Abstract
MicroRNAs (miRNA/miRs) are small, non-coding RNA molecules (19–25 nucleotides in length), which function to regulate gene expression. It has been reported that miR-128 serves an important role in regulating cancer cell growth; increasing evidence has indicated that the expression of miR-128 is decreased in pancreatic cancer (PC) cells. However, the specific mechanisms of miR-128 in regulating PC cell growth are unclear. In the present study, it was confirmed that the expression of miR-128 was significantly decreased within PC tissues compared with adjacent normal tissues via reverse transcription-quantitative polymerase chain reaction analysis. In addition, miR-128 mimics inhibited PC MIA-PaCa2 cell growth by enhancing cell apoptosis in a caspase-dependent manner. Furthermore, the results of the present study demonstrated that double minute 4 (MDM4) may be a direct target for miR-128 via a dual luciferase report assay; miR-128 may inhibit MDM4 expression, and increase p53 and cleaved caspase-3 protein expression levels. In summary, the present study indicated that miR-128 is downregulated in PC, and it may be a promising target for future PC diagnosis and treatment.
Keywords: microRNA, pancreatic cancer, cell growth, p53, double minute 4
Introduction
Pancreatic cancer (PC) is one of the most common fatal cancer types (1). The overall 5-year survival rate is <5%; annually, almost half of patients diagnosed with PC will succumb to the disease (2,3). At present, the options for chemotherapy only minimally prolong the life span of patients with PC (4–6). It is urgent to investigate and validate the mechanisms of PC growth (7,8). Extensive efforts have been made to identify biomarkers for this disease. Research has indicated that some miRNAs may exhibit variable expression levels within some cancer tissues (9–11).
MicroRNAs (miRNAs/miRs) are endogenous short non-coding RNAs, 19–25 nucleotides in length (11). miRNAs have been reported to serve an important role in regulating the expression and function of numerous genes and proteins (12). Previous studies have demonstrated that miRNA may facilitate cancer cell growth, invasion and migration via degradation of mRNA, by pairing with bases in its 3′-untranslated region (UTR) (13,14). Thus, miRNAs are considered to be key markers for the diagnosis and investigation of PC (15). In recent decades, a number of studies have been conducted to profile miRNA expression within PC cells, in order to identify differential miRNA expression between PC and adjacent normal tissues (16,17). Numerous miRNAs have been identified, including miR-128, miR-21-5p, miR-31-5p, miR-210, miR-217 and miR-375 (9,12,13,15,18–22); however, the specific mechanisms of these identified miRNAs require further investigation.
Double minute (MDM) family proteins are key regulators for the onco-suppressor p53. Marine and Jochemsen (23) identified that MDM4 was a p53 binding protein and knockdown of MDM4 could induce embryonic lethality within mice. The utero stage death could be rescued by deletion of the Trp53 gene.
In the present study, miR-128 expression levels within PC cells were significantly decreased compared with adjacent normal tissues, as confirmed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. Additionally, MDM4 may be associated with miR-128 expression and was demonstrated to be a target of miR-128 via a dual luciferase assay. Furthermore, the expression levels of p53 were upregulated by miR-128 by targeting MDM4 to suppress tumor growth. The results of the present study may contribute to the developments of PC treatment and diagnosis.
Materials and methods
Patient recruitment and tissue sample collection
A total of 30 patients (14 females and 16 males; age, 22–63) diagnosed with PC were recruited from The Third People's Hospital (Yancheng, China) from September 2015 to June 2017. In-hospital histories of patients were reviewed, including the diagnosis, American journal of critical care (AJCC, version 8) PC stage or metastasis (24). All PC and adjacent noncancerous tissues were obtained intraoperatively and fast frozen in liquid nitrogen, then stored at −80°C until experimental analysis. The present study was approved by the Institutional Review Board for Human Research of The Third People's Hospital. All recruited patients provided written informed consent.
Cell culture
Human pancreatic carcinoma cell line MIA PaCa-2 was provided by the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2 in a humidified cell culture incubator (Sanyo Electric Co., Ltd., Osaka, Japan).
Cell transfection
MIA PaCa-2 cells (1×105 cells/well) were plated in 6-well plates and transfected with 100 nM miR-128 mimics or scrambled sequence control (Shanghai GenePharma Co., Ltd., Shanghai, China) using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol, when MIA PaCa-2 cells reached 50–60% confluence within the plate. The micmic sequence was: 5′-UAUAUGGCUUUAGAUACUGUGAA-3′. The scramble sequence was: 5′-UAUAUGGCUUUAGAUGACGCUGA-3′. Subsequently, cells washed out following transfection 6 h and 2 ml Dulbecco's modified DMEM with 10% fetal bovine serum were added to each well. cells were harvested with trypsin at 37°C with 5% CO2 for 5 min for following experiments at certain time points after transfection, including: 24, 48 and 72 h.
Cell proliferation assay
Cell proliferation was determined with a Cell Counting kit (CCK)-8 (C0038; Beyotime Institute of Biotechnology, Shanghai, China) according to the manufacturer's protocol. Briefly, 2×103 cells in the logarithmic growth phase were plated in a 96-well plate and then were transfected with miR-128 mimics or scramble control. These cells were used for the cell proliferation assay at 24, 48 and 72 h by adding 20 µl CCK solution into each well and the cells were further incubated for 0.5 h at 37°C with 5% CO2 in the cell incubator. The number of living cells was measured using a microplate reader at 450 nm wavelength.
Colony formation assay
MIA PaCa-2 cells transfected with miR-128 mimics or scramble control vector were transferred into a 6-well plate (100 cells/well) and the medium was replaced after 6 h. Subsequently, the cells were cultured for another 72 h at 37°C with 5% CO2. When visible cell colonies formed, cells were stained with 0.1% crystal violet (Sigma Aldrich; Merck KGaA, Darmstadt, Germany) in DMEM for 15 min at room temperature and the colony was observed under light microscope, (magnification, ×40; Leica Microsystems, Inc., Buffalo Grove, IL, USA). The number of colony formation in each group was normalized to that in the mock vector (Shanghai GenePharma Co., Ltd.) transfection group.
Flow cytometry analysis of apoptotic cells
Apoptosis was measured using Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI)-staining kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, 1×106 MIA PaCa-2 cells were plated in 100 mm dishes (Corning Incorporated, Corning, NY, USA) and then transfected with miR-128 mimics or scrambled control After 72 h transfection, the cells were lysed and stained with Annexin V-FITC and PI prior to analysis with a flow cytometer. This assay was performed to sort and identify early apoptotic (Annexin V-FITC+/PI-) cells, primary necrotic (Annexin V-FITC-/PI+) cells and also late apoptotic (Annexin V-FITC+/PI+) cells (also known as secondary necrotic cells). For each group, the experiment was repeated in triplicate. Analyzation was performed using a FACScan flow cytometer with CellQuest software version 5.1 (BD Biosciences, Franklin Lakes, NJ, USA).
Predicted target analysis of miR-128
miRecords (http://c1.accurascience.com/miRecords/) is an online database for predicting target binding sites of miRNAs. It integrates the results of numerous online miRNA target prediction tools, including DIANAmicroT, miRanda, PicTar and TargetScan. Predicted miR-128 targets were identified by miRecords database.
3′UTR-luciferase reporter gene assay
All vectors were purchased from Ambion (Thermo Fisher Scientific, Inc.). Vectors carried the MDM4 sequence, which contained the predicted miR-128 binding sites with wild-type or mutant 3′UTR. MIA PaCa-2 cells were plated into 24-well plates (1×104 cells/well) and transfected with MDM4 vector or mutant vector using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. After 4 h, 50 nM of miR-128 mimics or scramble control was transfected into the cells respectively. After another 48 h, cells were lysed with 0.5% trypsin at 37°C containing 5% CO2 and luciferase activities were analyzed with a dual luciferase assay kit (Promega Corporation, Madison, WI, USA) along with the normalization to the control group treated with MDM4 wild-type and miR-128 scramble vectors.
RT-qPCR analysis
Total RNA was isolated from tumor and adjacent tissues using a PicoPure™ RNA Isolation kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Then, 2 µg of RNA was utilized for cDNA synthesis using SuperScript™ III first-strand synthesis system kit (Thermo Fisher Scientific, Inc.). The reverse transcription reaction was performed at 55°C for 30 min. Following reverse transcription reactions, qPCR was performed with TaqMan™ gene expression master mix and respective primers (Thermo Fisher Scientific, Inc.). The specific PCR primer for miR-128 was as follows: forward, 5′-AACACTCCAGCTGGGTCACAGTGAACCGGTCT-3′ and reverse, 5′-CTCAACTGGTGTCGTGGA-3′. The primer for GAPDH was as follows: Forward, 5′-AATGCATCCTGCACCACCAA-3′ and reverse, 5′-GTAGCCATATTCATTGTCATA-3′. The qPCR procedure was performed as follows: For hold stage: Step 1: Heating from 25 to 95°C at a rate of 1.6°C/sec, holding for 2 min at 50°C. Step 2: Heating from 50 to 95°C at a rate of 1.6°C/sec, holding for 10 min at 95°C. For qPCR stage: Step 1: Initial denaturation at 95°C for 15 sec. Step 2: Annealing extension at 60°C for 1 min. The temperature was reduced from 95 to 60°C at the rate of 1.6°C/sec; the denaturation and extension stages were repeated for 40 cycles. Cq was measured at the PCR stage. Based on the RT-qPCR results of Cq number, the mRNA expression levels were calculated using the 2−ΔΔCq method (25) and normalized to an internal reference control GAPDH.
Western blotting
MIA PaCa-2 cells were harvested 72 h post-transfection with miR-128 mimics or mock, then total proteins were extracted using 1X NuPAGE™ LDS sample buffer (Thermo Fisher Scientific, Inc.). Protein concentration was quantified using a Bicinchoninic Acid Protein Assay kit (Thermo Fisher Scientific, Inc.). A total of 30 µg of protein was loaded per lane and electrophoresis was performed in 10% Tris-SDS gel. Following electrophoresis, the proteins were transferred to polyvinylidene fluoride membranes (Thermo Fisher Scientific, Inc.). For subsequent blocking and antibody incubation, an iBind™ kit (Thermo Fisher Scientific, Inc.) was used according to the manufacturer's protocol. The prepared diluted primary antibodies, iBind™ Flex/iBind™ Flex FD solution, diluted secondary antibodies and iBind™ Flex/iBind™ Flex FD solution were sequentially added to each lane. Membranes were then incubated with primary antibodies overnight at room temperature. Following this, membranes were incubated with secondary antibodies for 1 h at room temperature. and were rinsed in water prior to immunodetection. The primary antibodies were as follows: Anti-p53 (cat. no. 2527, 1:300; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-MDM4 (cat. no. sc-374147, 1:300; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), anti-cleaved caspase-3 (cat. no. sc-98785, 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and anti-β-actin (cat. no. 7210, 1:1,000 Santa Cruz Biotechnology, Inc.). The secondary antibody (horseradish peroxidase conjugated anti-rabbit; cat. no. 7074) was used at 1:3,000 and was obtained from Cell Signaling Technology, Inc. The signal was detected with SuperSigal® west femto maximum sensitivity substrate (Thermo Fisher Scientific, Inc.), and the specific proteins were detected with ChemiDoc™ imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA), using Image Lab software version 4.0 (Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analysis between two groups was performed with two-tailed Student's t-test and statistical differences among multiple groups were analyzed by one-way analysis of variance followed by Dunnett's test using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference. All experiments were performed in triplicate unless otherwise stated and all data are presented as the mean ± standard deviation.
Results
Expression of miR-128 in PC tissues and normal adjacent tissues
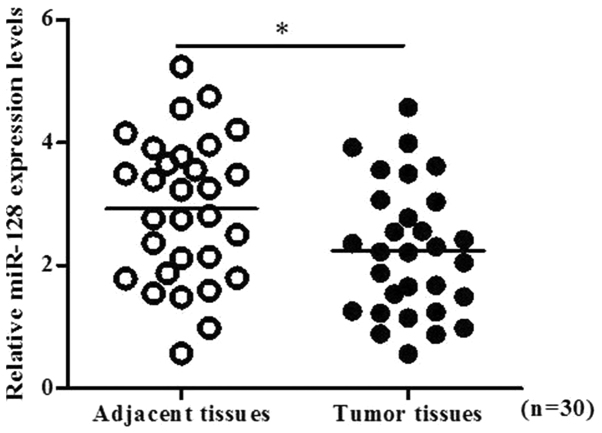
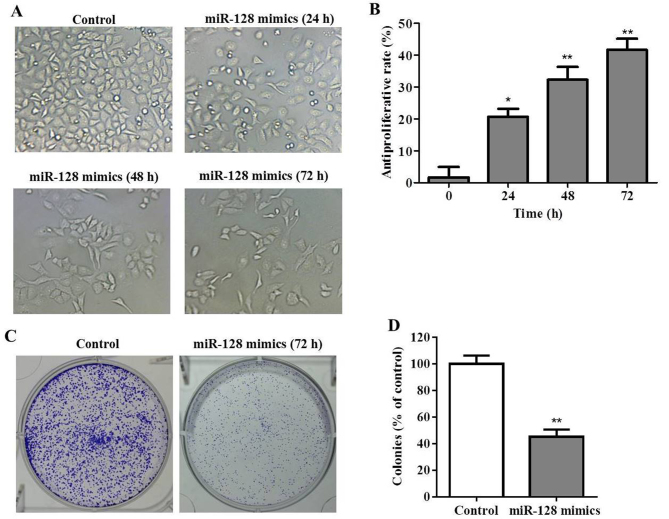
The expression of miR-128 within 30 pairs of PC tissues and normal adjacent tissues was determined by RT-qPCR. The results indicated that the levels of miR-128 were significantly decreased within PC tissues compared with in normal adjacent tissues (P<0.05; Fig. 1). In order to investigate the biological role of miR-128 in PC cells, miR-128 mimics or mock vectors were transiently transfected into MIA PaCa-2 cells. Subsequently, PC cell viability was determined with a CCK-8 assay at various time points post-transfection. As presented in Fig. 2A and B, miR-128 inhibited the growth of MIA PaCa-2 cells. The cell morphology and density observed under the microscope, reduced cell proliferation was observed within the miR-128 mimics group, particularly at 48 and 72 h post-transfection. In addition, cell morphologies were round instead of epithelial indicating that miR-128 may not only inhibit cell growth, but may also induce cell apoptosis. Furthermore, the result of the colony formation assay indicated that colony formation was significantly inhibited by miR-128 mimics transfection (P<0.01; Fig. 2C and D).
Figure 1.
miR-128 expression profile of pancreatic cancer tissues and normal adjacent tissues. Total RNA was extracted from tumor tissues and normal tissues of 30 patients with pancreatic cancer. The expression levels of miR-128 were analyzed by reverse transcription-quantitative polymerase chain reaction. For all samples, analysis was conducted in triplicate. *P<0.05. miR, microRNA.
Figure 2.
miR-128 mimics inhibits MIA PaCa-2 cell growth and colony formation. (A) Morphology alterations of MIA PaCa-2 cells transfected with miR-128 mimics for 24, 48 or 72 h (magnification, ×20). (B) Antiproliferative rate of miR-128 mimics at 24, 48 or 72 h (C) Colony formation assay at 72 h (magnification, ×10). (D) Quantification of colony formation assay. *P<0.05, **P<0.01 vs. control. miR, microRNA.
miR-128 induced apoptosis in PC cells
As observed in Fig. 2A, MIA PaCa-2 cells revealed apoptotic characteristics following transfection with miR-128 mimics. Thus, flow cytometry analysis was performed to investigate the proportion of apoptotic cells in the miR-128 mimics-transfected cells using Annexin V-FITC and PI staining. The results of the present study demonstrated that miR-128 mimics markedly increased the early apoptotic cell number (Annexin V-FITC+/PI-) and late apoptotic cell number (Annexin V-FITC+/PI+; Fig. 3A). The apoptotic rate following miR-128 mimics transfection was 32.1%, which was significantly increased compared with in the control (P<0.01; Fig. 3B). These results demonstrated that miR-128 may induce MIA PaCa-2 cell apoptosis.
Figure 3.
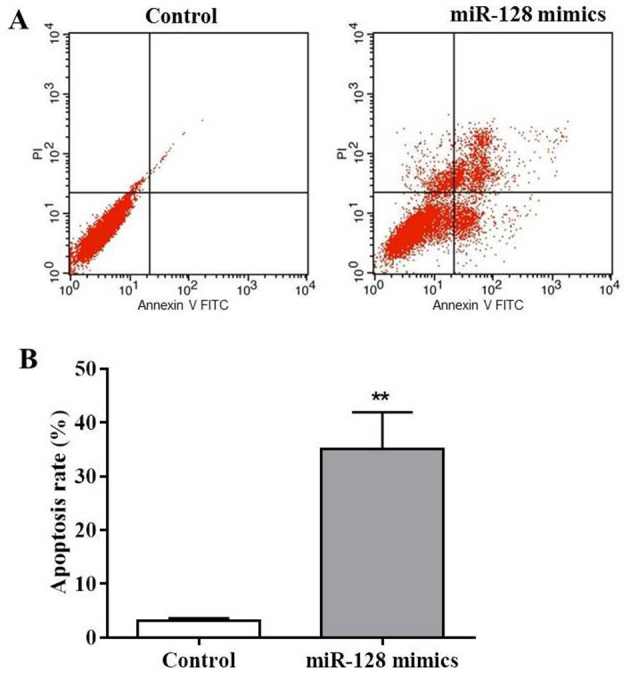
miR-128 mimics induces apoptosis in MIA PaCa-2 cells. MIA PaCa-2 cells were treated with miR-128 mimics or mock for 72 h followed by PI and Annexin V-FITC double staining of the apoptotic cells. (A) Flow cytometry analysis of apoptosis rate in MIA PaCa-2 cells. (B) Quantification of apoptotic rate. **P<0.01 vs. control. FITC, fluorescein isothiocyanate; miR, microRNA; PI, propidium iodide.
miR-128 directly targets MDM4
To investigate the underlying mechanisms of miR-128-mediated inhibition of MIA PaCa-2 cell growth and apoptosis induction, bioinformatics analysis was performed to identify the targets of miR-128 in humans. MDM4 was of particular interest among all the potential targets due to the high prediction scores in TargetScan (http://www.targetscan.org/vert_61/), and microRNA.org (http://www.microrna.org/microrna/) online bioinformatics methods. Therefore, the following experiments were performed to confirm whether MDM4 was a direct target of miR-128. In the present study, wild-type and mutant MDM4-3′-UTR expression plasmids were constructed, which were fused with a luciferase reporter gene (Fig. 4A). The results indicated that the miR-128 mimics significantly inhibited the luciferase activity of cells transfected with wild-type MDM4 (P<0.01). However, no significant variations between the control and miR-128 mimics treatment were observed with mutant MDM4 (Fig. 4B). Thus, the findings of the present study suggested that miR-128 may directly target MDM4.
Figure 4.
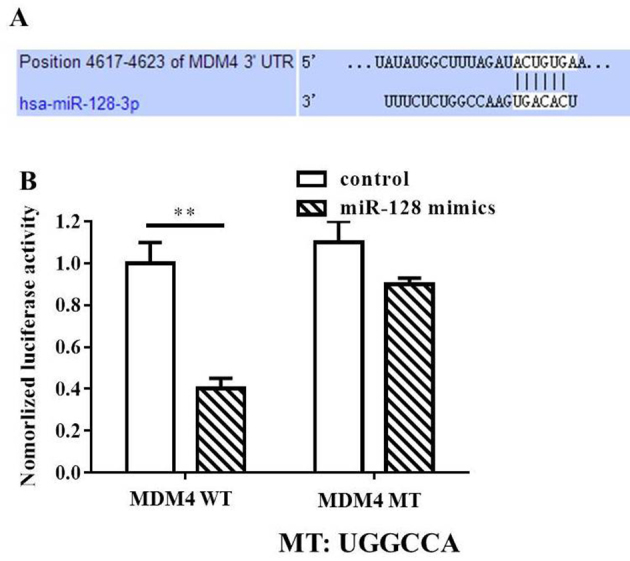
MDM4 is a direct target of miR-128. (A) Predicted target site of miR-128 in the 3′UTR of MDM4. (B) MIA PaCa-2 cells were transfected with a reporter gene containing WT (sequence UGACAC) or MT (sequence UGGCCA) MDM4 3′UTR along with miR-422a mimics or control as indicated. Luciferase activity was compared between groups. **P<0.01. MDM4, double minute 4; miR, microRNA; MT, mutant type; UTR, untranslated region; WT, wild type.
miR-128 induces apoptosis in PC cells via upregulation of p53 expression
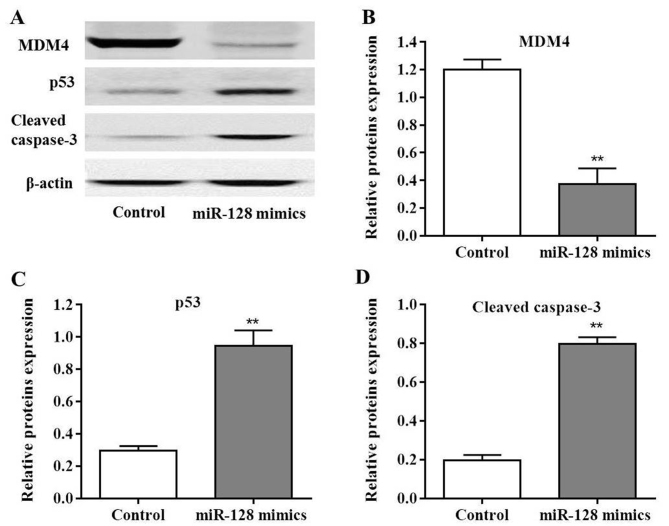
As MDM4 was considered to be the target of miR-128, MDM4 protein expression was measured in PC cells following transfection with miR-128 mimics. MDM4 levels were significantly decreased post-transfection compared with the control (P<0.01; Fig. 5A and B). Additionally, miR-128 may induce MIA PaCa-2 cell apoptosis; in order to further determine the possible mechanisms involved in this process, molecules were selected that have been reported to be involved in the caspase signaling pathway. The results of the present study indicated that the expression levels of p53 and cleaved caspase-3 protein were significantly upregulated in MIA PaCa-2 cells post-transfection with miR-128 mimics compared with the control (P<0.01; Fig. 5C and D). Collectively, these results indicated that miR-128 promoted MIA PaCa-2 cell apoptosis via the p53 and caspase-3 dependent pathway.
Figure 5.
Expression of p53 is enhanced by miR-128 mimics via inhibition of MDM4 expression. (A) Western blot analysis of MDM4, p53 and cleaved caspase-3 expression following transfection with miR-128 mimics. (B) Quantification of MDM4 expression. (C) Quantification of p53 expression. (D) Quantification of cleaved caspase-3 expression. **P<0.01 vs. control. MDM4, double minute 4; miR, microRNA.
Discussion
The role of miR-128 has been discussed in various types of cancer as its expression may be suppressed at the mRNA level (10,15). It has also been reported that miR-128 is decreased in PC and may inhibit PC cell proliferation (26); however, further investigation is required to understand the mechanisms of miR-128 in the regulation of PC cell growth (10,27,28).
The present study clarified one of the biological roles of miR-128 in regulating PC cell growth. Compared with in adjacent normal tissue, miR-128 expression levels were significantly downregulated in pancreatic cancer tissues. Additionally, miR-128 may significantly inhibit MIA PaCa-2 cell growth in functional experiments. miR-128 was indicated to induce cell apoptosis, as demonstrated by flow cytometry analysis. To the best of our knowledge, the present study is the first to report the induction of cell apoptosis by miR-128 in PC.
In the present study, MDM4 was identified as the direct target of miR-128 using bioinformatics analysis (29). miR-128 may bind to the MDM4 3′UTR via base pairing. Based on this knowledge, the dual luciferase assay demonstrated that restored miR-128 expression levels could decrease luciferase activity, whereas no alterations were observed in the luciferase activity with mutant MDM4 3′UTR. In addition, analysis of protein expression levels indicated that miR-128 decreased MDM4 protein expression. Furthermore, along with decreased MDM4 protein, significant increases in p53 and cleaved caspase-3 protein expression levels were observed. The results of the present study suggested that miR-128 may regulate the expression of MDM4 by directly binding to the 3′UTR of MDM4. Therefore, the increased p53 expression promoted the caspase-3 dependent cell apoptosis program.
In conclusion, the results of the present study demonstrate that miR-128 may inhibit the proliferation of pancreatic cancer cells by targeting MDM4 to induce cell apoptosis. These findings add to the current understanding of miR-128 in pancreatic cancer cells and may have potential applications in the development of treatment strategies for pancreatic cancer. However, the detailed mechanism of MDM4 in the pathogenesis of pancreatic cancer requires further investigation.
Acknowledgements
The authors would like to thank Dr Hongli Wang (The Third People's Hospital, Yancheng, China) for bioinformatics analysis.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HH managed the experimental design, tissue collection, the execution of experiments and was the major contributor in developing the first draft of the manuscript. LW analyzed and interpreted the patient data. JX reviewed and approved the final version of the manuscript. AW reviewed and approved the final draft of the manuscript prior to submission.
Ethics approval and consent to participate
Ethical approval for the present study was received from the Clinical Research and Ethics Committee at The Third People's Hospital (Yancheng, China).
Consent for publication
All patients provided written informed consent for the publication of all associated data in this study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Vaz J, Ansari D, Sasor A, Andersson R. SPARC: A potential prognostic and therapeutic target in pancreatic cancer. Pancreas. 2015;44:1024–1035. doi: 10.1097/MPA.0000000000000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spadi R, Brusa F, Ponzetti A, Chiappino I, Birocco N, Ciuffreda L, Satolli MA. Current therapeutic strategies for advanced pancreatic cancer: A review for clinicians. World J Clin Oncol. 2016;7:27–43. doi: 10.5306/wjco.v7.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin QJ, Yang F, Jin C, Fu DL. Current status and progress of pancreatic cancer in China. World J Gastroenterol. 2015;21:7988–8003. doi: 10.3748/wjg.v21.i26.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puleo F, Maréchal R, Demetter P, Bali MA, Calomme A, Closset J, Bachet JB, Deviere J, Van Laethem JL. New challenges in perioperative management of pancreatic cancer. World J Gastroenterol. 2015;21:2281–2293. doi: 10.3748/wjg.v21.i8.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis JL, Pandalai PK, Ripley RT, Langan RC, Avital I. Expanding surgical treatment of pancreatic cancer: The role of regional chemotherapy. Pancreas. 2014;41:678–684. doi: 10.1097/MPA.0b013e318249955a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rachagani S, Macha MA, Heimann N, Seshacharyulu P, Haridas D, Chugh S, Batra SK. Clinical implications of miRNAs in the pathogenesis, diagnosis and therapy of pancreatic cancer. Adv Drug Deliv Rev. 2015;81:16–33. doi: 10.1016/j.addr.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tempero MA, Arnoletti JP, Behrman SW, Ben-Josef E, Benson AB, III, Casper ES, Cohen SJ, Czito B, Ellenhorn JD, Hawkins WG, et al. Pancreatic Adenocarcinoma, version 2.2012: Featured updates to the NCCN Guidelines. J Natl Compr Canc Netw. 2012;10:703–713. doi: 10.6004/jnccn.2012.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herman JM, Wild AT, Wang H, Tran PT, Chang KJ, Taylor GE, Donehower RC, Pawlik TM, Ziegler MA, Cai H, et al. Randomized phase III multi-institutional study of TNFerade biologic with fluorouracil and radiotherapy for locally advanced pancreatic cancer: Final results. J Clin Oncol. 2013;31:886–894. doi: 10.1200/JCO.2012.44.7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao J, Zhang S, Zhou Y, Hu X, Shao C. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. FEBS Lett. 2011;585:207–213. doi: 10.1016/j.febslet.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 11.Khan S, Ansarullah DK, Kumar D, Jaggi M, Chauhan SC. Targeting microRNAs in pancreatic cancer: Microplayers in the big game. Cancer Res. 2013;73:6541–6547. doi: 10.1158/0008-5472.CAN-13-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Li A, Hong SM, Hruban RH, Giggins M. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res. 2012;18:981–992. doi: 10.1158/1078-0432.CCR-11-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali S, Saleh H, Sethi S, Sarkar FH, Philip PA. MicroRNA profiling of diagnostic needle aspirates from patients with pancreatic cancer. Br J Cancer. 2012;107:1354–1360. doi: 10.1038/bjc.2012.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, Liu J, Chen-Xiao K, Zhang X, Lee WN, Go VL, Xiao GG. Advance in microRNA as a potential biomarker for early detection of pancreatic cancer. Biomark Res. 2016;4:20. doi: 10.1186/s40364-016-0074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma MZ, Kong X, Weng MZ, Cheng K, Gong W, Quan ZW, Peng CH. Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: Meta-analysis, experimental validation and clinical significance. J Exp Clin Cancer Res. 2013;32:71. doi: 10.1186/1756-9966-32-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He H, Hao SJ, Yao L, Yang F, Di Y, Li J, Jiang YJ, Jin C, Fu DL. MicroRNA-218 inhibits cell invasion and migration of pancreatic cancer via regulating ROBO1. Cancer Biol Ther. 2014;15:1333–1339. doi: 10.4161/cbt.29706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, Zhou J, Wu J, Shao C. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- 19.Munding JB, Adai AT, Maghnouj A, Urbanik A, Zöllner H, Liffers ST, Chromik AM, Uhl W, Szafranska-Schwarzbach AE, Tannapfel A, Hahn SA. Global microRNA expression profiling of microdissected tissues identifies miR-135b as a novel biomarker for pancreatic ductal adenocarcinoma. Int J Cancer. 2012;131:E86–E95. doi: 10.1002/ijc.26466. [DOI] [PubMed] [Google Scholar]
- 20.Zhao C, Zhang J, Zhang S, Yu D, Chen Y, Liu Q, Shi M, Ni C, Zhu M. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncology Rep. 2013;30:276–284. doi: 10.3892/or.2013.2420. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Zhao CY, Zhang SH, Yu DH, Chen Y, Liu QH, Shi M, Ni CR, Zhu MH. Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma. Oncol Re. 2014;31:1157–1164. doi: 10.3892/or.2013.2960. [DOI] [PubMed] [Google Scholar]
- 22.Abue M, Yokoyama M, Shibuya R, Tamai K, Yamaguchi K, Sato I, Tanaka N, Hamada S, Shimosegawa T, Sugamura K, Satoh K. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. 2014;46:539–547. doi: 10.3892/ijo.2014.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marine JC, Jochemsen AG. Mdmx as an essential regulator of p53 activity. Biochem Biolphys Res Commun. 2005;331:750–760. doi: 10.1016/j.bbrc.2005.03.151. [DOI] [PubMed] [Google Scholar]
- 24.Gospodarowicz MK, Brierley JD, Wittekind C. TNM Classification of Malignant Tumours. 2017 [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delata Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, Chen C. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KH, Lee JK, Choi DW, Do IG, Sohn I, Jang KT, Jung SH, Heo JS, Choi SH, Lee KT. Postoperative prognosis prediction of pancreatic cancer with seven microRNAs. Pancreas. 2015;44:764–768. doi: 10.1097/MPA.0000000000000346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.