containing lyophilised RT-LAMP reagents; the authors state that the whole assay takes 40 minutes.
4.4. Sequencing
Directly sequencing sample, and comparing to a database, allows direct diagnosis of an infection [45]. As whole or partial fragments of the pathogen are amplified (average read length mid-2015 was 5 kb [46]) and sequenced, this can take longer than RT-qPCR. A major strength of this technique is that, in addition to providing a diagnosis, it allows tracking of pathogen spread, and monitoring for the development of virulence and potential resistance. While sequencing generally requires a well-resourced laboratory with both sequencing machinery and computer analytics, portable systems have been developed. During the 2014–2016 EBOV outbreak a sequencing device, MinION (Oxford Nanopore Technologies) was used in Guinea for sequencing and analysis of 142 EBOV samples [45,46], demonstrating that in-field use of sequencing is feasible. For general surveillance of circulating viruses in a region, next-generating sequencing has been proposed [47,48].
4.5. Novel and or secondary diagnostic nucleic acid methods
Novel filovirus nucleic acid-based diagnostics are presented in Table 6. Of these, only one was granted Emergency Use during the 2014–2016 Ebolavirus outbreak was the FilmArray Biothreat E test [49]. This test is similar to RT-qPCR in that it has a reverse transcription step followed by a multiplexed PCR step. However, the products of the initial PCR are distributed to an array of secondary PCRs which use nested (internal) primers in combination with an interchelating fluorescent dye. The final products are measured using a film array. The assay itself is within a self-contained pouch in which the RNA template is released by a combination of chemical and mechanical (bead beating) means prior to RT and PCR. This assay has been tested both in the UK [50] the USA [51] and in field conditions in Sierra Leone [50] and Guinea [52]. Aside from the Biothreat E test, those methods described in Table 6 were not tested during the outbreak but show promise in development of future diagnostics.
Table 6.
Novel NATs for detection of filoviruses.
| Test | Notes |
|---|---|
| FilmArray Biothreat-E test [[50], [51], [52],87] | Whole blood or urine sample. Estimated LoD: 6 × 105 PFU/ml. FDA EUA. |
| QuRapID platform [53] | In blood RT-qPCR (far red dyes), use in resource poor regions. 20 kg, table top device, car alternator/battery or mains capable. |
| Virocyt [88] | Flow based particle detection of virus. Fluorescent staining of both genome and protein. EXPERIMENTAL uses; not suited for clinical samples due to high levels of other protein. |
| Lab-on-chip Optofluidic detection [89] | LoD 0.2PFU/ml. Amplification free by using sample concentration before measurement by laser. |
| Circulating microRNA [90] | Measuring EBOV induced changes in miRNA in humans and NHP. Proof of principle assay. 36 differentially expressed miRNAs; 93.1% (27/29) accurate in acute cases |
| Padlock probe detection [91] | Rolling circle amplification (RCA) of EBOV L gene on magnetic beads followed by secondary circle to circle amplification. Combined Detection by biotin capture and magnetic bead for an electrochemical and magnetic actuation. LOD:33 cDNA molecules. |
| One step FRET-PCR [92] | Multiplex assay differentiating between RT-qPCR products by Tm; 6FAM and LCRed 640 probes. Products from different ebolavirus subtypes had both distinct Tm fluorescence and amplicon size which allows typing. |
| FILODIAG [62] | Filovirus Diagnostics. Ultra-fast laser amplification using laser-heated, primer coated, nanoparticles for rapid heating/cooling. Aim of 15 min sample-to-result. |
| Mofina [63] | Portable POC device for the detection of Ebolavirus or Marburg virus. Sample-to-result in 75 min |
Utilization of far red fluorophores has been examined with whole blood samples to overcome signal inhibition of blood constituents. The QuRapID system uses these dyes in addition to rapid freeze/thaw cycles to isolate and then amplify viral RNA. The 20 kg stand-alone system has been developed for field use [53]. Two bead-based PCR assays were developed to detect multiple RNA viruses from bat urine [54]. Briefly, a one-step RT-PCR is combined with primers with a 5′ tag (24 nt) and biotinylated dCTP nucleotides. Fluorescently labelled microbeads with an anti-tag sequence then bind to amplification products. A Bio-plex 200 flow cell instrument measures the bead and amplification product. This bead-based technology could be adapted for use in multiplex filovirus diagnostics for humans.
5. Point-of-care diagnosis
A significant goal for filovirus diagnostics is the development of point-of care (POC) diagnosis. The ASSURED criteria set out by the WHO for POC devices are:
Affordable, Sensitive, Selective, User-friendly, Rapid, Equipment-free, and Deliverable (to end users) [55]. After the start of the EVD outbreak in 2014, a target product profile for diagnostics for Ebolavirus was proposed [56].
While there are rapid diagnostic tests based on an antibody response to viral antigen(s), an early diagnosis of filoviral infection is preferable, ideally before the humoral response has developed, and nucleic acid testing can do this. Studies in non-human primates have shown that post exposure prophylaxis (PEP) using vaccines for filoviruses can increase rates of survival even 2 days post-exposure [57]. The recent phase III trial of the VSV-ZEBOV vaccine indicates that this may be the case for human infections with EBOV [28]. There is also evidence that PEP with antibodies can be effective in non-human primates [58,59] and murine models [60]; notably a definitive diagnosis of a patient would be required before administration of treatment. Evidence from animal models indicate the earlier the administration of either vaccine or antibodies the greater the survival odds.
Furthermore, in filovirus outbreak situations, POC devices could play a key role in the triage of patients presenting to a clinic with fever. Multiplexed devices could assess whether a patient has multiple infections; for example with a virus as well as malaria and thereby feed into the clinical and therapeutic pathway [61]. A key aspect of POC devices is that there is minimal sample handing and potentially pathogenic material does not require transport to distant sites, thereby improving the diagnostic turnaround time.
Two projects funded via the Innovative Medicine Initiative (IMI) are attempting to address the need for novel near-patient filovirus diagnostics. A device that uses a laser based ultra-fast PCR is being developed by the FILODIAG consortium [62]. This technology utilises primer coated nanoparticles that are rapidly heated by laser absorption and then cool down immediately. This is faster than conventional thermocyclers; the aim is to test for EBOV within 15 min. A POC diagnostic device is being developed by the Mofina consortium for Ebolaviruses and Marburg virus detection. It is small, portable and will deliver results within 75 min following skin prick blood sampling [63]. As such, it will be well suited for in field use during filovirus outbreaks.
6. Conclusions
Nucleic acid tests have the greatest potential for early detection of filovirus infection. Their main strength is that only a small amount of input material is required for both detection and typing (either by specific primers/probes or sequencing). These tests can also be used in live vaccine administration to assess viral replication.
While the main focus during an outbreak of filoviral infections is plasma viraemia, other sites of viral persistence have been identified [[64], [65], [66], [67], [68]]. Assessing the ability of the described NATs when starting with a different clinical sample matrix is important. This is even more critical for POC devices where sample is put into a device unprocessed rather than purified RNA. The majority of the tests described in this review have focused on filoviral infections, yet the ideal test would incorporate a number of likely pathogens for a region to allow discrimination between causes of fever. NATs that utilise multiplexing that are integrated with novel POC platforms are eminently suited to this objective and, ultimately, will revolutionise outbreak diagnostics.
Funding
HMS is supported by the Wellcome Trust Institutional Strategic Support Fund (204809/Z/16/Z) awarded to St. George’s University of London.
Competing interests statement
JT and ADS work for QuantuMDx, a company developing diagnostic devices. SK is a paid advisor and chairs the infectious diseases advisory board for QuantuMDx. SK and HMS are both shareholders in QuantuMDx. SK and HMS are in receipt of funds from QuantuMDx to develop diagnostic technologies and assays (that have supported DJC).
All authors approved the final manuscript.
Acknowledgements
None.
References
- 1.CDC, No Title (n.d.). https://www.cdc.gov/vhf/ebola/outbreaks/history/chronology.html.
- 2.CDC, No Title (n.d.). https://www.cdc.gov/vhf/virus-families/filoviridae.html.
- 3.Pattyn S., van der Groen G., Jacob W., Piot P., Courteille G. Isolation of Marburg-like virus from a case of haemorrhagic fever in Zaire. Lancet (London England). 1977;1:573–574. doi: 10.1016/s0140-6736(77)92002-5. http://www.ncbi.nlm.nih.gov/pubmed/65663 (Accessed 9 January 2018) [DOI] [PubMed] [Google Scholar]
- 4.Swanepoel R., Leman P.A., Burt F.J., Zachariades N.A., Braack L.E., Ksiazek T.G., Rollin P.E., Zaki S.R., Peters C.J. Experimental inoculation of plants and animals with ebola virus. Emerg. Infect. Dis. 1996;2:321–325. doi: 10.3201/eid0204.960407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biek R., Walsh P.D., Leroy E.M., Real L.A. Recent common ancestry of Ebola Zaire virus found in a bat reservoir. PLoS Pathog. 2006;2:e90. doi: 10.1371/journal.ppat.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Towner J.S., Amman B.R., Sealy T.K., Carroll S.A.R., Comer J.A., Kemp A., Swanepoel R., Paddock C.D., Balinandi S., Khristova M.L., Formenty P.B.H., Albarino C.G., Miller D.M., Reed Z.D., Kayiwa J.T., Mills J.N., Cannon D.L., Greer P.W., Byaruhanga E., Farnon E.C., Atimnedi P., Okware S., Katongole-Mbidde E., Downing R., Tappero J.W., Zaki S.R., Ksiazek T.G., Nichol S.T., Rollin P.E. Isolation of genetically diverse marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laing E.D., Mendenhall I.H., Linster M., Low D.H.W., Chen Y., Yan L., Sterling S.L., Borthwick S., Neves E.S., Lim J.S.L., Skiles M., Lee B.P.Y.-H., Wang L.-F., Broder C.C., Smith G.J.D. Serologic evidence of fruit bat exposure to filoviruses, Singapore 2011–2016. Emerg. Infect. Dis. 2018;24:114–117. doi: 10.3201/eid2401.170401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jayme S.I., Field H.E., de Jong C., Olival K.J., Marsh G., Tagtag A.M., Hughes T., Bucad A.C., Barr J., Azul R.R., Retes L.M., Foord A., Yu M., Cruz M.S., Santos I.J., Lim T.M.S., Benigno C.C., Epstein J.H., Wang L.-F., Daszak P., Newman S.H. Molecular evidence of Ebola Reston virus infection in Philippine bats. Virol. J. 2015;12:107. doi: 10.1186/s12985-015-0331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa H., Miyamoto H., Nakayama E., Yoshida R., Nakamura I., Sawa H., Ishii A., Thomas Y., Nakagawa E., Matsuno K., Kajihara M., Maruyama J., Nao N., Muramatsu M., Kuroda M., Simulundu E., Changula K., Hang’ombe B., Namangala B., Nambota A., Katampi J., Igarashi M., Ito K., Feldmann H., Sugimoto C., Moonga L., Mweene A., Takada A. Seroepidemiological prevalence of multiple species of filoviruses in fruit bats (Eidolon helvum) migrating in africa. J. Infect. Dis. 2015;212(Suppl):S101–S108. doi: 10.1093/infdis/jiv063. [DOI] [PubMed] [Google Scholar]
- 10.Hayman D.T.S., Emmerich P., Yu M., Wang L.-F., Suu-Ire R., Fooks A.R., Cunningham A.A., Wood J.L.N. Long-term survival of an urban fruit bat seropositive for ebola and lagos bat viruses. PLoS One. 2010;5:e11978. doi: 10.1371/journal.pone.0011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pourrut X., Souris M., Towner J.S., Rollin P.E., Nichol S.T., Gonzalez J.-P., Leroy E. Large serological survey showing cocirculation of Ebola and Marburg viruses in Gabonese bat populations, and a high seroprevalence of both viruses in Rousettus aegyptiacus. BMC Infect. Dis. 2009;9:159. doi: 10.1186/1471-2334-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leroy E.M., Epelboin A., Mondonge V., Pourrut X., Gonzalez J.-P., Muyembe-Tamfum J.-J., Formenty P. Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, democratic Republic of Congo. Vector-Borne Zoon. Dis. 2007;9(2009):723–728. doi: 10.1089/vbz.2008.0167. [DOI] [PubMed] [Google Scholar]
- 13.Weingartl H.M., Embury-Hyatt C., Nfon C., Leung A., Smith G., Kobinger G. Transmission of Ebola virus from pigs to non-human primates. Sci. Rep. 2012;2:811. doi: 10.1038/srep00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO, No Title (n.d.). http://www.who.int/mediacentre/news/statements/2014/ebola-20140808/en/.
- 15.WHO, No Title (n.d.). http://www.who.int/mediacentre/news/statements/2016/end-of-ebola-pheic/en/.
- 16.WHO Situation report: EBOLA VIRUS DISEASE 10 JUNE 2016, n.d. http://apps.who.int/iris/bitstream/10665/208883/1/ebolasitrep_10Jun2016_eng.pdf?ua=1 (Accessed 9 January 2018).
- 17.Agnandji S.T., Fernandes J.F., Bache E.B., Obiang Mba R.M., Brosnahan J.S., Kabwende L., Pitzinger P., Staarink P., Massinga-Loembe M., Krähling V., Biedenkopf N., Fehling S.K., Strecker T., Clark D.J., Staines H.M., Hooper J.W., Silvera P., Moorthy V., Kieny M.-P., Adegnika A.A., Grobusch M.P., Becker S., Ramharter M., Mordmüller B., Lell B., Krishna S., Kremsner P.G., Kremsner P.G. Safety and immunogenicity of rVSV(G-ZEBOV-GP Ebola vaccine in adults and children in Lambaränä, Gabon: a phase I randomised trial. PLoS Med. 2017;14:e1002402. doi: 10.1371/journal.pmed.1002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agnandji S.T., Huttner A., Zinser M.E., Njuguna P., Dahlke C., Fernandes J.F., Yerly S., Dayer J.-A., Kraehling V., Kasonta R., Adegnika A.A., Altfeld M., Auderset F., Bache E.B., Biedenkopf N., Borregaard S., Brosnahan J.S., Burrow R., Combescure C., Desmeules J., Eickmann M., Fehling S.K., Finckh A., Goncalves A.R., Grobusch M.P., Hooper J., Jambrecina A., Kabwende A.L., Kaya G., Kimani D., Lell B., Lemaître B., Lohse A.W., Massinga-Loembe M., Matthey A., Mordmüller B., Nolting A., Ogwang C., Ramharter M., Schmidt-Chanasit J., Schmiedel S., Silvera P., Stahl F.R., Staines H.M., Strecker T., Stubbe H.C., Tsofa B., Zaki S., Fast P., Moorthy V., Kaiser L., Krishna S., Becker S., Kieny M.-P., Bejon P., Kremsner P.G., Addo M.M., Siegrist C.-A. Phase 1 trials of rVSV Ebola vaccine in Africa and Europe. N. Engl. J. Med. 2016;374:1647–1660. doi: 10.1056/NEJMoa1502924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu F.-C., Wurie A.H., Hou L.-H., Liang Q., Li Y.-H., Russell J.B.W., Wu S.-P., Li J.-X., Hu Y.-M., Guo Q., Xu W.-B., Wurie A.R., Wang W.-J., Zhang Z., Yin W.-J., Ghazzawi M., Zhang X., Duan L., Wang J.-Z., Chen W. Safety and immunogenicity of a recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in Sierra Leone: a single-centre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2017;389:621–628. doi: 10.1016/S0140-6736(16)32617-4. [DOI] [PubMed] [Google Scholar]
- 20.Milligan I.D., Gibani M.M., Sewell R., Clutterbuck E.A., Campbell D., Plested E., Nuthall E., Voysey M., Silva-Reyes L., McElrath M.J., De Rosa S.C., Frahm N., Cohen K.W., Shukarev G., Orzabal N., van Duijnhoven W., Truyers C., Bachmayer N., Splinter D., Samy N., Pau M.G., Schuitemaker H., Luhn K., Callendret B., Van Hoof J., Douoguih M., Ewer K., Angus B., Pollard A.J., Snape M.D. Safety and immunogenicity of novel adenovirus type 26 – and modified vaccinia ankara – vectored Ebola vaccines. JAMA. 2016;315:1610. doi: 10.1001/jama.2016.4218. [DOI] [PubMed] [Google Scholar]
- 21.De Santis O., Audran R., Pothin E., Warpelin-Decrausaz L., Vallotton L., Wuerzner G., Cochet C., Estoppey D., Steiner-Monard V., Lonchampt S., Thierry A.-C., Mayor C., Bailer R.T., Mbaya O.T., Zhou Y., Ploquin A., Sullivan N.J., Graham B.S., Roman F., De Ryck I., Ballou W.R., Kieny M.P., Moorthy V., Spertini F., Genton B. Safety and immunogenicity of a chimpanzee adenovirus-vectored Ebola vaccine in healthy adults: a randomised, double-blind, placebo-controlled, dose-finding, phase 1/2a study. Lancet Infect. Dis. 2016;16:311–320. doi: 10.1016/S1473-3099(15)00486-7. [DOI] [PubMed] [Google Scholar]
- 22.Tapia M.D., Sow S.O., Lyke K.E., Haidara F.C., Diallo F., Doumbia M., Traore A., Coulibaly F., Kodio M., Onwuchekwa U., Sztein M.B., Wahid R., Campbell J.D., Kieny M.-P., Moorthy V., Imoukhuede E.B., Rampling T., Roman F., De Ryck I., Bellamy A.R., Dally L., Mbaya O.T., Ploquin A., Zhou Y., Stanley D.A., Bailer R., Koup R.A., Roederer M., Ledgerwood J., Hill A.V.S., Ballou W.R., Sullivan N., Graham B., Levine M.M. Use of ChAd3-EBO-Z Ebola virus vaccine in Malian and US adults, and boosting of Malian adults with MVA-BN-Filo: a phase 1, single-blind, randomised trial, a phase 1b, open-label and double-blind, dose-escalation trial, and a nested, randomised, double-blind, placebo-controlled trial. Lancet Infect. Dis. 2016;16:31–42. doi: 10.1016/S1473-3099(15)00362-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huttner A., Dayer J.-A., Yerly S., Combescure C., Auderset F., Desmeules J., Eickmann M., Finckh A., Goncalves A.R., Hooper J.W., Kaya G., Krähling V., Kwilas S., Lemaître B., Matthey A., Silvera P., Becker S., Fast P.E., Moorthy V., Kieny M.P., Kaiser L., Siegrist C.-A., Consortium VSV-Ebola. The effect of dose on the safety and immunogenicity of the VSV Ebola candidate vaccine: a randomised double-blind, placebo-controlled phase 1/2 trial. Lancet Infect. Dis. 2015;15:1156–1166. doi: 10.1016/S1473-3099(15)00154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regules J.A., Beigel J.H., Paolino K.M., Voell J., Castellano A.R., Hu Z., Muñoz P., Moon J.E., Ruck R.C., Bennett J.W., Twomey P.S., Gutiérrez R.L., Remich S.A., Hack H.R., Wisniewski M.L., Josleyn M.D., Kwilas S.A., Van Deusen N., Mbaya O.T., Zhou Y., Stanley D.A., Jing W., Smith K.S., Shi M., Ledgerwood J.E., Graham B.S., Sullivan N.J., Jagodzinski L.L., Peel S.A., Alimonti J.B., Hooper J.W., Silvera P.M., Martin B.K., Monath T.P., Ramsey W.J., Link C.J., Lane H.C., Michael N.L., Davey R.T., Thomas S.J. RVSV(G-ZEBOV-GP study group, a recombinant vesicular stomatitis virus Ebola vaccine. N. Engl. J. Med. 2017;376:330–341. doi: 10.1056/NEJMoa1414216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ewer K., Rampling T., Venkatraman N., Bowyer G., Wright D., Lambe T., Imoukhuede E.B., Payne R., Fehling S.K., Strecker T., Biedenkopf N., Krähling V., Tully C.M., Edwards N.J., Bentley E.M., Samuel D., Labbé G., Jin J., Gibani M., Minhinnick A., Wilkie M., Poulton I., Lella N., Roberts R., Hartnell F., Bliss C., Sierra-Davidson K., Powlson J., Berrie E., Tedder R., Roman F., De Ryck I., Nicosia A., Sullivan N.J., Stanley D.A., Mbaya O.T., Ledgerwood J.E., Schwartz R.M., Siani L., Colloca S., Folgori A., Di Marco S., Cortese R., Wright E., Becker S., Graham B.S., Koup R.A., Levine M.M., Volkmann A., Chaplin P., Pollard A.J., Draper S.J., Ballou W.R., Lawrie A., Gilbert S.C., Hill A.V.S. A monovalent chimpanzee adenovirus ebola vaccine boosted with MVA. N. Engl. J. Med. 2016;374:1635–1646. doi: 10.1056/NEJMoa1411627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kibuuka H., Berkowitz N.M., Millard M., Enama M.E., Tindikahwa A., Sekiziyivu A.B., Costner P., Sitar S., Glover D., Hu Z., Joshi G., Stanley D., Kunchai M., Eller L.A., Bailer R.T., Koup R.A., Nabel G.J., Mascola J.R., Sullivan N.J., Graham B.S., Roederer M., Michael N.L., Robb M.L., Ledgerwood J.E. RV 247 Study Team, Safety and immunogenicity of Ebola virus and Marburg virus glycoprotein DNA vaccines assessed separately and concomitantly in healthy Ugandan adults: a phase 1b, randomised, double-blind, placebo-controlled clinical trial. Lancet. 2015;385:1545–1554. doi: 10.1016/S0140-6736(14)62385-0. [DOI] [PubMed] [Google Scholar]
- 27.Ledgerwood J.E., DeZure A.D., Stanley D.A., Coates E.E., Novik L., Enama M.E., Berkowitz N.M., Hu Z., Joshi G., Ploquin A., Sitar S., Gordon I.J., Plummer S.A., Holman L.A., Hendel C.S., Yamshchikov G., Roman F., Nicosia A., Colloca S., Cortese R., Bailer R.T., Schwartz R.M., Roederer M., Mascola J.R., Koup R.A., Sullivan N.J., Graham B.S. VRC 207 study team, chimpanzee adenovirus vector Ebola vaccine. N. Engl. J. Med. 2017;376:928–938. doi: 10.1056/NEJMoa1410863. [DOI] [PubMed] [Google Scholar]
- 28.Henao-Restrepo A.M., Camacho A., Longini I.M., Watson C.H., Edmunds W.J., Egger M., Carroll M.W., Dean N.E., Diatta I., Doumbia M., Draguez B., Duraffour S., Enwere G., Grais R., Gunther S., Gsell P.-S., Hossmann S., Watle S.V., Kondé M.K., Kéïta S., Kone S., Kuisma E., Levine M.M., Mandal S., Mauget T., Norheim G., Riveros X., Soumah A., Trelle S., Vicari A.S., Røttingen J.-A., Kieny M.-P. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!) Lancet (London England) 2017;389:505–518. doi: 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WHO . WHO; 2014. How to Safely Collect Blood Samples by Phlebotomy from Patients Suspected to Be Infected with Ebola or Marburg. http://www.who.int/csr/resources/publications/ebola/blood-collect/en/(Accessed 11 January 2018) [Google Scholar]
- 30.Spengler J.R., Chakrabarti A.K., Coleman-McCray J.D., Martin B.E., Nichol S.T., Spiropoulou C.F., Bird B.H. Utility of oral swab sampling for Ebola virus detection in Guinea pig model. Emerg. Infect. Dis. 2015;21:1816–1819. doi: 10.3201/eid2110.150840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semper A.E., Broadhurst M.J., Richards J., Foster G.M., Simpson A.J.H., Logue C.H., Kelly J.D., Miller A., Brooks T.J.G., Murray M., Pollock N.R. Performance of the GeneXpert Ebola assay for diagnosis of ebola virus disease in Sierra Leone: a field evaluation study. PLoS Med. 2016;13:e1001980. doi: 10.1371/journal.pmed.1001980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrader C., Schielke A., Ellerbroek L., Johne R. 2012. PCR Inhibitors – occurrence, Properties and Removal. [DOI] [PubMed] [Google Scholar]
- 33.Haddock E., Feldmann F., Feldmann H. Effective chemical inactivation of Ebola virus. Emerg. Infect. Dis. 2016;22:1292–1294. doi: 10.3201/eid2207.160233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rieger T., Kerber R., El Halas H., Pallasch E., Duraffour S., Günther S., Ölschläger S. Evaluation of RealStar reverse transcription-polymerase chain reaction kits for filovirus detection in the laboratory and field. J. Infect. Dis. 2016;214:S243–S249. doi: 10.1093/infdis/jiw246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trombley A.R., Wachter L., Garrison J., Buckley-Beason V.A., Jahrling J., Hensley L.E., Schoepp R.J., Norwood D.A., Goba A., Fair J.N., Kulesh D.A. Comprehensive panel of real-time TaqMan polymerase chain reaction assays for detection and absolute quantification of filoviruses, arenaviruses, and New World hantaviruses. Am. J. Trop. Med. Hyg. 2010;82:954–960. doi: 10.4269/ajtmh.2010.09-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.FDA, Emergency Situations (Medical Devices) – Emergency Use Authorizations, (n.d.). https://www.fda.gov/MedicalDevices/Safety/EmergencySituations/ucm161496.htm#ebola (Accessed 9 January 2018).
- 37.Raftery P., Condell O., Wasunna C., Kpaka J., Zwizwai R., Nuha M., Fallah M., Freeman M., Harris V., Miller M., Baller A., Massaquoi M., Katawera V., Saindon J., Bemah P., Hamblion E., Castle E., Williams D., Gasasira A., Nyenswah T. Establishing Ebola Virus Disease (EVD) diagnostics using GeneXpert technology at a mobile laboratory in Liberia: impact on outbreak response, case management and laboratory systems strengthening. PLoS Negl. Trop. Dis. 2018;12:e0006135. doi: 10.1371/journal.pntd.0006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong Y.-P., Othman S., Lau Y.-L., Radu S., Chee H.-Y. Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2018;124:626–643. doi: 10.1111/jam.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Soud W.A., Jönsson L.J., Râdström P. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J. Clin. Microbiol. 2000;38:345–350. doi: 10.1128/jcm.38.1.345-350.2000. http://www.ncbi.nlm.nih.gov/pubmed/10618113 (Accessed 1 September 2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Soud W.A., Rådström P. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 2001;39:485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurosaki Y., Magassouba N., Bah H.A., Soropogui B., Doré A., Kourouma F., Cherif M.S., Keita S., Yasuda J. Deployment of a reverse transcription loop-mediated isothermal amplification test for ebola virus surveillance in remote areas in Guinea. J. Infect. Dis. 2016;214:S229–S233. doi: 10.1093/infdis/jiw255. [DOI] [PubMed] [Google Scholar]
- 42.Kurosaki Y., Magassouba N., Oloniniyi O.K., Cherif M.S., Sakabe S., Takada A., Hirayama K., Yasuda J. Development and evaluation of reverse transcription-loop-mediated isothermal amplification (RT-LAMP) assay coupled with a portable device for rapid diagnosis of ebola virus disease in Guinea. PLoS Negl. Trop. Dis. 2016;10:e0004472. doi: 10.1371/journal.pntd.0004472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurosaki Y., Takada A., Ebihara H., Grolla A., Kamo N., Feldmann H., Kawaoka Y., Yasuda J. Rapid and simple detection of Ebola virus by reverse transcription-loop-mediated isothermal amplification. J. Virol. Methods. 2007;141:78–83. doi: 10.1016/j.jviromet.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 44.Benzine J.W., Brown K.M., Agans K.N., Godiska R., Mire C.E., Gowda K., Converse B., Geisbert T.W., Mead D.A., Chander Y. Molecular diagnostic field test for point-of-care detection of ebola virus directly from blood. J. Infect. Dis. 2016;214:S234––S242. doi: 10.1093/infdis/jiw330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L., Bore J.A., Koundouno R., Dudas G., Mikhail A., Ouédraogo N., Afrough B., Bah A., Baum J.H.J., Becker-Ziaja B., Boettcher J.P., Cabeza-Cabrerizo M., Camino-Sánchez Á., Carter L.L., Doerrbecker J., Enkirch T., García-Dorival I., Hetzelt N., Hinzmann J., Holm T., Kafetzopoulou L.E., Koropogui M., Kosgey A., Kuisma E., Logue C.H., Mazzarelli A., Meisel S., Mertens M., Michel J., Ngabo D., Nitzsche K., Pallasch E., Patrono L.V., Portmann J., Repits J.G., Rickett N.Y., Sachse A., Singethan K., Vitoriano I., Yemanaberhan R.L., Zekeng E.G., Racine T., Bello A., Sall A.A., Faye O., Faye O., Magassouba N., Williams C.V., Amburgey V., Winona L., Davis E., Gerlach J., Washington F., Monteil V., Jourdain M., Bererd M., Camara A., Somlare H., Camara A., Gerard M., Bado G., Baillet B., Delaune D., Nebie K.Y., Diarra A., Savane Y., Pallawo R.B., Gutierrez G.J., Milhano N., Roger I., Williams C.J., Yattara F., Lewandowski K., Taylor J., Rachwal P., Turner D.J., Pollakis G., Hiscox J.A., Matthews D.A., O’Shea M.K., Johnston A.M., Wilson D., Hutley E., Smit E., Di Caro A., Wölfel R., Stoecker K., Fleischmann E., Gabriel M., Weller S.A., Koivogui L., Diallo B., Keïta S., Rambaut A., Formenty P., Günther S., Carroll M.W. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greninger A.L., Naccache S.N., Federman S., Yu G., Mbala P., Bres V., Stryke D., Bouquet J., Somasekar S., Linnen J.M., Dodd R., Mulembakani P., Schneider B.S., Muyembe-Tamfum J.-J., Stramer S.L., Chiu C.Y. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koehler J.W., Hall A.T., Rolfe P.A., Honko A.N., Palacios G.F., Fair J.N., Muyembe J.-J., Mulembekani P., Schoepp R.J., Adesokan A., Minogue T.D. Development and evaluation of a panel of filovirus sequence capture probes for pathogen detection by next-generation sequencing. PLoS One. 2014;9:e107007. doi: 10.1371/journal.pone.0107007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woolhouse M.E.J., Rambaut A., Kellam P. Lessons from Ebola: improving infectious disease surveillance to inform outbreak management. Sci. Transl. Med. 2015;7:307rv5. doi: 10.1126/scitranslmed.aab0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.FDA . 2014. Letter of Authorization for BioFire FilmArray Biothreat-E test. https://www.fda.gov/downloads/EmergencyPreparedness/Counterterrorism/MedicalCountermeasures/MCMLegalRegulatoryandPolicyFramework/UCM420421.pdf (Accessed 9 January 2018) [Google Scholar]
- 50.Weller S.A., Bailey D., Matthews S., Lumley S., Sweed A., Ready D., Eltringham G., Richards J., Vipond R., Lukaszewski R., Payne P.M., Aarons E., Simpson A.J., Hutley E.J., Brooks T. Evaluation of the biofire FilmArray bioThreat-E test (v2.5) for rapid identification of Ebola virus disease in heat-treated blood samples obtained in Sierra Leone and the United Kingdom. J. Clin. Microbiol. 2016;54:114–119. doi: 10.1128/JCM.02287-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Southern T.R., Racsa L.D., Albariño C.G., Fey P.D., Hinrichs S.H., Murphy C.N., Herrera V.L., Sambol A.R., Hill C.E., Ryan E.L., Kraft C.S., Campbell S., Sealy T.K., Schuh A., Ritchie J.C., Lyon G.M., Mehta A.K., Varkey J.B., Ribner B.S., Brantly K.P., Ströher U., Iwen P.C., Burd E.M. Comparison of FilmArray and quantitative real-time reverse transcriptase PCR for detection of zaire Ebolavirus from contrived and clinical specimens. J. Clin. Microbiol. 2015;53:2956–2960. doi: 10.1128/JCM.01317-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gay-Andrieu F., Magassouba N., Picot V., Phillips C.L., Peyrefitte C.N., Dacosta B., Doré A., Kourouma F., Ligeon-Ligeonnet V., Gauby C., Longuet C., Scullion M., Faye O., Machuron J.L., Miller M. Clinical evaluation of the BioFire FilmArray ® bioThreat-E test for the diagnosis of Ebola virus disease in Guinea. J. Clin. Virol. 2017;92:20–24. doi: 10.1016/j.jcv.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 53.Shah K., Bentley E., Tyler A., Richards K.S.R., Wright E., Easterbrook L., Lee D., Cleaver C., Usher L., Burton J.E., Pitman J.K., Bruce C.B., Edge D., Lee M., Nazareth N., Norwood D.A., Moschos S.A. Field-deployable, quantitative, rapid identification of active Ebola virus infection in unprocessed blood. Chem. Sci. 2017;8:7780–7797. doi: 10.1039/c7sc03281a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyd V., Smith I., Crameri G., Burroughs A.L., Durr P.A., White J., Cowled C., Marsh G.A., Wang L.-F. Development of multiplexed bead arrays for the simultaneous detection of nucleic acid from multiple viruses in Bat samples. J. Virol. Methods. 2015;223:5–12. doi: 10.1016/j.jviromet.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.TDR | Rapid diagnostic tests for sexually transmitted infections, WHO, (2015). http://www.who.int/tdr/publications/journal-supplements/sti-way-forward/en/(Accessed 9 January 2018).
- 56.Chua A.C., Cunningham J., Moussy F., Perkins M.D., Formenty P. The case for improved diagnostic tools to control ebola virus disease in West Africa and how to get there. PLoS Negl. Trop. Dis. 2015;9:e0003734. doi: 10.1371/journal.pntd.0003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geisbert T.W., Hensley L.E., Geisbert J.B., Leung A., Johnson J.C., Grolla A., Feldmann H. Postexposure treatment of marburg virus infection. Emerg. Infect. Dis. 2010;16:1119–1122. doi: 10.3201/eid1607.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pyankov O.V., Setoh Y.X., Bodnev S.A., Edmonds J.H., Pyankova O.G., Pyankov S.A., Pali G., Belford S., Lu L., La M., Lovrecz G., Volchkova V.A., Chappell K.J., Watterson D., Marsh G., Young P.R., Agafonov A.A., Farmer J.F., Volchkov V.E., Suhrbier A., Khromykh A.A. Successful post-exposure prophylaxis of Ebola infected non-human primates using Ebola glycoprotein-specific equine IgG. Sci. Rep. 2017;7:41537. doi: 10.1038/srep41537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dye J.M., Herbert A.S., Kuehne A.I., Barth J.F., Muhammad M.A., Zak S.E., Ortiz R.A., Prugar L.I., Pratt W.D. Postexposure antibody prophylaxis protects nonhuman primates from filovirus disease. Proc. Natl. Acad. Sci. 2012;109:5034–5039. doi: 10.1073/pnas.1200409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng X., Wong G., Zhao Y., Wang H., He S., Bi Y., Chen W., Jin H., Gai W., Chu D., Cao Z., Wang C., Fan Q., Chi H., Gao Y., Wang T., Feng N., Yan F., Huang G., Zheng Y., Li N., Li Y., Qian J., Zou Y., Kobinger G., Gao G.F., Qiu X., Yang S., Xia X. Treatment with hyperimmune equine immunoglobulin or immunoglobulin fragments completely protects rodents from Ebola virus infection. Sci. Rep. 2016;6:24179. doi: 10.1038/srep24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waxman M., Aluisio A.R., Rege S., Levine A.C. Characteristics and survival of patients with Ebola virus infection, malaria, or both in Sierra Leone: a retrospective cohort study. Lancet Infect. Dis. 2017;17:654–660. doi: 10.1016/S1473-3099(17)30112-3. [DOI] [PubMed] [Google Scholar]
- 62.IMI, FILODIAG | IMI – Innovative Medicines Initiative, (n.d.). http://www.imi.europa.eu/projects-results/project-factsheets/filodiag (Accessed 9 January 2018).
- 63.Mofina | IMI – Innovative Medicines Initiative, (n.d.). http://www.imi.europa.eu/projects-results/project-factsheets/mofina (Accessed 22 February 2018).
- 64.Deen G.F., Broutet N., Xu W., Knust B., Sesay F.R., McDonald S.L.R., Ervin E., Marrinan J.E., Gaillard P., Habib N., Liu H., Liu W., Thorson A.E., Yamba F., Massaquoi T.A., James F., Ariyarajah A., Ross C., Bernstein K., Coursier A., Klena J., Carino M., Wurie A.H., Zhang Y., Dumbuya M.S., Abad N., Idriss B., Wi T., Bennett S.D., Davies T., Ebrahim F.K., Meites E., Naidoo D., Smith S.J., Ongpin P., Malik T., Banerjee A., Erickson B.R., Liu Y., Liu Y., Xu K., Brault A., Durski K.N., Winter J., Sealy T., Nichol S.T., Lamunu M., Bangura J., Landoulsi S., Jambai A., Morgan O., Wu G., Liang M., Su Q., Lan Y., Hao Y., Formenty P., Ströher U., Sahr F. Ebola RNA persistence in semen of ebola virus disease survivors – Final report. N. Engl. J. Med. 2017;377:1428–1437. doi: 10.1056/NEJMoa1511410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mate S.E., Kugelman J.R., Nyenswah T.G., Ladner J.T., Wiley M.R., Cordier-Lassalle T., Christie A., Schroth G.P., Gross S.M., Davies-Wayne G.J., Shinde S.A., Murugan R., Sieh S.B., Badio M., Fakoli L., Taweh F., de Wit E., van Doremalen N., Munster V.J., Pettitt J., Prieto K., Humrighouse B.W., Ströher U., DiClaro J.W., Hensley L.E., Schoepp R.J., Safronetz D., Fair J., Kuhn J.H., Blackley D.J., Laney A.S., Williams D.E., Lo T., Gasasira A., Nichol S.T., Formenty P., Kateh F.N., De Cock K.M., Bolay F., Sanchez-Lockhart M., Palacios G. Molecular evidence of sexual transmission of Ebola virus. N. Engl. J. Med. 2015;373:2448–2454. doi: 10.1056/NEJMoa1509773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodriguez L.L., De Roo A., Guimard Y., Trappier S.G., Sanchez A., Bressler D., Williams A.J., Rowe A.K., Bertolli J., Khan A.S., Ksiazek T.G., Peters C.J., Nichol S.T. Persistence and genetic stability of ebola virus during the outbreak in Kikwit, Democratic Republic of the Congo. J. Infect. Dis. 1995;179(1999):S170–S176. doi: 10.1086/514291. [DOI] [PubMed] [Google Scholar]
- 67.Varkey J.B., Shantha J.G., Crozier I., Kraft C.S., Lyon G.M., Mehta A.K., Kumar G., Smith J.R., Kainulainen M.H., Whitmer S., Ströher U., Uyeki T.M., Ribner B.S., Yeh S. Persistence of Ebola virus in ocular fluid during convalescence. N. Engl. J. Med. 2015;372:2423–2427. doi: 10.1056/NEJMoa1500306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chughtai A.A., Barnes M., Macintyre C.R. Persistence of Ebola virus in various body fluids during convalescence: evidence and implications for disease transmission and control. Epidemiol. Infect. 2016;144:1652–1660. doi: 10.1017/S0950268816000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leroy E., Baize S., Lu C., McCormick J., Georges A., Georges-Courbot M., Lansoud-Soukate J., Fisher-Hoch S. Diagnosis of Ebola haemorrhagic fever by RT-PCR in an epidemic setting. J. Med. Virol. 2000;60:463–467. [PubMed] [Google Scholar]
- 70.Towner J.S., Rollin P.E., Bausch D.G., Sanchez A., Crary S.M., Vincent M., Lee W.F., Spiropoulou C.F., Ksiazek T.G., Lukwiya M., Kaducu F., Downing R., Nichol S.T. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J. Virol. 2004;78:4330–4341. doi: 10.1128/JVI.78.8.4330-4341.2004. http://www.ncbi.nlm.nih.gov/pubmed/15047846 (Accessed 7 June 2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park S.-W., Lee Y.-J., Lee W.-J., Jee Y., Choi W. One-step reverse transcription-polymerase chain reaction for ebola and marburg viruses. Osong Publ. Heal Res. Perspect. 2016;7:205–209. doi: 10.1016/j.phrp.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogawa H., Miyamoto H., Ebihara H., Ito K., Morikawa S., Feldmann H., Takada A. Detection of all known filovirus species by reverse transcription-polymerase chain reaction using a primer set specific for the viral nucleoprotein gene. J. Virol. Methods. 2011;171:310–313. doi: 10.1016/j.jviromet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bergqvist C., Holmström P., Lindegren G., Lagerqvist N., Leijon M., Falk K.I. Multiplex nucleic acid suspension bead arrays for detection and subtyping of filoviruses. J. Clin. Microbiol. 2015;53:1368–1370. doi: 10.1128/JCM.02787-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drosten C., Göttig S., Schilling S., Asper M., Panning M., Schmitz H., Günther S. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 2002;40:2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. http://www.ncbi.nlm.nih.gov/pubmed/12089242 (Accessed 8 January 2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanchez A., Ksiazek T.G., Rollin P.E., Miranda M.E., Trappier S.G., Khan A.S., Peters C.J., Nichol S.T. Detection and molecular characterization of Ebola viruses causing disease in human and nonhuman primates. J. Infect. Dis. 1999;179(Suppl. 1):S164–S169. doi: 10.1086/514282. [DOI] [PubMed] [Google Scholar]
- 76.Gibb T.R., Norwood D.A., Jr, Woollen N., Henchal E.A. Development and evaluation of a fluorogenic 5′-nuclease assay to identify Marburg virus. Mol. Cell. Probes. 2001;15:259–266. doi: 10.1006/mcpr.2001.0369. [DOI] [PubMed] [Google Scholar]
- 77.Weidmann M., Mühlberger E., Hufert F.T. Rapid detection protocol for filoviruses. J. Clin. Virol. 2004;30:94–99. doi: 10.1016/j.jcv.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 78.Cnops L., Van den Eede P., Pettitt J., Heyndrickx L., De Smet B., Coppens S., Andries I., Pattery T., Van Hove L., Meersseman G., Van Den Herrewegen S., Vergauwe N., Thijs R., Jahrling P.B., Nauwelaers D., Ariën K.K. Development, evaluation, and integration of a quantitative reverse-transcription polymerase chain reaction diagnostic test for ebola virus on a molecular diagnostics platform. J. Infect. Dis. 2016;214:S192–S202. doi: 10.1093/infdis/jiw150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pinsky B.A., Sahoo M.K., Sandlund J., Kleman M., Kulkarni M., Grufman P., Nygren M., Kwiatkowski R., Baron E.J., Tenover F., Denison B., Higuchi R., Van Atta R., Beer N.R., Carrillo A.C., Naraghi-Arani P., Mire C.E., Ranadheera C., Grolla A., Lagerqvist N., Persing D.H. Analytical performance characteristics of the cepheid GeneXpert ebola assay for the detection of Ebola virus. PLoS One. 2015;10:e0142216. doi: 10.1371/journal.pone.0142216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loftis A.J., Quellie S., Chason K., Sumo E., Toukolon M., Otieno Y., Ellerbrok H., Hobbs M.M., Hoover D., Dube K., Wohl D.A., Fischer W.A. Validation of the cepheid GeneXpert for detecting Ebola virus in semen. J. Infect. Dis. 2017;215:344–350. doi: 10.1093/infdis/jiw562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pettitt J., Higgs E., Fallah M., Nason M., Stavale E., Marchand J., Reilly C., Jensen K., Dighero-Kemp B., Tuznik K., Logue J., Bolay F., Hensley L. Assessment and optimization of the GeneXpert diagnostic platform for detection of ebola virus RNA in seminal fluid. J. Infect. Dis. 2016;215:jiw599. doi: 10.1093/infdis/jiw599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.LightMix ® Ebola Zaire rRT-PCR Test, (n.d.). https://www.fda.gov/downloads/MedicalDevices/Safety/EmergencySituations/UCM428004.pdf (Accessed 9 January 2018).
- 83.F.D.A., EZ1 EUO Authorisation, 2014 https://www.fda.gov/downloads/MedicalDevices/Safety/EmergencySituations/UCM418799.pdf (Accessed 9 January 2018).
- 84.WHO . 2015. WHO Emergency Use Assessment and Listing Procedure for EVD IVDs PUBLIC REPORT Product: Liferiver™ ?Ebola Virus (EBOV) Real Time RT-PCR Kit. http://www.who.int/diagnostics_laboratory/procurement/150427_liferiver_china_public_report.pdf (Accessed 9 January 2018) [Google Scholar]
- 85.Xu C., Wang H., Jin H., Feng N., Zheng X., Cao Z., Li L., Wang J., Yan F., Wang L., Chi H., Gai W., Wang C., Zhao Y., Feng Y., Wang T., Gao Y., Lu Y., Yang S., Xia X. Visual detection of Ebola virus using reverse transcription loop-mediated isothermal amplification combined with nucleic acid strip detection. Arch. Virol. 2016;161:1125–1133. doi: 10.1007/s00705-016-2763-5. [DOI] [PubMed] [Google Scholar]
- 86.Oloniniyi O.K., Kurosaki Y., Miyamoto H., Takada A., Yasuda J. Rapid detection of all known ebolavirus species by reverse transcription-loop-mediated isothermal amplification (RT-LAMP) J. Virol. Methods. 2017;246:8–14. doi: 10.1016/j.jviromet.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 87.Leski T.A., Ansumana R., Taitt C.R., Lamin J.M., Bangura U., Lahai J., Mbayo G., Kanneh M.B., Bawo B., Bockarie A.S., Scullion M., Phillips C.L., Horner C.P., Jacobsen K.H., Stenger D.A. Use of the FilmArray system for detection of zaire Ebolavirus in a small hospital in Bo, Sierra Leone. J. Clin. Microbiol. 2015;53:2368–2370. doi: 10.1128/JCM.00527-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rossi C.A., Kearney B.J., Olschner S.P., Williams P.L., Robinson C.G., Heinrich M.L., Zovanyi A.M., Ingram M.F., Norwood D.A., Schoepp R.J. Evaluation of ViroCyt® Virus Counter for rapid filovirus quantitation. Viruses. 2015;7:857–872. doi: 10.3390/v7030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cai H., Parks J.W., Wall T.A., Stott M.A., Stambaugh A., Alfson K., Griffiths A., Mathies R.A., Carrion R., Patterson J.L., Hawkins A.R., Schmidt H. Optofluidic analysis system for amplification-free, direct detection of Ebola infection. Sci. Rep. 2015;5:14494. doi: 10.1038/srep14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duy J., Koehler J.W., Honko A.N., Schoepp R.J., Wauquier N., Gonzalez J.-P., Pitt M.L., Mucker E.M., Johnson J.C., O’Hearn A., Bangura J., Coomber M., Minogue T.D. Circulating microRNA profiles of Ebola virus infection. Sci. Rep. 2016;6:24496. doi: 10.1038/srep24496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carinelli S., Kühnemund M., Nilsson M., Pividori M.I. Yoctomole electrochemical genosensing of Ebola virus cDNA by rolling circle and circle to circle amplification. Biosens. Bioelectron. 2017;93:65–71. doi: 10.1016/j.bios.2016.09.099. [DOI] [PubMed] [Google Scholar]
- 92.Lu G., Zhang J., Zhang C., Li X., Shi D., Yang Z., Wang C. One-step reverse-transcription FRET-PCR for differential detection of five ebolavirus species. PLoS One. 2015;10:e0126281. doi: 10.1371/journal.pone.0126281. [DOI] [PMC free article] [PubMed] [Google Scholar]