Abstract
Ovarian cancer is a markedly heterogeneous malignancy characterized by various histological subtypes. Molecular biomarkers have been indicated to serve significant functions in the early diagnosis and treatment of early-stage ovarian cancer. However, the detailed mechanism underlying the tumorigenesis of ovarian cancer remains unclear. The present study aimed to identify a novel long non-coding RNA in patients with ovarian cancer. Nicotinamide nucleotide transhydrogenase-antisense 1 (NNT-AS1) was markedly downregulated in patients with ovarian cancer and in cultured human ovarian cancer cells. Knockdown of NNT-AS1 in the human ovarian cancer cell lines HO-8910 and SK-OV-3 promoted colony formation and arrested the cell cycle at G0/G1 phase. Furthermore, Transwell demonstrated that the downregulation of NNT-AS1 increased cell migration and invasion by ~60 and 70%, respectively, in HO-8910 and SK-OV-3 cells. Furthermore, cell apoptosis was inhibited by the transfection of siNNT-AS1 in the two cell lines, whereas the relative activities of caspase-3 and caspase-9 were decreased. These results indicated a protective function of NNT-AS1 in human ovarian cancer, providing novel insights into the diagnosis and treatment of ovarian cancer in clinical settings.
Keywords: long non-coding RNA, nicotinamide nucleotide transhydrogenase-antisense RNA 1, ovarian cancer, proliferation, metastasis, apoptosis
Introduction
Ovarian cancer is one of the leading causes of gynecological cancer-associated mortality in developed countries, and often presents at an advanced stage (1). The current treatment strategy in the majority of patients with late-stage disease involves cytoreductive surgeries and platinum-based chemotherapy (2–4). However, the majority of patients are resistant to chemotherapy or radiotherapy (5,6). Therefore, it is urgent to diagnose ovarian cancer in the early stages of the disease, and to develop novel molecular biomarkers to measure the potential incidence rate in females worldwide.
Although the majority of the human genome may be transcribed into RNA, only a small fraction of RNAs has the potential to code proteins, accounting for ~3% of the human genome (7). The RNAs that lack these protein-coding abilities are termed non-coding RNAs, of which long non-coding RNAs (lncRNAs) have a length of >200 bp and they lack an open reading frame (8). LncRNAs are primarily transcriptionally regulated by RNA polymerase II and serve a significant function in various process, including the splicing of introns, 5′-end capping and 3′-end polyadenylation (8), and have been demonstrated to affect histone modifications, binding of transcription factors to targeted genes and chromatin remodeling (9). LncRNAs have been indicated to affect cancer cell proliferation (10), metastasis (11), apoptosis (12) and autophagy (13) in a number of types of malignancy.
The lncRNA nicotinamide nucleotide transhydrogenase-antisense RNA1 (NNT-AS1) is a newly identified lncRNA, and has been revealed to serve a regulatory function in human colorectal cancer (14). NNT-AS1 is overexpressed in human colorectal cancer, and is associated with lymph node metastasis, tumor-node-metastasis stage, vessel invasion and differentiation (14). Knockdown of NNT-AS1 in colorectal cancer cells suppressed tumor growth and metastasis by the mitogen-activated protein kinase/extracellular-signal-regulated kinase signaling pathway (14). However, the detailed mechanisms are not known.
The present study aimed to identify the function of NNT-AS1 in human ovarian cancer. To achieve this, specific small interfering (si)RNA was transfected into various ovarian cancer cells, and the effects of NNT-AS1 on cell proliferation, migration and apoptosis were investigated using cell proliferation, Transwell, wound-healing and cell apoptosis assays. The data indicated a protective function of NNT-AS1 in human ovarian cancer, providing novel insights into the diagnosis and treatment of early-stage ovarian cancer.
Materials and methods
Human tissues
The present study was approved by the Ethic Committee of the Third Affiliated Hospital of Soochow University (Changzhou, China). A total of 55 patients with ovarian cancer were included (age range, 45–75 years; median age, 58 years) between January 2015 and January 2016, and their tumor tissues and the adjacent non-cancerous counterparts were frozen in liquid nitrogen once removed from the body (surgical resection) and subjected to subsequent RNA extraction. All patients stated their full intention to participate in the present study, and written consent from each patient was also obtained.
Routine cell culture and transfection
HO-8910, and SK-OV-3 human ovarian cancer cells were purchased from the American Type Tissue Collection (Manassas, VA, USA). All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplied with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 incubator. Specific siRNAs against NNT-AS1 (GCAAGCUGACCCUGAAGUUCA) and siNC (CUACAUCGACAAGGUGCGCGU) were designed and synthesized by Shanghai GenePharma Co., Ltd (Shanghai, China) and the concentration of siRNA used for transfection was 1 µM. Cell transfection was performed using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 72 h, according to the manufacturer's protocol.
RNA extraction and quantitative polymerase chain reaction (qPCR)
Total RNA from clinical tissues and cultured HO-8910 and SK-OV-3 cells were extracted using TRIzol® reagent (Takara Biotechnology, Co., Ltd., Dalian, China), according to the manufacturers' protocol. The quality and quantity of RNA were determined using a NanoDrop 2000 instrument (NanoDrop Technologies; Thermo Fisher Scientific, Inc.) by measuring the absorbance at 260 and 280 nm. cDNA was reverse-transcribed from 500 ng RNA using Prime Script TM Master Mix (Takara Biotechnology, Co., Ltd.). qPCR was then performed using SYBR Green reagent (Takara Biotechnology, Co., Ltd.), according to the manufacturer's protocol. The thermocycling protocol was as follows: Initial denaturation at 95°C for 5 min, followed by 45 repeats of a three-step cycling program consisting of 10 sec at 95°C (denaturation), 10 sec at 60°C (primer annealing) and 10 sec at 72°C (elongation), and a final extension step for 10 min at 72°C. The results were normalized to the expression of GAPDH (15). The primers used were the following: NNT-AS1, 5′-CTCCGAACCAAAAGGCGAC-3′ (forward) and 5′-CTTTGTTCTGATGGGACCC-3′ (reverse); GAPDH, 5′-CGCTCTCTGCTCCTCCTGTTC-3′ (forward) and 5′-ATCCGTTGACTCCGACCTTCAC-3′ (reverse).
Colony formation assay
The HO-8910 and SK-OV-3 cells were seeded (1×104 cells) in 6-well plates, pre-treated with siNNT-AS1 for 72 h at 37°C and spread onto 12-well plates (100 cells/well) in triplicate. The plates were incubated at 37°C for 14 days continuously. Subsequently, the colonies were fixed with chilled 100% methanol and stained with crystal violet (1%) for 5 min at room temperature. Colonies that contained >50 cells were considered to be surviving cells under a light microscope (Nikon Corporation, Tokyo, Japan) at magnification, ×200.
Cell cycle analysis
Each group of HO-8910 and SK-OV-3 cells was seeded into 6-well plates at a concentration of 3×105 cells/well, and pre-treated with specific siRNA against NNT-AS1 when the cell lines reached 85% confluence. Subsequently, cells were collected following low-speed centrifugation (840 × g for 5 min) at 4°C, and the cell pellets were resuspended in 1 ml PBS, fixed with 75% ice-cold ethanol and stored at −20°C for 2 days. Prior to flow cytometry analysis, cells were lysed with NP-40 buffer (Beyotime Institute of Biotechnology, Haimen, China), centrifuged (840 × g for 5 min at 4°C) and resuspended in propidium iodide (PI; Thermo Fisher Scientific, Inc.) staining buffer containing 50 µl/ml PI and 250 µl/ml RNase A (Beyotime Institute of Biotechnology). Finally, the cell mixture was incubated at 4°C for 30 min in the dark, and analyzed using a fluorescence-activated cell sorting technique on a flow cytometer (Beckman Coulter, Inc., Brea, CA, USA).
Transwell assay
Cell migration and invasion assays were performed in 24-well Transwell plates (8 µm pore membrane, Corning, NY, USA). Following transfection with siRNAs, a total of 1×104 cells (HO-8910 and SK-OV-3 cells) were suspended with FBS-free DMEM and loaded into the upper chambers, while the lower chambers were filled with 600 µl DMEM supplemented with 10% FBS. The plates were incubated at 37°C for an additional 24 h. Subsequently, the cells were fixed with chilled 100% methanol for 5 min at room temperature and stained with 0.1% crystal violet for 5 min at room temperature. Cells on the upper surface of the chambers were removed using cotton swabs, and images of those on the lower surface were captured and counted using a light microscope (Nikon Corporation) at magnification ×200 for five random fields. For the invasion assay, the Transwell membranes were pre-incubated with Matrigel® (Corning Incorporated, Corning, NY, USA) for 6 h at 37°C.
Wound-healing assay
Wound-healing assays were performed in HO-8910 and SK-OV-3 cells by creating identical wounds using 10 µl sterile pipette tips. Briefly, cells were seeded (1×104) into 6-well plates and co-incubated with siNNT-AS1 for 72 h at 37°C. Subsequently, cells were washed with PBS twice and a cross was scraped in the center of each well, prior to washing twice and immediately being placed in fresh FBS-free DMEM. Images of the cells were captured under a light microscope (Nikon Corporation) at magnification, ×200 for each group and accounted for a time-point 0 h. Following 24 h incubation at 37°C, the two cell lines were observed and images were captured. A total of 5 random fields of view were selected, and the effects of NNT-AS1 were analyzed.
Cell apoptosis analysis
Flow cytometry (FACScan; BD Biosciences, Franklin Lakes, NJ, USA) was used to determine the apoptotic rate upon siNNT-AS1 treatment on HO-8910 and SK-OV-3 cells. Briefly, cells (1×104) cultured in 6-well plates were harvested, washed with PBS and collected following low-speed centrifugation (840 g for 5 min at 4°C). Subsequently, cells were resuspended in 100 µl staining buffer with 5 µl Annexin V-allophycocyanin (20 µg/ml) and 5 µl PI (10 mg/l) (BD Pharmingen; BD Biosciences). Cells were then subjected to flow cytometry (excitation, 488 nm; emission, 635 nm) and cells that were Annexin V-positive were considered to be apoptotic. CellQuest software Pro (version 1.0, BD Biosciences) was used to analyze the data, and each experiment was repeated at least three times in triplicate.
Relative caspase activity determination
The activities of caspase-3 and caspase-9 were determined using a Caspase-3 Activity kit and a Caspase-9 Activity kit (both from Beyotime Institute of Biotechnology), respectively, according to the manufacturer's protocols. In brief, cell lysates from HO-8910 and SK-OV-3 cells were collected using centrifugation at 840 × g for 5 min at 4°C following siRNA treatment. A total of 10 µl proteins from cell lysates from each group were added into 96-well plates and mixed with 80 µl reaction buffer, which contained caspase substrate (2 mM, Beyotime Institute of Biotechnology). Following incubation for 4 h at 37°C, caspase activities were determined using a microplate reader at an absorbance of 405 nm.
Statistical analysis
All results are presented as the means ± standard deviation. Each experiment was repeated at least three times in triplicate, unless otherwise stated. Paired student's t-test was performed to determine the difference between two groups. One-way analysis of variance followed by Student-Newman-Keuls test was used to compare the differences between multiple groups. P<0.05 was considered to indicate a statistically significant difference, All data were analyzed using SPSS software (version 19.0; IBM Corp., Armonk, NY, USA).
Results
LncRNA NNT-AS1 is downregulated in human ovarian cancer in vivo and in vitro
The relative expression of lncRNA NNT-AS1 in samples from 55 patients with ovarian cancer from The First People's Hospital of Changzhou was determined. It was demonstrated that the relative transcript level of NNT-AS1 was significantly decreased in tumor samples compared with adjacent non-tumor samples (Fig. 1A). Subsequently, four ovarian cancer cell lines were selected for RNA quantification of NNT-AS1. As presented in Fig. 1B, the relative expression of NNT-AS1 was significantly decreased in all four ovarian cancer cell lines used, compared with the expression of NNT-AS1 in 293T cells. Notably, HO-8910PM and OVCAR3 cells exhibited the lowest expression levels of NNT-AS1, whereas the RNA levels of HO-8910 and SK-OV-3 cells were slightly increased compared with the other two cell lines. These results indicated that the expression of NNT-AS1 was markedly decreased in human ovarian cancer cells in vivo and in vitro.
Figure 1.
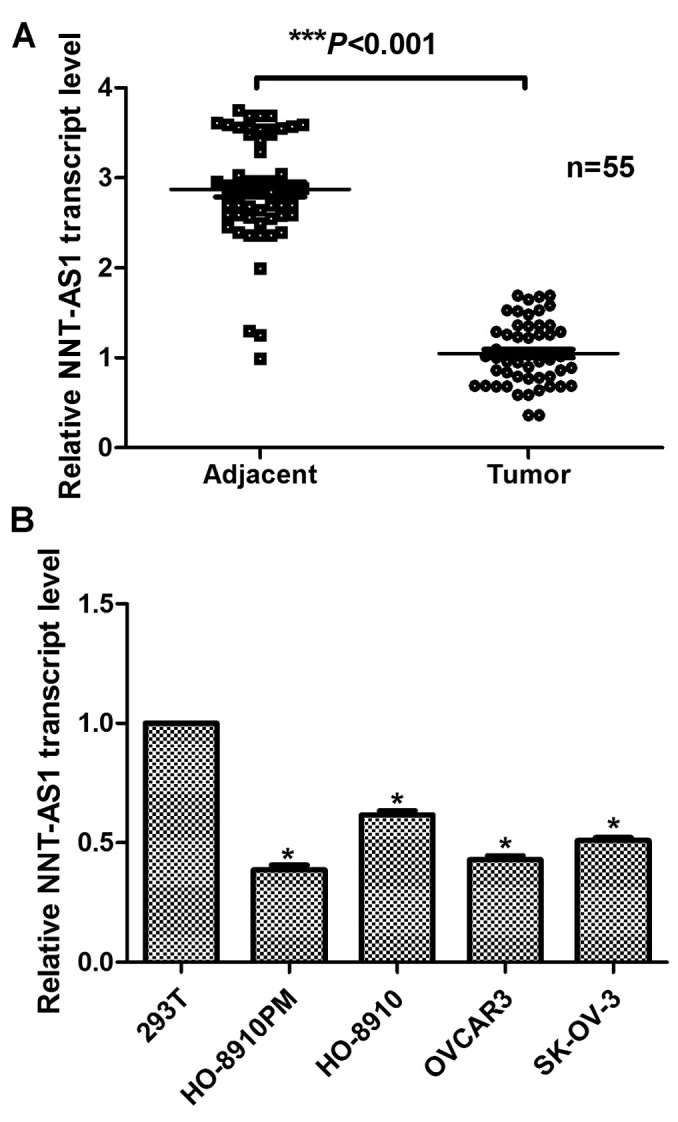
Long non-coding RNA NNT-AS1 is downregulated in human ovarian cancer in vivo and in vitro. (A) RT-qPCR analysis of the expression of NNT-AS1 in clinical human tissues from 55 patients. ***P<0.001 vs. adjacent non-cancerous tissues (Student's t-test). (B) RT-qPCR analysis of the expression of NNT-AS1 in cultured ovarian cancer cell lines. 293T cells were included as a control. *P<0.05 vs. 293T cells (analysis of variance and Student-Newman-Keuls test). RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NNT-AS1, nicotinamide nucleotide transhydrogenase-antisense RNA 1.
Knockdown of NNT-AS1 in ovarian cancer cells promotes colony formation and arrests the cell cycle at G0/G1 phase
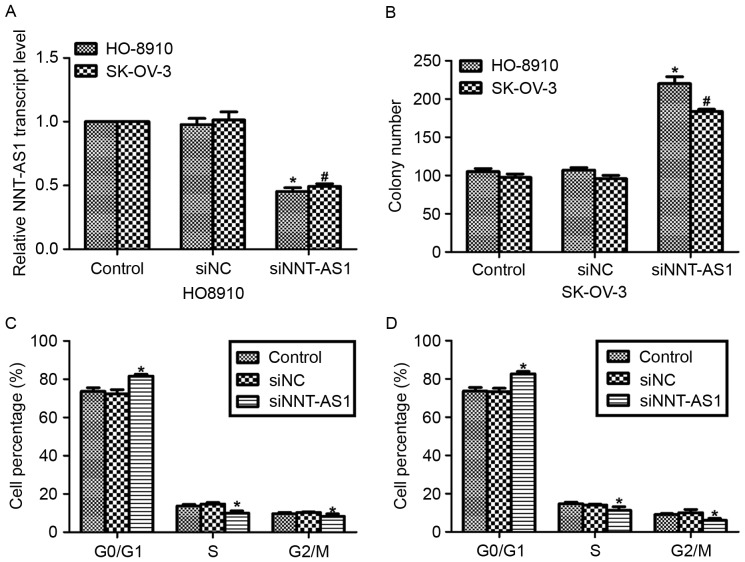
The effects of NNT-AS1 in human ovarian cancer were investigated. Specific siRNAs against NNT-AS1 were transfected into HO-8910 and SK-OV-3 cells, as these two cell lines exhibited the highest transcript levels of NNT-AS1 of the four ovarian cancer cell lines used. As presented in Fig. 2A, the expression levels of NNT-AS1 were significantly decreased in HO-8910 and SK-OV-3 cells when the cells were treated with siNNT-AS1 for 72 h. Subsequently, colony formation assays were performed in HO-8910 and SK-OV-3 cells with or without siNNT-AS1 treatment. Following transfection of the HO-8910 cells with siNNT-AS1, >200 colonies were counted, whereas only ~100 colonies were observed in the control cells (Fig. 2B). Furthermore, cell cycle analysis was also performed in the HO-8910 and SK-OV-3 cells. As presented in Fig. 2C and D, the cell cycle distribution of siNNT-AS1 cells was altered so that the highest proportion of cells were in the G0/G1 phase, compared with the other cells (control and siNC) in the two cell lines.
Figure 2.
Knockdown of NNT-AS1 in ovarian cancer cells promotes colony formation and cell cycle arrest at G0/G1 phase. (A) Reverse transcription-quantitative polymerase chain reaction analysis of the expression of NNT-AS1 in cultured ovarian cancer cell lines transfected with siNNT-AS1. (B) Colony formation assays were performed in HO-8910 and SK-OV-3 cells in the presence or absence of siNNT-AS1. *P<0.05 vs. control in HO-8910 cells (ANOVA and SNK test). #P<0.05 vs. control in SK-OV-3 cells (ANOVA and SNK test). (C) Cell cycle assays were performed in HO-8910 cells in the presence or absence of siNNT-AS1. (D) Cell cycle assays were performed in SK-OV-3 cells in the presence or absence of siNNT-AS1. *P<0.05 vs. control (ANOVA and SNK test). NNT-AS1, nicotinamide nucleotide transhydrogenase-antisense RNA 1; si, small interfering; NC, negative control; ANOVA, analysis of variance; SNK, Student-Newman-Keuls.
Knockdown of NNT-AS1 increases cell migration and invasion in HO-8910 and SK-OV-3 cells
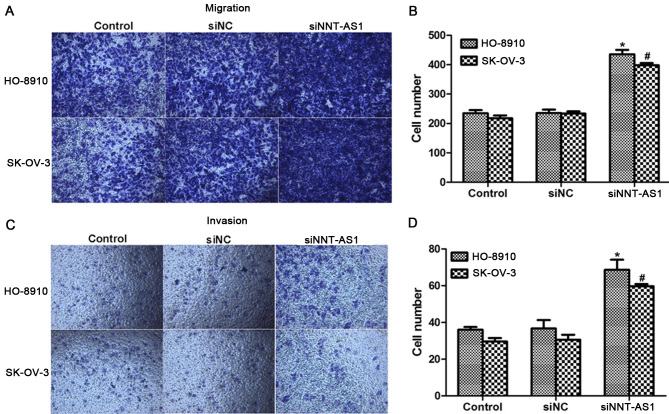
Cell migration are thetwo primary manifestations of malignancies in vivo and in vitro (16). Next, the function of NNT-AS1 in cell migration in HO-8910 and SK-OV-3 cells was investigated. As presentedin Fig. 3A and B, cell migratory capacities in the twocell lines were increased when siNNT-AS1 was transfected for 72 h, as was evidentfrom the >400 cells observed to migrate through the membrane in siNNT-AS1-stimulated cells, whereas only ~200 cells were counted on the lower surface of the membrane. Likewise, cell invasion was also upregulated in HO-8910 and SK-OV-3 cells when cells were treated with siNNT-AS1 (Fig. 3C and D).
Figure 3.
Knockdown of NNT-AS1 increases migration and invasion of HO-8910 and SK-OV-3 cells. (A) Representative images of migration assays for HO-8910 and SK-OV-3 cells (magnification, ×200). (B) Quantification of migration assays for HO-8910 and SK-OV-3 cells. (C) Representative images of cell invasion assays for HO-8910 and SK-OV-3 cells (magnification, ×200). (D) Quantification of cell invasion assays in the two cell lines. *P<0.05 vs. control in HO-8910 cells (ANOVA and SNK test). #P<0.05 vs. control in SK-OV-3 cells (ANOVA and SNK test). NNT-AS1, nicotinamide nucleotide transhydrogenase-antisense RNA 1; si, small interfering; NC, negative control; ANOVA, analysis of variance; SNK, Student-Newman-Keuls.
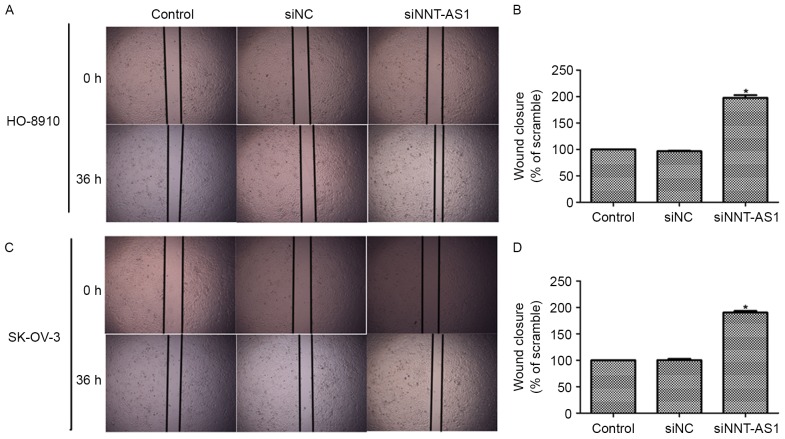
Wound-healing assays were also performed to assess the effects of NNT-AS1 on cell migration. As presentedin Fig. 4A and B, knockdown of NNT-AS1 in HO-8910 and SK-OV-3 cells led to increased cell migration and wound healing compared with the control cells without transfections. Quantification of the wound-healing assay also indicated that the wound closure in siNNT-AS1-treated cells was almost 2-fold that of the siNC-treated cells (Fig. 4C and D). Together with the results presented in Fig. 3, these results suggested that NNT-AS1 inhibited cell migration in human ovarian cancer cells in vitro.
Figure 4.
Downregulation of NNT-AS1 increases wound closure ability in HO-8910 and SK-OV-3 cells. (A) Representative images of the wound-healing assays for HO-8910 cells (magnification, ×200). (B) Quantification of wound-healing assays in HO-8910 cells relative to control. (C) Representative images of the wound-healing assays for SK-OV-3 cells (magnification, ×200). (D) Quantification of wound-healing assays in SK-OV-3 cells relative to control. *P<0.05 vs. control (analysis of variance and Student-Newman-Keuls test). NNT-AS1, nicotinamide nucleotide transhydrogenase-antisense RNA 1; si, small interfering; NC, negative control.
Knockdown of NNT-AS1 inhibits cell apoptosis in human ovarian cancer cells
Subsequently, the function of NNT-AS1 in cell apoptosis was assessed. As presented in Fig. 5A, transfection with NNT-AS1 inhibited cell apoptosis by 7% in HO-8910 cells and by 5% in SK-OV-3 cells. Next, the relative activity of caspase-3 and caspase-9 was determined in the two cell lines. It was demonstrated that the caspase-3 activity was suppressed by >50% in HO-8910 and SK-OV-3 cells following siNNT-AS1 treatment (Fig. 5B). Furthermore, the relative caspase-9 activity was also inhibited by NNT-AS1 knockdown in the two cell lines (Fig. 5C). These data suggested that the downregulation of NNT-AS1 in human ovarian cancer inhibited cell apoptosis in vitro.
Figure 5.
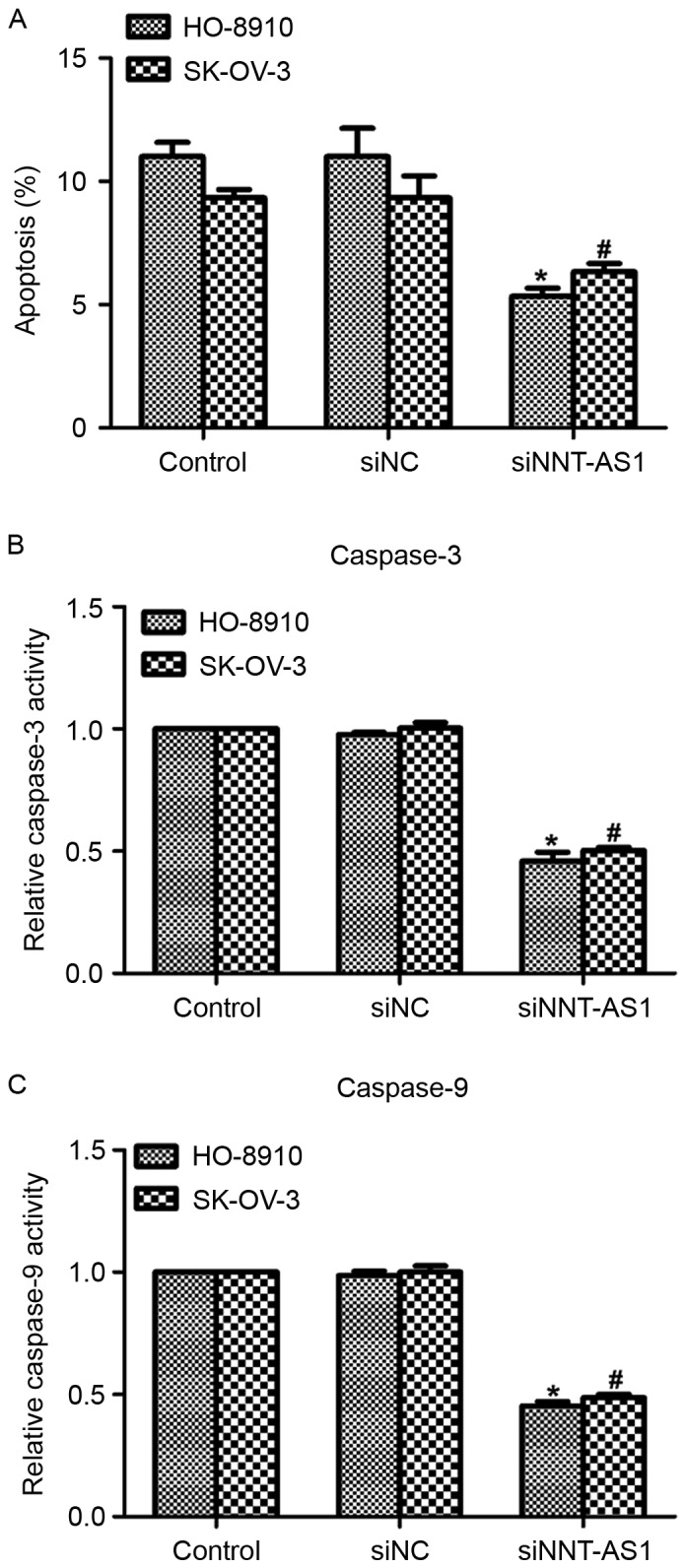
Knockdown of NNT-AS1 inhibits cell apoptosis in human ovarian cancer cells. (A) Cell apoptosis was assessed in HO-8910 and SK-OV-3 cells in the presence or absence of siNNT-AS1. (B) Relative caspase-3 activity was determined in HO-8910 and SK-OV-3 cells in the presence or absence of siNNT-AS1. (C) Relative caspase-9 activity was determined in HO-8910 and SK-OV-3 cells in the presence or absence of siNNT-AS1. *P<0.05 vs. control in HO-8910 cells (ANOVA and SNK test). #P<0.05 vs. control in SK-OV-3 cells (ANOVA and SNK test). NNT-AS1, nicotinamide nucleotide transhydrogenase-antisense RNA 1; si, small interfering; NC, negative control; ANOVA, analysis of variance; SNK, Student-Newman-Keuls.
Discussion
Ovarian cancer is a histologically, clinically and molecularly diverse malignancy that occurs worldwide (17). A total of >21,980 cases of ovarian cancer were diagnosed and 14,270 mortalities occurred in 2015 in the USA, making it the gynecological tumor with the highest mortality rate in developed countries (18). Therefore, an increasing number of molecular biomarkers have become the focus of previous studies.
The present study revealed that the newlyidentified lncRNA NNT-AS1, previously demonstrated to be overexpressed in human colorectal cancer (19), was downregulated in human ovarian cancer, asindicated using in vivo and in vitro experiments. Of the fourovarian cancer cell lines used, HO-8910PM and OVCAR3 exhibited the lowest expression of NNT-AS1. Therefore, NNT-AS1 was overexpressed in these two cell lines in subsequent experiments. However, construction ofan NNT-AS1-expressing plasmid with the whole genomic DNA failed (data not shown), and there isno known commercial source from which to obtain this plasmid. Consequently, a second methodof knocking down the expression of NNT-AS1 was selected, and this demonstrated lower NNT-AS1 transcript levels in HO-8910 and SK-OV-3 cells compared with in 293T cells, and increased expression in HO-8910 and SK-OV-3 cells compared with in HO-8910PM and OVCAR3 cells. This issue is currently being addressedas part of the ongoing research carried out by the present authors, so that HO-8910PM and OVCAR3 cells may be transfected with anNNT-AS1 expression plasmid. Another limitation of the present study was that commercial normal ovarian cells were not used as the internal control.
The results of the present study indicated anassociation between the expression of NNT-AS1 and cell migration (and therefore metastasis), as the lowest expression of NNT-AS1 was demonstrated in the HO-8910PM cells, which exhibited the highest potential for metastasis. It was also identified that knockdown of NNT-AS1 in HO-8910 and SK-OV-3 cells promoted cell metastasis in vitro. However, epithelial-mesenchymal transition markers were not included in the present study. The protein level of epithelial cadherin, neural cadherin and cyclin B1 by western blot analysis in HO-8910 and SK-OV-3 in the presence or absence of siNNT-AS1 are being investigated.
There have been twosignaling pathways demonstrated to be involved in the induction of cell apoptosis: the intrinsic and extrinsic pathways (20). The initiation of the intrinsic pathway is associated with pro-apoptotic factors, including B-cell lymphoma 2 (Bcl-2)-associated X protein and Bcl-2-associated death promoter, which leads to an increased permeability of the mitochondria membrane, loss of membrane potential and the release of cytochrome c into the cytosol (20). The intrinsic pathway is associated with activated caspase-3, whereas the extrinsic pathway is associated with the activation of caspase-8 (21). In the results of the present study, it was indicated that the relative activities of caspase-8 remained unchanged (data not shown), therefore it may be that the intrinsic pathway was involved in the process, which is a focus of future study.
Taken together, the results of the present study suggested that the relative transcript level of NNT-AS1 was downregulated in human ovarian cancer. HO-8910 and SK-OV-3 cells with NNT-AS1 knocked downexhibited a greater potential to form colonies compared with the control cells. Cell cycle was arrested at G0/G1 phase upon siNNT-AS1 treatment in the two cell lines. Downregulation of NNT-AS1 promoted cell migration and invasion in HO-8910 and SK-OV-3 cells. In addition, depletion of NNT-AS1 in human ovarian cancer cells inhibited cell apoptosis by decreasing the relative activities of caspase-3 and caspase-9. The results of the present study indicated the protective function of NNT-AS1 in human ovarian cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YH and YX constructed the experiments and organized the data, JS assisted in the analysis of data and YX wrote the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Third Affiliated Hospital of Soochow University. All patients stated their full intention to participate in the present study, and written consent from each patient was also obtained.
Consent for publication
All patients provided permission to participate in the present study, and written consent was obtained from each patient.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Vergote I, Oaknin A, Baurain JF, Ananda S, Wong S, Su X, Wu B, Zhong Z, Warner D, Casado A. A phase 1b, open-label study of trebananib in combination with paclitaxel and carboplatin in patients with ovarian cancer receiving interval or primary debulking surgery. Eur J Cancer. 2014;50:2408–2416. doi: 10.1016/j.ejca.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Vergote I, Amant F, Kristensen G, Ehlen T, Reed NS, Casado A. Primary surgery or neoadjuvant chemotherapy followed by interval debulking surgery in advanced ovarian cancer. Eur J Cancer. 2011;47(Suppl 3):S88–S92. doi: 10.1016/S0959-8049(11)70152-6. [DOI] [PubMed] [Google Scholar]
- 4.Reed NS, Mangioni C, Malmström H, Scarfone G, Poveda A, Pecorelli S, Tateo S, Franchi M, Jobsen JJ, Coens C, et al. Phase III randomised study to evaluate the role of adjuvant pelvic radiotherapy in the treatment of uterine sarcomas stages I and II: An European Organisation for Research and Treatment of Cancer Gynaecological Cancer Group Study (protocol 55874) Eur J Cancer. 2008;44:808–818. doi: 10.1016/j.ejca.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Pinsky PF, Yu K, Kramer BS, Black A, Buys SS, Partridge E, Gohagan J, Berg CD, Prorok PC. Extended mortality results for ovarian cancer screening in the PLCO trial with median 15 years follow-up. Gynecol Oncol. 2016;143:270–275. doi: 10.1016/j.ygyno.2016.08.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, Amso NN, Apostolidou S, Benjamin E, Cruickshank D, et al. Ovarian cancer screening and mortality in the UK collaborative trial of ovarian cancer screening (UKCTOCS): A randomised controlled trial. Lancet. 2016;387:945–956. doi: 10.1016/S0140-6736(15)01224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melissari MT, Grote P. Roles for long non-coding RNAs in physiology and disease. Pflugers Arch. 2016;468:945–958. doi: 10.1007/s00424-016-1804-y. [DOI] [PubMed] [Google Scholar]
- 10.Lin CY, Kleinbrink EL, Dachet F, Cai J, Ju D, Goldstone A, Wood EJ, Liu K, Jia H, Goustin AS, et al. Primate-specific oestrogen-responsive long non-coding RNAs regulate proliferation and viability of human breast cancer cells. Open Biol. 2016;6:150262. doi: 10.1098/rsob.150262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Sehgal L, Jain N, Khashab T, Mathur R, Samaniego F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J Transl Med. 2016;14:346. doi: 10.1186/s12967-016-1100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei S, Wang K. Long noncoding RNAs: Pivotal regulators in acute myeloid leukemia. Exp Hematol Oncol. 2016;5:30. doi: 10.1186/s40164-016-0059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuo C, Jiang R, Lin X, Shao M. LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget. 2017;8:1429–1437. doi: 10.18632/oncotarget.13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Yang L, Hu X, Jiang Y, Hu Y, Liu Z, Liu J, Wen T, Ma Y, An G, Feng G. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget. 2017;8:3441–3453. doi: 10.18632/oncotarget.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 17.Yeung TL, Leung CS, Li F, Wong SS, Mok SC. Targeting stromal-cancer cell crosstalk networks in ovarian cancer treatment. Biomolecules. 2016;6:3. doi: 10.3390/biom6010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 19.Liu T, Zhang X, Gao S, Jing F, Yang Y, Du L, Zheng G, Li P, Li C, Wang C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–85563. doi: 10.18632/oncotarget.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spencer SL, Sorger PK. Measuring and modeling apoptosis in single cells. Cell. 2011;144:926–939. doi: 10.1016/j.cell.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Y, Gong J, Tian X, Yan X, Guo T, Huang M, Zhang B, Hu X, Liu H, Wang Y, et al. Japonicone A inhibits the growth of non-small cell lung cancer cells via mitochondria-mediated pathways. Tumour Biol. 2015;36:7473–7482. doi: 10.1007/s13277-015-3439-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.