Abstract
We present association results from a large genome-wide association study of tooth agenesis (TA) as well as selective TA, including 1,944 subjects with congenitally missing teeth, excluding third molars, and 338,554 controls, all of European ancestry. We also tested the association of previously identified risk variants, for timing of tooth eruption and orofacial clefts, with TA. We report associations between TA and 9 novel risk variants. Five of these variants associate with selective TA, including a variant conferring risk of orofacial clefts. These results contribute to a deeper understanding of the genetic architecture of tooth development and disease. The few variants previously associated with TA were uncovered through candidate gene studies guided by mouse knockouts. Knowing the etiology and clinical features of TA is important for planning oral rehabilitation that often involves an interdisciplinary approach.
Keywords: genetics, molecular genetics, hypodontia, oligodontia, orofacial cleft(s), odontogenesis
Introduction
Tooth agenesis (TA) is a developmental anomaly defined as the absence of ≥1 permanent teeth. Based on severity, TA is classified as hypodontia and oligodontia—that is, missing ≤5 and ≥6 teeth, respectively (Polder et al. 2004). The prevalence of TA, excluding the third molars (wisdom teeth), is reported to range between 3% and 10% (Magnússon 1977; Johannsdottir et al. 1997; Polder et al. 2004) and is considerably lower for primary teeth (Magnússon 1984). Agenesis of permanent teeth has a slightly higher prevalence in women than men (Magnússon 1977; Magnússon 1979). The worldwide prevalence of agenesis of the third molars is 23% (Carter and Worthington 2015). The most commonly congenitally missing permanent teeth, after the third molars, are the mandibular second premolars, followed by the maxillary lateral incisors and maxillary second premolars (Magnússon 1977; Polder et al. 2004).
There are several syndromes accompanied by increased prevalence of dental anomalies. These include orofacial clefts (OFCs; Howe et al. 2015) and ectodermal dysplasia (Adaimy et al. 2007). Hence, genes involved in odontogenesis may play a role in other developmental processes. Individuals with OFC have an especially high risk of TA in the maxillary cleft region (Rullo et al. 2015). The risk for TA outside the cleft area is also higher for those with OFC than for population controls (Howe et al. 2015).
The investigation of the genetic regulation of tooth development with mouse models has identified more than a hundred genes involved in odontogenesis. These studies showed that 4 conserved pathways are of major importance during odontogenesis: bone morphogenetic proteins, fibroblast growth factors, sonic hedgehog, and wingless-related pathways (Lan et al. 2014). Analyses of humans, as guided by the animal models, have yielded a small number of mutations associated with TA, mainly with oligodontia (Lan et al. 2014). These include variants disrupting WNT10A, where variants have been associated with hypodontia and oligodontia (Adaimy et al. 2007), and in other genes in the WNT pathway, such as LRP6 (Ockeloen et al. 2016).
A well-powered genome-wide association study (GWAS) for TA has not been reported. It is notable that a relatively small GWAS for agenesis of the third molars (Haga et al. 2013) failed to show association at a genome-wide significant level. However, GWASs for the dental traits of sequence and timing of eruption of permanent (Geller et al. 2011) and primary (Pillas et al. 2010; Fatemifar et al. 2013) teeth identified several risk loci.
Here we describe a GWAS for TA, excluding third molars. We found 9 novel TA-associated variants, including 5 variants associated with agenesis of specific teeth. One variant also associates with OFC.
Subjects and Methods
Study Populations
Icelandic Discovery Sample
The measures of missing teeth used in this study were subjects with TA (n = 1,944) missing up to 5 teeth, defined as hypodontia (n = 1,755), and subjects missing ≥6 teeth, defined as oligodontia (n = 189). In this study, we did not have information regarding the third molars (the wisdom teeth); thus, TA was defined as missing ≥1 teeth, excluding the third molars. In total, 338,554 subjects were included as controls.
The TA cases included were identified in an epidemiologic study by Magnússon (1977; n = 85), at 2 orthodontic clinics in Iceland (n = 158 and n = 110), and from the Icelandic Health Insurance registry (n = 1,591). The Icelandic Health Insurance registry comprises all individuals who have, over the last 20 y, received a refund for dental service necessitated by agenesis of teeth. Since this refund is categorically related to TA, the sample represents all forms of hypodontia occurring in the population, missing only a few individuals neglecting or not needing dental treatment. All cases were diagnosed with TA via radiography.
The Icelandic study was approved by the Data Protection Authority and the National Bioethics Committee. Participants giving samples also gave written informed consent.
Follow-up Samples
The follow-up samples are described in the Appendix Subjects and Methods.
Genotyping, Imputation, and Association Testing in the Icelandic Sample
We carried out chip typing of 151,677 Icelanders using Illumina arrays from the HumanHap and Omni series as described previously (Gudbjartsson et al. 2015). The chip-typed individuals were long range phased (Kong et al. 2008), and variants from whole-genome sequencing of Icelanders (n = 15,220) were imputed into the chip-typed individuals as well as 282,894 close relatives (Gudbjartsson et al. 2015).
Association testing was carried out through logistic regression with 1) the TA phenotype as the response and 2) the genotype counts and a set of nuisance variables as predictors (i.e., sex, county of birth, current age or age at death [first- and second-order terms included], blood sample availability, and an indicator of the overlap of the lifetime of the individual with the time span of phenotype collection; Steinthorsdottir et al. 2014). The intercept from LD score regression (Bulik-Sullivan et al. 2015) was used to estimate correction factors for inflation of test statistics due to relatedness and population stratification. The estimated correction factors were 1.14 for TA, 1.14 for agenesis of maxillary lateral incisors, 1.07 for agenesis of maxillary second premolars, and 1.10 for mandibular second premolars. Information about the functional impact of classes of variants was used for a class-specific Bonferroni procedure used for the threshold for genome-wide significance as described in detail in the study by Sveinbjornsson et al. (2016). Combination of results from the follow-up samples, as well as joint analysis of the discovery and follow-up samples, was carried out with METAL software (Willer et al. 2010).
Variants Known to Confer Risk of TA-Related Phenotypes Tested in the Icelandic Sample
In total, 40 known OFC variants and 17 variants associated with primary and permanent tooth eruption were tested in our TA sample, as described in the Appendix Subjects and Methods.
Gene Set Enrichment Analysis
We performed gene set enrichment analysis for the 11 TA-associated markers using the GO database through INRICH, as described in the Appendix Subjects and Methods.
Results
Our TA sample consists of an Icelandic discovery sample and follow-up samples from Denmark, the Netherlands, Portugal, and the United States, as described in the study design flowchart in Figure 1. First, we conducted a GWAS for TA as well as for selective TA of the most frequently absent teeth, excluding third molars (Fig. 2), in the Icelandic sample (Fig. 3 and Appendix Figs. 1-11). Second, we tested previously reported risk variants for TA-related phenotypes in the Icelandic sample (Appendix Tables 1 and 2). Finally, variants significantly associated with TA or selective TA (11 from the GWASs and 1 from the testing of risk variants for TA-related phenotypes) were included in joint analyses with data from the follow-up samples (Table 1). Following the joint analysis, 11 of the 12 variants taken forward were significantly associated with TA, of which 9 have not previously been associated with TA. Five of the 9 variants associate with agenesis of specific teeth, including 1 variant also conferring risk of the TA-related phenotype OFC.
Figure 1.
Study design flowchart. GWAS, genome-wide association study; TA, tooth agenesis.
Figure 2.
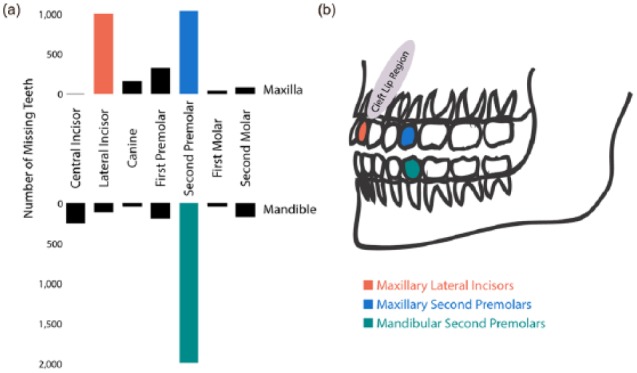
The Icelandic tooth agenesis sample. (a) The number of absent teeth at the specific dental positions, excluding the third molars, in an Icelandic sample of 1,944 subjects in which 5,423 teeth were missing. In total, 1,196 subjects were missing 1,987 mandibular second premolars; 600 subjects, 1,034 maxillary second premolars; and 600 subjects, 998 maxillary lateral incisors. (b) Illustration of the teeth in the mandible and the maxilla, as well as the cleft lip region. The 3 most commonly missing teeth analyzed in the 3 genome-wide association studies of selective tooth agenesis are indicated as red (maxillary lateral incisors), blue (maxillary second premolars), and green (mandibular second premolars).
Figure 3.
Manhattan plots for tooth agenesis (TA). (a) Genome-wide association study (GWAS) for TA in the Icelandic discovery sample. (b) GWAS for selective TA in the Icelandic discovery sample. The new signals (FOXP1 and EDA) from the GWAS of maxillary lateral incisors are indicated in red. (c) The new signal (LEF1) from the GWAS of the mandibular second premolars is indicated in green. (d) The new signal (NOL11) from the GWAS of maxillary second premolars is indicated in blue. Pthreshold = 2.30 × 10-9 for other variants within DNase I hypersensitivity sites (DHSs). Genome-wide significance thresholds after correction for multiple testing are based on type of variant as described in the study by Sveinbjornsson et al. (2016) per the following thresholds: Pthreshold = 2.60 × 10-7 for high-impact variants (stop-gain and stop-loss, frameshift indel, donor and acceptor splice-site, and initiator codon variants), Pthreshold = 5.10 × 10-8 for moderate-impact variants (missense, in-frame indel, and splice-region variants), Pthreshold = 4.60 × 10-9 for low-impact variants (synonymous, 3′ and 5′ UTR, and upstream and downstream variants), Pthreshold = 2.30 × 10-9 for other variants within DHSs, and Pthreshold = 7.90 × 10-10 for other variants not within DHSs.
Table 1.
Sequence Variants Conferring Risk of Tooth Agenesis.
| celandic Discovery Sample | Combined Follow-up Samples a | Combined Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype/Marker | Gene Region | EA/OA | EAF, % | OR (95% CI) | P Value b | OR (95% CI) | P Value | OR (95% CI) | P Value b |
| Tooth agenesis | n = 1,944 | n = 301 | |||||||
| rs4498834 | ASCL5/CACNA1S | C/T | 43.0 | 1.41 (1.29 to 1.54) | 2.9 × 10-14 | 1.37 (1.14 to 1.65) | 0.0016 | 1.40 (1.29 to 1.52) | 1.8 × 10-16 |
| rs35822372 | FOXI3 | T/C | 21.0 | 1.46 (1.32 to 1.62) | 3.4 × 10-13 | 1.34 (1.10 to 1.65) | 0.0046 | 1.44 (1.31 to 1.57) | 5.8 × 10-15 |
| chr2:108,896,996c | EDAR | A/G | 0.02 | 23.7 (8.16 to 68.8) | 5.9 × 10-9 | — | — | 23.7 (8.16 to 68.8) | 5.9 × 10-9 |
| rs2034604 d | ARHGAP15 | T/C | 52.7 | 1.41 (1.29 to 1.54) | 2.1 × 10-14 | 1.18 (0.98 to 1.41) | 0.010 | 1.36 (1.26 to 1.47) | 8.3 × 10-16 |
| rs121908120 e | WNT10A | A/T | 2.60 | 3.42 (2.85 to 4.10) | 6.1 × 10-40 | 2.25 (1.39 to 3.65) | 0.0040 | 3.25 (2.74 to 3.85) | 1.7 × 10-40 |
| rs121908119 c,e | WNT10A | A/C | 0.14 | 5.56 (3.00 to 10.3) | 4.9 × 10-8 | — | — | 5.56 (3.00 to 10.3) | 4.9 × 10-8 |
| rs371555610 f,g | ZFHX4 | delCTT/CTT | 85.7 | 1.61 (1.40 to 1.86) | 4.4 × 10-11 | 0.98 (0.73 to 1.32) | 1.00 | 1.47 (1.25 to 1.61) | ns |
| Mandibular second premolars | n = 1,196 | n = 148 | |||||||
| rs917412 | LEF1 | T/C | 9.20 | 1.71 (1.45 to 2.02) | 2.5 × 10-10 | 1.23 (0.87 to 1.72) | 0.24 | 1.60 (1.38 to 1.86) | 5.6 × 10-10 |
| Maxillary second premolars | n = 600 | — | |||||||
| rs758468472 c | NOL11 | G/T | 0.04 | 27.4 (8.47 to 88.7) | 3.3 × 10-8 | — | — | 27.4 (8.47 to 88.7) | 3.3 × 10-8 |
| Maxillary lateral incisors | n = 600 | n = 134 | |||||||
| rs35956082 d | FOXP1 | A/G | 74.8 | 2.08 (1.68 to 2.57) | 1.1 × 10-11 | 1.42 (0.94 to 2.15) | 0.097 | 1.92 (1.59 to 2.32) | 1.0 × 10-11 |
| rs55846652 f | EDA | T/C | 67.2 | 1.92 (1.58 to 2.34) | 6.7 × 10-11 | 1.57 (1.10 to 2.23) | 0.013 | 1.83 (1.54 to 2.17) | 4.8 × 10-12 |
EA, effect allele for which OR is shown; EAF, effect allele frequency (%); n, number of TA cases; ns, not significant; OA, other allele; OR, odds ratio (allelic); TA, tooth agenesis.
The follow-up samples included subjects from four locations as described in Figure 1. Since only direction of effect could be tested in the U.S. sample, we present ORs in the combined analysis not including the U.S. sample and P values from analysis with only direction of effect for all samples combined.
Genome-wide significance after correction for multiple testing is based on type of variant as described in the study by Sveinbjornsson et al. (2016) per the following thresholds: Pthreshold = 2.60 × 10-7 for high-impact variants (stop-gain and stop-loss, frameshift indel, donor and acceptor splice-site, and initiator codon variants), Pthreshold = 5.10 × 10-8 for moderate-impact variants (missense, in-frame indel, and splice region variants), Pthreshold = 4.60 × 10-9 for low-impact variants (synonymous, 3′ and 5′ UTR, and upstream and downstream variants), Pthreshold = 2.30 × 10-9 for other variants within DNase I hypersensitivity sites, and Pthreshold = 7.90 × 10-10 for other variants not within DNase I hypersensitivity sites.
Variant was too rare to be tested in follow-up samples.
To test if there were multiple independent signals within each locus, we conducted association analysis of all variants in each region ±500 kb surrounding each signal peak, conditioned on the effect of the top single-nucleotide polymorphism. Two second signals were identified: rs354700 in 2q22.2 ARHGAP15 (allele frequency [G] = 34.0, OR = 1.24, P = 7.1 × 10-7) and rs9825432 in 3p13 FOXP1 region (allele frequency [G] = 66.0, OR = 1.49, P = 7.5 × 10-7).
Known TA sequence variant.
A different variant was tested in follow-up samples. Variant rs17348154-C (minor allele frequency = 14.0) in linkage disequilibrium with rs371555610,rs529942527 (r2 = 0.87) was included in the follow-up samples. The 2 rs numbers (rs371555610, rs529942527) represent 1 variant; thus, only 1 P value and 1 OR are presented. Variant rs2520378-T in linkage disequilibrium with rs55846652 (r2 = 0.69), was included from the follow-up sample from Portugal (Alves-Ferreira et al. 2014).
The following rs name is reported for this variant: rs371555610 and rs529942517.
The 2 previously reported TA risk variants are rs121908120-A (p.Phe228Ile) and rs121908119-A (p.Cys107Ter) in WNT10A (Table 1). These variants confer high risk of TA and are independent signals in our sample (Padjusted = 7.8 × 10-9 and Padjusted = 2.4 × 10-40 for rs121908119 and rs121908120, respectively, in the Icelandic sample). We found that the rs121908120-A association has a recessive component (the multiplicative model is rejected in favor of the full model, P = 5.2 × 10-6; Appendix Table 3) with estimated odds ratios (ORs) of 3.0 for AT heterozygotes and 51.3 for AA homozygotes.
Of the 9 previously unreported associations with TA, 5 are located within or close to genes implicated in odontogenesis and/or affecting other ectodermal structures. One is a rare missense variant in EDAR (chr2:108896996-A, p.Arg420Trp) that confers a high risk of TA (Table 1). This variant is not found in gnomAD (Lek et al. 2016), is probably specific to the Icelandic population, and was not tested for association in the follow-up samples. The second variant, rs35822372-T, is a common variant located 9.3 kb downstream of FOXI3 and confers modest risk of TA (Table 1). The third variant, rs55846652-T, is on chromosome X downstream of EDA and associates with agenesis of maxillary lateral incisors (Table 1). The ORs for rs55846652-T did not differ significantly between men and women in the Icelandic sample (ORwomen = 2.04, 95% CI = 1.62 to 2.56, P = 9.1 × 10-10; ORmen = 1.66, 95% CI = 1.18 to 2.34, P = 0.0033). The fourth, rs917412-T, is located upstream of LEF1 and associates with agenesis of mandibular second premolars, and the fifth, rs35956082-A, is in the third intron of FOXP1 and associates with agenesis of the maxillary lateral incisors (Table 1).
The other novel TA risk variants map to novel TA risk genes. The first, rs4498834-C is 3.8 kb downstream of ASCL5 and in the first intron of CACNA1S and confers modest risk of TA (Table 1). The second, rs2034604-T, is located in the second intron of ARHGAP15 and confers modest risk of TA (Table 1). The third, rs758468472-T, is a rare splice region variant of the first intron of NOL11 and associates with agenesis of maxillary second premolars (Table 1). Like the EDAR p.Arg420Trp variant, the rs758468472-T has not been reported in gnomAD (Lek et al. 2016), is likely specific to the Icelandic population, and was therefore not tested in follow-up samples. Finally, in our test of 40 known variants conferring risk of the TA-related phenotype OFC, rs5829552-TA associates with agenesis of the maxillary lateral incisors (Table 2, Appendix Table 1). This variant is located 3.8 kb upstream of FAM49A.
Table 2.
Association between a Known Orofacial Cleft Locus and Agenesis of the Maxillary Lateral Incisors.
| Markera: Locus/Gene Region | Position (hg38) | EA/OA | EAF, % | OR (95% CI) | P Valueb |
|---|---|---|---|---|---|
| rs5829552c: 2p24.2 FAM49A | |||||
| Icelandic discovery sample (n = 600) | 16,545,695 | TA/T | 22.5 | 1.55 (1.30 to 1.85) | 1.4 × 10-6 |
| Combined follow-up samples (n = 134)d | 1.13 (0.83 to 1.55) | 0.44 | |||
| All samples combined | 1.44 (1.23 to 1.68) | 4.9 × 10-6 |
EA, effect allele for which OR is shown; EAF, effect allele frequency; n, number of TA cases; OA, other allele; OR, odds ratio (allelic); TA, tooth agenesis.
In total, 40 orofacial cleft variants were tested in our TA data (see Appendix Table 1 for results of all tested variants).
Significant association after correction for multiple testing based on Pthreshold = 0.00031 (correcting for 40 variants and 4 phenotypes).
The following rs names are reported for this variant: rs5829552, rs397784935, and rs869032736. The variant rs7552-G (r2 = 0.97 with rs5829552-TA) was tested in the replication samples.
The follow-up samples for agenesis of maxillary lateral incisors included the Dutch sample and the Portuguese sample.
We also tested variants associating with the sequence and timing of tooth eruption (Pillas et al. 2010; Geller et al. 2011; Fatemifar et al. 2013), including primary and permanent teeth, for association in the Icelandic sample (Fig. 1). Two variants associate with TA in our sample (Appendix Table 2) after Bonferroni correction for multiple testing (Pthreshold = 0.00074). The first variant, rs2281845-T, previously associated with fewer permanent teeth erupted between age 6 and 14 y (Geller et al. 2011), is in linkage disequilibrium (LD) with the ASCL5/CACNA1S rs4498834-C that associates with TA in our study (r2 = 0.90). The second variant, rs11796357-A at EDA, previously associated with slower primary tooth eruption and fewer primary teeth erupted (Pillas et al. 2010; Fatemifar et al. 2013), is in LD with rs55846652-T, which associates with agenesis of maxillary lateral incisors in our study (r2 = 0.77; Appendix Table 2).
One variant located close to ZFHX4 on 8q21.13 (rs371555610, rs529942527-delCTT) was associated with TA in the Icelandic sample (Table 1) but did not survive the threshold for genome-wide significance in the combined analysis including all samples (P = 1.1 × 10-9) and was not regarded as a significantly TA-associated variant.
We tested if the risk variants for TA associate with gene expression (Genotype- Tissue Expression [GTEx] project, V6p; GTEx Consortium 2015). The rs4498834-C associates with lower expression of ASCL5 in several tissues, with the strongest association seen in thyroid tissue (β = −0.87, P = 3.5 × 10-39) and is in strong LD with the strongest GTEx expression signal for ASCL5, rs7516023 (r2 = 0.87). The rs4498834-C allele is also associated with expression of the neighboring genes TMEM9 and IFGN1 (P = 1.7 × 10-5 and 1.4 × 10-14, respectively) but is not in high LD with the strongest expression signals for either gene (r2 = 0.07 and 0.002 with rs1010502 and rs831745, respectively) and does not associate with the expression of CACNA1S. We did not find significant gene expression associations for the other variants.
Gene set enrichment analysis of the TA loci revealed association with odontogenesis of dentine-containing tooth (GO:0042475, P = 1.0 × 10-5), trachea gland development (GO:0061153, P = 1.0 × 10-5), positive regulation of gene expression (GO:0010628, P = 2.0 × 10-5), salivary gland cavitation (GO:0060662, P = 2.0 × 10-5), odontogenesis (GO:0042476, P = 3.0 × 10-5), and positive regulation of NF-kB import into nucleus (GO:0042346, P = 4.0 × 10-5; Appendix Table 4).
Discussion
Here we present a GWAS for TA and agenesis of the 3 most commonly congenitally missing teeth, apart from the third molars. The TA variants previously reported are rare and confer a high risk of oligodontia (Lan et al. 2014). Our sample consists largely of hypodontia cases (90.3%), and we find association with common and rare sequence variants. Of the 9 novel risk variants, 4 associate with TA, and 5 associate with selective TA, including 1 OFC variant. Consistent with previous reports (Polder et al. 2004), the most commonly missing teeth in our data are the mandibular second premolars, followed by the maxillary second premolars and maxillary lateral incisors (Fig. 2).
Of the 9 novel TA variants identified in our study, 5 are located in or close to genes previously implicated in odontogenesis and/or affect other ectodermal structures. These genes are WNT pathway genes EDA, EDAR, FOXI3, FOXP1, and LEF1. We also find an association with 2 known TA variants in WNT10A, including a missense variant (rs121908120-A, p.Phe228Ile) and a nonsense mutation (rs121908119-A, p.Cys107Ter; Adaimy et al. 2007). Mutations in WNT10A were first associated with odonto-onycho-dermal dysplasia (MIM 257980; Adaimy et al. 2007). Gene set enrichment analysis of the associated TA genes revealed, not unexpectedly, association with odontogenesis. In addition, we identified enriched gene sets related to abnormal skin pigmentation and defects in the salivary gland that have been reported for ectodermal dysplasia. The enrichment of the NF-kB signaling genes is important for ectodermal development, including that of teeth.
Although the variants close to EDA (rs55846652-T) and its receptor EDAR (p.Arg420Trp) are novel, these genes have been associated with TA (Arte et al. 2013) and primary tooth development (Pillas et al. 2010; Fatemifar et al. 2013). Analysis of a previously reported missense variant in EDAR (p.Arg420Gln) showed that the reported mutation affects activation of the NF-kB signaling pathway involved in ectodermal differentiation (Kumar et al. 2001). How the missense mutation in EDAR (p.Arg420Trp) associated with TA in this study confers risk is not known, although it does affect the same amino acid and therefore may affect activation of the NF-kB signaling pathway. The EDA locus is known to associate with primary tooth eruption (Pillas et al. 2010; Fatemifar et al. 2013), suggesting an involvement of EDA in tooth development and tooth eruption. The “Tabby” mouse model of EDA includes symptoms of ectodermal dysplasia, including TA or abnormal teeth (Srivastava et al. 1997). FOXI3, of which EDA is an upstream regulator, is expressed in dental tissues during development in animal models (Jussila et al. 2014). A variant in FOXI3 associates with canine ectodermal dysplasia, which includes abnormally shaped or absent teeth from dogs (Drögemüller et al. 2008). Expression of FOXP1 was shown to be greater in human deciduous tooth germs of middle cap stage as compared with lip tissue (Huang et al. 2014). Functional analyses also showed that FOXP1 is involved in the regulation of the WNT signaling pathway (Walker et al. 2015). LEF1, associated with agenesis of mandibular second premolars, was reported to have important functions during odontogenesis. Tooth development arrests at the bud stage in Lef1 homozygous mouse mutants in which anomalies in other ectodermal structures are also present (Van Genderen et al. 1994). LEF1 forms transcriptional units with CTNNB1 in the Wnt signaling pathway (Lan et al. 2014).
Of the 9 novel TA risk variants, 4 are located in or close to the genes ASCL5/CACNA1S, ARHGAP15, NOL11, and FAM49A, which have not been implicated in TA. These genes do not appear to be in previously described gene networks. The function of ASCL5 is not fully understood; however, ASCL5 is a part of the achaete-scute complex-like family, which may be important in tumorigenesis (Wang et al. 2017). For the other gene in this region, CACNA1S, the Cacna1sMDG/MDG mouse model often includes secondary cleft palate as well as micrognathia (Chaudhari 1992). The ASCL5/CACNA1S locus has been associated with timing of eruption of the primary dentition (Geller et al. 2011). Gene expression data support ASCL5, while some literature supports CACNA1S as the TA gene in this region. ARHGAP15 is involved in several signaling mechanisms as a Rac-specific GTPase-activating protein (Radu et al. 2013). A known TA gene, PITX2 affects the Rac-specific GTPase signaling pathway (Wei and Adelstein 2002), suggesting a related function for ARHGAP15. Nol11 is required for craniofacial development in vertebrates (Griffin et al. 2015); however, it has not been associated with TA. Nol11 is of importance in ribosome biogenesis, a process that is disrupted in, for example, Treacher-Collins syndrome (MIM 154500), characterized by orofacial dysmorphies, often including OFC.
The FAM49A locus confers risk of OFC (Leslie et al. 2016; Yu et al. 2017) and agenesis of maxillary lateral incisors in our sample. The function of FAM49A is not well described; however, a recent study showed expression of Fam49a during mouse development in the palatal mesenchymal cells and epithelium cells (Yu et al. 2017). High prevalence of various dental anomalies in children with OFC has been reported (Howe et al. 2015). The maxillary lateral incisors are more commonly missing from subjects with OFC than from controls (Howe et al. 2015; Rullo et al. 2015). A few TA variants (e.g., MSX1) also confer risk of oral clefts (Van Den Boogaard et al. 2000), the formation of which occurs in parallel with that of tooth buds (Hovorakova et al. 2006). These TA genes are expressed at the same time in tissue at neighboring anatomic positions (Hovorakova et al. 2006). Hence, it may not be surprising that dental anomalies, including TA, within and outside the cleft area have been reported to be more frequent in persons with nonsyndromic OFC (Howe et al. 2015; Rullo et al. 2015).
Many genes that affect tooth development have been found through gene expression and functional studies in mice. However, only a few genes have been associated with TA in humans (Lan et al. 2014). Homozygous carriers of high-impact TA mutations are at high risk of syndromes, such as odonto-onycho-dermal dysplasia (Adaimy et al. 2007).
Here we report findings from the first GWAS for TA uncovering 9 novel TA-associated variants—common variants conferring low risk and rare variants conferring modest to high risk. Of the 9 novel variants, 5 are implicated in known TA gene networks, while the remaining 4 provide new information on the genetic background for TA in humans. The reported TA genes affect several stages of odontogenesis, including the emergence of the dental placode as well as the bud, cap, and bell stages and the timing of eruption (Lan et al. 2014). Of the novel TA variants, 1 is a known OFC variant conferring risk of agenesis of the maxillary lateral incisors located in the cleft. This result adds support to the growing evidence for an overlapping genetic etiology of TA and OFC. Some of the variants conferring high risk of TA also associate with development of other associated anomalies (Adaimy et al. 2007; Howe et al. 2015). Thus, novel variants conferring risk of TA can uncover associations with other phenotypes and help in defining new syndromes. Oral rehabilitation of patients with TA may require a multidisciplinary approach. Better understanding of the etiology and clinical features of TA is important for planning and treatment.
Author Contributions
L. Jonsson, H. Stefansson, K. Stefansson, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; T.E. Magnusson, contributed to conception, design, data acquisition, and interpretation, drafted and critically revised the manuscript; A. Thordarson, T. Jonsson, contributed to conception, design, data acquisition, and interpretation, critically revised the manuscript; F. Geller, B. Feenstra, M. Melbye, E.A. Nohr, S. Vucic, B. Dhamo, F. Rivadeneira, E.M. Ongkosuwito, E.B. Wolvius, E.J. Leslie, M.L. Marazita, B.J. Howe, L.M. Moreno Uribe, I. Alonso, M. Santos, T. Pinho, contributed to data acquisition and analysis, critically revised the manuscript; R. Jonsson, G. Audolfsson, contributed to data acquisition, critically revised the manuscript; L. Gudmundsson, M.S. Nawaz, S. Olafsson, O. Gustafsson, A. Ingason, U. Unnsteinsdottir, G. Bjornsdottir, G.B. Walters, M. Zervas, A. Oddsson, D.F. Gudbjartsson, S. Steinberg, contributed to data analysis and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
The authors are grateful to the individuals who participated in this study. We also thank the staff and researchers at the deCODE genetics core facilities.
Footnotes
A supplemental appendix to this article is available online.
L.J. received support from the Swedish Society of Medicine and the Swedish Brain Foundation. The financial support for the Icelandic study from the National Institutes of Health (NIH) (National Institute of Dental and Craniofacial Research; R0IDE022905) is acknowledged. The Danish National Birth Cohort was established with a significant grant from the Danish National Research Foundation. Additional support was obtained from the Danish Regional Committees, the Pharmacy Foundation, the Egmont Foundation, the March of Dimes Birth Defects Foundation, the Health Foundation, and other minor grants. The Danish National Birth Cohort biobank is a part of the Danish National Biobank resource, which is supported by the Novo Nordisk Foundation. B.F. was supported by an Oak Foundation fellowship and by a grant from the Novo Nordisk Foundation (12955). Additional support was provided by grants from the NIH, including R00-DE025060 [E.J.L.], R01-DE016148 [M.L.M.], X01-HG007485 [M.L.M.]. L.G., M.S.N., S.O., O.G., A.I., U.U., G.B., G.B.W., M.Z., A.O., D.F.G., S.S, H.S., and K.S are employees of deCODE genetics/Amgen.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iD: L. Jonsson  https://orcid.org/0000-0002-3175-103X
https://orcid.org/0000-0002-3175-103X
S. Vucic  https://orcid.org/0000-0003-3352-3900
https://orcid.org/0000-0003-3352-3900
B. Dhamo  https://orcid.org/0000-0002-5537-067X
https://orcid.org/0000-0002-5537-067X
E. M. Ongkosuwito  https://orcid.org/0000-0001-9250-027X
https://orcid.org/0000-0001-9250-027X
E. J. Leslie  https://orcid.org/0000-0002-5735-1712
https://orcid.org/0000-0002-5735-1712
B. J. Howe  https://orcid.org/0000-0002-6737-9735
https://orcid.org/0000-0002-6737-9735
G. B. Walters  https://orcid.org/0000-0002-5415-6487
https://orcid.org/0000-0002-5415-6487
References
- Adaimy L, Chouery E, Megarbane H, Mroueh S, Delague V, Nicolas E, Belguith H, De Mazancourt P, Megarbane A. 2007. Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia. Am J Hum Genet. 81(4):821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Ferreira M, Pinho T, Sousa A, Sequeiros J, Lemos C, Alonso I. 2014. Identification of genetic risk factors for maxillary lateral incisor agenesis. J Dent Res. 93(5):452–458. [DOI] [PubMed] [Google Scholar]
- Arte S, Parmanen S, Pirinen S, Alaluusua S, Nieminen P. 2013. Candidate gene analysis of tooth agenesis identifies novel mutations in six genes and suggests significant role for WNT and EDA signaling and allele combinations. PLoS One. 8(8):e73705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N, Daly MJ, Price AL, Neale BM. 2015. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter K, Worthington S. 2015. Morphologic and demographic predictors of third molar agenesis: a systematic review and meta-analysis. J Dent Res. 94(7):886–894. [DOI] [PubMed] [Google Scholar]
- Chaudhari N. 1992. A single nucleotide deletion in the skeletal muscle-specific calcium channel transcript of muscular dysgenesis (mdg) mice. J Biol Chem. 267(36):25636–25639. [PubMed] [Google Scholar]
- Drögemüller C, Karlsson EK, Hytonen MK, Perloski M, Dolf G, Sainio K, Lohi H, Lindblad-Toh K, Leeb T. 2008. A mutation in hairless dogs implicates FOXI3 in ectodermal development. Science. 321(5895):1462. [DOI] [PubMed] [Google Scholar]
- Fatemifar G, Hoggart CJ, Paternoster L, Kemp JP, Prokopenko I, Horikoshi M, Wright VJ, Tobias JH, Richmond S, Zhurov AI, et al. 2013. Genome-wide association study of primary tooth eruption identifies pleiotropic loci associated with height and craniofacial distances. Hum Mol Genet. 22(18):3807–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller F, Feenstra B, Zhang H, Shaffer JR, Hansen T, Esserlind AL, Boyd HA, Nohr EA, Timpson NJ, Fatemifar G, et al. 2011. Genome-wide association study identifies four loci associated with eruption of permanent teeth. PLoS Genet. 7(9):e1002275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JN, Sondalle SB, Del Viso F, Baserga SJ, Khokha MK. 2015. The ribosome biogenesis factor Nol11 is required for optimal rDNA transcription and craniofacial development in Xenopus. PLoS Genet. 11(3): e1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. 2015. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans; Science. 348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, Besenbacher S, Magnusson G, Halldorsson BV, Hjartarson E, et al. 2015. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 47(5):435–444. [DOI] [PubMed] [Google Scholar]
- Haga S, Nakaoka H, Yamaguchi T, Yamamoto K, Kim YI, Samoto H, Ohno T, Katayama K, Ishida H, Park SB, et al. 2013. A genome-wide association study of third molar agenesis in Japanese and Korean populations. J Hum Genet. 58(12):799–803. [DOI] [PubMed] [Google Scholar]
- Hovorakova M, Lesot H, Peterkova R, Peterka M. 2006. Origin of the deciduous upper lateral incisor and its clinical aspects. J Dent Res. 85(2):167–171. [DOI] [PubMed] [Google Scholar]
- Howe BJ, Cooper ME, Vieira AR, Weinberg SM, Resick JM, Nidey NL, Wehby GL, Marazita ML, Moreno Uribe LM. 2015. Spectrum of dental phenotypes in nonsyndromic orofacial clefting. J Dent Res. 94(7):905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Hu X, Lin C, Chen S, Huang F, Zhang Y. 2014. Genome-wide analysis of gene expression in human embryonic tooth germ. J Mol Histol. 45(6):609–617. [DOI] [PubMed] [Google Scholar]
- Johannsdottir B, Wisth PJ, Magnusson TE. 1997. Prevalence of malocclusion in 6-year-old Icelandic children. Acta Odontol Scand. 55(6):398–402. [DOI] [PubMed] [Google Scholar]
- Jussila M, Crespo Yanez X, Thesleff I. 2014. Initiation of teeth from the dental lamina in the ferret. Differentiation. 87(1–2):32–43. [DOI] [PubMed] [Google Scholar]
- Kong A, Masson G, Frigge ML, Gylfason A, Zusmanovich P, Thorleifsson G, Olason PI, Ingason A, Steinberg S, Rafnar T, et al. 2008. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet. 40(9):1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Eby MT, Sinha S, Jasmin A, Chaudhary PM. 2001. The ectodermal dysplasia receptor activates the nuclear factor-kappaB, JNK, and cell death pathways and binds to ectodysplasin A. J Biol Chem. 276(4):2668–2677. [DOI] [PubMed] [Google Scholar]
- Lan Y, Jia S, Jiang R. 2014. Molecular patterning of the mammalian dentition. Semin Cell Dev Biol. 25–26:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, Mchenry T, et al. 2016. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet. 25(13):2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnússon TE. 1977. Prevalence of hypodontia and malformations of permanent teeth in Iceland. Community Dent Oral Epidemiol. 5(4):173–178. [DOI] [PubMed] [Google Scholar]
- Magnússon TE. 1979. Prevalence of congenitally missing and malformations of permanent teeth in Iceland. Hardjaxl. 2(16):53–64. [Google Scholar]
- Magnússon TE. 1984. Hypodontia, hyperodontia, and double formation of primary teeth in Iceland: an epidemiological study. Acta Odontol Scand. 42(3):137–139. [DOI] [PubMed] [Google Scholar]
- Ockeloen CW, Khandelwal KD, Dreesen K, Ludwig KU, Sullivan R, van Rooij IA, Thonissen M, Swinnen S, Phan M, Conte F, et al. 2016. Novel mutations in LRP6 highlight the role of WNT signaling in tooth agenesis. Genet Med. 18(11):1158–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillas D, Hoggart CJ, Evans DM, O’Reilly PF, Sipila K, Lahdesmaki R, Millwood IY, Kaakinen M, Netuveli G, Blane D, et al. 2010. Genome-wide association study reveals multiple loci associated with primary tooth development during infancy. PLoS Genet. 6(2):e1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polder BJ, Van’t Hof MA, Van Der Linden FP, Kuijpers-Jagtman AM. 2004. A meta-analysis of the prevalence of dental agenesis of permanent teeth. Community Dent Oral Epidemiol. 32(3):217–226. [DOI] [PubMed] [Google Scholar]
- Radu M, Rawat SJ, Beeser A, Iliuk A, Tao WA, Chernoff J. 2013. ArhGAP15, a Rac-specific GTPase-activating protein, plays a dual role in inhibiting small GTPase signaling. J Biol Chem. 288(29):21117–21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rullo R, Festa VM, Rullo R, Addabbo F, Chiodini P, Vitale M, Perillo L. 2015. Prevalence of dental anomalies in children with cleft lip and unilateral and bilateral cleft lip and palate. Eur J Paediatr Dent. 16(3):229–232. [PubMed] [Google Scholar]
- Srivastava AK, Pispa J, Hartung AJ, Du Y, Ezer S, Jenks T, Shimada T, Pekkanen M, Mikkola ML, Ko MS, et al. 1997. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc Natl Acad Sci U S A. 94(24):13069–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinthorsdottir V, Thorleifsson G, Sulem P, Helgason H, Grarup N, Sigurdsson A, Helgadottir HT, Johannsdottir H, Magnusson OT, Gudjonsson SA, et al. 2014. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 46(3):294–298. [DOI] [PubMed] [Google Scholar]
- Sveinbjornsson G, Albrechtsen A, Zink F, Gudjonsson SA, Oddson A, Masson G, Holm H, Kong A, Thorsteinsdottir U, Sulem P, et al. 2016. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat Genet. 48(3):314–317. [DOI] [PubMed] [Google Scholar]
- Van Den Boogaard MJ, Dorland M, Beemer FA, Van Amstel HK. 2000. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 24(4):342–343. Erratum in Nat Genet. 2000. 25(1):125. [DOI] [PubMed] [Google Scholar]
- Van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. 1994. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 8(22):2691–2703. [DOI] [PubMed] [Google Scholar]
- Walker MP, Stopford CM, Cederlund M, Fang F, Jahn C, Rabinowitz AD, Goldfarb D, Graham DM, Yan F, Deal AM, et al. 2015. FOXP1 potentiates Wnt/β-catenin signaling in diffuse large B cell lymphoma. Sci Signal. 8(362):ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Shahi P, Huang JT, Phan NN, Sun Z, Lin YC, Lai MD, Werb Z. 2017. Systematic analysis of the achaete-scute complex-like gene signature in clinical cancer patients. Mol Clin Oncol. 6(1):7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Adelstein RS. 2002. Pitx2a expression alters actin-myosin cytoskeleton and migration of HeLa cells through Rho GTPase signaling. Mol Biol Cell. 13(2):683–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. 2010. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Zuo X, He M, Gao J, Fu Y, Qin C, Meng L, Wang W, Song Y, Cheng Y, et al. 2017. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat Commun. 8:14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




