Abstract
Hepatocellular carcinoma (HCC) can result from hepatitis B or C infection, fibrosis or cirrhosis. Transforming growth factor-β (TGF-β) is one of the main growth factors associated with fibrosis or cirrhosis progression in the liver, but its role is controversial in hepatocarcinogenesis. In the present study, the effect of TGF-β on the HCC Huh-7 and Huh-Bat cell lines was evaluated. To study the effect of TGF-β, Huh-7 and Huh-Bat cells were treated with TGF-β and a TGF-β receptor inhibitor (SB431542). Cell survival, cell cycle, numbers of side population (SP) cells and expression of the cancer stem cell marker cluster of differentiation (CD)133, epithelial-mesenchymal transition markers (E-cadherin, α-smooth muscle actin and vimentin) and TGF-β-regulated proteins [phospho-c-Jun N-terminal kinase (p-JNK), p-c-Jun and p-smad2] were investigated. TGF-β treatment resulted in decreased cell survival with a targeted effect on SP cells. Expression of CD133 and vimentin was upregulated by treatment with the TGF-β receptor antagonist SB431542, but not with TGF-β. By contrast, TGF-β induced accumulation of cells at G0/G1, and upregulated expression of p-JNK, p-c-Jun and p-smad2. However, these effects were blocked when cells were treated with TGF-β plus SB431542, indicating the specificity of the TGF-β effect. The present results indicated that TGF-β has anticancer effects mediated by survival inhibition of cancer stem cells, which may be developed as a novel therapy for HCC.
Keywords: hepatocellular carcinoma, transforming growth factor-β, cancer stem cells, side population, drug resistance, c-Jun N-terminal kinase signaling
Introduction
Hepatocellular carcinoma (HCC) is the most fatal cancer worldwide (1). Known causes of liver cancer include hepatitis B or C infection, alcohol-induced cirrhosis (2) and chronic diseases, including obesity and diabetes, which lead to liver fibrosis (3). HCC is characterized by mixed cell types, strong resistance to chemotherapy and local (rather than distant) metastasis, all of which present challenges to effective treatment.
Transforming growth factor-β (TGF-β) has been suggested as a useful biomarker for fibrosis, cirrhosis and epithelial-mesenchymal transition (EMT) in patients with HCC (4). EMT is a predisposing factor for the development of HCC (5) and has been shown to be induced by several cytokines, including interleukins, TGF-β and interferons (6).
TGF-β induces differentiation of embryonic stem cells, somatic stem cells and cancer stem cells (7). In addition, TGF-β induces differentiation of quiescent hepatic stellate cells into activated cells in the liver (8), fibrosis through extracellular matrix (ECM) deposition and HCC (9). TGF-β binds several proteins, activates several signaling pathways and is involved in HCC progression (10). TGF-β enhances tumor initiating ability through EMT of cancer stem cells (11). Tumor initiating ability is a known characteristic of cancer stem cells (12).
Cancer stem cells are characterized by resistance to anticancer drugs (13), high sphere forming efficacy (14), EMT (15) and specific protein expression that promotes transformation (16). HCC stem cells express cluster of differentiation (CD)90, CD133 and CD24 (17), and retain side population (SP) characteristics, as well as resistance to anticancer drugs (18). In addition, liver cancer typically exhibits strong resistance to anticancer drugs, since detoxification is one of its functions (19).
The present study focused on the effect of TGF-β on cancer stem cells in vitro. The proliferation, fraction of SP cells and expression of relevant signaling molecules were assessed following TGF-β and SB431542 treatment using Huh-7 and Huh-Bat cells as a model for HCC.
Materials and methods
Cell culture
Huh-7 and Huh-Bat [Huh-7 transfected with human Na+-taurocholate co-transporting polypeptide (bile acid transporter) cDNA] cell lines were obtained from the Korean Cell Line Bank (Seoul, Korea). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.). For adherent cultures, 5×105 cells were seeded onto tissue culture dishes (BD Biosciences, San Jose, CA, USA). All cultures were maintained at 37°C in a humidified 5% CO2 atmosphere.
Cell survival rate following SB431542 and TGF-β treatment
Cells were cultured (5×105 cells) in DMEM containing 10% FBS at 37°C. After 24 h, the cells were washed twice with PBS, and fresh medium was added. Cells were treated for 72 h at 37°C with 0.01% dimethyl sulfoxide (vehicle control), 1 µM SB431542 (TGF-β inhibitor; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), 1 ng/ml TGF-β (LC Laboratories, Woburn, MA, USA) or 1 µM SB431542 plus 1 ng/ml TGF-β. Following treatment for 72 h, cells were harvested for subsequent analysis. Cell survival rates were estimated by the number of viable cells, counted by positive staining with 0.4% trypan blue dye (Sigma-Aldrich; Merck KGaA) in a Neubauer chamber.
Cell cycle analysis
To determine the effect of treatment on cell cycle progression, at 72 h following treatment, cell samples were harvested and fixed in 70% ethanol for 25 min. Subsequent to washing in PBS, samples were treated with 100 µg/ml RNaseA for 90 min at 37°C, and stained with 10 µg/ml propidium iodide (PI). Flow cytometry was performed in triplicate for each experiment using a FACSCalibur (BD Biosciences).
SP cell analysis
SP cell analysis was performed as reported previously (20). Briefly, cells were detached, transferred to microcentrifuge tubes, washed in PBS and resuspended at 1×106 cells/ml. Samples were stained with Hoechst 33342 dye (5 µg/ml; Sigma-Aldrich; Merck KGaA) or Hoechst dye and 50–100 µM verapamil (an efflux blocker), in DMEM containing 10% FBS for 90 min at 37°C. Following staining, samples were centrifuged for 3 min at 150 × g and 4°C and collected for analysis of the SP fraction. PI (1 µg/ml) was added prior to analysis to identify and exclude dead cells. Samples were analyzed using a FACSAria (BD Biosciences).
Expression of CD133, E-Cadherin, α-smooth muscle actin (SMA) and vimentin
To determine the phenotype of the cell populations following TGF-β and/or SB431542 treatment, cells were harvested and stained for CD133, E-cadherin, α-SMA and vimentin expression. Briefly, cell samples were washed twice with PBS, then fixed and permeabilized according to the manufacturer's protocol (BD Biosciences). Following centrifugation for 3 min at 150 × g and 4°C, cells were washed and incubated for 45 min at 4°C with anti-human CD133-APC (cat. no. 130-090-826; 1:50; Miltenyi Biotec Ltd., Bergisch Gladbach, Germany), PE anti-human E-cadherin (cat. no. 562870; 1:50; BD Biosciences), anti-human α-SMA-fluorescein isothiocyanate (cat. no. ab8211; 1:50; Abcam, Cambridge, UK) and anti-human vimentin-Alexa488 (cat. no. 562338; 1:50; BD Biosciences). Following staining, cells were washed with PBS three times, and flow cytometry analysis was performed in triplicate using FACSCalibur (BD Biosciences).
Immunoblotting
Total cell lysates were prepared in 100 µl lysis buffer (Cell Signaling Technology, Inc., Danvers, MA, USA). Protein concentrations were measured using Bio-Rad protein assay kit I, according to the manufacturer's protocol (cat. no. 5000001; Bio-Rad Laboratories, Inc., Hercules, CA, USA). Cell lysates were separated using 10% SDS-PAGE, and proteins were electrotransferred onto Hybond-enhanced chemiluminescence nitrocellulose membranes (GE Healthcare Life Sciences, Chalfont, UK). Blots were blocked for 1 h with blocking buffer (5% skim milk in TBS-tween 20) and incubated overnight at 4°C with mouse anti-phospho-SAPK/JNK (Thr183/Tyr185; cat. no. 9251; 1:1,000; Cell Signaling Technology, Inc.), rabbit anti-phospho-c-Jun (ser63; cat. no. 2361; 1:1,000; Cell Signaling Technology, Inc.), rabbit anti-phospho-smad2 (cat. no. 8828; 1:1,000; Cell Signaling Technology), and mouse anti-β-actin (cat. no. sc47778; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Blots were washed and incubated for 1 h at room temperature with peroxidase-conjugated AffiniPure rabbit anti-mouse IgG (cat. no. 315-005-003; 1:2,500; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) or peroxidase-conjugated AffiniPure mouse anti-rabbit IgG (cat. no. 211-005-109; 1:2,500; Jackson ImmunoResearch Laboratories, Inc.). Labeled proteins were detected using an enhanced chemiluminescence detection system (GE Healthcare Life Sciences).
Statistical analysis
At least three replicate experiments were performed for all analyses. Data are expressed as the mean ± standard error. Student's t-tests were used to compare the results of treated and control cells. P<0.05 was considered to indicate a statistically significant difference.
Results
TGF-β treatment inhibits proliferation of Huh-7 and Huh-Bat cells
To determine the effect of TGF-β on the survival rate of Huh-7 and Huh-Bat cells, cultures were treated with 1 µM SB431542, 1 ng/ml TGF-β or 1 µM SB431542 plus 1 ng/ml TGF-β for 72 h at 37°C. The survival of Huh-7 and Huh-Bat cells was reduced by TGF-β treatment and this effect was blocked by 1 µM SB431542 (Fig. 1). These results revealed that TGF-β specifically inhibited the survival of HCC cells in vitro.
Figure 1.
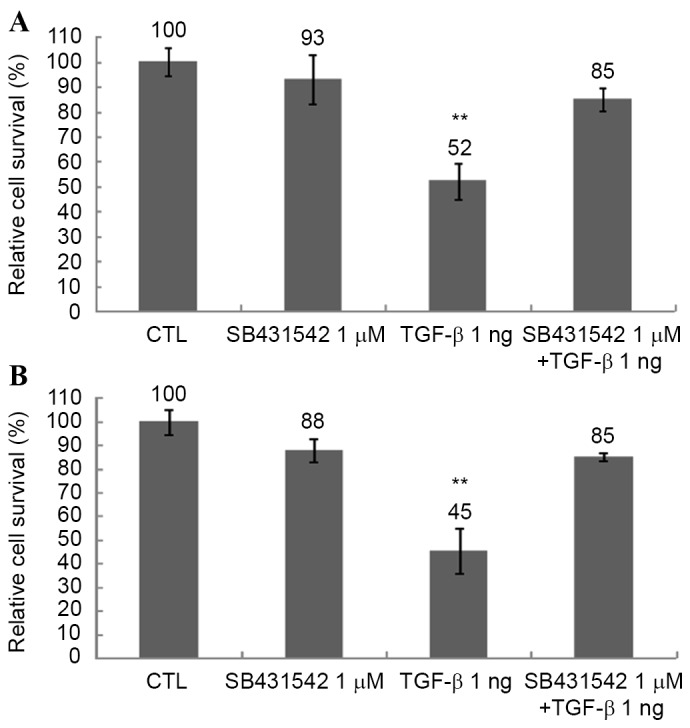
Cell survival of Huh-7 and Huh-Bat cells following SB431542 and TGF-β treatment. Cells (5×105) were seeded onto plates in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cultures were incubated for 72 h with dimethyl sulfoxide (control), 1 µM SB431542, 1 ng/ml TGF-β or 1 µM SB431542 plus 1 ng/ml TGF-β. Cell survival was assessed by cell counting using trypan blue dye exclusion. (A) Huh-7 cells. (B) Huh-Bat cells. The relative cell survival rate is shown as survival percentage vs. control cells. Values are presented as the mean ± standard error from at least three independent experiments. *P≤0.05; **P≤0.01. TGF-β, transforming growth factor-β; CTL, control.
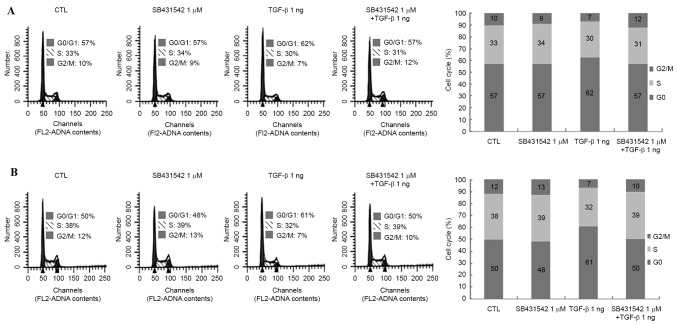
TGF-β treatment induces cell cycle arrest in Huh-7 and Huh-Bat cells
TGF-β is known to induce cell cycle arrest in several types of cancer (21). TGF-β was found to decrease the survival rate of Huh-7 and Huh-Bat cells after 72 h (Fig. 1). To confirm the inhibitory effects of TGF-β on cell survival rate, the cell cycle was analyzed following TGF-β and SB431542 treatment in Huh-7 and Huh-Bat cells. TGF-β induced a 5% increase of Huh-7 cells in G0/G1 and an 11% increase of Huh-Bat cells in G0/G1 compared with the control; cell cycle arrest was blocked by SB431542 (Fig. 2). These findings indicated that TGF-β reduces survival of HCC cells by induction of cell cycle arrest.
Figure 2.
Cell cycle analysis of Huh-7 and Huh-Bat cells following SB431542 and TGF-β treatment. Huh-7 or Huh-Bat cells (5×105) were seeded onto Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cells were incubated for 72 h with dimethyl sulfoxide (control), 1 µM SB431542, 1 ng/ml TGF-β or 1 µM SB431542 plus 1 ng/ml TGF-β. Cells were stained with propidium iodide and analyzed by flow cytometry. (A) Huh-7 cells. (B) Huh-Bat cells. TGF-β, transforming growth factor-β; CTL, control.
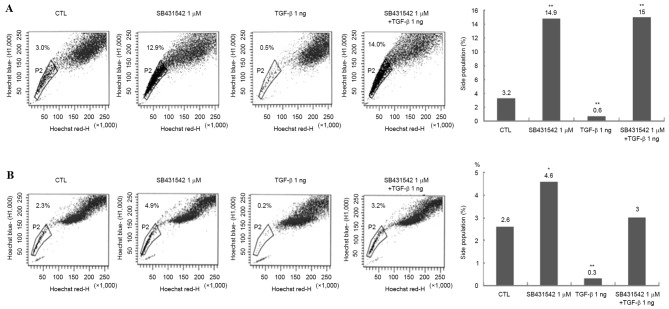
TGF-β treatment reduces the number of SP cells in Huh-7 and Huh-Bat cultures
To determine whether TGF-β treatment targeted cancer stem cells in Huh-7 and Huh-Bat cultures, SP cells were analyzed using Hoechst dye staining following 1 µM SB431542, 1 ng/ml TGF-β or 1 µM SB431542 plus 1 ng/ml TGF-β treatment for 72 h. TGF-β treatment reduced the proportion of SP cells, whereas SB431542 treatment increased the proportion of SP cells, and TGF-β plus SB431542 blocked the effect of TGF-β on SP cells (Fig. 3). These results indicated that the effect of TGF-β may be specific for cancer stem cells.
Figure 3.
SP cells in Huh-7 and Huh-Bat cells following SB431542 and TGF-β treatment. Huh-7 or Huh-Bat cells (5×105) were seeded onto Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The cultures were incubated for 72 h with dimethyl sulfoxide (control), 1 µM SB431542, 1 ng/ml TGF-β or 1 µM SB431542 plus 1 ng/ml TGF-β. (A) Huh-7 cells. (B) Huh-Bat cells. SP cells were assayed using a FACSAria with a 515 nm SP filter (Hoechst blue). *P<0.05; **P<0.01, compared with control and treated cells. SP, side population; TGF-β, transforming growth factor-β; CTL, control.
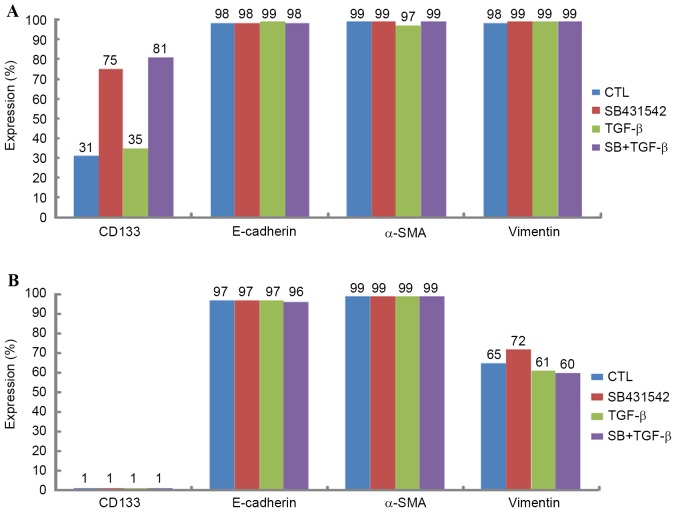
Expression of CD133, E-cadherin, α-SMA and vimentin in Huh-7 and Huh-bat cells following TGF-β treatment
To confirm the effect of TGF-β on SP cells, the expression levels of the cancer stem cell markers CD133 and epithelial mesenchymal transition markers (EMT, E-cadherin, α-SMA and vimentin) were analyzed. Expression of the cancer stem cell marker CD133 and EMT markers (E-cadherin, α-SMA and vimentin) differed depending on cell type. Huh-7 cells expressed high levels of CD133 and vimentin compared with Huh-Bat cells (Fig. 4). However, following SB431542 treatment, expression of CD133 and vimentin was upregulated in Huh-Bat cells. By contrast, expression of these markers was similar in control and TGF-β-treated cells (Fig. 4). The present results revealed that the expression of cancer stem cell and EMT markers was dependent on cell type, and was upregulated by blockade of TGF-β signaling.
Figure 4.
Expression of CD133, E-cadherin, α-SMA and vimentin in Huh-7 and Huh-Bat cells following SB431542 and TGF-β treatment. Huh-7 or Huh-Bat cells (5×105 cells) were seeded in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cells were incubated for 72 h with dimethyl sulfoxide (control), 1 µM SB431542, 1 ng/ml TGF-β or 1 µM SB431542 plus 1 ng/ml TGF-β. Expression of CD133, E-cadherin, α-SMA and vimentin was determined by fluorescence-activated cell sorting analysis. (A) Huh-7 cells, (B) Huh-Bat cells. α-SMA, α-smooth muscle actin; TGF-β, transforming growth factor-β; CTL, control; CD133, cluster of differentiation 133.
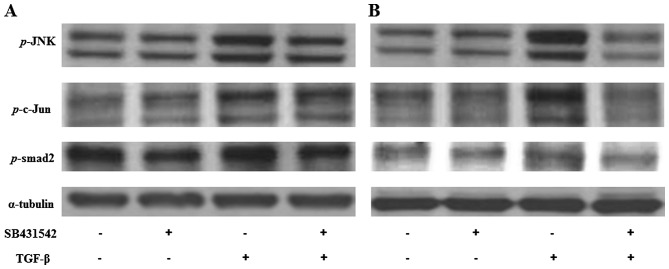
Expression of p-JNK, p-c-Jun and p-smad2 in Huh-7 and Huh-Bat cells following TGF-β treatment
The expression of JNK signaling molecules, including p-SAPK/JNK, p-c-Jun and p-smad2, which are all associated with protein regulation by TGF-β in Huh-7 and Huh-Bat cells following SB431542 and TGF-β treatment, was examined. p-SAPK/JNK, p-c-Jun and p-smad2 were upregulated by TGF-β treatment, which was blocked by TGF-β plus SB431542 treatment (Fig. 5). These results indicated that TGF-β specifically upregulates proteins associated with growth inhibition that targeted the SP subpopulation.
Figure 5.
Expression of p-SAPK/JNK, p-c-Jun, and p-smad2 in Huh-7 and Huh-Bat cells following SB431542 and TGF-β treatment. Huh-7 or Huh-Bat cells (5×105 cells) were seeded in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cells were incubated for 72 h with DMSO (control), 1 µM SB431542, 1 ng/ml TGF-β or 1 µM SB431542 plus 1 ng/ml TGF-β. (A) Huh-7 cells. (B) Huh-bat cells. Expression of p-JNK, p-c-Jun and p-smad2 was detected by immunoblotting. Cells were treated with distilled water and DMSO (lane 1), 1 µM SB431542 (lane 2), 1 ng/ml TGF-β (lane 3) or 1 µM SB431542 plus 1 ng/ml TGF-β (lane 4). TGF-β, transforming growth factor-β; SAPK, stress-activated protein kinase; JNK, c-Jun N-terminal kinase; DMSO, dimethyl sulfoxide.
Discussion
TGF-β performs a role in liver fibrosis through activation of stellate cells and TGF-β promotion of ECM accumulation. TGF-β also plays an important role in differentiation of stem cells and has been used as an agent for differentiation of stem cells. In relevance to cancer, TGF-β has been shown to induce metastatic ability through EMT or suppress cancer cell proliferation through cell cycle arrest. In the present study, the anticancer effects of TGF-β on cancer stem cells were studied in HCC cell lines. It was hypothesized that TGF-β treatment reduces the proliferation of cancer stem cells. Therefore, proliferation, SP cell numbers and expression of cancer stem cell markers, EMT markers and TGF-β signaling molecules were evaluated following TGF-β treatment of Huh-7 and Huh-Bat cells.
TGF-β treatment decreased survival cell (specifically of the SP fraction) and upregulated expression of JNK signaling molecules. These effects were blocked by the TGF-β receptor inhibitor SB431542. Furthermore, the effect of TGF-β was through cell cycle arrest. These results are similar to those of previous studies investigating the tumor suppressive effects of TGF-β in liver cancer. Baek et al reported that TGF-β inhibited cell proliferation through cell cycle arrest (22). Senturk et al demonstrated that TGF-β induced downregulation of p21 (Cip1) and p15 (Ink4b) and inhibited cell proliferation through cell cycle arrest in HCC cell lines (23). In addition, Hashimoto et al identified that TGF-β inhibited cell proliferation through G2 arrest in HCC cells (24).
With regard to TGF-β signaling molecules, TGF-β-induced upregulation of p-JNK, p-c-Jun and p-smad2 expression was observed. Hu et al reported that upregulated TGF-β/smad signaling was associated with suppression of hepatocarcinogenesis (25). Dzieran et al also reported that TGF-β inhibited proliferation and induced apoptosis through increased smad3 activity in human HCC cell lines (26). Park et al reported that TGF-β1 induced apoptosis through the activation of p38, JNK and caspases (27). Suzuki et al reported that TGF-β regulated autophagy activation, which is involved in cancer progression through the smad and JNK pathways (28). Kim et al reported that TGF-β induces apoptosis through cleavage of Bcl-2-associated death promoter in a smad3-dependent mechanism in FaO HCC cells (29). The present results indicated that TGF-β inhibited proliferation through the activation of JNK and smad signaling.
SPs have been proposed as cancer stem cells due to their tumor initiation ability and drug resistance (18). TGF-β was found to decrease the SP fraction through inhibition of cell proliferation. Therefore, the present results demonstrated that TGF-β treatment specifically targeted cancer stem cells in HCC cell lines. Ehata et al reported that TGF-β decreased the number of SP cells through repression of ATP binding cassette subfamily G member 2 (ABCG2) transcription, which prevented the direct binding of smad2/3 to its promoter/enhancer in gastric carcinoma cells (30).
TGF-β is a known EMT-inducing agent in several cancers (31). However, in the present study, TGF-β did not induce upregulation of the liver cancer stem cell marker (CD133) and EMT markers (E-cadherin, α-SMA and vimentin). This may be due to the high level of expression of EMT markers (E-cadherin, α-SMA and vimentin) in Huh-7 and Huh-bat cells. Relevant to EMT, Yin et al reported that TGF-β induced EMT but decreased SP cells and expression of ABCG2 in the breast cancer MCF7 cell line (32). In contrast to the present results, You et al reported that TGF-β upregulated CD133 expression and increased tumor initiation ability in an HCC cell line (33). Martin et al reported that TGF-β increased the proportion of CD133+ cells in liver cancer cell lines (34). Nishimura et al reported that TGF-β induced increased proliferative capacity and drug resistance in SP cells in the hepatic tumor cell line K-251 (35). These findings were different from the present results, and may be due to differences in culture environment and drug treatment methods.
In conclusion, the present results indicated that TGF-β may be used to specifically target SP cells through the induction of JNK signaling. This effect of TGF-β on liver cancer stem cells indicated that TGF-β requires investigation as a novel method to treat liver cancer.
Acknowledgements
The present study was supported by grants from the Seoul National University Hospital [grant no. 0320140200 (2014–1290)] and the ILDONG pharmaceutical corporation [grant no. 0620133000 (2013–1890)].
References
- 1.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.Armengol C, Bartolí R, Sanjurjo L, Serra I, Amézaga N, Sala M, Sarrias MR. Role of scavenger receptors in the pathophysiology of chronic liver diseases. Crit Rev Immunol. 2013;33:57–96. [PubMed] [Google Scholar]
- 3.Petta S, Craxi A. Hepatocellular carcinoma and non-alcoholic fatty liver disease: From a clinical to a molecular association. Curr Pharm Des. 2010;16:741–752. doi: 10.2174/138161210790883787. [DOI] [PubMed] [Google Scholar]
- 4.Song BC, Chung YH, Kim JA, Choi WB, Suh DD, Pyo SI, Shin JW, Lee HC, Lee YS, Suh DJ. Transforming growth factor-beta1 as a useful serologic marker of small hepatocellular carcinoma. Cancer. 2002;94:175–180. doi: 10.1002/cncr.10170. [DOI] [PubMed] [Google Scholar]
- 5.Reichl P, Haider C, Grubinger M, Mikulits W. TGF-β in epithelial to mesenchymal transition and metastasis of liver carcinoma. Curr Pharm Des. 2012;18:4135–4147. doi: 10.2174/138161212802430477. [DOI] [PubMed] [Google Scholar]
- 6.Saito T, Yoshida K, Matsumoto K, Saeki K, Tanaka Y, Ong SM, Sasaki N, Nishimura R, Nakagawa T. Inflammatory cytokines induce a reduction in E-cadherin expression and morphological changes in MDCK cells. Res Vet Sci. 2014;96:288–291. doi: 10.1016/j.rvsc.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Selesniemi K, Reedy M, Gultice A, Guilbert LJ, Brown TL. Transforming growth factor-beta induces differentiation of the labyrinthine trophoblast stem cell line SM10. Stem Cells Dev. 2005;14:697–711. doi: 10.1089/scd.2005.14.697. [DOI] [PubMed] [Google Scholar]
- 8.Kim JB, Ann YH, Park SY, Jee HG, Kim HR, Lee JH, Yu SJ, Lee HS, Kim YJ. Side population in LX2 cells decreased by transforming growth factor-β. Hepatol Res. 2014;44:229–237. doi: 10.1111/hepr.12106. [DOI] [PubMed] [Google Scholar]
- 9.Giannelli G, Mazzocca A, Fransvea E, Lahn M, Antonaci S. Inhibiting TGF-β signaling in hepatocellular carcinoma. Biochim Biophys Acta. 2011;1815:214–223. doi: 10.1016/j.bbcan.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Meindl-Beinker NM, Matsuzaki K, Dooley S. TGF-β signaling in onset and progression of hepatocellular carcinoma. Dig Dis. 2012;30:514–523. doi: 10.1159/000341704. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Zhu F, Xiao L, Wang M, Tian R, Shi C, Qin R. Side population cells in human gallbladder cancer cell line GBC-SD regulated by TGF-β-induced epithelial-mesenchymal transition. J Huazhong Univ Sci Technolog Med Sci. 2011;31:749–755. doi: 10.1007/s11596-011-0671-1. [DOI] [PubMed] [Google Scholar]
- 12.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 13.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 14.Cioce M, Gherardi S, Viglietto G, Strano S, Blandino G, Muti P, Ciliberto G. Mammosphere-forming cells from breast cancer cell lines as a tool for the identification of CSC-like- and early progenitor-targeting drugs. Cell Cycle. 2010;9:2878–2887. doi: 10.4161/cc.9.14.12371. [DOI] [PubMed] [Google Scholar]
- 15.Pinto CA, Widodo E, Waltham M, Thompson EW. Breast cancer stem cells and epithelial mesenchymal plasticity-implications for chemoresistance. Cancer Lett. 2013;341:56–62. doi: 10.1016/j.canlet.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Kai K, Arima Y, Kamiya T, Saya H. Breast cancer stem cells. Breast Cancer. 2010;17:80–85. doi: 10.1007/s12282-009-0176-y. [DOI] [PubMed] [Google Scholar]
- 17.Michishita M, Ezaki S, Ogihara K, Naya Y, Azakami D, Nakagawa T, Sasaki N, Arai T, Shida T, Takahashi K. Identification of tumor-initiating cells in a canine hepatocellular carcinoma cell line. Res Vet Sci. 2014;96:315–322. doi: 10.1016/j.rvsc.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, Nakauchi H, Taniguchi H. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 19.Dietrich CG, Geier A, Wasmuth HE, Matern S, Gartung C, de Waart DR, Elferink RP. Influence of biliary cirrhosis on the detoxification and elimination of a food derived carcinogen. Gut. 2004;53:1850–1855. doi: 10.1136/gut.2003.037507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damdinsuren B, Nagano H, Kondo M, Natsag J, Hanada H, Nakamura M, Wada H, Kato H, Marubashi S, Miyamoto A, et al. TGF-beta1-induced cell growth arrest and partial differentiation is related to the suppression of Id1 in human hepatoma cells. Oncol Rep. 2006;15:401–408. [PubMed] [Google Scholar]
- 22.Baek HJ, Pishvaian MJ, Tang Y, Kim TH, Yang S, Zouhairi ME, Mendelson J, Shetty K, Kallakury B, Berry DL, et al. Transforming growth factor-β adaptor, β2-spectrin, modulates cyclin dependent kinase 4 to reduce development of hepatocellular cancer. Hepatology. 2011;53:1676–1684. doi: 10.1002/hep.24128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senturk S, Mumcuoglu M, Gursoy-Yuzugullu O, Cingoz B, Akcali KC, Ozturk M. Transforming growth factor-beta induces senescence in hepatocellular carcinoma cells and inhibits tumor growth. Hepatology. 2010;52:966–974. doi: 10.1002/hep.23769. [DOI] [PubMed] [Google Scholar]
- 24.Hashimoto O, Ueno T, Kimura R, Ohtsubo M, Nakamura T, Koga H, Torimura T, Uchida S, Yamashita K, Sata M. Inhibition of proteasome-dependent degradation of Wee1 in G2-arrested Hep3B cells by TGF beta 1. Mol Carcinog. 2003;36:171–182. doi: 10.1002/mc.10111. [DOI] [PubMed] [Google Scholar]
- 25.Hu X, Rui W, Wu C, He S, Jiang J, Zhang X, Yang Y. Compound astragalus and salvia miltiorrhiza extracts suppress hepatocarcinogenesis by modulating transforming growth factor-β/Smad signaling. J Gastroenterol Hepatol. 2014;29:1284–1291. doi: 10.1111/jgh.12490. [DOI] [PubMed] [Google Scholar]
- 26.Dzieran J, Fabian J, Feng T, Coulouarn C, Ilkavets I, Kyselova A, Breuhahn K, Dooley S, Meindl-Beinker NM. Comparative analysis of TGF-β/Smad signaling dependent cytostasis in human hepatocellular carcinoma cell lines. PLoS One. 2013;8:e72252. doi: 10.1371/journal.pone.0072252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SS, Eom YW, Kim EH, Lee JH, Min DS, Kim S, Kim SJ, Choi KS. Involvement of c-Src kinase in the regulation of TGF-beta1-induced apoptosis. Oncogene. 2004;23:6272–6281. doi: 10.1038/sj.onc.1207856. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki HI, Kiyono K, Miyazono K. Regulation of autophagy by transforming growth factor-β (TGF-β) signaling. Autophagy. 2010;6:645–647. doi: 10.4161/auto.6.5.12046. [DOI] [PubMed] [Google Scholar]
- 29.Kim BC, Mamura M, Choi KS, Calabretta B, Kim SJ. Transforming growth factor beta 1 induces apoptosis through cleavage of BAD in a Smad3-dependent mechanism in FaO hepatoma cells. Mol Cell Biol. 2002;22:1369–1378. doi: 10.1128/MCB.22.5.1369-1378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehata S, Johansson E, Katayama R, Koike S, Watanabe A, Hoshino Y, Katsuno Y, Komuro A, Koinuma D, Kano MR, et al. Transforming growth factor-β decreases the cancer-initiating cell population within diffuse-type gastric carcinoma cells. Oncogene. 2011;30:1693–1705. doi: 10.1038/onc.2010.546. [DOI] [PubMed] [Google Scholar]
- 31.Tirino V, Camerlingo R, Bifulco K, Irollo E, Montella R, Paino F, Sessa G, Carriero MV, Normanno N, Rocco G, Pirozzi G. TGF-β1 exposure induces epithelial to mesenchymal transition both in CSCs and non-CSCs of the A549 cell line, leading to an increase of migration ability in the CD133+ A549 cell fraction. Cell Death Dis. 2013;4:e620. doi: 10.1038/cddis.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin L, Castagnino P, Assoian RK. ABCG2 expression and side population abundance regulated by a transforming growth factor beta-directed epithelial-mesenchymal transition. Cancer Res. 2008;68:800–807. doi: 10.1158/0008-5472.CAN-07-2545. [DOI] [PubMed] [Google Scholar]
- 33.You H, Ding W, Rountree CB. Epigenetic regulation of cancer stem cell marker CD133 by transforming growth factor-beta. Hepatology. 2010;51:1635–1644. doi: 10.1002/hep.23544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M, Ancey PB, Cros MP, Durand G, Le Calvez-Kelm F, Hernandez-Vargas H, Herceg Z. Dynamic imbalance between cancer cell subpopulations induced by transforming growth factor beta (TGF-β) is associated with a DNA methylome switch. BMC Genomics. 2014;15:435. doi: 10.1186/1471-2164-15-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura T, Azuma T, Yokoyama A, Ochiai H, Saito H, Hibi T. New mechanism of transforming growth factor-beta signaling in hepatoma: Dramatic up-regulation of tumor initiating cells and epidermal growth factor receptor expression. Hepatol Res. 2009;39:501–509. doi: 10.1111/j.1872-034X.2008.00480.x. [DOI] [PubMed] [Google Scholar]