Abstract
Lung cancer is a major cause of death worldwide, and non-small cell lung cancer (NSCLC) is the most common type of lung cancer. The aim of this study was to investigate whether miR-96 mediated the invasion and metastasis of NSCLC by targeting glypican-3 (GPC3). Reverse transcription-quantitative PCR (RT-qPCR) was employed to detect the level of miR-96 and GPC3 mRNA. We applied western blot analysis to measure the protein expression level of GPC3 gene. The luciferase reporter assay was employed to confirm that GPC3 was a target gene of miR-96. The Transwell assay was used to detect migration and invasion. The results revealed that miR-96 was upregulated in NSCLC tissues and lung cancer cells (A549 and H460) compared with corresponding paracancerous tissues and normal epidermic MRC-5 cells. Overexpression of miR-96 promoted invasion and migration in A549 cells. GPC3 was a direct target of miR-96 and regulated by miR-96. GPC3 could reverse partial fuction of miR-96 on proliferation. In conclusion, miR-96 was able to promote the migration and invasion of lung cancer cells by targeting GPC3 gene. The newly identified miR-96/GPC3 axis may provide a therapeutic method for the treatment of NSCLC.
Keywords: miR-96, glypican-3, migration, invasion
Introduction
Cancer is caused by the genetic mutations induced by various carcinogenic factors (1). Lung cancer is the leading cause of death worldwide (2). The incidence of lung cancer is related to smoking, and it is reported that 85% of male patients and 47% of female lung cancer patients are smokers (3,4). Non-small cell lung cancer (NSCLC) is the most common type of lung cancer which accounts for >75% of all lung cancer cases (5). Lung cancer is a highly complicated and heterogeneous disease with intricate genetic mutations (6). Therefore, further investigation of the underlying mechanism of NSCLC is urgently needed.
MicroRNAs (miRNAs) are 21–28 nucleotides in length and are highly conserved small non-coding RNAs that bind to mRNAs of their target gene at their 3′-UTR to regulate target gene expression at post-transcriptional (7,8). miRNAs play an important role in various biological processes, such as proliferation, metastasis, and apoptosis (9). In NSCLC, multiple miRNAs have been identified as tumor suppressors, including miR-16, miR-23b, and miR-143 (10–12). miR-96 is a member of the miR-183 family, which constituted a polycistronic paralogous miRNA cluster (13). In breast cancer, miR-96 promoted cell invasion, migration, and proliferation in vitro by silencing PTPN9 (14). Ress et al reported that miR-96 promoted cell proliferation and predicted poor prognosis in colorectal cancer (15). Recently, miR-96 has been reported to be upregulated and acts as an oncogene in many cancers, including NSCLC (16). The reversion-inducing-cysteine-rich protein with kazal motifs (RECK) is the functional target of miR-96 (17). The majority of human miRNAs were imprecisely bound to mRNA of target genes (8). Thus, we considered there could be other unknown targets of miR-96 in NSCLC. Jalvy-Delvaille et al have shown that glypican-3 (GPC3) is a potential target of miR-96 in hepatocellular carcinoma (HCC) cells (18).
GPC3, heparan sulfate proteoglycans (HSPGs) which are the major part of extracellular matrix, play a pivotal role in the regulation of heparin-binding growth factor and various intracellular signaling pathways (19,20). It has been found that GPC3 functions as a tumor suppressor in various tumors. GPC3 was significantly downregulated in human malignant mesothelioma cell lines and the ectopic expression of GPC3 suppressed the colony formation of human MM cells in vitro (21). White et al reported that GPC3 may participate in the tumorigenesis of some WT cases (22).
In the present study, we investigated the role of miR-96 and GPC3 in NSCLC. We found that miR-96 was upregulated in lung cancer cells and NSCLC tissues compared with normal epithelium cells and paracancerous tissues. The miR-96 overexpression significantly promoted the migratory and invasive ability of lung cancer cell lines. Moreover, we identified that GPC3 was a direct target of miR-96 and miR-96 regulated the expression of GPC3. GPC3 was decreased in lung cancer cell lines and a low expression of GPC3 suppressed the proliferation of lung cancer cells.
Materials and methods
Tissue samples and cell lines
In total, 57 human NSCLC samples and corresponding paracancerous tissues from the patients who underwent surgery at the First Affiliated Hospital of Jiamusi University (Jiamusi, China) between January, 2012 and December, 2016 were collected. The above experimental samples were obtained with informed consent of the patients and verified by the Ethics Committee of the First Affiliated Hospital of Jiamusi University.
Human lung cancer cells A549 and H460 and normal lung epithelium cells MRC-5 were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured at 37°C in a humidified atmosphere with 5% CO2 using RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA), and supplemented with 10% fetal bovine serum.
Western blot analysis
Total proteins were extracted from the cells and tissues using the cell lysis buffer (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's instructions. The proteins were quantified using the Bradford assay kit (Takara Biotechnology Co., Ltd.). The proteins were separated by SDS-PAGE, and then transferred onto a PVDF membrane (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membrane was blocked in Tween-20 containing 5% skimmed milk for 1 h and then incubated with primary antibody (GPC3 rabbit polyclonal antibody, P51654; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at 4°C overnight. After the membranes were washed three times (10 min each) with PBST, the anti-rabbit secondary antibody (1:3,000; sc-362280; Santa Cruz Biotechnology, Inc.) was incubated with the membranes for 1 h at room temperature. The protein bands were visualized using the Bio-Rad Gel Doc XR instrument (Bio-Rad Laboratories, Inc.). Each experiment was performed three times in duplicate.
Plasmid construction and cell transfection
miRNA vectors, including miR-96 mimic and LNA anti-miR-96 and their control vectors were designed and purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China), which were employed to overexpress or knock down miR-96. pcDNA3.1-GPC3 and siRNA-GPC3 plasmids (purchased from Shanghai GenePharma Co., Ltd.) were utilized to overexpress or knock down GPC3.
A549 cells were seeded onto 6-well plates and when the confluence was ~70%, the plasmids were transfected using Lipofectamine 2000 and RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) according to the manufacturer's instructions. Plasmids or Lipofectamine 2000 were diluted with Opti-MEM Reduced-Serum Medium and set aside. After mixing the two dilutions, and allowing to stand for 20 min, the dilutions were added onto the 6-well plates containing cells. After transfection for 4–6 h, fresh RPMI-1640 medium was added.
Total RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
The total RNA was extracted using the RNAprep Pure Tissue kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer's instructions. The cDNA was synthesized using the FastKing RT kit (with gDNase) (Tiangen Biotech Co., Ltd.). GAPDH mRNA levels were used for the normalization of GPC3. The primer sequences were as follows: miR-96 forward, 5′-GCCCGCTTTGGCACTAGCACATT-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; GPC3 forward, 5′-CAGACTCGAGCTGCCTGGTGCCCAGC-3′ and reverse, 5′-GAGAGGTACCCAAAGAAATCCATGCAAAGAG-3′; GAPDH forward, 5′-CCACTCCTCCACCTTTGAC-3′ and reverse, 5′-ACCCTGTTGCTGTAGCCA-3′; and U6 forward, 5′-CTTCGGCAGCACATATACT-3′ and reverse, 5′-AAAATATGGAACGCTTCACG-3′. RT-PCR was performed using the Roche LightCycler 480 instrument (Roche Diagnostics, Basel, Switzerland). The U6 snRNA levels were used for the normalization of miR-96.
Luciferase reporter assay
TargetScan website (www.targetscan.org) was used to predict the target gene of miR-96, GPC3 was identified to be a potential target of miR-96 and the binding site of GPC3 for miR-96 was at the 3′-UTR. The 3′-UTR sequence of GPC3 was amplified and cloned into the pGL3 vector (pGL3-GPC3-WT; WT). The binding site of GPC3 for miR-96 was mutated and cloned into the pcDNA3.1 vector (pGL3-GPC3-MT; MT). Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to perform the luciferase activity assay.
Migration and invasion assays
The migration of lung cancer A549 cell lines was tested using Transwell chamber (8 µm pore; Corning, Inc., Corning, NY, USA). Approximately 1×106 cells were added into the upper chamber and cell medium was added into the lower chamber. Cells were incubated for 24 h at 37°C. Crystal violet was used to stain the migrated cells, which were visualized using an inverted microscope. The upper chamber was filled with Matrigel (BD Biosciences, San Jose, CA, USA) for the invasion test.
Cell proliferation assay
The 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) was employed to investigate cell proliferation. Before the experiment, the cells were seeded onto 96-well plates, cultivated with RPMI-1640 medium for 24 h to adherence and then MTT solution was added and incubated for 4 h at 37°C. After the supernatants were removed, the formazan crystals were dissolved using DMSO (200 µl/well). The absorbance at 490 nm was tested using the Thermo Scientific Evolution 300 instrument (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Statistical analysis
SPSS 18.0 software was used for statistical analysis. Student's t-test or ANOVA and Scheffe's test were employed to carry out the comparison between means of two groups or multiple groups. Results were considered significant at P<0.05.
Results
miR-96 is significantly upregulated in NSCLC, the opposite of GPC3
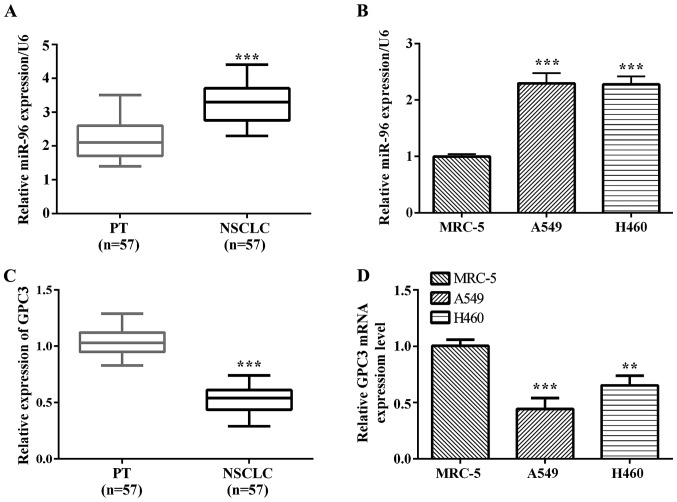
In this study, we investigated the miR-96 expression level in tissues and cell lines by RT-qPCR. The results showed that miR-96 was significantly upregulated in NSCLC tissues compared with the corresponding paracancerous tissues (P<0.0001; Fig. 1A). We also found that miR-96 was upregulated in A549 (P=0.0003) and H460 (P=0.0001) lung cancer cell lines compared with normal lung epithelium MRC-5 cells (Fig. 1B). GPC3 mRNA expression was observed in 53 cases of NSCLC samples and their matched normal paracancerous tissues. We noted a trend of lower expression in NSCLC samples compared to matched paracancerous tissues (Fig. 1C). Fig. 1D shows normal lung epithelium MRC-5 cells with the A549 and H460 lung cancer cells, depicting decreased GPC3 mRNA in A549 (P=0.0009) and H460 (P=0.0039) lung cancer cells compared to matched normal lung epithelium MRC-5 cells.
Figure 1.
miR-96 is significantly upregulated in NSCLC, the opposite of GPC3. (A) miR-96 expression levels of NSCLC tissues and corresponding paracancerous tissues were confirmed by RT-qPCR. (B) miR-96 expression levels of lung cancer cells A549, H460 and normal lung MRC-5 cell line were confirmed by RT-qPCR. (C) The expression of GPC3 mRNA in NSCLC tissues and corresponding paracancerous tissues were determined by RT-qPCR. (D) GPC3 levels in A549, H460 and MRC-5 were quantified by RT-qPCR. NSCLC, non-small cell lung cancer; GPC3, glypican-3; RT-qPCR, reverse transcription-quantitative PCR. **P<0.01; ***P<0.0001.
Overexpression of miR-96 promotes cell migration and invasion in vitro
In order to investigate the influence of miR-96 on cell migration and invasion, we transfected the miR-96 mimic, LNA anti-miR-96 or negative control plasmid into A549 cell lines. The effect of transfection of miR-96 mimic (P<0.0001) and anti-miR-96 (P=0.0022) into the A549 cell line was confirmed by RT-qPCR (Fig. 2A). The effect of miR-96 on lung cancer cell migration and invasion was analyzed by Transwell assay. The migratory and invasive ability of the transfected cells was measured and compared with that of NC at 24, 48, 72 and 96 h post-transfection. As shown in Fig. 2B, upregulation of miR-96 significantly promoted the migration (P=0.0043) and invasion (P=0.0023) of A549 cells, conversely, downregulation of miR-96 inhibited the cell migration (P=0.0018) and invasion (P=0.0010).
Figure 2.
Overexpression of miR-96 significantly promotes cell migration and invasion of NSCLC cells in vitro. (A) A549 cells were transfected with miR-96 mimic or LAN-anti-miR-96 and the expression of miR-96 was confirmed by RT-qPCR. (B) Transwell assay was used to analyze migratory and invasive ability of A549 cells. NSCLC, non-small cell lung cancer. **P<0.01; ***P<0.001.
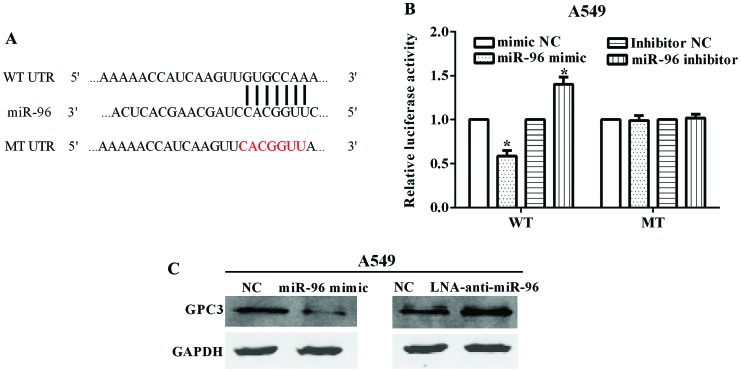
GPC3 is a target of miR-96 and is downregulated by miR-96
By the TargetScan online tool (www.targetscan.org/vert_71/), we identified GPC3 as a potential target of miR-96. The binding site on GPC3 of miR-96 locates at its 3′-UTR with binding site 5′-GUGCCAA-3′ (Fig. 3A). After mutated the 3′-UTR from 5′-GUGCCAAA-3′ (WT) to 5′-CACGGAA-3′ (MT), we constructed the Psicheck™-2-GPC3-WT and Psicheck™-2-GPC3-MT plasmids. The luciferase reporter assay demonstrated that the luciferase activity was significantly inhibited when the A549 cell line was co-transfected with miR-96 mimic and Psicheck™-2-GPC3-WT (P=0.0116), while there was no obvious change in transfection of miR-96 mimic and Psicheck™-2-GPC3-MT (P=0.826) (Fig. 3B). By contrast, the luciferase activity was increased when the cell lines were co-transfected with miR-96 inhibitor and Psicheck™-2-GPC3-WT (P=0.0218), whereas there was no obvious change in transfection of miR-96 inhibitor and Psicheck™-2-GPC3-MT (P=0.574) (Fig. 3B). These results indicated that the GPC3 was a target of miR-96.
Figure 3.
GPC3 is a target of miR-96 and downregulated by miR-96. (A) The binding site of GPC3 for miR-96 is located at its 3′-UTR. The mutated nucleotides are indicated in red. (B) miR-153 significantly suppressed the luciferase activity that carried WT but not MT 3′-UTR of SNAI1. Anti-miR-153 led to a marked increase in luciferase activity of WT 3′-UTR of SNAI1. (C) GPC3 protein level was determined by western blot analysis, when A549 cell line was transfected with miR-96 mimic or LNA anti-miR-96. GPC3, glypican-3. *P<0.05.
We also investigated the GPC3 protein levels when changed by the expression of miR-96. The results showed that GPC3 was significantly decreased when A549 cell line was transfected with miR-96 mimic. By contrast, the GPC3 protein level was increased when the A549 cell line was transfected with LNA anti-miR-96 (Fig. 3C).
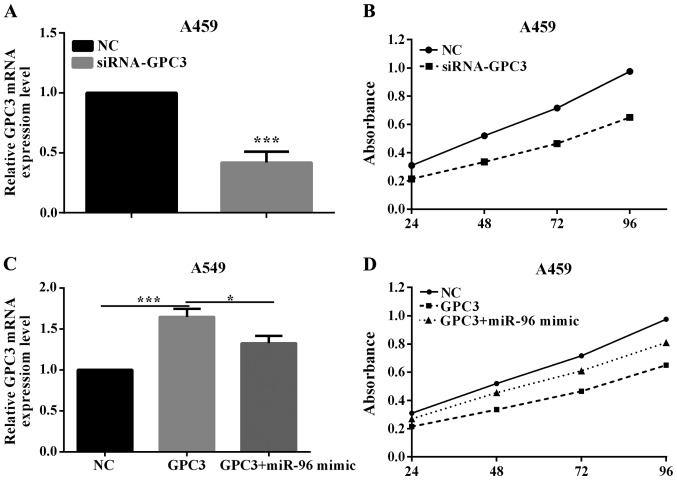
GPC3 reverses partial fuction of miR-96 on proliferation
To investigate whether miR-96 may promote NSCLC tumorigenesis by silencing GPC3, we assessed the role of PTPN9 on cell proliferation. In order to knock down GPC3 gene, the siRNA-GPC3 or negative control (siRNA-NC) was transfected into A549 cell lines. The efficient knockdown of GPC3 mRNA in A549 cells (P=0.0048) is shown in Fig. 4A. The proliferation assay demonstrated that knockdown of GPC3 could significantly promote the proliferation of A549 cell lines (P=0.0352; Fig. 4B). This indicated that GPC3 was a tumor suppressor in NSCLC.
Figure 4.
GPC3 reverses partial fuction of miR-96 on proliferation. (A) The expression of GPC3 was detected by RT-qPCR when transfected siRNA-GPC3 or siRNA-NC into A549 cells. (B) The proliferative ability was measured after A549 cells were transfected with siRNA or negative control. (C) The cells transfected with both miR-96 mimic and pcDNA3.1-GPC3 exhibited a lower GPC3 mRNA level compared with the cells transfected with pcDNA3.1-GPC3. (D) The proliferative ability was measured after A549 cells were co-transfected with miR-96 mimic and pcDNA3.1-GPC3 or only transfected with pcDNA3.1-GPC3. GPC3, glypican-3; RT-qPCR, reverse transcription-quantitative PCR. *P<0.05; ***P<0.001.
To explore whether miR-96 regulation of cell proliferation is executed in a GPC3-dependent manner, we co-transfected A549 cells with miR-96 mimic and pcDNA3.1-GPC3 plasmid. Compared with cells transfected with pcDNA3.1-GPC3, the cells transfected with both miR-96 mimic and pcDNA3.1-GPC3 exhibited a lower GPC3 mRNA level (P=0.0003; Fig. 4C). Consequently, the cells transfected with both miR-96 mimic and pcDNA3.1-GPC3 exhibited an obviously lower proliferation (P=0.0131; Fig. 4D), suggesting that miR-96-resistant GPC3 can attenuate the proliferative effect of GPC3 on lung cells. The results indicate that miR-96 may regulate the proliferation of lung cancer cells in a GPC3-dependent manner.
Discussion
Lung cancer is the leading cause of mortality in men, and the second leading cause of mortality in women worldwide. NSCLC is one of the most common histological subtypes of lung cancer, which can be divided into three subtypes, which are large cell carcinoma (LCC), lung adenocarcinoma (LAD), and lung squamous cell carcinoma (LSCC) (23). Although cancer treatments have improved in recent years, the outcomes of patients with GC remain unsatisfactory. Thus, to find new and effective therapeutic methods for treatment of NSCLC is imperative.
miRNAs have been found to play an important role in the development of diverse diseases, including cancer. As reported in previous studies, several miRNAs play an important role in tumorigenesis of human NSCLC due to their overexpression in tumor tissues. miR-96 is a member of the miR-183 family, which constituted a polycistronic paralogous miRNA cluster (13). miR-96 was found to be highly upregulated in different kinds of tumors, including breast cancer (14), and colorectal cancer (15). Similarly, in our study, miR-96 was found to be upregulated in 53 pairs of NSCLC tissues and lung cancer cells. Upregulation of miR-96 could promote lung cancer cell A549 migration and invasion, while downregulation suppressed A549 cell migratory and invasive ability. In addition, GPC3 was found to be downregulated in NSCLC tissues and lung cancer cells. Hong et al reported that miR-96 promoted cell invasion, migration, and proliferation in breast cancer (14). Considering these results, we strongly believe that the impact of miR-96 on migration and invasion may be through direct inhibition of GPC3.
GPC3 is an HSPG playing a pivotal role in the regulation of heparin-binding growth factor and various intracellular signaling pathways (24). The glypican proteins use a glycosylphosphatidylinositol anchor to link to the cytoplasmic membrane (25). GPC3 was significantly downregulated in human malignant mesothelioma cell lines and ectopic expression of GPC3 suppressed colony formation of human MM cells (21). In accordance with these reports, we identified that knockdown of GPC3 by siRNA promoted proliferative ability in lung cancer A549 cell line. In HCC, GPC3 is a transcriptor target of c-Myc and GPC3 can also regulate the expression of c-Myc (26). In the present study, we found that GPC3 was a direct target of miR-96 and regulated by miR-96. Moreover, when GPC3 was overexpressed, the cell proliferative ability increased. In co-transfection of GPC3 and miR-96 mimic, proliferative ability was reduced compared with only transfection with GPC3. Thus, we propose that miR-96 and GPC3 had a relationship with NSCLN proliferation.
There still exist some disadvantages in our research. For example, the number of patient samples was small. Our research did not refer to the clinical experiments. In our further study, we will perform the above experiments.
Taken together, we identified that miR-96 promoted migration and invasion of lung cancer cells by targeting GPC3. miR-96 functioned as an oncogene in NSCLC and GPC3 could reverse partial fuction of miR-96 on proliferation. The identified miR-96/GPC3 axis may provide a meaningful therapeutic method for the treatment of NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request
Authors' contributions
SL contributed to the conception of the study. XF contributed significantly in performing the experiment and helped to write the manuscript. JZ wrote the manuscript and helped to perform the experiment. YZ performed the data analyses. MS and HZ helped perform the analysis with constructive discussions. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All the samples were obtained with informed consent of the patients and verified by the Ethics Committee of the First Affiliated Hospital of Jiamusi University (Jiamusi, China).
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
References
- 1.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Islami F, Torre LA, Jemal A. Global trends of lung cancer mortality and smoking prevalence. Transl Lung Cancer Res. 2015;4:327–338. doi: 10.3978/j.issn.2218-6751.2015.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramanian J, Govindan R. Lung cancer in never smokers: A review. J Clin Oncol. 2007;25:561–570. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Wang DC, Shi L, Zhu B, Min Z, Jin J. Genome analyses identify the genetic modification of lung cancer subtypes. Semin Cancer Biol. 2017;42:20–30. doi: 10.1016/j.semcancer.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Sittka A, Schmeck B. MicroRNAs in the lung. Adv Exp Med Biol. 2013;774:121–134. doi: 10.1007/978-94-007-5590-1_7. [DOI] [PubMed] [Google Scholar]
- 10.Ke Y, Zhao W, Xiong J, Cao R. Downregulation of miR-16 promotes growth and motility by targeting HDGF in non-small cell lung cancer cells. FEBS Lett. 2013;587:3153–3157. doi: 10.1016/j.febslet.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Han H, Yang J, Wang Y, Chen W, Chen J, Yang Y, Li Q. Nucleobase-modified polyamidoamine-mediated miR-23b delivery to inhibit the proliferation and migration of lung cancer. Biomater Sci. 2017;5:2268–2275. doi: 10.1039/C7BM00599G. [DOI] [PubMed] [Google Scholar]
- 12.Ma Q, Jiang Q, Pu Q, Zhang X, Yang W, Wang Y, Ye S, Wu S, Zhong G, Ren J, et al. MicroRNA-143 inhibits migration and invasion of human non-small-cell lung cancer and its relative mechanism. Int J Biol Sci. 2013;9:680–692. doi: 10.7150/ijbs.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 14.Hong Y, Liang H, Uzair-Ur-Rehman, Wang Y, Zhang W, Zhou Y, Chen S, Yu M, Cui S, Liu M, et al. miR-96 promotes cell proliferation, migration and invasion by targeting PTPN9 in breast cancer. Sci Rep. 2016;6:37421. doi: 10.1038/srep37421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ress AL, Stiegelbauer V, Winter E, Schwarzenbacher D, Kiesslich T, Lax S, Jahn S, Deutsch A, Bauernhofer T, Ling H, et al. MiR-96-5p influences cellular growth and is associated with poor survival in colorectal cancer patients. Mol Carcinog. 2015;54:1442–1450. doi: 10.1002/mc.22218. [DOI] [PubMed] [Google Scholar]
- 16.Wu L, Pu X, Wang Q, Cao J, Xu F, Xu LI, Li K. miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9. Oncol Lett. 2016;11:945–952. doi: 10.3892/ol.2015.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo H, Li Q, Li W, Zheng T, Zhao S, Liu Z. MiR-96 downregulates RECK to promote growth and motility of non-small cell lung cancer cells. Mol Cell Biochem. 2014;390:155–160. doi: 10.1007/s11010-014-1966-x. [DOI] [PubMed] [Google Scholar]
- 18.Jalvy-Delvaille S, Maurel M, Majo V, Pierre N, Chabas S, Combe C, Rosenbaum J, Sagliocco F, Grosset CF. Molecular basis of differential target regulation by miR-96 and miR-182: The Glypican-3 as a model. Nucleic Acids Res. 2012;40:1356–1365. doi: 10.1093/nar/gkr843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Z, Grobe K, Zhang X. Role of heparan sulfate proteoglycans in optic disc and stalk morphogenesis. Dev Dyn. 2014;243:1310–1316. doi: 10.1002/dvdy.24142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernfield M, Götte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 21.Murthy SS, Shen T, De Rienzo A, Lee WC, Ferriola PC, Jhanwar SC, Mossman BT, Filmus J, Testa JR. Expression of GPC3, an X-linked recessive overgrowth gene, is silenced in malignant mesothelioma. Oncogene. 2000;19:410–416. doi: 10.1038/sj.onc.1203322. [DOI] [PubMed] [Google Scholar]
- 22.White GR, Kelsey AM, Varley JM, Birch JM. Somatic glypican 3 (GPC3) mutations in Wilms' tumour. Br J Cancer. 2002;86:1920–1922. doi: 10.1038/sj.bjc.6600417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filmus J. Glypicans in growth control and cancer. Glycobiology. 2001;11:19R–23R. doi: 10.1093/glycob/11.3.19R. [DOI] [PubMed] [Google Scholar]
- 25.Ho M, Kim H. Glypican-3: A new target for cancer immunotherapy. Eur J Cancer. 2011;47:333–338. doi: 10.1016/j.ejca.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Jin R, Zhang X, Lv F, Liu L, Liu D, Liu K, Li N, Chen D. Oncogenic activation of glypican-3 by c-Myc in human hepatocellular carcinoma. Hepatology. 2012;56:1380–1390. doi: 10.1002/hep.25891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request