Abstract
The last few decades have witnessed an increasing interest in studying the human microbiome and its role in health and disease. The focus of those studies was mainly the characterization of changes in the composition of the microbial communities under different conditions. As a result of those studies, we now know that imbalance in the composition of the microbiome, also referred to as microbial dysbiosis, is directly linked to developing certain conditions. Dysbiosis of the oral microbiome is a prime example of how this imbalance leads to disease in the case of periodontal disease. However, there is considerable overlap in the phylogenetic profiles of microbial communities associated with active and inactive lesions, suggesting that the difference in periodontal status of those sites may not be explained solely by differences in the subgingival microbial composition. These findings suggest that differences in functional activities may be the essential elements that define the dysbiotic process. Researchers have recently begun to study gene expression of the oral microbiome in situ with the goal of identifying changes in functional activities that could explain the transition from health to disease. These initial results suggest that, rather than a specific composition, a better understanding of oral dysbiosis can be obtained from the study of functional activities of the microbial community. In this review, we give a summary of these initial studies, which have opened a new door to our understanding of the dynamics of the oral community during the dysbiotic process in the oral cavity.
Keywords: dysbiosis, periodontitis, caries, metagenome, microbiota, transcriptome
Introduction
The term microbiome was coined by Joshua Lederberg to signify the “ecological community of commensal, symbiotic, and pathogenic microorganisms that share our body space” (Lederberg and McCray 2001). Since then, most of the effort to understand the role of the microbiome in health and disease has been placed in characterizing changes in community composition under different conditions.
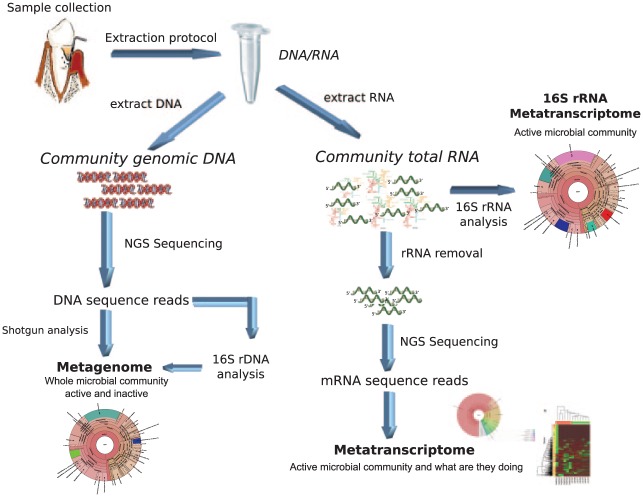
The assessment of community composition gives just a partial picture of the molecular processes by which dysbiosis occurs. A more comprehensive approach is needed to study the host-pathogen interactions in vivo that reveal the links between microbial community crosstalk with the host and microbial activity in the oral microbiome. The metatranscriptome reveals the taxonomic composition and active functions of a complex microbial community in contrast with the metagenome, which shows only the microbial composition of the community (Fig. 1).
Figure 1.
Laboratory work flow for metatranscriptome analysis. A general overview of the different kinds of analyses that can be performed to study the oral microbiome metatranscriptome.
The National Institutes of Health recently established the Integrative Human Microbiome Project (iHMP) as the second phase of the original Human Microbiome Project. iHMP will focus on the study of host-microbiome interactions by analyzing microbiome and host activities via different “omics” (iHMP Research Network Consortium 2014).
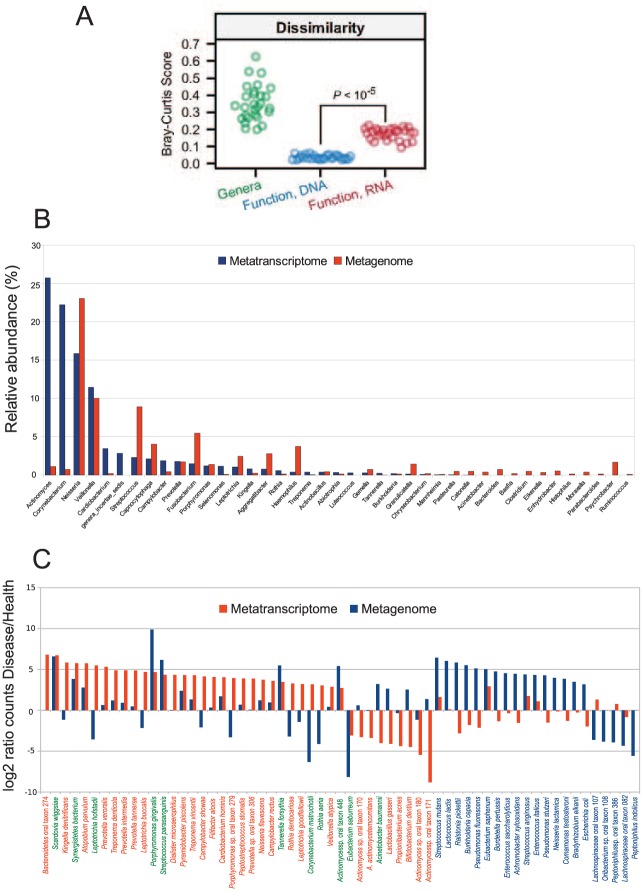
Community composition does not necessarily reflect the active members of the community (Fig. 2), thereby supporting the need for functional analysis of the microbiome. Previous studies showed that metagenomic (microbial composition) and metatranscriptomic (global gene expression) profiles from the same sample differ in phylogenetic composition (Benítez-Páez et al. 2014; Duran-Pinedo et al. 2014; Franzosa et al. 2014; Yost et al. 2015). One direct consequence of this is that members of the community that are present at low numbers in the metagenome may still be critical in carrying on metabolic activities essential to the community. For example, at low colonization numbers, Porphyromonas gingivalis modulates the behavior of the oral community, showing its role as a “keystone pathogen” in disease (Hajishengallis et al. 2012).
Figure 2.
Comparison of metagenome and metatranscriptome in the oral and gut microbiome. (A) Metagenome and metatranscriptome analysis of the gut microbiome. Comparison of between-sample diversity for taxonomic and functional profiles with the Bray-Curtis metric (adapted with permissions from Franzosa et al. 2014). (B) Microbiota composition in the human oral biofilm from a 24-h dental plaque sample. Relative abundance of bacterial genera from metagenomic data differs from that obtained from metatranscriptomic data (adapted from Benítez-Páez et al. 2014). (C) Rank distribution of statistically significant relative increase in number of hits for the metagenome and metatranscriptome results in samples from healthy patients and patients with severe periodontitis. In green, species with statistical differences in both metagenome and metatranscriptome. In blue, species with statistical differences in metagenomic counts. In red, species with statistical differences in metatranscriptome counts (adapted from Duran-Pinedo et al. 2014).
Metagenomic and metatranscriptomic analyses treat the microbial community as a whole, overcoming problems associated with polymerase chain reaction amplification. Moreover, they are not limited to analysis of a predetermined assembly of bacteria, such as that used in checkerboard hybridization (Socransky et al. 1994), microarray-based techniques (Colombo et al. 2009), or a specific gene such as 16S rRNA (Kumar et al. 2003). However, these techniques need complete genomes to be performed. Fortunately, the oral community is one of the best characterized in the human microbiome, which significantly facilitates metatranscriptome analysis.
Metatranscriptomic analysis characterizes gene expression profiles of the entire microbial community based on the set of transcripts being synthesized under diverse environmental conditions. Given the complexity of the oral microbiome, it would not have been possible to perform this type of analysis without the arrival of next-generation sequencing technologies. For this reason, metatranscriptomic analyses have just recently begun to be used in studying the human microbiome (Booijink et al. 2010; Duran-Pinedo et al. 2014; Jorth et al. 2014).
One of the challenges faced when measuring bacterial gene expression in situ is the limited amount of biomass present in oral samples. To overcome this problem, researchers have followed 2 strategies. The first involves pooling samples with similar clinical characteristics to reach the amount of RNA needed for analysis (Jorth et al. 2014), including an amplification step where the final amplified RNA is a direct representation of the relative abundance of mRNA in the original sample (Duran-Pinedo et al. 2014; Yost et al. 2015). The second involves linear RNA amplification methods that have been successfully applied in deep sequencing, rendering coverage that is indistinguishable from that of nonamplified libraries (Hoeijmakers et al. 2011).
Although metatranscriptomics is emerging as a powerful technology for the functional characterization of microbiomes, there is still no consensus on what is the best approach to perform these kinds of studies.
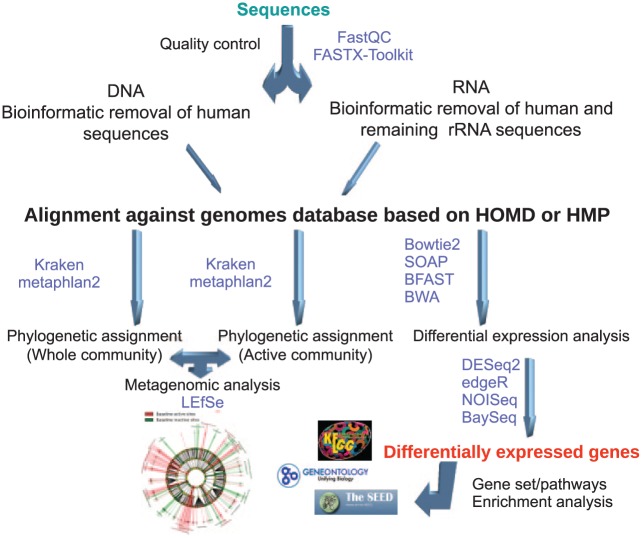
Bioinformatic analyses of the metatranscriptome involve managing large data sets through a series of steps, called a pipeline or a work flow. The first step is to remove low-quality sequences, followed by an alignment step against the genomes of interest, then a differential expression analysis to identify changes in metabolic activities, and, finally, a phylogenetic analysis of the transcripts to identify active members of the community (Fig. 3). One of the crucial elements when optimizing metatranscriptomic experimental designs is sample size estimation with a particular statistical power. In estimating sample size, we should consider sequencing depth, power wanted, single- or paired-end reads, the coefficient of variation (CV), effect size, and budget constraints (Hart et al. 2013; Ching et al. 2014). Increasing sample size and sequencing depth and using paired-end reads significantly enhance the statistical power (Ching et al. 2014; Liu et al. 2014). However, increasing sample size is more potent than sequencing depth to increase power, especially when the sequencing depth reaches 20 million reads (Ching et al. 2014). In most cases, a large number of replicates is not possible. Nonetheless, the statistical power of deep sequencing provides a robust output that can provide highly informative statistically significant data. This in vivo–derived information can then be followed by studies of clinical samples with quantitative polymerase chain reaction to further validate the findings. Interestingly, in model organisms, it seems that there is no need for a large number of replicates to reach the desired statistical power. For instance, in a study by Ching et al. (2014) comparing 5 differential expression analysis packages and evaluating their performance by power and other metrics, the authors found that when the sequencing depth reached 20 million paired-end reads, all packages reached a power >0.8 with only 10 replicates per condition. Hart et al. (2013) performed a similar analysis and found that the biological CV of the data sets was a significant driver in calculating sample size, but with low CVs, the number of samples per group was also small. With a CV of 0.4, they needed only 10 samples per condition and 10 million sequences to reach a power of 0.8. However, with a CV of 1.2, they needed 40 samples per condition to achieve the same statistical power.
Figure 3.
Overview of a general bioinformatic work flow for metatranscriptome analysis. Common steps in the bioinformatic analysis of microbial transcriptomes: quality control of the sequences obtained by next-generation sequencing, alignment against the genomes of interest, phylogenetic assignment of the transcripts and metagenome, differential expression analysis and gene set or pathway enrichment analysis based on the obtained differentially expressed genes. In purple are software packages widely used for metatranscriptome analysis. HMP, Human Microbiome Project; HOMD, Human Oral Microbiome Database; LEfSe, linear discriminant analysis effect size.
Microbial Metatranscriptome in Health
To understand the dynamics of gene expression of any microbiome, there is a need to characterize what the transcriptome looks like under healthy conditions. Thus far, there is a significant void in the metatranscriptomic studies under healthy conditions. Only 1 study focused on the metatranscriptome during biofilm formation and after meal ingestion (Benítez-Páez et al. 2014). The authors characterized the gene activity repertoire of the microbial community during development in supragingival dental plaque samples and determined the taxonomic identity of the active microbiome before and after ingestion of a meal. Changes in bacterial activity during plaque development and after meal ingestion were person-specific. In some cases, >80% of active bacteria corresponded to only 3 genera (Actinomyces, Corynebacterium, and Rothia), whereas other individuals did not show any dominant genera in the active microbial community.
The authors (Benítez-Páez et al. 2014) minimized potential errors in taxonomic assignment in the active community by assigning reads at the family and genus taxonomic levels only, selecting matches against a 16S rRNA database of 100% sequence identity and keeping only hypervariable informative regions of the 16S rRNA gene. The predominant genera of active members of the community were Streptococcus (12% to 19%) and Actino-myces (3% to 12%). Actinomyces showed higher frequencies in early plaque samples, in agreement with its known role as an early colonizer. Other frequent active genera were the Actinobacteria Rothia, Angustibacter, and Kineococcus; the Proteobacteria Neisseria, Kingella, and Alysiella; the Firmicutes Gemella, Paeni-bacillus, and Veillonella; and, finally, Cap-nocytophaga and Fusobacterium. Kineo-coccus, Alysiella, and Paenibacillus are genera commonly found in environmental samples. Nonetheless, members of those genera have been identified in oral samples, although generally at low numbers (Kuhn 1981; Kraal et al. 2014; Tetz et al. 2015; Tønjum 2015; Zheng et al. 2015; Tetz et al. 2016).
In early samples, genes involved in the metabolism of carbohydrates, energy, amino acids, cofactor/vitamins, and xenobiotic degradation were predominantly upregulated. In the late stages, the researchers (Benítez-Páez et al. 2014) observed upregulation of genes involved in quorum sensing response—in particular, genes identified as belonging to type II secretion systems. Since type II secretion systems promote secretion of folded periplasmic proteins that typically play a role in survival, this finding indicates that the community is adapting as it develops. Finally, regarding changes in the active community after a meal, some individuals demonstrated microbial communities that were very resilient to changes in gene expression profile, while others had more apparent differences in the proportions of active bacteria; however, no specific pattern was common to all individuals. Actinomyces was the only genus found in a percentage of >10% in all samples.
Among the significant findings of this work (Benítez-Páez et al. 2014) is that the metatranscriptomic profiles during biofilm formation and after meal ingestion were person-specific. Some individuals showed virtually no changes in the active bacterial population after food ingestion, which suggests that their microbiota is not affected by food ingestion, thereby potentially reducing the risk of acidic pH and promoting dental health. The authors also showed that expression of genes linked to translation machinery is higher in early biofilm stages, whereas more specialized genes are expressed in the mature biofilm. Among them, genes involved in competence, quorum sensing, mutacin production, and DNA uptake were overexpressed in late biofilm, indicating a more complex level of interactions in mature biofilm than at earlier stages of biofilm formation.
Microbial Metatranscriptome in Caries
For a long time, acidogenic species of the genus Streptococcus (e.g., S. mutans) have been considered the causative agent of dental caries. Indeed, numerous 16S rRNA studies showed that certain acidogenic and aciduric species, such as S. mutans and Lactobacillus spp., are highly correlated with active caries (McLean 2014). Many other species are likely to be relevant, as evidenced by the diverse microbial populations present in caries. They include members of the genera Actinomyces, Fusobacterium, Porphyromonas, Selenomonas, Bifidobacteria, Scardovia, and Haemophi-lus (Tanner et al. 2011), supporting the idea that, as in periodontitis, a complex consortium formed by multiple microorganisms act collectively to initiate and expand the disease (Simón-Soro and Mira 2014).
In a metatranscriptomic study that focused on the active bacterial communities in caries lesions by pyrosequencing of 16S rRNA, Simón-Soro et al. (2014) found that active caries lesions contained between 70 and 400 metabolically active species of bacteria. They discovered that noncavitated (“white spot” lesions), open dentin, or enamel-dentin caries presented different active communities. While members of the genera Streptococcus and Veillonella were highly active in all 3 types of caries, Lactobacillus spp. were highly active only in the enamel-dentin caries sites. Enamel-dentin caries, also known as hidden caries, is a phenomenon that leads to formation of highly mineralized, strengthened enamel surfaces, under which the loss of mineral may progress gradually and the carious lesion might extend into dentin without a clinically visible crack at the enamel surface. Interestingly, among members of the genus Streptococcus, S. sanguinis was the most active species in all 3 kinds of lesions, while S. mutans was active only in noncavitated and open dentin lesions, indicating an association of S. mutans with open lesions rather than unexposed, hidden caries, in agreement with the idea that oxygen availability is a crucial factor in the interspecies competition between S. mutans and S. sanguinis in the oral biofilm (Kreth et al. 2008).
When looking at community-wide expression profiles of dental plaque samples from dental caries, Peterson et al. (2014) found that transcripts of the community were produced by a limited number of species. Again, S. sanguinis was the most active member of the community with 16% of the transcripts, followed by Streptococcus mitis (10%), Veillonella parvula (9%), Capnocytophaga sp. (9%), Streptococcus oralis (8%), Streptococcus spp. (7%), Gemella haemolysans (5%), Strepto-coccus gordonii (4%), and Neisseria sp. (3%). Despite interpersonal variation at the level of specific genes, distinct patterns of expression emerged in terms of functional categories. A large number of activities were associated with oxidative stress, with high expression of proteins that metabolize superoxides and peroxides (superoxide dismutase, peroxiredoxins, and ferroxidase; Peterson et al. 2013).
May et al. (2016) recently performed an analysis of already-existing metatranscriptomic libraries to identify functional differences between health and caries. They determined that deregulated metabolic subnetworks were indeed significantly different. The disease-associated parts of the caries subnetwork included 9 KEGG Orthology groups (KOs) from the pathways of the phosphotransferase system and fructose and mannose metabolism. Among them are sugar phosphotransferases involved in the beta-glucoside metabolism, which are critical elements in the process of colonization (Loo et al. 2003; Kiliç et al. 2004). Another set of disease-associated pathways was a pathway that converts sorbitol to fructose 6-phosphate. Unlike many other oral species, S. mutans can metabolize sorbitol as a carbon source (Janda and Kuramitsu 1978; Rölla et al. 1981). In contrast, only 1 KO was downregulated in disease in the same pathway. This KO converts fructose to fructose 6-phosphate, suggesting that while in health fructose may be the primary carbon source, in dental caries sorbitol might be used as an additional source of carbon (May et al. 2016). Interestingly, these results seem to contradict previous laboratory studies where sorbitol inhibited sugar metabolism of S. mutans (Takahashi-Abbe et al. 2001). These contradictory results may reflect the different physiologies of S. mutans, when living as part of a complex microbiome as opposed to being grown isolated under laboratory conditions.
Microbial Metatranscriptome of Subgingival Plaque in Chronic Periodontitis
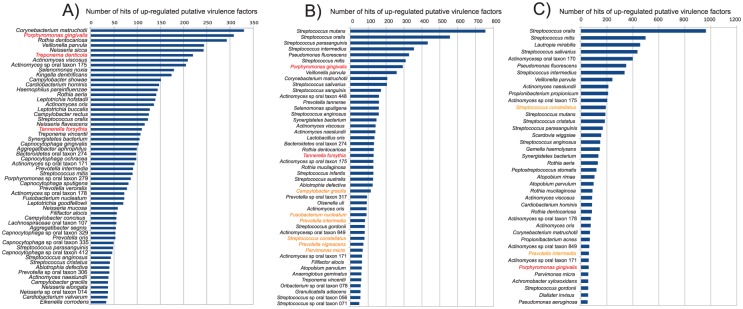
As in the case of dental caries, only recently has metatranscriptomic analysis been applied to study the functional activities associated with periodontal disease (Duran-Pinedo et al. 2014; Jorth et al. 2014; Duran-Pinedo et al. 2015; Szafrański et al. 2015; Yost et al. 2015). However, with even a limited number of studies, we have started to obtain some general conclusions associated with the functional profiles of the microbiome in health and disease. The first conclusion is that despite high interpatient variability in microbiome composition, the disease-associated communities displayed conserved functional activities, while the health-associated communities exhibited a more diverse range of those functional activities (Duran-Pinedo et al. 2014; Jorth et al. 2014). As a corollary of this observation, although the activities associated with periodontitis are conserved, the species linked to those activities are different among patients (Duran-Pinedo et al. 2014; Jorth et al. 2014; Yost et al. 2015). These results suggest that rather than focusing on the activity of a specific pathogen or pathogens, we should consider changes in the behavior of the community as a whole if we want to explain those observations. One recent hypothesis that tries to explain the role of some prominent members of the community in disease is that specific low-abundance microbial pathogens could act as keystone pathogens and modulate the composition and behavior of the oral community (Darveau 2009; Hajishengallis et al. 2012). The periodontopathogen P. gingivalis has been proposed as a keystone pathogen in periodontal disease via host modulation (Hajishengallis et al. 2012). It does so through the expression of the virulence factor gingipain, which cleaves the complement component 5 (C5), generating high levels of C5a and acting as a highly inflammatory peptide (Hajishengallis et al. 2012). Jorth et al. (2014) observed that Fusobacterium nucleatum was the only species degrading lysine to butyrate in all patients, and they proposed that production of butyrate may contribute to the stability of the community by preventing the proliferation of human gingival fibroblasts. However, Szafrański et al. (2015) observed that expression of genes for the synthesis of butyrate occurred under all conditions, not only during disease. Moreover, their study determined that 4 additional species with alternative pathways contributed to butyrate synthesis. Therefore, they concluded that the proinflammatory role of F. nucleatum might not be related to butyrate biosynthesis. Another exciting aspect of the ecology of periodontitis is that it results in changes not only in community composition but also in functional dysbiosis. Jorth et al. (2014) showed that the proportions of F. nucleatum did not significantly change between healthy and periodontitis samples, but its metabolism was utterly different under those 2 conditions. Unexpectedly, in a separate study, Duran-Pinedo et al. (2014) observed that the majority of virulence factors upregulated in subjects with periodontitis came from organisms not considered major periodontal pathogens (Fig. 4), in agreement with the idea of the “pathogenic microbial community” (Berezow and Darveau 2011) or “the community as pathogen” (Relman 2012), which postulates that the integrated actions of the components of the microbial community would result in disease.
Figure 4.
Ranked species by the number of upregulated putative virulence factors in the metatranscriptome. Putative virulence factors were identified by alignment of the protein sequences from the different genomes against the Virulence Factors of Pathogenic Bacteria Database. The numbers in the graph are the absolute number of hits for the different species of the upregulated putative virulence factors identified. In red are the members of the “red complex.” In orange are members of the “orange complex.” (A) Comparison of health vs. severe periodontitis. (B) Comparison of baseline vs. progressing sites in periodontitis progression (adapted from Yost et al. 2015). (C) Comparison of baseline nonprogressing vs. baseline progressing in periodontitis progression. (Adapted from Duran-Pinedo et al. 2014 and Yost et al. 2015.)
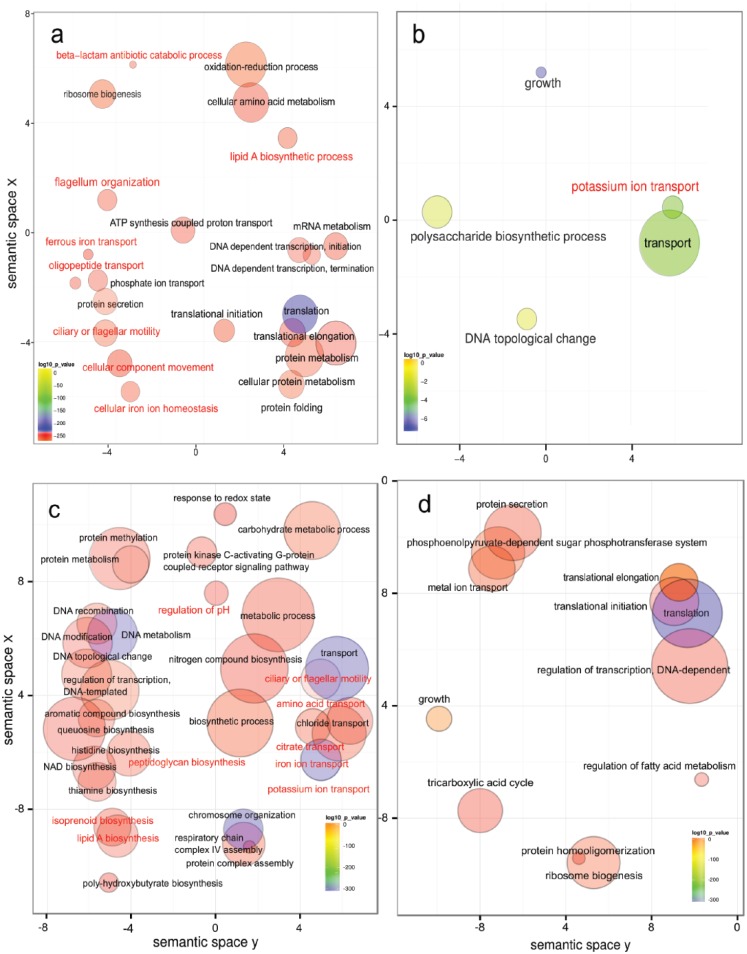
In terms of functional changes during disease, the overall picture of metabolic activities showed that Gene Ontology (GO) biological processes related to flagellar motility, peptide transport, iron acquisition, and beta-lactam degradation were overrepresented in disease, as was biosynthesis of the lipid A component of endotoxins from Gram-negative bacteria (Fig. 5a). GO biological processes underrepresented in disease included potassium transport and polysaccharide biosynthesis (Fig. 5b). In agreement with those observations, Szafrański et al. (2015) found that flagellin and transcripts assigned to chemotaxis, iron acquisition, antimicrobial resistance, secretion, and iron transport were significantly more abundant and could be directly linked to pathogenesis. Interestingly, a metatranscriptome analysis of caries samples also found transcripts encoding for resistance to antibiotics, including transcripts encoding the acriflavine resistance complex (Peterson et al. 2014).
Figure 5.
Gene Ontology (GO) enrichment analysis in severe periodontitis and periodontitis progression. Enriched terms obtained with goseq were summarized and visualized as a scatter plot via REVIGO. Summarized GO terms related to biological processes in (A) severe periodontitis (adapted from Duran-Pinedo et al. 2014) and (B) health (adapted from Duran-Pinedo et al. 2014). GO enrichment analysis comparison of baselines from progressing and nonprogressing sites: Summarized GO terms related to biological processes in baselines of (C) progressing and (D) nonprogressing sites (adapted from Yost et al. 2015). In red are activities that have been associated with pathogenesis in periodontitis. Circle size is proportional to the frequency of the GO term, whereas color indicates the log10 P value (red higher, blue lower).
One of the advantages of using metatranscriptome analysis is that we can also zoom into specific members of the microbiome to assess their activities. The “red complex,” which appears later in biofilm development, comprises 3 species that are considered the primary periodontal pathogens: P. gingivalis, Treponema denticola, and Tannerella forsythia (Socransky et al. 1998). Despite a good understanding of the association between red complex species and periodontitis, we have only limited information on the in situ activity of these organisms. In a study of microbial metatranscriptome on chronic periodontitis, the 3 members of the red complex showed high expression of metalloproteases, peptidases, and genes associated with motility and invasion. Additionally, proteins involved in iron metabolism represented a significant fraction of upregulated putative virulence factors in these periodontal pathogens (Duran-Pinedo et al. 2014).
Microbial Metatranscriptome in Periodontitis Progression
One still unanswered question regarding periodontal disease is why in some cases teeth with clinical symptoms of periodontitis progress to tooth loss and in other cases remain stable despite the lack of treatment (Goodson et al. 1982). Most effort regarding this question has focused on identifying markers that would distinguish between active and nonactive sites. While there have been many studies associating markers that might distinguish between active and inactive sites—including bacterial profiles (Byrne et al. 2009; Charalampakis et al. 2013), genetic markers (Heitz-Mayfield 2005; Ricci et al. 2011), protein activities (Eley and Cox 1996), cytokines (Khalaf et al. 2014), and clinical markers (Charalampakis et al. 2013)—none of these associations shed light on the mechanisms of progression. Periodontal disease progresses through periods of acute exacerbation (activity), followed by periods of remission (Socransky et al. 1984; Haffajee and Socransky 1986). Goodson et al. (1982) found that among untreated patients whose attachment levels were measured every month for 1 y, 5.7% of the sites became significantly deeper, while 11.5% became markedly shallower during that period. Among sites with increased pocket depth, approximately half showed spontaneous recovery to their original depth, and half exhibited cyclic deepening, followed by spontaneous recovery to their initial depth (Goodson et al. 1982). Patterns of “exacerbation” and “remission” were also described by others (Jeffcoat and Reddy 1991; Clerehugh et al. 1995). It has been postulated that changes in the composition of subgingival biofilms could explain these periods of disease activity. In fact, a few studies found differences in the levels of subgingival species when comparing progressing and nonprogressing sites through cultural approaches (Dzink et al. 1988) and molecular approaches (Tanner et al. 2007; Teles et al. 2008; Byrne et al. 2009). These studies also demonstrated a considerable overlap in the composition of the microbial communities associated with progressing and nonprogressing lesions, suggesting that the difference in the periodontal status of the sites could not be explained solely by the reported differences in the subgingival microbial composition.
Yost et al. (2015) compared metatranscriptomic profiles of subgingival plaque from active and inactive sites in patients with chronic periodontitis, trying to identify functional signatures that could explain the initial stages of dysbiosis. In this study, the microbiome of the progressing sites was already dysbiotic at the initiation of the study. When the baselines of active versus nonactive sites were compared, the differences in the composition of the active communities were more significant than they were when active communities were compared at baseline and after progression (Yost et al. 2015). In the baseline of progressing sites, GO enrichment analysis showed an overrepresentation of terms related to cell motility, transport (iron, potassium, chloride, citrate, and amino acids transport), lipid A and peptidoglycan biosynthesis, and protein kinase C-activating G protein–coupled receptor signaling pathway, as well as synthesis of aromatic compounds (Fig. 5c). However, in the baseline samples from nonprogressing sites, there was an overrepresentation of GO terms related to tricarboxylic acid cycle, metal ion transport, phosphoenolpyruvate-dependent sugar phosphotransferase system, and protein secretion (Fig. 5d). Interestingly, histidine biosynthesis was overrepresented at the initial stages of the progressing sites (Fig. 5c), while Jorth et al. (2014) found that histidine catabolism was upregulated in the disease sites of their study.
One of the key altered metabolic activities in periodontitis and its progression seems to be potassium ion transport (Duran-Pinedo et al. 2014; Yost et al. 2015). Its importance was confirmed by demonstrating that potassium levels increased the virulence of the oral community as a whole, while altering the immune response of gingival epithelium, increasing the production of TNF-α, and reducing the expression of IL-6 and the antimicrobial peptide human β-defensin 3 (Yost et al. 2017).
Conclusions and Future Directions
We present recent studies that focused on the metatranscriptome of the oral community. These studies are the starting point for the identification of critical environmental signals that modify the behavior of the community from commensal to dysbiotic. These studies give insight into how microbial communities behave—information that was unimaginable only a few years ago. In the case of caries, sugar metabolism was identified as a central element that distinguishes health and disease, but new observations were made, such as the potential use of sorbitol as an additional source of carbon by S. mutans in dental caries. In the case of periodontal disease, the fact that levels of extracellular potassium are an important signal in disease is just one among other dysbiotic signals identified by metatranscriptome analysis.
Longitudinal studies are needed to identify the real markers of the initial stages of the disease. There are no studies yet on the transcriptome of the fungal fraction of the oral microbiome, which could be an important element influencing the dynamics of the whole microbial community. Thus far, the study of oral metatranscriptomics has been restricted to only 2 diseases: caries and periodontal disease. Naturally, other oral diseases will lengthen the list of conditions that should be studied. Moreover, the metatranscriptome does not necessarily represent the final metabolic products generated by the microbial community. Improvements in proteomics and metabolomics should allow us to determine the origin of proteins and metabolites produced by the microbiome under different conditions. Finally, integrating the expression profiles of the host with the patterns from the microbiome should be one of the directions to take. The immune response plays a crucial role in the outcome of the progression of the disease. Thus, understanding how the host interacts with the microbiome is essential to have a complete picture of health and disease.
Author Contributions
J. Frias-Lopez, J. Solbiati, contributed to conception and design, drafted and critically revised the manuscript. Both authors gave final approval and agree to be accountable for all aspects of the work.
Acknowledgments
The authors thank Dr. Mary-Ellen Davey and Susan Yost for reviewing the manuscript and their useful suggestions and comments.
Footnotes
This work was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health (award 2R01DE021553).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iD: J. Frias-Lopez  https://orcid.org/0000-0002-2097-3171
https://orcid.org/0000-0002-2097-3171
References
- Benítez-Páez A, Belda-Ferre P, Simón-Soro A, Mira A. 2014. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genomics. 15:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezow AB, Darveau RP. 2011. Microbial shift and periodontitis. Periodontol 2000. 55(1):36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booijink CCGM, Boekhorst J, Zoetendal EG, Smidt H, Kleerebezem M, de Vos WM. 2010. Metatranscriptome analysis of the human fecal microbiota reveals subject-specific expression profiles, with genes encoding proteins involved in carbohydrate metabolism being dominantly expressed. Appl Environ Microbiol. 76(16):5533–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SJ, Dashper SG, Darby IB, Adams GG, Hoffmann B, Reynolds EC. 2009. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol Immunol. 24(6):469–477. [DOI] [PubMed] [Google Scholar]
- Charalampakis G, Dahlén G, Carlén A, Leonhardt A. 2013. Bacterial markers vs. clinical markers to predict progression of chronic periodontitis: a 2-yr prospective observational study. Eur J Oral Sci. 121(5):394–402. [DOI] [PubMed] [Google Scholar]
- Ching T, Huang S, Garmire LX. 2014. Power analysis and sample size estimation for RNA-Seq differential expression. RNA. 20(11):1684–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerehugh V, Worthington HV, Lennon MA, Chandler R. 1995. Site progression of loss of attachment over 5 years in 14- to 19-year-old adolescents. J Clin Periodontol. 22(1):15–21. [DOI] [PubMed] [Google Scholar]
- Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, et al. 2009. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 80(9):1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP. 2009. The oral microbial consortium’s interaction with the periodontal innate defense system. DNA Cell Biol. 28(8):389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Pinedo AE, Chen T, Teles R, Starr JR, Wang X, Krishnan K, Frias-Lopez J. 2014. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 8(8):1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Pinedo AE, Yost S, Frias-Lopez J. 2015. Small RNA transcriptome of the oral microbiome during periodontitis progression. Appl Environ Microbiol. 81(19):6688–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzink JL, Socransky SS, Haffajee AD. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 15(5):316–323. [DOI] [PubMed] [Google Scholar]
- Eley BM, Cox SW. 1996. A 2-year longitudinal study of elastase in human gingival crevicular fluid and periodontal attachment loss. J Clin Periodontol. 23(7):681–692. [DOI] [PubMed] [Google Scholar]
- Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, et al. 2014. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci U S A. 111(22):E2329–E2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson JM, Tanner AC, Haffajee AD, Sornberger GC, Socransky SS. 1982. Patterns of progression and regression of advanced destructive periodontal disease. J Clin Periodontol. 9(6):472–481. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS. 1986. Attachment level changes in destructive periodontal diseases. J Clin Periodontol. 13(5):461–475. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat Rev Microbiol. 10(10):717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher JP. 2013. Calculating sample size estimates for RNA sequencing data. J Comput Biol. 20(12):970–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz-Mayfield LJ. 2005. Disease progression: identification of high-risk groups and individuals for periodontitis. J Clin Periodontol. 32 Suppl 6:196–209. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers WAM, Bártfai R, Françoijs KJ, Stunnenberg HG. 2011. Linear amplification for deep sequencing. Nat Protoc. 6(7):1026–1036. [DOI] [PubMed] [Google Scholar]
- iHMP Research Network Consortium. 2014. The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe. 16(3):276–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda WM, Kuramitsu HK. 1978. Production of extracellular and cell-associated glucosyltransferase activity by Streptococcus mutans during growth on various carbon sources. Infect Immun. 19(1):116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffcoat MK, Reddy MS. 1991. Progression of probing attachment loss in adult periodontitis. J Periodontol. 62(3):185–189. [DOI] [PubMed] [Google Scholar]
- Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. 2014. Metatranscriptomics of the human oral microbiome during health and disease. mBio. 5(2):e01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf H, Lönn J, Bengtsson T. 2014. Cytokines and chemokines are differentially expressed in patients with periodontitis: possible role for TGF-β1 as a marker for disease progression. Cytokine. 67(1):29–35. [DOI] [PubMed] [Google Scholar]
- Kiliç AO, Tao L, Zhang Y, Lei Y, Khammanivong A, Herzberg MC. 2004. Involvement of Streptococcus gordonii beta-glucoside metabolism systems in adhesion, biofilm formation, and in vivo gene expression. J Bacteriol. 186(13):4246–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraal L, Abubucker S, Kota K, Fischbach MA, Mitreva M. 2014. The prevalence of species and strains in the human microbiome: a resource for experimental efforts. PLoS One. 9(5):e97279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 190(13):4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn DA. 1981. The genera Simonsiella and Alysiella. In: The prokaryotes. 2nd ed. Berlin (Germany): Springer; p. 390–399. [Google Scholar]
- Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. 2003. New bacterial species associated with chronic periodontitis. J Dent Res. 82(5):338–344. [DOI] [PubMed] [Google Scholar]
- Lederberg J, McCray A. 2001. ’Ome sweet ’omics—a genealogical treasury of words. Scientist. 15(7):8. [Google Scholar]
- Liu Y, Zhou J, White KP. 2014. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics. 30(3):301–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo CY, Mitrakul K, Voss IB, Hughes CV, Ganeshkumar N. 2003. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J Bacteriol. 185(21):6241–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A, Brandt BW, El-Kebir M, Klau GW, Zaura E, Crielaard W, Heringa J, Abeln S. 2016. metaModules identifies key functional subnetworks in microbiome-related disease. Bioinformatics. 32(11):1678–1685. [DOI] [PubMed] [Google Scholar]
- McLean JS. 2014. Advancements toward a systems level understanding of the human oral microbiome. Front Cell Infect Microbiol. 4:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SN, Meissner T, Su AI, Snesrud E, Ong AC, Schork NJ, Bretz WA. 2014. Functional expression of dental plaque microbiota. Front Cell Infect Microbiol. 4:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SN, Snesrud E, Liu J, Ong AC, Kilian M, Schork NJ, Bretz W. 2013. The dental plaque microbiome in health and disease. PLoS One. 8(3):e58487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relman DA. 2012. The human microbiome: ecosystem resilience and health. Nutr Rev. 70 Suppl 1:S2–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci M, Garoia F, Tabarroni C, Marchisio O, Barone A, Genovesi A, Covani U. 2011. Association between genetic risk score and periodontitis onset and progression: a pilot study. Arch Oral Biol. 56(12):1499–1505. [DOI] [PubMed] [Google Scholar]
- Rölla G, Oppermann RV, Waaler SM, Assev S. 1981. Effect of aqueous solutions of sorbitol-xylitol on plaque metabolism and on growth of Streptococcus mutans. Scand J Dent Res. 89(3):247–250. [DOI] [PubMed] [Google Scholar]
- Simón-Soro A, Guillen-Navarro M, Mira A. 2014. Metatranscriptomics reveals overall active bacterial composition in caries lesions. J Oral Microbiol. 6:25443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Soro A, Mira A. 2014. Solving the etiology of dental caries. Trends Microbiol. 23(2):76–82. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol. 25(2):134–144. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Goodson JM, Lindhe J. 1984. New concepts of destructive periodontal disease. J Clin Periodontol. 11(1):21–32. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. 1994. “Checkerboard” DNA-DNA hybridization. BioTechniques. 17(4):788–792. [PubMed] [Google Scholar]
- Szafrański SP, Deng ZL, Tomasch J, Jarek M, Bhuju S, Meisinger C, Kühnisch J, Sztajer H, Wagner-Döbler I. 2015. Functional biomarkers for chronic periodontitis and insights into the roles of Prevotella nigrescens and Fusobacterium nucleatum: a metatranscriptome analysis. NPJ Biofilms Microbiomes. 1:15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi-Abbe S, Abbe K, Takahashi N, Tamazawa Y, Yamada T. 2001. Inhibitory effect of sorbitol on sugar metabolism of Streptococcus mutans in vitro and on acid production in dental plaque in vivo. Oral Microbiol Immunol. 16(2):94–99. [DOI] [PubMed] [Google Scholar]
- Tanner AC, Kent R, Jr, Kanasi E, Lu SC, Paster BJ, Sonis ST, Murray LA, Van Dyke TE. 2007. Clinical characteristics and microbiota of progressing slight chronic periodontitis in adults. J Clin Periodontol. 34(11):917–930. [DOI] [PubMed] [Google Scholar]
- Tanner AC, Kent RL, Jr, Holgerson PL, Hughes CV, Loo CY, Kanasi E, Chalmers NI, Johansson I. 2011. Microbiota of severe early childhood caries before and after therapy. J Dent Res. 90(11):1298–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles RP, Patel M, Socransky SS, Haffajee AD. 2008. Disease progression in periodontally healthy and maintenance subjects. J Periodontol. 79(5):784–794. [DOI] [PubMed] [Google Scholar]
- Tetz G, Tetz V, Vecherkovskaya M. 2015. Complete genome sequence of Paenibacillus sp. strain VT 400, isolated from the saliva of a child with acute lymphoblastic leukemia. Genome Announc. 3(4):e00894-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetz G, Tetz V, Vecherkovskaya M. 2016. Genomic characterization and assessment of the virulence and antibiotic resistance of the novel species Paenibacillus sp. strain VT-400, a potentially pathogenic bacterium in the oral cavity of patients with hematological malignancies. Gut Pathog. 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tønjum T. 2015. Alysiella. In: Bergey’s manual of systematics of archaea and bacteria. New York (NY): John Wiley & Sons, Ltd; p. 1–18. [Google Scholar]
- Yost S, Duran-Pinedo AE, Krishnan K, Frias-Lopez J. 2017. Potassium is a key signal in host-microbiome dysbiosis in periodontitis. PLoS Pathog. 13(6):e1006457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost S, Duran-Pinedo AE, Teles R, Krishnan K, Frias-Lopez J. 2015. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 7(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhang Z, Liu C, Qiao Y, Zhou D, Qu J, An H, Xiong M, Zhu Z, Zhao X. 2015. Metagenomic sequencing reveals altered metabolic pathways in the oral microbiota of sailors during a long sea voyage. Sci Rep. 5:9131. [DOI] [PMC free article] [PubMed] [Google Scholar]