Abstract
Background
Polytene chromosome banding patterns have long been used by Drosophila evolutionists to infer degree of relatedness among taxa. Recently, nucleotide sequences have preempted this traditional method. We place the classical Drosophila evolutionary biology tools of polytene chromosome inversion analysis in a phylogenetic context and assess their utility in comparison to nucleotide sequences.
Results
A simultaneous analysis framework was used to examine the congruence of the chromosomal inversion data with more recent DNA sequence data in four Drosophila species groups – the melanogaster, virilis, repleta, and picture wing. Inversions and nucleotides were highly congruent with one another based on incongruence length difference and partitioned Bremer support values. Inversion phylogenies were less resolved because of fewer numbers of characters. Partitioned Bremer supports, corrected for the number of characters in each matrix, were higher for inversion matrices.
Conclusions
Polytene chromosome data are highly congruent with DNA sequence data and, when placed in a simultaneous analysis framework, are shown to be more information rich than nucleotide data.
Background
Species in the family Drosophilidae have been premier research subjects in evolutionary biology since T. H. Morgan first used Drosophila melanogaster as a genetic tool in the early part of the 20th Century. Polytene chromosome phylogenies have become commonplace in the examination of this family of flies. The chromosomal analyses have been used in two ways in evolutionary studies. The first use is as genetic markers [1,2] in which the chromosomal inversions are considered alleles and are utilized to examine gene flow and other population genetic parameters. The second use of polytene chromosomes in evolutionary studies is as tracers of phylogeny [3-7]. This approach has resulted in important and detailed chromosomal phylogenies for several groups of flies in the genus Drosophila. Lists of species for which polytene chromosome maps have been produced are available [8,9], and over 300 species of Drosophila have been examined at this level. In particular, extensive chromosomal phylogenies for flies in the picture wing[4],melanogaster[5], virilis[6], and repleta[7] species groups exist. Cladistic analyses of the chromosomal data for these groups exist for the picture wing[10] and melanogaster species groups [11]. More recently, large amounts of DNA sequence information have been collected for these and many other species groups, yet no detailed analysis of the overall utility of chromosomal inversion data or of their congruence with DNA sequence data has been carried out.
The main objective of the present study is to place the chromosomal inversion data into a phylogenetic framework. We examine the congruence of DNA sequence characters and chromosomal inversion data in four different species groups in the genus Drosophila (picture wing, repleta, melanogaster and virilis) to assess the relative contribution to phylogenetic hypotheses that the two different sources of character information make.
Results and Discussion
Chromosomal inversions
Table 1 lists the sources of the data used in this study. Table 2 shows the results of phylogenetic analysis of the inversion and DNA sequence character partitions separately and in combination. Tree topologies of the chromosomal and DNA sequence character partitions were very similar (Figure 1). The major difference upon direct examination of the molecular and inversion cladograms in Figure 1 was the lack of resolution for inversions compared to DNA sequences. The similarity in topology of these trees was borne out by the ILD analyses (picture wing = 0.007 [NS], melanogaster = 0.000 [NS], repleta = 0.022 [NS] and virilis = 0.046 [NS], where none of the ILD measures was greater than 0.05. These results indicated that there was less than a 5% increase in length of the simultaneous analysis cladogram due to combining the DNA and inversion partitions. Figure 2 shows the simultaneous analysis of the four data sets with Bremer support indices and bootstrap values on nodes.
Table 1.
Data Sources for the five data matrices used in this study.
| References | ||||
| Species Group | DNA | No. of bp | Molecular | Chromosome |
| picture wing | Yp-1 | 849 | [22] | [3,10] |
| melanogaster | Adh | 771 | [23,24,25,26,27,28] | [5] |
| Sry | 1498 | [29] | ||
| nullo | 566 | [29] | ||
| 28S | 342 | [30,31] | ||
| 5S | 187 | [32,33] | ||
| Amy | 1568 | [34] | ||
| virilis | G6pd | 1063 | [34,35] | [6] |
| Adh | 988 | [37] | ||
| repleta | COII | 442 | [38] | [7] |
| 16S | 521 | [38] | ||
| hb | 526 | [38] | ||
| ND1 | 129 | [38] | ||
Gene Abbreviations:Yp-1 = Yolk protein, Adh = Alcohol dehydrogenase, Sry = Serendipity, nullo = nullo, 28S = 28S rDNA, 5S = 5S rDNA, Amy = α-amylase, G6pd = glycerol-6-phosphate dehydrogenase, COII = cytochrome oxidase subunit II, 16S = 16S rDNA, hb = hunchback, ND1 = NADH dehydrogenase subunit 1, bp = base pairs, Chromo = chromosome.
Table 2.
Tree statistics for the four analyses.
| picture wing group | repleta group | ||||||
| Total | Molecular | Inversions | Total | Molecular | Inversions | ||
| CI | 0.62 | 0.59 | 0.95 | CI | 0.31 | 0.30 | 0.89 |
| RI | 0.85 | 0.81 | 0.99 | RI | 0.52 | 0.51 | 0.93 |
| PI | 177 | 141 | 36 | PI | 501 | 461 | 40 |
| ST | 542 | 443 | 95 | ST | 2774 | 2647 | 80 |
| # T | 36 | 60 | 2 | # T | 1 | 57 | 4 |
| nch | 942 | 849 | 93 | nch | 1737 | 1618 | 119 |
| ntx | 35 | 35 | 35 | ntx | 54 | 54 | 54 |
| melanogaster group | virilis group | ||||||
| Total | Molecular | Inversions | Total | Molecular | Inversions | ||
| CI | 0.80 | 0.77 | 1.00 | CI | 0.83 | 0.84 | 0.84 |
| RI | 0.81 | 0.77 | 1.00 | RI | 0.90 | 0.90 | 0.94 |
| PI | 258 | 221 | 37 | PI | 112 | 91 | 21 |
| ST | 790 | 729 | 61 | ST | 260 | 209 | 49 |
| # T | 1 | 1 | 1 | # T | 6 | 24 | 33 |
| nch | 4991 | 4930 | 61 | nch | 2071 | 2024 | 47 |
| ntx | 8 | 8 | 8 | ntx | 11 | 11 | 11 |
Abbreviations: Total = combined data set, Molecular = only DNA sequence data, Inversions = only inversion data, CI = consistency index, RI = retention index, PI = number of parsimony informative characters in the data partition, ST = length of shortest tree, #T = number of most parsimonious trees obtained in the analysis, nch = number of characters in the data set, ntx = number of taxa.
Figure 1.
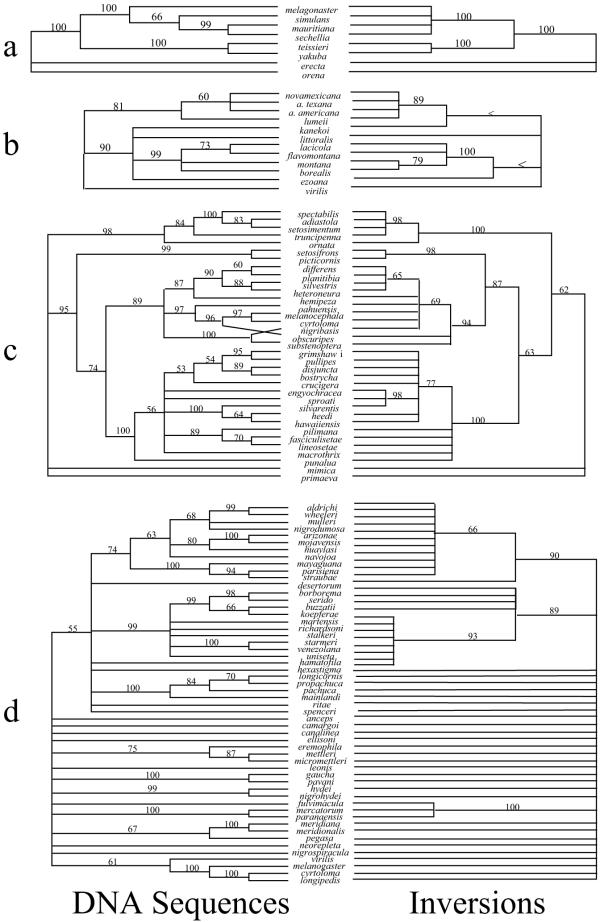
Cladograms as described in the text for the four species groups used in this study – a) the melanogaster species group; b) the virilis species group; c) the picture wing species group and d) the repleta species group. Numbers above branches indicate the bootstrap values for the nodes to the right of the number. The trees on the left are for DNA sequences only and the trees on the right are for chromosomal inversion data. < indicates a bootstrap value less than 50%.
Figure 2.
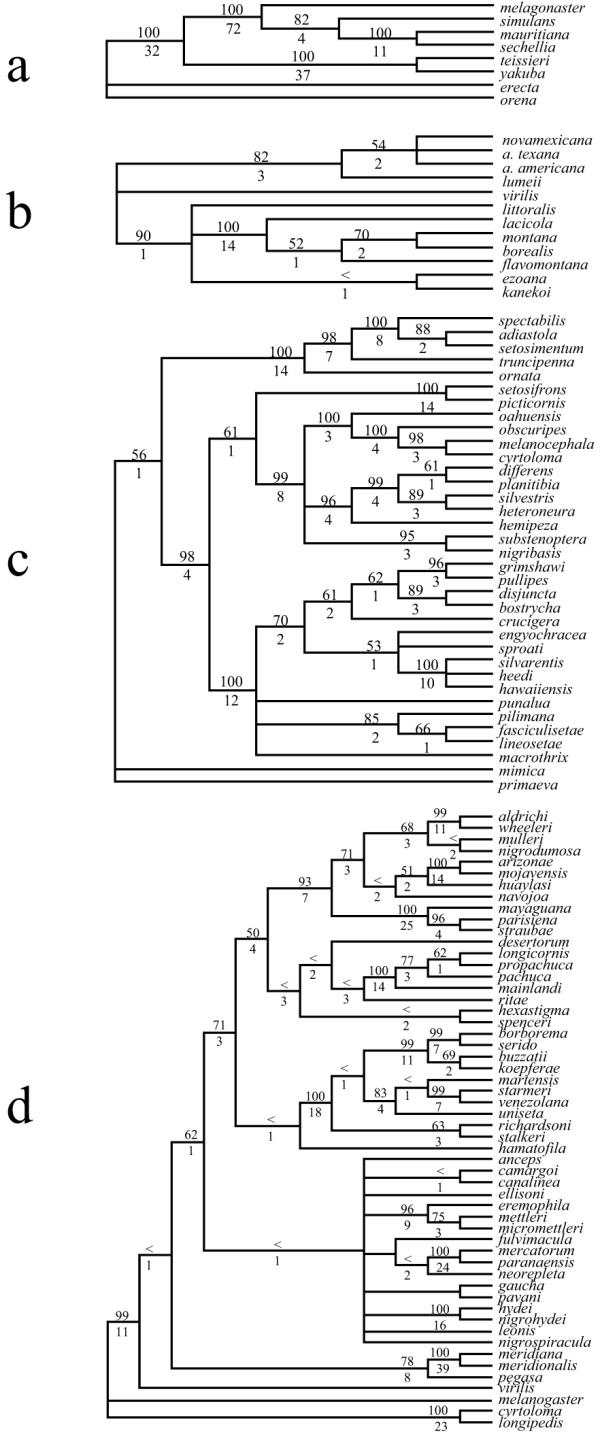
Cladograms showing the total evidence hypotheses for combined analyses of DNA sequences and chromosomes. a) the melanogaster species group; b) the virilis species group; c) the picture wing species group and d) the repleta species group. The numbers above nodes indicates the bootstrap value and the number below indicates the Bremer index. < indicates a bootstrap value less than 50%.
In general, the agreement of chromosomal inversion topology with DNA sequence topology was extremely good. The number of nodes in the trees that disagreed (as assessed by a negative partitioned Bremer support value) for both data partitions was extremely low (Table 3). In all species groups examined here there was at least one node that was negative for partitioned Bremer support of the molecular partition, indicating that the molecular data are in conflict with the simultaneous analysis hypothesis for these nodes. Both the melanogaster and the virilis group chromosomal data showed complete agreement with the simultaneous analysis tree, while three nodes in the repleta group tree and one node in the picture wing tree had negative partitioned Bremer supports for the inversion partition. While there were many nodes that had zero partitioned Bremer support for the inversion partition (Table 3), the total support rendered by the inversion data to the simultaneous analysis tree was relatively large ("total BS" column in Table 3). To standardize the total partitioned Bremer support contribution of each partition to the simultaneous analysis tree we divided the total Bremer support by the number of phylogenetically informative characters in that character partition and by the minimum steps of the simultaneous analysis tree ("corrected total BS" column in Table 3). Using both kinds of correction (Table 3) resulted in inversions showing higher corrected values relative to the DNA sequence characters in seven out of eight comparisons. This result is most likely due to the considerably higher consistency of the chromosomal inversion data. Only when the total Bremer support values of both the inversion data and the molecular data were standardized by the minimum steps of the simultaneous analysis for the repleta group did we find that both data partitions contributed equally to the simultaneous analysis tree.
Table 3.
Results of partitioning Bremer support to molecular and inversion character sources.
| corrected total BS | |||||||
| total | negative | zero | % zero or | # PI | min. # | ||
| Dataset | nodes | nodes | nodes | negative | total BS | chars. | steps |
| picture wing | 32 | 1/1 | 21/6 | 69/22 | 34/91 | 0.98/0.63 | 0.37/0.29 |
| melanogaster | 5 | 0/1 | 2/0 | 40/20 | 47/109 | 1.5/0.5 | 0.77/0.17 |
| repleta | 44 | 3/1 | 25/3 | 64/9 | 30.5/269.5 | 1.6/0.6 | 0.37/0.37 |
| virilis | 9 | 0/1 | 4/4 | 44/56 | 11/12 | 0.5/0.2 | 0.16/0.06 |
Each entry in the table is listed as values calculated for inversions / molecular data. The number of nodes (# of nodes), the umber of negative nodes (negative nodes) and the # number of zero nodes (zero nodes) are given in the first three columns. The percent of zero or negative nodes refers to nodes that have either zero or negative Bremer support values. Total BS is the Bremer support summed over all nodes in the cladogram. This value has been corrected by either dividing by the number of parsimony informative characters or the minimum steps in each partition (see text).
We also computed the consistency indices of the inversion and DNA partitions for the four data matrices using the simultaneous analysis trees for each as a constraint. The consistency indices for the inversion partitions were considerably higher than those for the DNA partitions. Previous surveys of the patterns and distributions of consistency indices with taxon number indicate that in general the CI decreases with the number of taxa in a study [12]. Figure 3 shows a plot of the CI versus number of taxa for both the inversion and the DNA partitions for all four data matrices. This figure demonstrates that inversions were highly consistent in all four cladograms and that they did not show the characteristic lowering of consistency index with increasing number of taxa that most character data show. However, the DNA partitions did show the depression of consistency index value with number of taxa. Together these analyses suggest that there is a high degree of agreement among classical chromosomal data and more recent DNA sequence data and that inversion data are extremely consistent with simultaneous analysis hypotheses of relationship in these groups of Drosophila.
Figure 3.
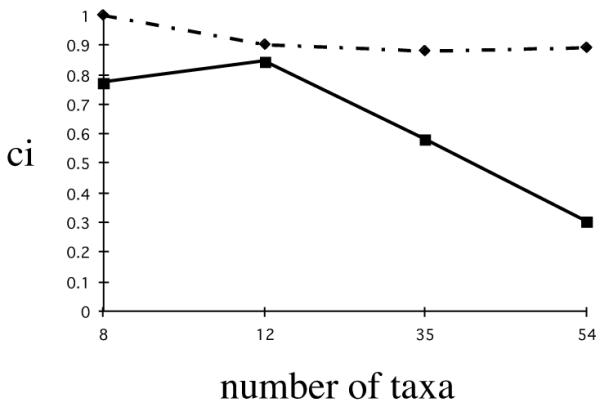
Plot of the consistency index of chromosomal inversion partition (dotted line) and the DNA sequence partition (solid line) when forced to fit the parsimony tree versus the number of taxa in the data set. The melanogaster species group analysis has eight taxa, the virilis species group analysis has 12 taxa, the picture wing species group has 35 taxa and the repleta species group has 54 taxa.
Conclusion
Classical Drosophila studies have used chromosomal inversions to understand phylogeny and speciation. More recent DNA sequence information can be combined with these classical data to make inferences about phylogeny and species divergence. The approach we have taken here is to combine the chromosomal inversion data with DNA sequence data to examine some of the classical notions of Drosophila evolution. Our results suggest that there is a great deal of congruence among DNA sequence data and chromosomal inversion data. Although this result is reassuring, there are still some areas of the phylogenies of the four species groups examined here that are not congruent. These areas are indicative of poor phylogenetic signal from one or both of the kinds of data – DNA and chromosomal inversion data. Chromosomal inversion data are much more information rich as assessed in the present study, and this is probably due to the higher consistency of these characters. This higher consistency stems from that fact that, while nucleotide characters have only four possible states to change to and from, chromosome breaks can occur at many different locations. Therefore, it is much simpler to change from an A to a T in two unrelated lineages than it is to have the exact same chromosome break in those same lineages.
Materials and Methods
Data matrices
Four data matrices were constructed using DNA sequences and chromosomal inversion data from the literature (Table 1). The four species groups for which we have obtained inversion data represent the four major species groups for which such data exist. Chromosome inversion information for other smaller groups such as the antopocerus species group (Hawaiian Drosophila) is published [13], however DNA sequence data are not yet available for these groups.
Phylogenetic Trees
Phylogenetic trees were generated using PAUP 4.0b7 [14]. In order to assess the relative contribution of chromosomal inversion and DNA sequence data we placed our analysis in a simultaneous analysis framework [15-17]. Bootstrap values were computed using PAUP 4.0b7. Decay indices were computed using AUTODECAY [18] and using the methods described in Baker et al. [19]. Significance of Incongruence Length Differences (ILDs [20,21]) were calculated using the Partition Homogeneity Option in PAUP 4.0b7 [14]. Partitioned Bremer supports for the inversion partition and the DNA partition were calculated using the approaches outlined in Baker et al. [19].
Phylogenetic Measures
Here we include definitions of several phylogenetic measures used in this paper. The consistency index (CI; [39]) is used to determine how much homoplasy (i.e., how many times a character evolves on a tree) a given character displays. The CI of a character is the mimimum number of steps for that character on a given tree divided by the total number of steps reconstructed for that character on the same tree. Those characters which are highly consistent, or without homoplasy, would have a consistency index of 1.
The decay index or Bremer support value is used to measure support at a node of interest in a phylogenetic tree. To obtain the decay index, the treelength of a tree constrained not to contain a node of interest is substracted from the unconstrained (most parsimonious) treelength. Higher numbers generally indicate greater support at a node. Partitioned Bremer support (PBS; [19]) measures the amount of support provided by each individual partition to the DI for every node in the combined analysis phylogenies. PBS is an extension of the decay index in that it shows the contribution of each partition to the decay index of every node on the combined analysis tree. To obtain the PBS value for a given node on a combined tree, the length of the partition on the unconstrained combined tree is subtracted from the length of a partition on a tree constrained to not contain the node of interest. If the partition supports a relationship represented by a node in the combined tree, then the PBS value will be positive. If, on the other hand, a partition supports an alternative relationship, the PBS value will be negative. The magnitude of PBS values indicate the level of support for, or disagreement with, a node. The sum of all partition lengths for any given node will always equal the decay index for that node on the total evidence tree.
Contributor Information
Patrick M O'Grady, Email: durando@amnh.org.
Richard H Baker, Email: richardbaker@hotmail.com.
Celeste M Durando, Email: ogardy@amnh.org.
William J Etges, Email: wetges@uark.edu.
Robert DeSalle, Email: desalle@amnh.org.
References
- Powell JR. Progress and Prospects in Evolutionary Biology: The Drosophila Model Oxford, Oxford University Press, 1997.
- Krimbas CB, Powell JR. Drosophila Inversion Polymorphism. Boca Raton, CRC Press, 1992.
- Carson HL. Chromosomal tracers of evolution. Science. 1970;168:1414–1418. doi: 10.1126/science.168.3938.1414. [DOI] [PubMed] [Google Scholar]
- Carson HL. Tracing ancestry with chromosomal sequences. TREE. 1987;2:203–207. doi: 10.1016/0169-5347(87)90021-8. [DOI] [PubMed] [Google Scholar]
- Lemunier F, David JR, Tsacas L. The melanogaster species group. The Genetics and Biology of Drosophila Vol 3e (Edited by Ashburner, M, Carson, H. L., Thompson, J. N) New York, Academic Press, 1986. pp. 148–256.
- Throckmorton LH. The virilis species group. The Genetics and Biology of Drosophila Vol 3b (Edited by Ashburner, M, Carson, H. L., Thompson, J. N) New York, Academic Press, 1982. pp. 227–296.
- Wasserman M. Cytological evolution of the Drosophila repleta group. In: Krimbas, C. B., Powell, J. R., editor. Drosophila Inversion Polymorphism. Boca Raton, CRC Press; 1992. pp. 455–552. [Google Scholar]
- Ashburner M. Drosophila : A Laboratory Handbook. New York, Cold Spring Harbor Press, 1989.
- Ashburner M, Carson HL. A checklist of maps of polytene chromosomes of drosophilids. Dros Inf Serv. 1983;59:148–151. [Google Scholar]
- Kaneshiro KY, Gillespie R, Carson HL. Chromosomes and male genitalia of Hawaiian Drosophila : tools for interpreting phylogeny and geography. Hawaiian Biogeography: Evolution on a Hotspot Archipelago (Edited by, Wagner, W.L., Funk, V. A.) Washington, DC, Smithsonian Institution Press, 1995. pp. 52–72.
- Lemunier F, Ashburner M. Relationships in the melanogaster species subgroup of the genus Drosophila (Sophophora). II. Phylogenetic relationships of six species based upon polytene banding sequences. Proc R Soc Lond B. 1976;193:257–294. doi: 10.1098/rspb.1976.0046. [DOI] [PubMed] [Google Scholar]
- Sanderson M, Donoghue M. Patterns of variation in levels of homoplasy. Evolution. 1992;43:1781–1795. doi: 10.1111/j.1558-5646.1989.tb02626.x. [DOI] [PubMed] [Google Scholar]
- Yoon JS, Richardson R. Evolution of the Hawaiian Drosophilidae II. Patterns and rates of chromosome evolution in the Antopocerus phylogeny. Genetics. 1976;83:827–843. doi: 10.1093/genetics/83.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. PAUP*: Phylogenetic Analysis Using Parsimony* (and other methods), 4.0b7 beta version. Sunderland, MA, Sinauer Associates, 2001.
- Nixon K, Carpenter J. On simultaneous analysis. Cladistics. 1996;12:221–241. doi: 10.1006/clad.1996.0016. [DOI] [PubMed] [Google Scholar]
- Kluge A. A concern for evidence and a phylogenetic hypothesis of relationships among Epicrates (Boidae, Serpentes). Syst Zool. 1989;38:7–25. [Google Scholar]
- Brower AVZ, DeSalle R, Vogler AP. Gene trees, species trees, and systematics: a cladistic perspective. Annu Rev Ecol Syst. 1996;27:423–450. doi: 10.1146/annurev.ecolsys.27.1.423. [DOI] [Google Scholar]
- Eriksson T. Auto-Decay. Software and documentation, version 2.9.5. Department of Botany, Stockholm University, 1996.
- Baker RH, Yu X, DeSalle R. Assessing the relative contribution of molecular and morphological characters in simultaneous analysis trees. Mol Phyl Evol. 1998;9:427–436. doi: 10.1006/mpev.1998.0519. [DOI] [PubMed] [Google Scholar]
- Farris JS, Kallersjo M, Kluge AG, Bult C. Testing significance of congruence. Cladistics. 1994;10:315–320. doi: 10.1006/clad.1994.1021. [DOI] [Google Scholar]
- Farris JS, Kallersjo M, Kluge AG, Bult C. Constructing a significance test for incongruence. Syst Biol. 1995;44:570–572. [Google Scholar]
- Kambysellis MP, Ho KF, Craddock EM, Piano F, Parisi M, Cohen J. Patterns of ecological shifts in the diversification of Hawaiian Drosophila inferred from a molecular phylogeny. Current Biology. 1995;5:1129–1139. doi: 10.1016/s0960-9822(95)00229-6. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Kreitman M. Evolutionary genetics of two sibling species, Drosophila simulans and D. sechellia. Evolution. 1986;40:673–691. doi: 10.1111/j.1558-5646.1986.tb00530.x. [DOI] [PubMed] [Google Scholar]
- Cohn VH, Moore GP. Organization and evolution of the alcohol dehydrogenase gene in Drosophila. Mol Biol Evol. 1988;5:154–166. doi: 10.1093/oxfordjournals.molbev.a040487. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Genbank accession number X54118. 1990.
- Moses K, Heberlein U, Ashburner M. The Adh gene promoters of Drosophila melanogaster and Drosophila orena are functionally conserved and share features of sequence structure and nuclease-protected sites. Mol Cell Biol. 1990;10:539–548. doi: 10.1128/mcb.10.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Jeffs PS, Holmes EC, Ashburner M. The molecular evolution of the alcohol dehydrogenase and alcohol dehydrogenase-related genes in the Drosophila melanogaster species subgroup. Mol Biol Evol. 1994;11:287–304. doi: 10.1093/oxfordjournals.molbev.a040110. [DOI] [PubMed] [Google Scholar]
- Caccone A, Moriyama EN, Gleason JM, Nigro L, Powell JR. A molecular phylogeny for the Drosophila melanogaster subgroup and the problem of polymorphism data. Mol Biol Evol. 1996;13:1224–1232. doi: 10.1093/oxfordjournals.molbev.a025688. [DOI] [PubMed] [Google Scholar]
- Ruiz Linares A, Hancock JM, Dover GA. Secondary structure constraints on the evolution of Drosophila 28S ribosomal RNA expansion segments. J Mol Biol. 1991;219:381–390. doi: 10.1016/0022-2836(91)90178-9. [DOI] [PubMed] [Google Scholar]
- Pelandakis M, Solignac M. Molecular phylogeny of Drosophila based on ribosomal RNA sequences. J Mol Evol. 1993;37:525–543. doi: 10.1007/BF00160433. [DOI] [PubMed] [Google Scholar]
- Samson ML, Wegnez M. The 5S ribosomal genes in the Drosophila melanogaster species subgroup: nucleotide sequence of a 5S unit from Drosophila simulans and Drosophila teissieri. Nucleic Acids Res. 1984;12:1003–1014. doi: 10.1093/nar/12.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Samson ML, Jordan P, Wegnez M. Structural evolution of the Drosophila 5S ribosomal genes. J Mol Evol. 1995;41:615–21. doi: 10.1007/BF00175820. [DOI] [PubMed] [Google Scholar]
- Shibata H, Yamazaki T. Molecular evolution of the duplicated Amy locus in the Drosophila melanogaster species subgroup: concerted evolution only in the coding region and an excess of nonsynonymous substitutions in speciation. Genetics. 1995;141:223–236. doi: 10.1093/genetics/141.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga H, Narise S. Sequence evolution of the Gpdh gene in the Drosophila virilis species group. Genetica. 1995;96:293–302. doi: 10.1007/BF01439583. [DOI] [PubMed] [Google Scholar]
- Kitagawa H. Genbank accession numbers AB019507 AB019546-AB019550, 1998.
- Nurminsky DI, Moriyama EN, Lozovskaya ER, Hartl DL. Molecular phylogeny and genome evolution in the Drosophila virilis species group duplications of the alcohol dehydrogenase gene. Mol Biol Evol. 1996;13:132–149. doi: 10.1093/oxfordjournals.molbev.a025551. [DOI] [PubMed] [Google Scholar]
- Durando CM, Baker RH, Etges WJ, Heed WB, Wasserman M, DeSalle R. Phylogenetic analysis of the repleta species group of the genus Drosophila using multiple sources of characters. Mol Phyl Evol. 2000;16:296–307. doi: 10.1006/mpev.2000.0824. [DOI] [PubMed] [Google Scholar]
- Kulge AG, Farris JS. Quantitative phyletics and the phylogeny of the anurans. Syst Zool. 1969;18:1–32. [Google Scholar]


