Abstract
Mutations in the mitochondrial 12S ribosomal RNA gene have been identified to be associated with deafness. Among these, the A to G transition at position 1555 is one of the most common pathogenic mutations associated with hearing loss. In order to evaluate the allele frequency of this mutation in infants with hearing loss, the A1555G mutation was screened in 300 deaf children and 100 age- and sex-matched healthy subjects. Consequently, 5 patients with this mutation were identified, whereas the mutation was absent in healthy controls. Among the patients with the mutation, only one had an obvious family history of hearing impairment. Notably, this pedigree manifested a high penetrance of deafness. In particular, the penetrance of deafness was 80 and 40%, when the aminoglycoside antibiotics (AmAn) was included or excluded, respectively. Clinical evaluation of this family exhibited a wide degree of hearing loss. Furthermore, screening for the complete mitochondrial genes revealed the occurrence of A1555G and transfer (t)RNAThr T15943C mutations, together with other genetic variations belonging to East Asian haplogroup C. Notably, the T15943C mutation, located at the T arm of tRNAThr, could disrupt the 63T-55A base-pairing and impair tRNA metabolism. Therefore, it was hypothesized that the combination of A1555G and T15943C mutations may result in mitochondrial dysfunction that is responsible for deafness. Screening for A1555G, as well as other potential pathogenic mutations in the mitochondrial genome, is critical for clinical diagnosis and prevention of hearing impairment.
Keywords: infant, deafness, mitochondrial mutations, A1555G, T15943C, transfer RNA metabolism
Introduction
Deafness is a serious health problem with unknown etiology, and it is estimated that 1 in 1,000 newborns suffer from the condition (1). It may be associated with numerous risk factors, including the administration of aminoglycoside antibiotics (AmAn) (2). Genetic alterations in human mitochondrial (mt)DNA are also thought to be associated with deafness (3). Human mtDNA is a double-stranded, circular molecule, encoding 13 protein subunits, 2 ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs). In particular, the well-known 12S rRNA A1555G mutation has been demonstrated to be an important molecular basis of hearing loss (4,5). Furthermore, the A to G transition at position 1555 is located in the region of small ribosomal RNA that is highly conserved in bacteria and mammals (6). The corresponding region in Escherichia coli forms an essential part of the decoding site of the ribosome (7) and is crucial for subunit association either by RNA-protein or the RNA-RNA interaction (8). It is interesting to note that among individuals who carry this mutation, the use of AmAn for clinical treatment of fever is likely to result in irreversible hearing impairment (9). However, individuals who carry the A1555G mutation exhibit a wide range of clinical phenotypes, from profound hearing loss to normal hearing (4). Thus, if an individual carrying the A1555G mutation cannot use aminoglycosides, screening for the A1555G mutation is crucial for prevention and early diagnosis of maternally inherited deafness. In addition, lymphoblastoid cell lines derived from an Arab-Israeli family demonstrated more severe biochemical defects in those that were symptomatic to those that were not (10). Therefore, it is hypothesized that other unidentified risk factors, including polymorphisms in mtDNA, nuclear gene mutations, AmAn and epigenetic modification, may contribute to hearing impairment (10).
To explore the potential association between mtDNA polymorphisms and deafness, a mutational screening was conducted for the presence of A1555G in 300 infants with hearing impairment and 100 healthy controls. Consequently, 5 patients with the mutation were identified. Among these infants carrying the A1555G mutation, only one had a family history of exposure to AmAn.
Materials and methods
Subjects and clinical evaluations
From January 2015 to January 2017, a total of 300 infants with hearing loss (180 females and 120 males), aged from 1–3 years (with an average age of 2 years), as well as 100 age- and sex-matched control subjects (60 females and 40 males) were recruited from Fujian Provincial Hospital (Fuzhou, China). A physical examination was performed for each participant that was enrolled. In addition, the level of hearing impairment was tested using pure-tone audiometry. Furthermore, the level of hearing loss was classified as follows: Normal, ≤25 dBHL; mild, 26–40 dBHL; moderate, 41–70 dBHL; severe, 71–90 dBHL; profound, >90 dBHL. Written informed consent was obtained from the patients' parents or guardians, and the study was approved by the Ethics Committee of Fujian Provincial Hospital. Patients were included if the infants possessed the sole clinical phenotype. Those patients who carried other mitochondrial disorders, including Leigh syndrome, exercise intolerance and developmental delay were excluded. In addition, healthy patients exhibited normal hearing, and did not carry any diseases.
Screening for the mitochondrial A1555G mutation
Genomic DNA was extracted from blood samples obtained from each participant using Puregene DNA Isolation kits (Gentra; Qiagen, Inc., Valencia, CA, USA). The mitochondrial 12S rRNA gene was amplified using polymerase chain reaction (PCR; Takara, Bio, Inc., Otsu, Japan), the primer sequence for amplification the 12S rRNA gene was as follows: (5): Forward, 5′-CGATCAACCTCACCACCTCT-3′ and reverse, 5′-TGGACAACCAGCTATCACCA-3′. The thermocycling conditions for PCR were as follows: 95°C for 5 min, 94°C for 10 sec, 60°C for 30 sec, 30 cycles at 72°C for 60 sec and 72°C for 10 min for extension. Following amplification, the PCR product was digested with the restriction enzyme BsmAI to screen for the occurrence of the A1555G mutation. Equal amounts (5 µl) of various digested samples were then analyzed using electrophoresis with 1.5% agarose gel. Following electrophoresis, a BandPeeper (MaestroGen Inc., Hsinchu, Taiwan) instrument was utilized to detect the results using the Invitrogen™ E-Gel™ Imager software (version 2.2.4; Invitrogen, Thermo Fisher Scientific Inc., Waltham, MA, USA).
Molecular features of the pedigree harboring mitochondrial A1555G mutation
A Chinese pedigree was collected in Fujian Provincial Hospital. The complete mitochondrial genomes of the matrilineal individuals (I-2, II-1, II-5, II-8, II-10, III-9, III-10 and III-12) were amplified using 24 primers, as previously described (6). The PCR reagents (all obtained from Takara Bio, Inc.,) were as follows: 200 µM dNTPs, 2 µl 10X PCR buffer (10X0.2 µl Taq DNA polymerase and 15 mmol/l Mg2+). Subsequently, the PCR products were purified and sequenced using the ABI PRISM™ 3700 machine (Applied Biosystems; Thermo Fisher Scientific, Inc.) (11). A total of 24 PCR product sequences were compared with the reverse Cambridge sequence, using DNA Star software version 5.01 (DNASTAR Inc., Madison, WI, USA) to detect variations in mitochondrial genes (GenBank accession no. NC_012920) (12).
Phylogenetic conservation analysis
To discern the potential pathogenic mtDNA mutations, the phylogenetic approach was utilized to assess the conservation index (CI) of each mtDNA mutation. A total of 17 species were used to analysis the CIs. Notably, CI>75% was regarded as exhibiting functional potential (13).
Detecting GJB2 mutations
In order to identify whether GJB2 mutations were involved in hearing impairment, variants of GJB2 were screened using PCR and direct sequencing. The primers for amplification of the GJB2 coding region were as follows: Forward, 5′-TATGACACTCCCCAGCACAG-3′ and reverse, 5′-GGGCAATGCTTAAACTGGC-3′ (14). The PCR reagents (all obtained from Takara Bio, Inc.) were as follows: 200 µM dNTPs, 2 µl 10X PCR buffer (10×0.2 µl Taq DNA polymerase) and 15 mmol/l Mg2+. PCR was performed under the following thermocycling conditions: 95°C for 5 min, 94°C for 10 sec, 60°C for 30 sec, 30 cycles at 72°C for 60 sec, and 72°C for 10 min for extension Then, the PCR products were sequenced using the ABI PRISM™ 3700 machine (Applied Biosystems; Thermo Fisher Scientific, Inc.) and compared with the wild-type GJB2 sequence using DNA Star software version 5.01 (GenBank accession no. M86849) to detect potential pathogenic mutations.
Screening for the TRMU A10S variant
Nuclear modified gene TRMU was previously identified to be associated with hearing loss (15). To understand the role of TRMU in hearing impairment, the TRMU A10S variant was screened in the matrilineal relatives using PCR amplification of exon 1 of this gene. The PCR reagents (all obtained from Takara Bio, Inc.,) were as follows: 200 µM dNTPs, 2 µl 10X PCR buffer (10×0.2 µl Taq DNA polymerase) and 15 mmol/l Mg2+. The primer sequence for exon 1 was as described previously (15). PCR was performed under the following thermocycling conditions: 95°C for 5 min, 94°C for 10 sec, 58°C for 30 sec, 30 cycles at 72°C for 60 sec, and 72°C for 10 min for extension. Then, the PCR product was sequenced using the ABI PRISM™ 3700 machine (Applied Biosystems; Thermo Fisher Scientific, Inc.) and compared with the wild-type version of TRMU and DNA Star software version 5.01 (GenBank accession no. AF448221) (16).
Bioinformatics analysis
To investigate whether T15943C mutation altered the structure of tRNAThr, the RNA Fold web server was employed to analyze the secondary structure of tRNAThr with and without the T15943C mutation (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) (17). The wild-type version of tRNAThr gene was: 5′-GTCCTTGTAGTATAAACTAATACACCAGTCTTGTAAACCGGAGATGAAAACCTTTTTCCAAGGACA-3′. The sequence of tRNAThr gene carrying the T15943C mutation was: 5′-GTCCTTGTAGTATAAACTAATACACCAGTCTTGTAAACCGGAGATGAAAACCTTTCTCCAAGGACA-3′.
Results
Frequency of mitochondrial A1555G mutation in infants with deafness
To detect the allele frequency of the A1555G mutation, 300 infants were enrolled who were confirmed to have non-syndromic hearing loss by a clinical physician in Fujian Provincial Hospital. The restriction enzyme BsmAI was used to screen the mitochondrial A1555G mutation following PCR amplification of the 12S rRNA gene. As indicated in Fig. 1, following electrophoresis, it was observed that samples carrying the A1555G mutation exhibited only one 797 bp band, while the samples without this mutation resulted in two specific bands: 472 and 325 bp. Consequently, 5 patients with this mutation were identified (5/300; 1.67%). The mutation was not detected in the 100 healthy controls. Further clinical evaluation indicated that 1 patient with the mutation had an obvious family history of using AmAn.
Figure 1.
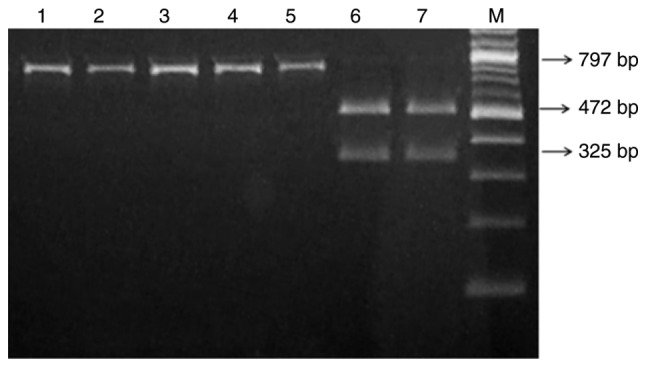
Detection of the A1555G mutation using BsmAI. Lanes 1–5 indicate samples with the A1555G mutation. Lanes 6 and 7 indicate samples without the A1555G mutation. M, DNA marker.
Molecular characterization of the pedigree carrying mitochondrial A1555G mutation
The proband's mother (II-10) was 26 years old and came from the Fuzhou area of Fujian Province, China. She received AmAn (gentamycin, 5 mg/kg/dose) for clinical treatment of a high fever when she was 21 years old. Unfortunately, 2 weeks following the antibiotic treatment, she suffered from bilateral hearing loss and clinical examination indicated that she had severe deafness (75 dB in left ear, 88 dB in right ear; see Table I). Further physical and audiological examinations were performed for the matrilineal relatives in this family. It was observed that I-2, II-5 and II-8 had a history of exposure to AmAn.
Table I.
Summary of clinical and molecular data for members of a Han Chinese family with AmAn-induced hearing loss.
| Subject | Sex | Age at onset (years) | Age at test (years) | Use of AmAn | PTA (dB), left ear | PTA (dB), right ear | Level of hearing loss |
|---|---|---|---|---|---|---|---|
| I-2 | Female | 55 | 62 | Yes | 90 | 90 | Profound |
| II-1 | Male | 18 | 30 | No | 75 | 80 | Severe |
| II-5 | Male | 25 | 32 | Yes | 35 | 40 | Moderate |
| II-8 | Female | 23 | 28 | Yes | 88 | 85 | Severe |
| II-10 | Female | 21 | 26 | Yes | 75 | 88 | Severe |
| III-9 | Male | 3 | 5 | No | 70 | 80 | Severe |
| III-10 | Female | 1 | 2 | No | 95 | 90 | Profound |
| III-12 | Female | 1 | 1 | No | 55 | 65 | Severe |
| III-1 | Male | N/A | 5 | No | 18 | 20 | Normal |
PTA, pure tone audiometry.
As indicated in Fig. 2, this pedigree had an obvious maternal transmission pattern. It was noted that the pedigree manifested a high penetrance and expressivity of deafness: The penetrance of deafness was 80 and 40% when patients who were exposed to AmAn were included and excluded, respectively. Furthermore, it was identified that the age at onset of deafness varied from 1 to 55 years (Table I). Other members of this family did not exhibit any other mitochondrial disorders, including type 2 diabetes, cardiovascular events, vision loss or neurological diseases.
Figure 2.
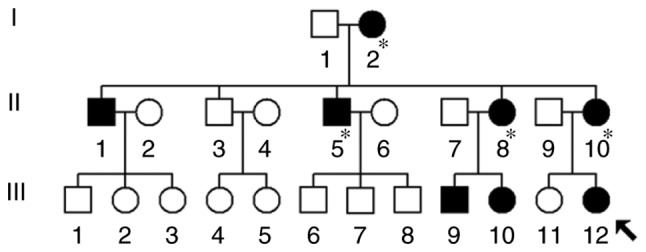
A Han Chinese family with AmAn-induced hearing loss. Males are indicated with squares and females are indicated with circles. Subjects with hearing loss are indicated by filled symbols. The arrow denotes the proband. Asterisks denote subjects with a history of AmAn exposure. AmAn, aminoglycoside antibiotic.
Screening for mutations in mitochondrial genes
The maternally inherited pattern of this family indicated that mitochondrial dysfunction may be involved in the pathogenesis of hearing loss. In order to investigate this, the variants in mtDNA were screened using PCR and direct sequencing. The results indicated that matrilineal relatives (I-2, II-1, II-5, II-8, II-10, III-9, III-10 and III-12) in this family harbored 22 mtDNA polymorphisms, as well as the well-known A1555G mutation (Table II). Of these, six variants were identified in the D-loop, three variants in 12S rRNA, one variant in 16S rRNA and one mutation in tRNAThr. The other sequence alterations were primarily in oxidative phosphorylation-associated genes. Furthermore, there were six missense mutations, including ND2 C5178A (L237M) and A5301G mutations (I278V), ATP6 A8860G mutation (T112A), ND3 A10398G mutation (T114A), and CytB C14766T (T7I) and A15326G (T194A) mutations. In addition, evolutionary conservation analysis was performed for these variants between 17 vertebrates, including the mouse (18), bovine (19) and Xenopus laevis (20). However, it was identified that none of these variants were conserved, with the exception of A1555G and T15943C mutations (Fig. 3 and Table III). In addition, the A1555G and T15943C mutations were absent in the control subjects. This suggested that the A1555G and T15943C mutations were involved in the pathogenesis of deafness in this family.
Table II.
Mitochondrial DNA sequence variants in a Han Chinese family with hearing loss.
| Gene | Position | Nucleotide change | Conservation (H/B/M/X)a |
|---|---|---|---|
| D-loop | 73 | A to G | |
| 263 | A to G | ||
| 310 | T to CTC | ||
| 16189 | T to C | ||
| 16355 | C to T | ||
| 16519 | T to C | ||
| 12S rRNA | 750 | A to G | A/G/G/- |
| 1438 | A to G | A/A/A/G | |
| 1555 | A to G | A/A/A/A | |
| 16S rRNA | 2706 | A to G | A/G/A/A |
| ND2 | 4769 | A to G | |
| 5178 | C to A (Leu to Met) | L/T/T/T | |
| 5301 | A to G (Ile to Val) | I/I/M/L | |
| CO1 | 7028 | C to T | |
| ATP6 | 8860 | A to G (Thr to Ala) | T/A/A/T |
| ND3 | 10398 | A to G (Thr to Ala) | T/T/T/A |
| ND4 | 11719 | G to A | |
| ND5 | 12705 | C to T | |
| 13344 | A to G | ||
| CytB | 14766 | C to T (Thr to Ile) | T/S/T/S |
| 15301 | G to A | ||
| 15326 | A to G (Thr to Ala) | T/M/I/I | |
| tRNAThr | 15943 | T to C | T/T/T/T |
Conservation of amino acid (for polypeptides) or nucleotide (for rRNAs) in H/B/M/X. H, human; B, bovine; M, mouse; X, Xenopus laevis; rRNA, ribosomal RNA; tRNA, transfer RNA; ND, NADH dehydrogenase; CO1, cytochrome c oxidase 1; ATP6, adenosine triphosphate 6; CytB, cytochrome B.
Figure 3.
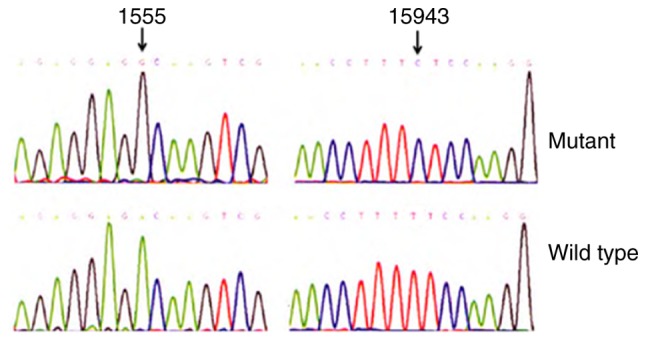
Identification of the mitochondrial A1555G and T15943C mutations using polymerase chain reaction and Sanger sequencing.
Table III.
Sequence alignment of tRNAThr gene from different species.
| Organism | Acc-stem | D-stem | D-loop | D-stem | Ac-Stem | Anticd-loop | Ac-stem | V-region | T-stem | T-loop | T-stema | Acc-stem |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Homo sapiens | GTCCTTGTA | GTAT | AAACTA | ATACA | CCAGT | CTTGTAA | ACCGG | AGAT | GAAAA | CCT | TTTTC | CAAGGACA |
| Mus musculus | GTCTTGATA | GTAT | AAACA | TTACT | CTGGT | CTTGTAA | ACCTG | AAAT | GAAGA | TCTTC | TCTTC | TCAAGACA |
| Myoxus glis | GTCCTGGTA | GTAT | AAAGTTA | TTACT | CTGGT | CTTGTAA | ACCAG | AAAT | GGAAA | TCTCAAT | TTTCC | CCAGGACA |
| Colobus guereza | GCCCTCGTA | GTAC | AAACTA | GTATA | CCGGT | CTTGTAA | ACCGA | AGAT | GGAGA | CT | TCTCC | CTAGGACA |
| Pan paniscus | GCCCTTGTA | GTAT | AAGCTA | ATACA | CCGGT | CTTGTAA | ACCGG | AAAC | GAAAA | CTT | TATTC | CAAGGACA |
| Ceratotherium simum | GTCTTTGTA | GTAT | ATGAA | TTACT | CTGGT | CTTGTAA | ACCAG | AAAA | GGAAA | GCACCAC | TTTCC | CCAAGACT |
| Arctocephalus forsteri | GTCTTCGTA | GTAT | AACAA | TTACC | TTGGT | CTTGTAA | ACCAA | AAAT | GGAGA | ACACCACGAC | TCTCC | CTAAGACT |
| Elephas maximus | ACCCCTATA | GTAT | AAGACA | TTACA | ATGGT | CTTGTAA | GCCAT | AAAT | GAAAA | CCA | CTTTC | TAAGGGTA |
| Bradypus tridactylus | GCCCTCGTA | GTAT | AACAAA | TTACA | CTAGC | CTTGTAA | ACCAG | AAAT | GAAAT | ACGC | ATTTC | CAAGGGCA |
| Gorilla gorilla | GCCCTTGTA | GTAC | AGACCA | ATACA | CCAGT | CTTGTAA | ACCGG | AAAC | GAAGA | CCT | CCTTC | CAAGGGCA |
Third nucleotide in this sequence represents position 63, corresponding to the T15943C mutation.
Genotyping analysis of GJB2 gene
To evaluate the contribution of GJB2 gene variants to deafness expression, GJB2 mutations were analyzed in the matrilineal relatives (I-2, II-1, II-5, II-8, II-10, III-9, III-10 and III-12). However, no functional variants were observed in the GJB2 gene.
Mutational analysis of TRMU gene
A previous study indicated that the TRMU A10S variant may contribute to the clinical expression of hearing loss associated with mitochondrial A1555G mutation (15). Therefore, the TRMU A10S variant was screened in matrilineal members from the current pedigree (I-2, II-1, II-5, II-8, II-10, III-9, III-10 and III-12). However, the variant was not detected in these subjects.
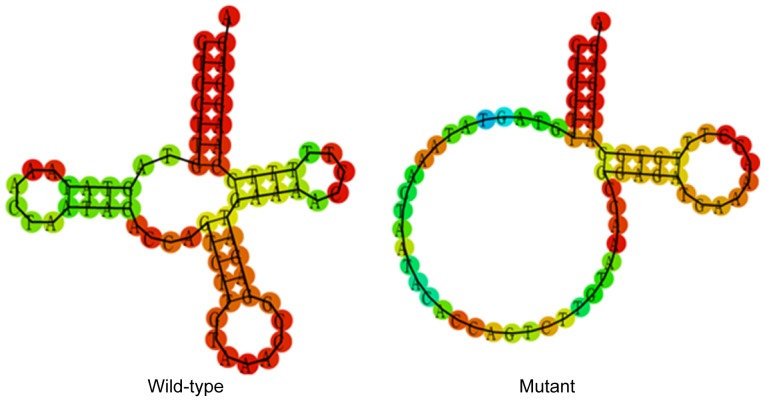
T15943C mutation affects the secondary structure of tRNAThr
To evaluate the effect of T15943C mutation on tRNAThr, a bioinformatics tool was used to predict the tRNAThr secondary structure with and without this mutation (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) (17). As indicated in Fig. 4, the T15943C mutation notably altered the tRNAThr secondary structure, strongly indicating that this mutation was pathogenic.
Figure 4.
Prediction of the secondary structure of transfer RNAThr with and without the T15943C mutation.
Discussion
The current study screened the occurrence of mitochondrial A1555G mutation in pediatric subjects with hearing loss. As early as 1993, the A1555G mutation was reported in an Arab-Israeli family with non-syndromic hearing impairment (4). In fact, this mutation occurred in the A-site of 12S rRNA gene, which was strongly conserved between multiple species and was critical for the binding of AmAn (21,22). Therefore, patients carrying A1555G mutation were more vulnerable to developing hearing loss when using AmAn.
Using PCR and direct sequencing, 5 infants with hearing loss were identified to be carrying the A1555G mutation in the current study. Its frequency was 1.67% in patients, but the mutation was not detected in the control subjects. The frequency of the A1555G mutation varies worldwide. For patients with hearing loss who used AmAn, the frequency of the A1555G mutation was reported to be 33% in a Japanese population (23), whereas the incidence was reported to be 13, 10.4 and 5% in three Han Chinese populations (24–26). These data are important, since children who carry this mutation should not be exposed to AmAn. Thus, screening for the occurrence of the A1555G mutation is advisable, particularly for those who have a family history of using AmAn.
In the current study, 5 infants with hearing loss were identified to be carrying the homoplasmic A1555G mutation. However, only 1 of these patients had an obvious family history of using AmAn. Following comprehensive genetic counseling, it was identified that the family members only exhibited the deafness phenotype, without other common diseases, including type 2 diabetes mellitus, cardiovascular events and neurological disorders. In addition, the pedigree manifested a high expressivity of deafness. PCR-Sanger sequencing was performed to identify the occurrence of the homoplasmic A1555G mutation, together with a further 22 genetic variations belonging to the East Asian haplogroup C (20). Among these, the T15943C mutation in tRNAThr was of particular interest. This mutation was identified in this Chinese family with hearing loss (Patients: I-2, II-1, II-5, II-8, II-10, III-9, III-10 and III-12) but absent in 100 healthy subjects. In addition, this mutation occurred at an evolutionary conserved position of tRNAThr (conventional position 63), which could disrupt 63T-55A base pairing and may result in failures in tRNA metabolism. In fact, RNA Fold results also indicated that this mutation caused a structural change of tRNAThr. Thus, it was speculated that the T15943C mutation affects tRNAThr function via the alteration of its secondary structure.
The phenotypic variability of matrilineal relatives within the pedigree suggested that nuclear genes or mtDNA genetic modifiers may serve key functions in deafness expression. For example, the TRMU A10S variant has previously been reported to increase the penetrance and expressivity of deafness-associated mitochondrial A1555G mutation (15). Mutations in the GJB2 gene have been regarded as important causes for non-syndromic hearing loss (14). However, an absence of functional sequence variants in TRMU and GJB2 genes in the current study indicated that these genes may not play active roles in deafness expression. Hence, other unidentified nuclear genes may be involved in the clinical manifestation of deafness in this pedigree.
In conclusion, the tRNAThr T15943C may be a secondary mutation for the phenotypic expression of mitochondrial deafness, and should be regarded as a risk factor for hearing impairment. Furthermore, screening for the presence of mitochondrial A1555G mutation is advisable, particularly for those who have a family history of using AmAn. The primary limitation of this study was the small sample size; further studies including larger cohorts of patients with hearing impairment should be performed in the future.
Acknowledgements
The authors would like to thank the patients and control subjects for participating in the present study. They would also like to thank the members in our otolaryngology department for discussing the project and responding to reviewer's comments.
Funding
The present study was supported by Fujian Provincial Clinical Priority Specialty Construction Project [Fujian Medical Health (2015) 593].
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LW, RL and JC designed the project, YC, MY and QW performed the experiments and LW wrote the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics of Fujian Provincial Hospital (Fuzhou, China) and all parents/guardians of participants provided written informed consent to participate.
Consent for publication
The patients' parents provided written informed consent for the publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Morton ME. Genetic epidemiology of hearing impairment. Ann NY Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 2.Bitner-Glindzicz M, Rahman S, Chant K, Marlow N. Gentamicin, genetic variation and deafness in preterm children. BMC Pediatr. 2014;14:66. doi: 10.1186/1471-2431-14-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischel-Ghodsian N. Genetic factors in aminoglycoside toxicity. Pharmacogenomics. 2005;6:27–36. doi: 10.1517/14622416.6.1.27. [DOI] [PubMed] [Google Scholar]
- 4.Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 5.Li R, Greinwald JH, Jr, Yang L, Choo DI, Wenstrup RJ, Guan MX. Molecular analysis of mitochondrial 12S rRNA and tRNASer(UCN) genes in paediatric subjects with non-syndromic hearing loss. J Med Genet. 2004;41:615–620. doi: 10.1136/jmg.2004.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neefs JM, Van de Peer Y, De Rijk P, Goris A, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1991;19(Suppl):S1987–S2015. doi: 10.1093/nar/19.suppl.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Connor M, Thomas CL, Zimmermann RA, Dahlberg AE. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 1997;25:1185–1193. doi: 10.1093/nar/25.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zwieb CD, Jemiolo DK, Jacob WF, Wagner R, Dahlberg AE. Characterization of a collection of deletion mutants at the 3′ end of 16S ribosomal RNA of Escherichia coli. Mol Gen Genet. 1986;203:256–264. doi: 10.1007/BF00333963. [DOI] [PubMed] [Google Scholar]
- 9.Qian Y, Guan MX. Interaction of aminoglycosides with human mitochondrial 12S rRNA carrying the deafness-associated mutation. Antimicrob Agents Chemother. 2009;53:4612–4618. doi: 10.1128/AAC.00965-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan MX, Fischel-Ghodsian N, Attardi G. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet. 1996;5:963–971. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- 11.Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: Analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz-Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of human mitochondrial DNA. Hum Mutat. 2006;27:1072–1081. doi: 10.1002/humu.20378. [DOI] [PubMed] [Google Scholar]
- 14.Dai ZY, Sun BC, Huang SS, Yuan YY, Zhu YH, Su Y, Dai P. Correlation analysis of phenotype and genotype of GJB2 in patients with non-syndromic hearing loss in China. Gene. 2015;570:272–276. doi: 10.1016/j.gene.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 15.Guan MX, Yan Q, Li X, Bykhovskaya Y, Gallo-Teran J, Hajek P, Umeda N, Zhao H, Garrido G, Mengesha E, et al. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am J Hum Genet. 2006;79:291–302. doi: 10.1086/506389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Y, Li Y, You J, Yang L, Chen B, Lu J, Guan MX. Mitochondrial tRNA(Glu) A14693G variant may modulate the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in a Han Chinese family. J Genet Genomics. 2009;36:241–250. doi: 10.1016/S1673-8527(08)60111-3. [DOI] [PubMed] [Google Scholar]
- 17.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The vienna RNA website. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. (Web Server Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 19.Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisà E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: Cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- 20.Roe A, Ma DP, Wilson RK, Wong JF. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- 21.Purohit P, Stern S. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature. 1994;370:659–662. doi: 10.1038/370659a0. [DOI] [PubMed] [Google Scholar]
- 22.Hamasaki K, Rando RR. Specific binding of aminoglycosides to a human rRNA construct based on a DNA polymorphism, which causes aminoglycoside-induced deafness. Biochemistry. 1997;36:12323–12328. doi: 10.1021/bi970962r. [DOI] [PubMed] [Google Scholar]
- 23.Usami S, Abe S, Akita J, Namba A, Shinkawa H, Ishii M, Iwasaki S, Hoshino T, Ito J, Doi K, et al. Prevalence of mitochondrial gene mutations among hearing impaired patients. J Med Genet. 2000;37:38–40. doi: 10.1136/jmg.37.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutchin T, Haworth I, Higashi K, Fischel-Ghodsian N, Stoneking M, Saha N, Arnos C, Cortopassi G. A molecular basis for human hypersensitivity to aminoglycoside antibiotics. Nuclear Acids Res. 1993;21:4174–4179. doi: 10.1093/nar/21.18.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Li R, Chen J, Liao Z, Zhu Y, Qian Y, Xiong S, Heman-Ackah S, Wu J, Choo DI, Guan MX. Mutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside-induced and non-syndromic hearing loss. Hum Genet. 2005;117:9–15. doi: 10.1007/s00439-005-1276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, Li Z, Zhu Y, Yang A, Li R, Zheng J, Cai Q, Peng G, Zheng W, Tang X, et al. Mitochondrial 12S rRNA variants in 1642 Han Chinese pediatric subjects with aminoglycoside-induced and nonsyndromic hearing loss. Mitochondrion. 2010;10:380–390. doi: 10.1016/j.mito.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.