Abstract
The aim of the present study was to examine the expression of phosphoglycerate mutase 1 (PGAM1) in astrocytomas, and to investigate its role in the progression of astrocytomas. The expression of PGAM1 mRNA in rat C6 glioma cells and normal astrocytes was determined using the reverse transcription-semi-quantitative polymerase chain reaction, and immunohistochemistry was used to detect the expression of PGAM1 protein in human astrocytomas and adjacent brain tissue. These data suggested that the expression of PGAM1 in rat C6 glioma cells was significantly increased compared with that of normal astrocytes (P<0.05), and the expression of PGAM1 protein in human astrocytoma tissue was significantly increased compared with that of the brain tissue surrounding the tumor (P<0.05). In addition, PGAM1 protein was more frequently expressed in high-grade astrocytomas compared with low-grade astrocytomas. These data indicate that the expression of PGAM1 is increased in C6 cells and human astrocytomas, and PGAM1 is probably involved in the tumorigenesis and progression of glioma, which may be a potential target for glioma treatment.
Keywords: phosphoglycerate mutase l, glioma, astrocytoma, semi-quantitative reverse transcription polymerase chain reaction, immunohistochemistry
Introduction
Gliomas are the most common primary malignant brain tumor with poor clinical outcome and account for 40–50% of primary intracranial neoplasms (1). The etiology and pathogenesis of gliomas remain to be elucidated. Despite the advances in therapeutic approaches, treatment offers limited aid in prolonging survival (2). In view of the overall poor outcome with current therapies, a better understanding of glioma etiology is crucial for future development of more effective treatments to cure this rapidly progressing disease.
It has been demonstrated that the growth of cancer cells is primarily dependent on the glycolysis pathway (3). Phosphoglycerate mutase 1 (PGAM1) catalyzes the conversion of 3-phosphoglycerate (3-PG) to 2-phosphoglycerate (2-PG) to release energy and is one of the key enzymes in the glycolytic pathway. A previous study by Ramanathan et al (3) suggested that knocking out the glycolysis pathway may cause extreme damage to tumor cells. However, the role of PGAM1 in glioma is poorly investigated.
In the present study, PGAM1 mRNA expression in rat C6 glioma cells and astrocytes, and the protein expression in human infiltrating astrocytomas of different World Health Organization (WHO) grades (4) and peritumoral brain tissue were investigated, and the role of PGAM1 in glioma proliferation and progression was discussed.
Materials and methods
Animals and cells
A total of 40 male adult Sprague-Dawley rats about 3 months old, weight 250±30 g, were provided by the Experimental Animal Center of Shandong University (Jinan, China), and maintained in a climate-controlled and sound isolated room (21°C) under a 12:12-h light:dark cycle, with 40–60% relative humidity and fed on laboratory chow and water ad libitum. Following 1-week acclimation, rats were sacrificed by intraperitoneal injection of sodium pentobarbital (45 mg/kg). Brain tissue was obtained for the experiments. The glioma C6 cell lines were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). Ethical approval was provided by the Ethics Committee of Qilu Hospital (Shandong, China).
Patient specimens
A total of 102 cases of astrocytomas treated in the Department of Neurosurgery, Shandong Cancer Hospital (Jinan, China) and Qilu Hospital of Shandong University (Jinan, China) between December 2011 and June 2013 were collected (59 males and 43 females; median age, 47 years; age range, 28–66 years). According to WHO 2007 criteria, 38 cases were WHO grade II, 30 cases were WHO grade III and 34 were WHO grade IV (4). A total of 11 cases of peritumoral brain tissue were also collected. No patients had ever received chemotherapy or radiation therapy prior to surgery.
Reagents
TRIzol was purchased from Invitrogen (Thermo Fisher Scientific, Inc. Waltham, MA, USA). Chloroform, isopropanol and diethyl pyrocarbonate-treated water were purchased from Shanghai Sangon Biological Engineering Technology Services Ltd. (Shanghai, China). Ethidium bromide (EB) was purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany); the reverse transcription-polymerase chain reaction (RT-PCR) PrimeScript™ RT reagent kit (cat no. RR037A) was purchased from Takara Biotechnology Co., Ltd. (Dalian, China), deoxynucleotide triphosphates (dNTPs), Taq DNA polymerase and CASsuper 7Kb DNA Marker were purchased from Promega Corporation (Madison, WI, USA). Poly-L-lysine, immunohistochemical hypersensitivity kit, goat anti-human PGAM1 antibody (sc-376638), 3,3′-diaminobenzidine (DAB) dye (sc-24982) and antibody dilutions (sc-2023) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
Culture, passage and identification of astrocytes
Bilateral brain cortex tissue (~0.5 g) from 1-week-old SD rats was collected and washed with Ca2+/Mg2+-free Hanks solution (sc-391061; Santa Cruz Biotechnology, Inc.) twice. The brain tissue was shredded and kept at room temperature for 10 min with trypsin (sc-391055; 1:250; Santa Cruz Biotechnology, Inc.). The resulting brain tissue was transferred into the centrifuge tube and centrifuged at 1,000 × g for 3 min at 4°C for cell separation. Cell suspension was collected and transferred into a new tube and centrifuged at 1,500 × g for 5 min at 4°C. The second cell suspension was seeded into a culture flask that was previously coated (4°C, overnight) with poly-L-lysine (sc-286689; 0.1%; Santa Cruz Biotechnology, Inc.). Cells were cultured with Dulbecco's modified Eagle's medium (DMEM)/Ham's F12 with 15% fetal bovine serum (FBS; 16250086; Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 incubator. Finally, astrocytes were identified with glial fibrillary acidic protein (GFAP; sc-33673; Santa Cruz Biotechnology, Inc.) immunofluorescence (5).
The immunostaining steps were as follows: Coverslips with their attached cells were removed from the culture medium and washed three times in phosphate buffered saline (PBS; sc-286634; Santa Cruz Biotechnology, Inc.) before being fixed in 4% paraformaldehyde (sc-281692; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. The coverslips were then washed three times in cold PBS (5 min each). Prior to staining, cells were permeabilised in PBS containing 0.01% Triton for 10 min at room temperature and then washed three times in cold PBS (3 min each). The coverslips were blocked with 10% normal goat serum for 1 h at 37°C. Each coverslip was incubated with 20 µl mouse anti-GFAP primary antibody (cat no. sc-33673; Santa Cruz Biotechnology, Inc.) 1:200 at 4°C overnight. Negative controls were incubated identically but without primary antibody. Coverslips were then washed three times in PBS, then incubated with a goat anti-rabbit secondary antibody conjugated to FITC (sc-2010; 1:100; Santa Cruz Biotechnology, Inc.) for 30 min at room temperature and washed three times in cold PBS 5 min each. Then the coverslips were incubated with 20 µl DAPI (sc-3598; Santa Cruz Biotechnology, Inc.) for 5 min at room temperature in the dark. Images were captured using a confocal microscope (PerkinElmer, Inc., Waltham, MA, USA) at a wavelength of 520 nm.
Culture of C6 glioma cells
C6 glioma cells were cultured in DMEM/Ham's F12 containing 15% FBS at 37°C in a 5% CO2 incubator. Following cells reaching exponential phase, they were subcultured into two culture flasks.
Total RNA extraction and detection of PGAM1 mRNA expression in rat astrocytes and C6 glioma cells
Extraction of total RNA from C6 and astrocytes
Once the cultured cells were confluent in the culture flasks, the culture medium was removed and cells were washed for 30 sec with PBS twice. Total RNA was extracted using a TRIzol RNA extraction kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA concentration and quality were determined using a NanoDrop ND-1000 spectrophotometer (A260/A280 values were 1.80–2.0; Thermo Fisher Scientific, Inc.) and gel electrophoresis.
Reverse transcription (first-strand cDNA synthesis)
A total of 2 µg total RNA, 2.0 µl 10 µmol/l oligo dNTPs, 2.0 µl 10X PCR buffer, 2.0 µl 5 µmol/l dNTPmix, 1.0 µl dNTP mixture, RNasin® Ribonuclease Inhibitor (4 U/µl; Invitrogen; Thermo Fisher Scientific, Inc.), and an appropriate amount of diethyl pyrocarbonate-treated water was added to a total volume of 20 µl, and thenplaced into a 0.5 ml Eppendorf tube. The mixture was incubated at 37°C for 60 min, and the reaction was stopped by heating at 70°C for 15 min. Then, the reaction tube was transferred to ice, and 20 µl Tris-EDTA buffer (Invitrogen; Thermo Fisher Scientific, Inc.) was added.
PCR and electrophoresis of PCR products
PCR primers were synthesized by Beyotime Institute of Biotechnology (Shanghai, China). The PGAM1 forward primer sequence was 5′-CCTCCTGTGAGAGCCTGAAG-3′ (nt452-nt471) and the PGAM1 reverse primer sequence was 5′-CTTCTTCACCTTGCCCTGAG-3′ (nt762-nt743). β-actin was used as an internal reference. A total of 2 µl cDNA, 2.5 µl 10X PCR buffer, 1.5 µl MgCl2, 1.0 µl dNTP mixture (2 mmol/l), 0.5 µl forward primer (10 µmol/l), 0.5 µl reverse primer (10 µmol/l), 0.5 µl Taq enzyme (2 U/µl) and 16.5 µl double-distilled water were added into a 0.2 ml Eppendorf PCR tube on ice. Following PCR (pre-denaturation at 94°C for 7 min, denaturation at 94°C for 30 sec, annealing at 56°C for 30 sec, extension at 72°C for 30 sec), 5 µl PCR product was analyzed on 1.5% agarose gel electrophoresis at 100 V for 15 min. The image was captured under UV light (WFH-204A; JK Technology, Shanghai, China). The intensity of these bands was semi-quantified using an imaging analyzer (ImageJ version 1.48; National Institutes of Health, Bethesda, MD, USA).
Immunohistochemistry on paraffin-embedded tissue sections
Sections of paraffin tissue 5-µm thick were deparaffinized by using xylene and rehydrated through a graded alcohol series to water. The dewaxed slides underwent antigen retrieval in citrate buffer (Beyotime Institute of Biotechnology, Shanghai, China) for 30 min by microwaving (Midea, Guangdong, China) and then cooled to room temperature. Peroxidase blocking solution (Beyotime Institute of Biotechnology, Shanghai, China) was used to block endogenous peroxidase activity at room temperature for 10 min. Tissue sections were incubated with anti-PGAM1 antibody (1:100 dilution) at 4°C overnight, and then incubated with biotinylated rabbit anti-Goat antibody (1:1,000; cat no. sc-2768; Santa Cruz Biotechnology, Inc.) at room temperature for 10 min. A drop of streptavidin-biotin-peroxidase solution Beyotime Institute of Biotechnology, Shanghai, China was added onto each section and incubated at room temperature for 10 min. Tissue sections were incubated with freshly prepared DAB chromogenic reagent for 1–2 min at room temperature, then counterstained with hematoxylin (1 µM; Beyotime Institute of Biotechnology, Shanghai, China) for 3 min at room temperature. Ten low power fields (magnification, ×100) were selected randomly and immunostaining was detected at a high power (magnification, ×400) using a light microscope (BX43; Olympus Corporation, Tokyo, Japan). The immunohistochemical stain results were divided into negative (<10% of cells with marked cytoplasmic staining), positive (10–70% cells with marked cytoplasmic staining) and strongly positive (>70% cells with marked cytoplasmic staining). Positive and strongly positive results were statistically counted as positive.
Statistical analysis
Data are expressed as mean ± standard deviation. Statistical analysis was performed using SPSS software version 14.0 (SPSS, Inc., Chicago, IL, USA). The relative absorbance of the astrocytes and C6 glioma cells was analyzed by unpaired Student's t-test. For the positive expression rates of PGAM1 in astrocytoma tissues and peritumoral brain tissue, the χ2 test was used, and P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of PGAM1 mRNA in C6 glioma cells and normal astrocytes
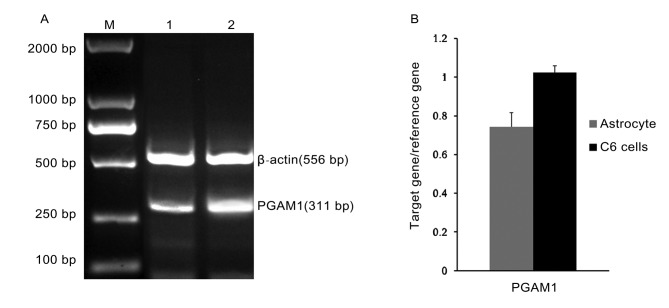
The RT-PCR PGAM1 products from astrocytes and C6 glioma were visualized by gel electrophoresis and the intensity of these bands was semi-quantified (Fig. 1A and B). Using SPSS version 14.0 statistical software, 2 samples in triplicate (C6 glioma cells vs. normal astrocytes) were compared. It was identified that the expression of PGAM1 in C6 glioma cells (1.26±0.05) was significantly increased compared with that of normal astrocytes (0.75±0.07). The difference was statistically significant (P<0.05; Table I).
Figure 1.
PGAM1 mRNA expression level in C6 glioma cells and normal astrocytes. (A) Electrophoresis of PCR products of PGAM1. M, Marker; 1, Astrocytes; 2, C6 glioma cells. (B) Semi-quantitative analysis of PCR products of PGAM1 in C6 glioma cells and normal astrocytes. PGAM1, phosphoglycerate mutase 1; PCR, polymerase chain reaction.
Table I.
Comparison of phosphoglycerate mutase 1 expression in C6 glioma cells and astrocytes.
| Group | No. of cases | Expression quantity (mean ± SD) |
|---|---|---|
| C6 glioma cells | 30 | 1.26±0.05 |
| Astrocytes | 10 | 0.75±0.07 |
t=3.89, P<0.05. SD, standard deviation.
PGAM1 immunohistochemistry of astrocytoma and peritumoral brain tissue
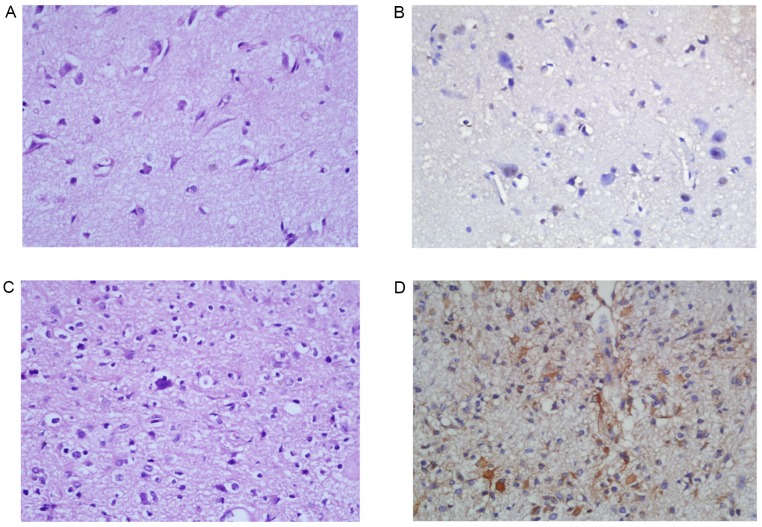
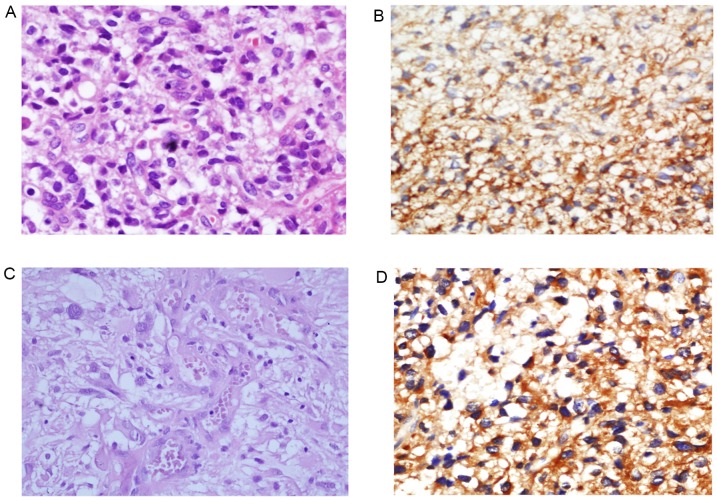
A total of 8/11 (73.8%) peritumoral brain tissues were negative for PGAM1 (Fig. 2A and B). Only 3 peritumoral brain tissues were positive for PGAM1 (3/11; 27.2%). In 102 cases of astrocytoma tissues, 74 cases (70.6%) were positive for PGAM1 and 28 cases were negative for PGAM1. There was a statistically significant difference in PGAM1 positive rates between the astrocytoma group and the peritumoral brain tissue group (χ2=8.35; P<0.01; Table II). The expression of PGAM1 in the low-grade diffuse astrocytomas (WHO Grade II; Fig. 2C and D) and high-grade astrocytomas (WHO Grade III–IV; Fig. 3) was compared. PGAM1 was predominantly confined to the cytoplasm of tumor cells (Figs. 2 and 3). The results demonstrated that 18/38 (47.4%) cases of WHO Grade II astrocytoma were positive for PGAM1. A total of 54/64 cases (84.4%) of high-grade astrocytomas were positive for PGAM1. The PGAM1 positive rate was significantly different between low-grade astrocytomas (WHO grade II) and high-grade astrocytomas (WHO grade III–IV) (χ2=15.73; P<0.01; Table III).
Figure 2.
PGAM1 immunohistochemical staining in peritumoral brain tissue and diffuse astrocytoma (WHO grade II). (A) Hematoxylin and eosin stain of peritumoral brain tissue. Magnification, ×400. (B) Peritumoral brain tissue is negative for PGAM1. Magnification, ×400. (C) Hematoxylin and eosin stain demonstrates brain tissue infiltrated by mildly hypercellular glial cells with irregular and atypical nuclei and inconspicuous mitotic activity, diagnostic of diffuse astrocytoma (WHO grade II). Magnification, ×200. (D) Diffuse astrocytoma is positive for PGAM1 with a cytoplasmic staining pattern. Magnification, ×200. PGAM1, phosphoglycerate mutase 1.
Table II.
Comparison of phosphoglycerate mutase 1 protein in glioma tissue and peritumoral brain tissue.
| Group | No. of cases | Negative | Positive | Positive rate (%) |
|---|---|---|---|---|
| Normal brain tissue | 11 | 8 | 3 | 27.2 |
| Glioma tissue | 102 | 30 | 72 | 70.6 |
χ2=8.35, P<0.01.
Figure 3.
PGAM1 immunohistochemical stains in high-grade astrocytoma. (A) Hematoxylin and eosin stain demonstrates hypercellular astrocytoma with occasional mitoses, diagnostic of anaplastic astrocytoma (WHO grade III). Magnification, ×400. (B) Anaplastic astrocytoma is positive for PGAM with a marked cytoplasmic staining pattern. Magnification, ×200. (C) Hematoxylin and eosin staining demonstrated high-grade astrocytoma with microvascular proliferation, diagnostic of glioblastoma (WHO grade IV). Magnification, ×400. (D) Glioblastoma is strongly and diffusely positive for PGAM1. Magnification, ×200. PGAM1, phosphoglycerate mutase 1.
Table III.
Association between phosphoglycerate mutase 1 expression and the grades of gliomas.
| Group | No. of cases | Negative | Positive | Positive rate (%) |
|---|---|---|---|---|
| Grade II | 38 | 20 | 18 | 47.4 |
| Grade III–IV | 64 | 10 | 54 | 84.4 |
χ2=15.73, P<0.01.
Discussion
As one of the key enzymes in the glycolytic pathway, PGAM1 primarily catalyzes the conversion of 3-phosphoglycerate into 2-phosphoglycerate. PGAM1 may be in muscle-derived form (m-type) and brain-derived form (b-type) in mammals (6). B-type PGAM1 has been identified previously (7). Tumor cells are distinct from normal cells by their unique feature of the glucose metabolism: Tumor cells obtain energy primarily through the glycolysis pathway, whereas normal cells primarily metabolize glucose through aerobic oxidation (8). Bustamante et al (9) identify that the hexokinase activity is markedly low in normal tissue, whereas the activity of this enzyme in tumor cells is significantly increased. Previous studies have indicated that certain enzymes which inhibit glycolysis may effectively kill liver cancer cells and other cancer cells (10–12).
PGAM1 is the only enzyme in the glycolytic pathway that is regulated at the transcriptional level by the tumor suppressor gene p53 (13), In 2005, Evans et al (14) suggest that PGAM1 may be a novel target for cancer treatment. PGAM1 is significantly upregulated in 66.7% of hepatocellular carcinomas, and markedly associated with the poorer survival of patients and poor differentiation of cancer cells (15). PGAM1 protein is overexpressed in breast cancer and prostate cancer (16,17). The expression of PGAM1 is upregulated in esophageal cancer (18) and glioma cells (19,20). In the present study, PGAM1 is expressed in astrocytes and C6 glioma cells using RT-semi-quantitative PCR, but that the expression level in C6 glioma cells is significantly increased compared with that of astrocytes. Using immunohistochemical staining, it is identified that the level of PGAM1 protein in astrocytoma tissue is significantly increased compared with that in peritumoral brain tissue. Furthermore, high-grade astrocytomas exhibit higher levels of PGAM1 positivity compared with low-grade astrocytomas. Therefore, PGAM1 may play an important role in the tumorigenesis and progression of malignant gliomas. The results indicate that PGAM1 inhibitors may be potential anticancer drugs, but extensive future studies are required.
In summary, the expression of PGAM1 is upregulated in glioma cell lines and astrocytoma tissues. This suggests that PGAM1 may be involved in the tumorigenesis and progression of malignant gliomas. PGAM1 may become a novel target for glioma treatment in the future.
Acknowledgements
The authors would like to thank Mr. Kuan Zhen (Department of Neurosurgery, Second Hospital of Shandong University), Mr. Yan Song (Department of Neurosurgery, Qilu Hospital of Shandong University, Brain Science Research Institute of Shandong University) and Mr. Baibin Bi (Department of Neurosurgery, Qilu Hospital of Shandong University, Brain Science Research Institute of Shandong University) for administrative and operational support.
Funding
The present study was supported by intradepartmental funding.
Availability of data and materials
All data generated during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
ZGL, JD and CD conducted the immunohistochemistry and immunofluorescence experiments. ZGL drafted the manuscript. NX, ELW and YYW participated in PCR experiments. YYW proofread the manuscript. ZGL and JD participated in data analysis, and statistical analysis. JYL made contributions to analysis and interpretation of data for histopathology and immunostain results, made the pathology figures and proofread the manuscript. JMY participated in experimental design and coordination of the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Ethical approval was provided by the Ethics Committee of Qilu Hospital (Shandong, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhou L. Editor in chief: Modern Neurosurgery. Shanghai: Fudan University Press; 2004. pp. 376–377. Front Page. [Google Scholar]
- 2.Ma LN, Song B, Cai JY. Research of the killing effects of cinobufacini on MCF-7cells. Chin Pharmacol Bull. 2011;27:37–40. (In Chinese) [Google Scholar]
- 3.Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci USA. 2005;102:5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schildge S, Bohrer C, Beck K, Schachtrup C. Isolation and culture of mouse cortical astrocytes. J Vis Exp. 2013;19 doi: 10.3791/50079. pii: 50079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grisolia S, Cleland WW. Influence of salt, substrate, and cofactor concentrations on the kinetic and mechanistic behavior of phosphoglycerate mutase. Biochemistry. 1968;7:1115–1121. doi: 10.1021/bi00843a032. [DOI] [PubMed] [Google Scholar]
- 7.Scatena R, Bottoi P, Pontoglio A, Mastrototaro L, Giardina B. Glycolytic enzyme inhibitors in cancer treatment. Expert Opin Investig Drugs. 2008;17:1533–1545. doi: 10.1517/13543784.17.10.1533. [DOI] [PubMed] [Google Scholar]
- 8.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: A recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustamante E, Morris HP, Pedersen PL. Energy metabolism of tumor cell. Requirement for a form of hexokinase with a propensity for mitochondrial binding. J Biol Chem. 1981;256:8699–8704. [PubMed] [Google Scholar]
- 10.Ko YH, Pedersen PL, Geschwind JF. Glucose catabolism in the rabbit VX2 tumor model for liver cancer: Characterization and targeting hexokinase. Cancer Lett. 2001;173:83–91. doi: 10.1016/S0304-3835(01)00667-X. [DOI] [PubMed] [Google Scholar]
- 11.Xu RH, Pelicano H, Zhou Y, Carew JS, Feng L, Bhalla KN, Keating MJ, Huang P. Inhibition of glycolysis in cancer cells: A novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res. 2005;65:613–621. [PubMed] [Google Scholar]
- 12.Gwak GY, Yoon JH, Kim KM, Lee HS, Chung JW, Gores GJ. Hypoxia stimulates proliferation of human hepatoma cells through the induction of hexokinase II expression. J Hepatol. 2005;42:358–364. doi: 10.1016/j.jhep.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 13.Cheung EC, Vousden KH. The role of p53 in glucose metabolism. Curr Opin Cell Biol. 2010;22:186–191. doi: 10.1016/j.ceb.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Evans MJ, Saghatelian A, Sorensen EJ, Cravatt BF. Target discovery in small molecule cell-based screens by in situ proteome reactivity profiling. Nat Biotechnol. 2005;23:1303–1307. doi: 10.1038/nbt1149. [DOI] [PubMed] [Google Scholar]
- 15.Ren F, Wu H, Lei Y, Zhang H, Liu R, Zhao Y, Chen X, Zeng D, Tong A, Chen L, et al. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol Cancer. 2010;9:81. doi: 10.1186/1476-4598-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortesi L, Barchetti A, De Matteis E, Rossi E, Della Casa L, Marcheselli L, Tazzioli G, Lazzaretti MG, Ficarra G, Federico M, Iannone A. Identification of protein clusters predictive of response to chemotherapy in breast cancer patients. J Proteome Res. 2009;8:4916–4933. doi: 10.1021/pr900239h. [DOI] [PubMed] [Google Scholar]
- 17.Zhang XB, Tang ZY, Zu XB, Qi L, Ruan JD. Modified serum-guided immunoblotting for differential study of prostate cancer. Zhonghua Nan Ke Xu. 2010;16:438–444. (In Chinese) [PubMed] [Google Scholar]
- 18.Saeki H, Kitao H, Yoshinaga K, Nakanoko T, Kubo N, Kakeji Y, Morita M, Maehara Y. Copy-neutral loss of heterozygosity at the p53 locus in carcinogenesis of esophageal squamous cell carcinomas associated with p53 mutations. Clin Cancer Res. 2011;17:1731–1740. doi: 10.1158/1078-0432.CCR-10-1996. [DOI] [PubMed] [Google Scholar]
- 19.Gao H, Yu B, Yan Y, Shen J, Zhao S, Zhu J, Qin W, Gao Y. Correlation of expression levels of ANXA2, PGAM1, and CALR with glioma grade and prognosis. J Neurosurg. 2013;118:846–853. doi: 10.3171/2012.9.JNS112134. [DOI] [PubMed] [Google Scholar]
- 20.Jain R, Kulkarni P, Dhali S, Rapole S, Srivastava S. Quantitative proteomic analysis of global effect of LLL12 on U87 cell's proteome: An insight into the molecular mechanism of LLL12. J Proteomics. 2015;113:127–142. doi: 10.1016/j.jprot.2014.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated during the present study are available from the corresponding author upon reasonable request.