Abstract
E2Fs, as a family of pivotal transcription factors, have been implicated in multiple biological functions in human cancer; however, the expression and prognostic significance of E2Fs in breast cancer remains unknown. In the present study, the mRNA expression patterns of E2Fs in breast cancer were investigated with Oncomine and The Cancer Genome Atlas data. Prognostic values of E2Fs for patients with breast cancer were determined using the Kaplan-Meier plotter database. The results strongly indicated that E2F1, E2F2, E2F3, E2F5, E2F7 and E2F8 were overexpressed in patients with breast cancer, whereas E2F4 and E2F6 exhibited no expression difference between patients with cancer and healthy controls. In survival analyses, elevated E2F1, E2F3, E2F5, E2F7 and E2F8 expression levels were significantly associated with lower overall survival, relapse-free survival (RFS), distant metastasis-free survival (DMFS) or post-progression survival for patients with breast cancer. Furthermore, high expression of E2F4 indicated improved RFS but reduced DMFS. Subgroup analyses based on four clinicopathological factors further revealed that E2Fs were associated with the prognosis of patients with breast cancer in an estrogen receptor-, progesterone receptor-, human epidermal growth factor 2- and lymph node status-specific manner. These data indicated that E2Fs may serve as promising biomarkers and therapeutic targets for breast cancer.
Keywords: E2F, breast cancer, bioinformatics, the cancer genome atlas, oncomine, Kaplan-Meier plotter, prognosis
Introduction
Breast cancer is the most common malignancy in females and remains a major cause of cancer-associated mortality for females globally, particularly in less developed countries (1). As of yet, the risk factors for breast cancer remain uncertain, but have been indicated to be associated with complex and heterogeneous processes involving reproductive, hormonal and numerous other potential factors, including being overweight, menopausal hormone therapy, physical inactivity and alcohol intake (2,3). The incidence rate of breast cancer remains at a relatively high level (4). Despite improved diagnostics, advanced surgical techniques and growing numbers of anticancer drugs and targeted therapies that have largely improved the clinical outcomes of breast cancer, the recurrence or metastasis frequently occurs and the long-term survival of patients with breast cancer is not optimistic (4–6); therefore, it is necessary to further investigate the underling mechanisms of initiation and development of breast cancer. Furthermore, novel biomarkers that may serve as therapeutic targets or prognostic indicators are also urgently required.
E2Fs are a group of transcription factors, including ≥10 members encoded by eight distinct genes (7). The majority of studies have divided E2Fs into two subgroups: Transcriptional activators (E2F1-E2F3) and repressors (E2F4-E2F8) based on their structures and functions (7,8). At present, E2Fs have been well characterized as central regulators of cell cycle progression (9). During G0 and early G1 phase, unphosphorylated pRB binds to certain E2Fs and negatively regulates their transcriptional activity (10). Subsequently, cyclin-dependent kinase complexes mediating phosphorylation of pRB in late G1 phase enable E2Fs to activate target genes, resulting in DNA and protein synthesis that are necessary for S-phase entry (10). Furthermore, an increasing number of studies have revealed the roles of E2Fs beyond simply participating in the regulation of the cell cycle (11,12). Numerous other physiological processes, including proliferation, apoptosis, DNA damage repair, senescence and autophagy, which were known to be crucial for tumor progression, have also been determined to heavily rely on the involvement of E2Fs (11,12).
In human malignances, E2Fs are frequently deregulated. Expression of E2F1 was reported to be elevated in lung cancer, compared with normal tissues, and a high level of E2F1 was significantly associated with a poorer prognosis (13,14). In hepatocellular carcinoma (HCC), E2F1, E2F3, E2F4 and E2F8 are overexpressed in tumor specimens (15–17). Overexpression of E2F8 contributes to HCC cell proliferation via promoting cells to entry into S-phase, which may be mediated by the transcriptional effect of E2F8 on cyclin D1 (16). Previous studies have determined that several E2Fs were upregulated in ovarian cancer, and high expression levels of E2F4 and E2F7 were associated with an improved prognosis, while E2F8 indicated a reduced overall survival (OS) (18–20). Recent studies have also provided evidence demonstrating that E2Fs family may act as promising biomarkers in breast cancer (21–23). A study based on 165 lymph node-negative breast carcinomas demonstrated that patients with E2F1-positive tumors would exhibit a reduced disease-free survival (DFS) or overall survival (OS) rate than those with E2F1-negative tumors (21). Similarly, increased nuclear expression of E2F4 demonstrated reduced survival outcomes for patients with breast cancer (22). Fujiwara et al (23) determined that E2F2 expression was associated with relapse-free survival (RFS) rate.
Although these data indicated that E2Fs may serve as reliable markers for breast cancer, the different expression levels, various biological functions, detailed molecular mechanisms and prognostic significance of the majority of E2Fs members remain elusive. A comprehensive study of all eight E2F genes is required.
Materials and methods
Oncomine database and the cancer genome atlas (TCGA) data
Oncomine (http://www.oncomine.org), an online microarray database, was utilized to examine the mRNA expression levels of E2Fs in breast cancer. The thresholds were restricted as follows: P-value=0.0001; fold-change=2; gene rank=10%; and data type, mRNA. For each gene, comparison by cancer vs. normal analysis was performed. Cancer type, fold change, Student's t-test value, P-value and sample size were abstracted from comparisons with statistical significance. Integrin mRNA HiSeq expression data of TCGA were downloaded from the Cancer Genomics Browser of University of California Santa Cruz (version 2015-02-24; https://genome-cancer.ucsc.edu/).
Kaplan-Meier database analysis
Kaplan-Meier plotter (KM plotter; http://kmplot.com/analysis/) (24) was used to determine the prognostic values of E2Fs in breast cancer. KM plotter is an online database containing microarray gene expression data and survival information derived from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/), European Genome-Phenome Archive (https://ega.crg.eu/) and TCGA containing a total of 4,142 patients with breast cancer with survival data. For each gene symbol, the desired probe ID was identified according to the file of probe sets provided by KM plotter. Patients were divided into high and low expression groups by median values of mRNA expression level and survival analyses were performed without follow-up restrictions. In brief, the desired probe IDs representing eight genes were separately entered into the database to perform Kaplan-Meier survival analysis for OS, RFS, distant metastasis-free survival (DMFS) and post-progression survival (PPS) Kaplan-Meier Plots, which were automatically generated by the database. Subgroup analyses were performed via separating patients based on the factors of expression of: Estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor 2 (HER-2) and lymph node status. Factors were defined as either positive or negative, with the status information being included in the database. The number of cases, hazard ratios (HRs), 95% confidence intervals (CIs) and log rank P-values were obtained from the webpage of the KM plotter.
Statistical analysis
An un-paired Student's t-test was performed to examine the mRNA expression difference between tumor and normal tissues from TCGA using SPSS 20.0 (IBM Corp., Armonk, NY, USA). The boxplots were created using GraphPad software 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Data are expressed as mean ± standard error of the mean. P<0.05 was considered to indicate a statistically significant significance.
Results
Expression levels of E2Fs in breast cancer
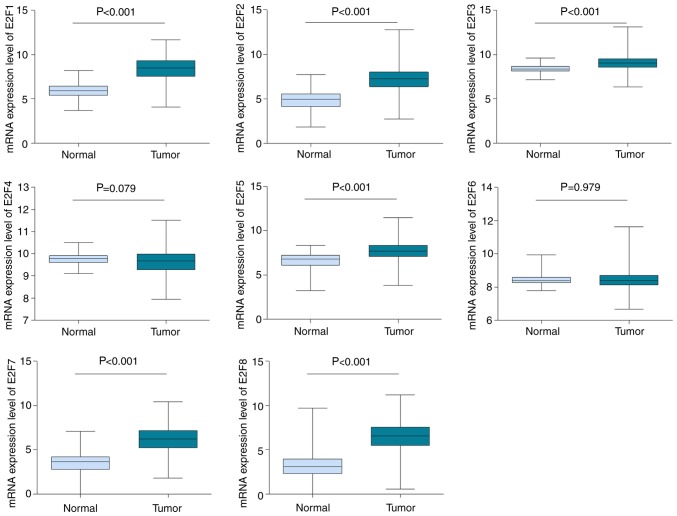
The mRNA expression levels of E2Fs via cancer vs. normal analysis were firstly investigated using the Oncomine database, which contains publicly available microarray data from multiple cancer types, including breast carcinoma. With the following thresholds: P-value=0.0001; fold change=2; gene rank=10%, E2F1 was determined to be overexpressed in breast cancer tissues, compared with normal samples, according to datasets from TCGA and Gluck et al (25). A total of nine comparisons, including datasets from Curtis et al (26), Gluck et al (25), TCGA, Zhao et al (27) and Richardson et al (28), revealed that the mRNA expression level of E2F2 was higher in breast cancer samples than in healthy controls. By contrast, the dataset by Radvanyi et al (29) demonstrated a lower expression level of E2F2 in breast cancer, but caution should be taken due to the limited sample size, with only six normal controls against two invasive lobular breast carcinomas. In datasets by Curtis et al (26) and Richardson et al (28), E2F3 was significantly upregulated in breast cancer, compared with normal tissues. However, all 13 datasets available for E2F4 indicated no expression difference between tumor and normal groups. Based on datasets by Richardson et al (28) and TCGA, it was determined that the transcription levels of E2F5 in ductal breast carcinoma and invasive breast carcinoma were higher than in normal breast tissues. As for E2F6, there were seven datasets in Oncomine, but none of these revealed a significant statistical difference between tumor and normal samples. The mRNA expression level of E2F7 was notably increased in breast cancer when datasets by Richardson et al (28) and TCGA were analyzed. Similarly, the mRNA expression level of E2F8 was increased in breast carcinomas, compared with normal tissues in datasets by Gluck et al (25) and TCGA. All of the results are summarized in Table I. Furthermore, the mRNA HiSeq expression data involving 1,095 tumors and 113 normal samples from TCGA database was utilized to further investigate and confirm the expression difference of E2Fs in breast cancer and normal tissue. As depicted in Fig. 1, consistent with the Oncomine data, the mRNA expression levels of E2F1, E2F2, E2F3, E2F5, E2F7 and E2F8 were determined to be upregulated in breast cancer (P<0.001), compared with normal tissues. There was no difference in transcription levels of E2F4 and E2F6 between tumor tissues and normal tissues (Fig. 1).
Table I.
Analyses of E2Fs in breast cancer.
| Gene symbol | Dataset | Reporter | Normal (no. of cases) | Tumor (no. of cases) | Fold-change | T-value | P-value |
|---|---|---|---|---|---|---|---|
| E2F1 | TCGA Breast | A_23_P80032 | Breast (61) | Invasive breast carcinoma (76) | 2.734 | 11.690 | 1.68×10−22 |
| A_23_P80032 | Breast (61) | Invasive ductal breast carcinoma (389) | 3.216 | 18.607 | 4.37×10−35 | ||
| A_23_P80032 | Breast (61) | Invasive lobular breast carcinoma (36) | 2.088 | 6.425 | 1.56×10−8 | ||
| Gluck Breast | 26634 | Breast (4) | Invasive breast carcinoma (154) | 2.545 | 10.681 | 2.29×10−5 | |
| E2F2 | Curtis Breast | ILMN_1777233 | Breast (144) | Medullary breast carcinoma (32) | 5.025 | 14.502 | 1.28×10−16 |
| ILMN_1777233 | Breast (144) | Invasive ductal breast carcinoma (1,556) | 2.767 | 33.780 | 3.89×10−93 | ||
| ILMN_1777233 | Breast (144) | Invasive breast carcinoma (21) | 2.315 | 6.844 | 3.56×10−7 | ||
| Gluck Breast | 20301 | Breast (4) | Invasive breast carcinoma (154) | 2.637 | 11.788 | 7.06×10−7 | |
| TCGA Breast | A_23_P408957 | Breast (61) | Invasive ductal breast carcinoma (389) | 3.790 | 18.457 | 7.38×10−35 | |
| A_23_P408957 | Breast (61) | Invasive breast carcinoma (76) | 3.094 | 11.308 | 1.62×10−21 | ||
| A_23_P408955 | Breast (61) | Invasive lobular breast carcinoma (36) | 2.243 | 7.520 | 6.13×10−11 | ||
| Zhao Breast | IMAGE: 293331 | Breast (3) | Invasive ductal breast carcinoma (37) | 2.222 | 6.084 | 7.11×10−6 | |
| Richardson Breast 2 | 228361_at | Breast (7) | Ductal breast carcinoma (40) | 3.077 | 6.237 | 5.25×10−5 | |
| E2F3 | Curtis Breast | ILMN_1669502 | Breast (144) | Medullary breast carcinoma (32) | 2.522 | 11.118 | 2.89×10−13 |
| Richardson Breast 2 | 203692_s_at | Breast (7) | Ductal breast carcinoma (40) | 3.558 | 8.624 | 9.00×10−9 | |
| E2F4 | Not available | ||||||
| E2F5 | Richardson Breast 2 | 221586_s_at | Breast (7) | Ductal breast carcinoma (40) | 2.573 | 6.253 | 9.00×10−8 |
| TCGA Breast | A_23_P31713 | Breast (61) | Invasive breast carcinoma (76) | 2.077 | 7.228 | 1.75×10−11 | |
| E2F6 | Not available | ||||||
| E2F7 | Richardson Breast 2 | 228033_s_at | Breast (7) | Ductal breast carcinoma (40) | 4.535 | 7.879 | 4.65×10−10 |
| TCGA Breast | A_23_P336178 | Breast (61) | Invasive breast carcinoma (76) | 5.193 | 12.912 | 1.43×10−25 | |
| A_23_P336178 | Breast (61) | Invasive ductal breast carcinoma (389) | 7.456 | 22.097 | 5.96×10−40 | ||
| A_23_P336178 | Breast (61) | Invasive lobular breast carcinoma (36) | 4.262 | 9.860 | 8.61×10−15 | ||
| E2F8 | Gluck Breast | 20493 | Breast (4) | Invasive breast carcinoma (154) | 2.489 | 13.421 | 4.37×10−6 |
| TCGA Breast | A_23_P35871 | Breast (61) | Invasive lobular breast carcinoma (36) | 5.188 | 9.033 | 2.05×10−14 | |
| A_23_P35871 | Breast (61) | Invasive breast carcinoma (76) | 7.581 | 11.979 | 5.70×10−23 | ||
| NM_024680_1_1600 | Breast (61) | Invasive ductal breast carcinoma (389) | 2.416 | 17.311 | 2.20×10−34 |
P<0.05. TCGA, The Cancer Genome Atlas.
Figure 1.
The mRNA expression levels of E2Fs in breast cancer. The mRNA expression levels of E2Fs were investigated with The Cancer Genome Atlas mRNA HiSeq expression data including 1,095 breast cancer tissues and 113 cases of normal tissues.
Association of the expression of E2Fs and OS rates in patients with breast cancer
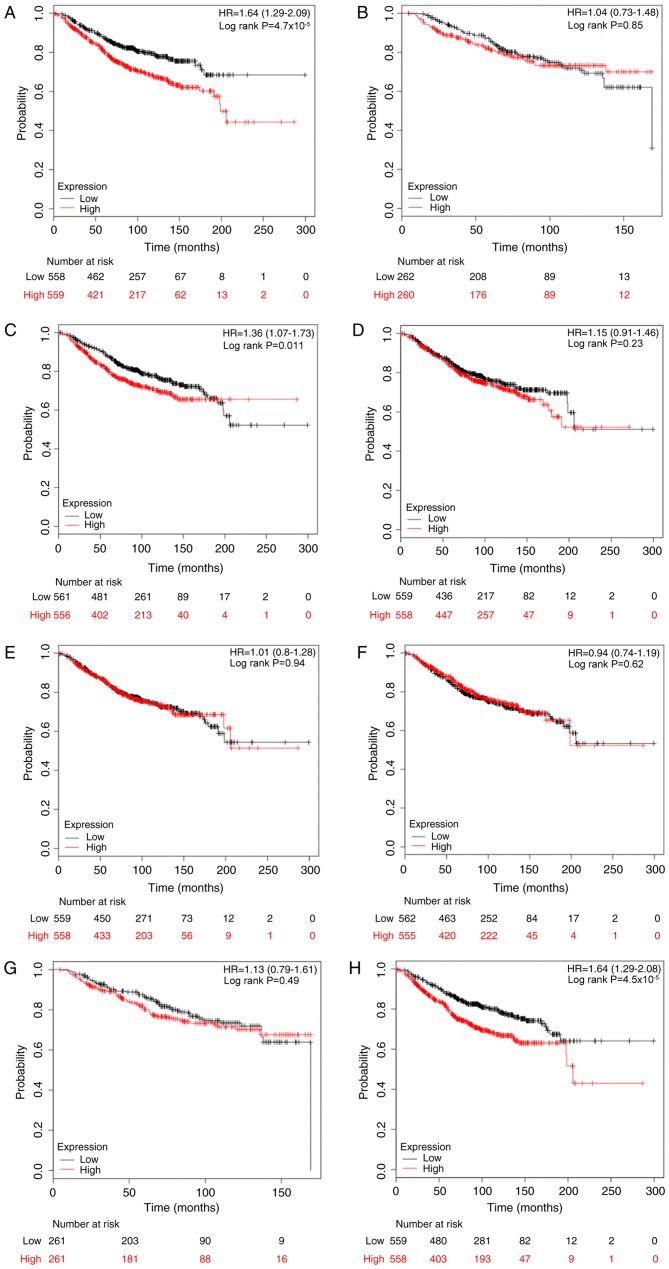
The association between E2Fs and OS rates was determined using the KM plotter database. The desired Affymetrix IDs were as follows: 204947_at, E2F1; 228361_at, E2F2; 203693_s_at, E2F3; 202248_at, E2F4; 221586_s_at, E2F5; 203957_at, E2F6; 228033_at, E2F7; and 219990_at, E2F8. As depicted in Fig. 2, it was determined that high mRNA expression of E2F1, E2F3 and E2F8 was significantly associated with reduced OS rates for patients with breast cancer, with HR=1.64 (1.29–2.09) and P<0.001; HR=1.36 (1.07–1.73) and P=0.011; and HR=1.64 (1.29–2.08) and P<0.001, compared with the low expression group, respectively. However, as for the other five members, E2F2 and E2F4-7, there was no clear association with OS (Fig. 2).
Figure 2.
The prognostic effects of E2Fs on overall survival. Kaplan-Meier survival curves are presented: (A) E2F1 (204947_at, n=1117); (B) E2F2 (228361_at, n=522); (C) E2F3 (203693_s_at, n=1117); (D) E2F4 (202248_at, n=1117); (E) E2F5 (221586_s_at, n=1117); (F) (203957_at, n=1117); (G) E2R7 (228033_at, n=522); and (H) E2F8 (219990_at, n=1117). HR, hazard ratio.
Following this, the prognostic values of E2Fs were examined in patients with breast cancer based on clinicopathological features, including ER, PR, HER-2 and lymph node status (Table II). The results demonstrated that high expression of E2F1 (HR, 1.82; 95% CI, 1.18–2.81; P=0.006), E2F3 (HR, 1.92; 95% CI, 1.25–2.95; P=0.003) and E2F8 (HR, 2.94; 95% CI, 1.87–4.63; P<0.001) indicated reduced OS rates in ER-positive patients, but not in ER-negative patients. Notably, high expression of E2F2, E2F5 and E2F6 were determined to be significantly associated with improved OS rates in ER-negative patients, with HR=0.29 (95% CI, 0.09–0.92) and P=0.025; HR=0.39 (95% CI, 0.21–0.71) and P=0.001; HR=0.52 (95% CI, 0.29–0.94) and P=0.027, respectively. Since there were a limited number of cases with PR information, analysis of the prognostic significance of E2Fs stratifying by PR status in KM plotter was not conducted. Although E2F1 and E2F5 were associated with OS in HER-2-positive patients, the results should be treated with caution due to a small sample size (n=28). Furthermore, increased E2F5 predicted an improved OS rate in lymph node-positive patients (HR, 0.60; 95% CI, 0.36–1.00; P=0.048), whilst E2F1 (HR, 2.15; 95% CI, 1.39–3.32; P<0.001) and E2F8 (HR, 2.14; 95% CI, 1.40–3.28; P<0.001) were significantly associated with reduced OS rates in lymph node-negative patients.
Table II.
The association between E2Fs and overall survival for patients with breast cancer based on clinicopathological features.
| Positive status | Negative status | ||||||
|---|---|---|---|---|---|---|---|
| Clinicopathological factor | Gene symbol | Cases | HR (95% CI) | P-value | Cases | HR (95% CI) | P-value |
| ER | E2F1 | 377 | 1.82 (1.18–2.81) | 0.006a | 142 | 0.83 (0.47–1.46) | 0.512 |
| E2F2 | 42 | 1.33 (0.36–5.00) | 0.669 | 45 | 0.29 (0.09–0.92) | 0.025a | |
| E2F3 | 377 | 1.92 (1.25–2.95) | 0.003a | 142 | 0.77 (0.44–1.35) | 0.362 | |
| E2F4 | 377 | 1.20 (0.79–1.82) | 0.403 | 142 | 0.68 (0.38–1.22) | 0.192 | |
| E2F5 | 377 | 1.03 (0.68–1.56) | 0.897 | 142 | 0.39 (0.21–0.71) | 0.001a | |
| E2F6 | 377 | 1.41 (0.93–2.15) | 0.107 | 142 | 0.52 (0.29–0.94) | 0.027a | |
| E2F7 | 42 | 0.84 (0.23–3.15) | 0.801 | 45 | 0.74 (0.27–1.98) | 0.543 | |
| E2F8 | 377 | 2.94 (1.87–4.63) | <0.001a | 142 | 0.95 (0.54–1.67) | 0.866 | |
| PR | N/A | ||||||
| HER-2 | E2F1 | 28 | 0.22 (0.06–0.81) | 0.013a | 62 | 1.04 (0.36–2.96) | 0.945 |
| E2F2 | 26 | 0.36 (0.10–1.39) | 0.125 | 62 | 1.38 (0.48–3.97) | 0.554 | |
| E2F3 | 28 | 0.50 (0.16–1.55) | 0.221 | 62 | 0.68 (0.24–1.98) | 0.481 | |
| E2F4 | 28 | 0.70 (0.22–2.18) | 0.534 | 62 | 1.39 (0.48–4.00) | 0.544 | |
| E2F5 | 28 | 0.27 (0.08–0.88) | 0.020a | 62 | 0.39 (0.12–1.24) | 0.097 | |
| E2F6 | 28 | 0.56 (0.18–1.78) | 0.320 | 62 | 0.53 (0.18–1.59) | 0.251 | |
| E2F7 | 26 | 0.79 (0.24–2.61) | 0.704 | 62 | 1.02 (0.36–2.91) | 0.969 | |
| E2F8 | 28 | 0.62 (0.20–1.91) | 0.404 | 62 | 1.02 (0.36–2.91) | 0.975 | |
| Lymph node | E2F1 | 197 | 1.27 (0.77–2.11) | 0.342 | 425 | 2.15 (1.39–3.32) | <0.001a |
| E2F2 | 118 | 0.77 (0.36–1.66) | 0.504 | 77 | 0.62 (0.19–2.07) | 0.433 | |
| E2F3 | 197 | 1.34 (0.81–2.21) | 0.255 | 425 | 1.10 (0.73–1.66) | 0.655 | |
| E2F4 | 197 | 1.39 (0.84–2.30) | 0.199 | 425 | 0.71 (0.47–1.08) | 0.107 | |
| E2F5 | 197 | 0.60 (0.36–1.00) | 0.048a | 425 | 1.00 (0.66–1.51) | 0.995 | |
| E2F6 | 197 | 0.63 (0.38–1.06) | 0.079 | 425 | 0.71 (0.46–1.09) | 0.112 | |
| E2F7 | 118 | 0.84 (0.40–1.77) | 0.650 | 77 | 1.52 (0.48–4.78) | 0.474 | |
| E2F8 | 197 | 0.78 (0.47–1.30) | 0.342 | 425 | 2.14 (1.40–3.28) | <0.001a | |
P<0.05. HR, hazard radio; CI, confidence interval; N/A, not available; HER-2, human epidermal growth factor; ER, estrogen receptor; PR, progesterone receptor.
Association between E2F expression and RFS rates in patients with breast cancer
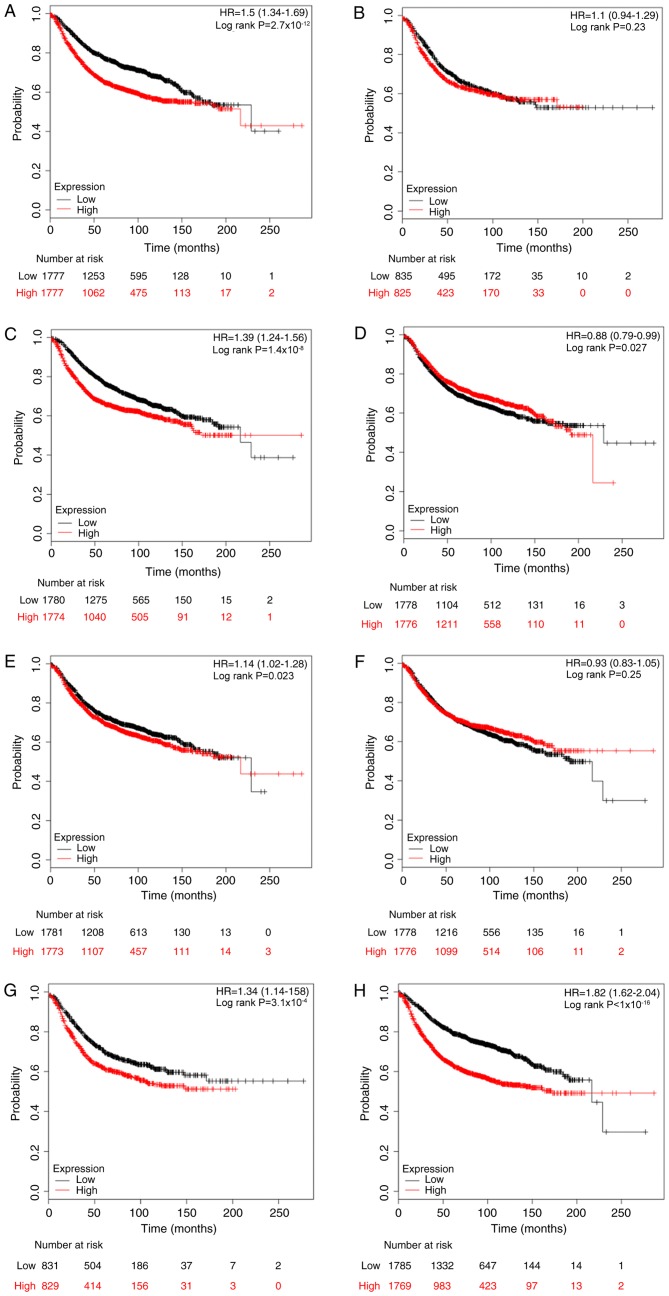
The prognostic values of E2Fs for RFS rates were then investigated using the KM plotter database, with the desired Affymetrix IDs of each gene symbol. Kaplan-Meier analyses indicated that high mRNA expression levels of E2F1, E2F3, E2F5, E2F7 and E2F8 were all significantly associated with reduced RFS rates (E2F1: HR, 1.50, 95% CI, 1.34–1.69, P<0.001; E2F3: HR, 1.39, 95% CI, 1.24–1.56, P<0.001; E2F5: HR, 1.14, 95% CI, 1.02–1.28, P=0.023; E2F7: HR, 1.34, 95% CI, 1.14–1.58, P<0.001; and E2F8: HR, 1.82, 95% CI, 1.62–2.04, P<0.001), while E2F4 was associated with improved RFS rates (HR, 0.88; 95% CI, 0.79–0.99; P=0.027). By contrast, E2F2 and E2F6 were not associated with RFS rates. The Kaplan-Meier curves are presented in Fig. 3.
Figure 3.
The prognostic effects of E2Fs on relapse-free survival. Kaplan-Meier survival curves are presented: (A) E2F1 (204947_at, n=3554); (B) E2F2 (228361_at t, n=1660); (C) E2F3 (203693_s_at t, n=3554); (D) E2F4 (202248_at t, n=3554); (E) E2F5 (221586_s_at t, n=3554); (F) (203957_at t, n=3554); (G) E2R7 (228033_at t, n=1660); and (H) E2F8 (219990_at t, n=3554). HR, hazard ratio.
When analyses were performed by stratifying patients into subgroups based on the clinicopathological features, it was determined that E2F1, E2F7 and E2F8 were significantly associated with reduced RFS rates in patients with ER-positive breast cancer (E2F1: HR, 1.49, 95% CI, 1.25–1.77, P<0.001; E2F7: HR, 1.50, 95% CI, 1.09–2.05, P=0.011; and E2F8: HR, 1.75, 95% CI, 1.47–2.09, P<0.001), but not in the ER-negative cohort (Table III). By contrast, high expression of E2F5 and E2F6 predicted improved RFS rates in ER-negative patients but not in ER-positive patients. With regards to PR status, E2F1, E2F7 and E2F8 indicated a reduced RFS rate in PR-positive patients, while E2F2 and E2F4 predicted a reduced RFS rate in the PR-negative group (Table III). In the HER-2-positive subgroup, only E2F2 was marginally associated with RFS rate (HR, 0.57; 95% CI, 0.33–0.99; P=0.045). However, high expression of E2F2 indicated an opposite association with RFS in the HER-2-negative subgroup (HR, 1.80; 95% CI, 1.33–2.44; P<0.001). In addition, E2F1, E2F3, E2F7 and E2F8 were also significantly associated with reduced RFS rates in HER-2-negative patients (Table III). E2F1, E2F3, E2F7 and E2F8 were associated with reduced RFS rates in lymph node-positive and HER-2-negative patients (Table III). E2F2 was determined to be associated with reduced RFS rates in the lymph node-positive subgroup (HR, 1.52; 95% CI, 1.16–2.00; P=0.003).
Table III.
The association between E2Fs and relapse-free survival for patients with breast cancer based on clinicopathological features.
| Positive status | Negative status | ||||||
|---|---|---|---|---|---|---|---|
| Clinicopathological factor | Gene symbol | Cases | HR (95% CI) | P-value | Cases | HR (95% CI) | P-value |
| ER | E2F1 | 1802 | 1.49 (1.25–1.77) | <0.001a | 671 | 1.13 (0.88–1.44) | 0.332 |
| E2F2 | 695 | 1.35 (0.99–1.85) | 0.056 | 313 | 0.99 (0.69–1.41) | 0.941 | |
| E2F3 | 1802 | 1.18 (0.99–1.40) | 0.060 | 671 | 0.91 (0.71–1.17) | 0.461 | |
| E2F4 | 1802 | 1.14 (0.96–1.36) | 0.131 | 671 | 1.05 (0.82–1.35) | 0.674 | |
| E2F5 | 1802 | 1.13 (0.95–1.34) | 0.174 | 671 | 0.75 (0.58–0.96) | 0.021a | |
| E2F6 | 1802 | 1.19 (1.00–1.41) | 0.052 | 671 | 0.75 (0.58–0.96) | 0.021a | |
| E2F7 | 695 | 1.50 (1.09–2.05) | 0.011a | 313 | 1.14 (0.79–1.62) | 0.487 | |
| E2F8 | 1802 | 1.75 (1.47–2.09) | <0.001a | 671 | 1.16 (0.91–1.49) | 0.227 | |
| PR | E2F1 | 525 | 1.84 (1.27–2.68) | 0.001a | 483 | 1.10 (0.81–1.49) | 0.550 |
| E2F2 | 489 | 1.34 (0.92–1.96) | 0.130 | 372 | 1.44 (1.00–2.05) | 0.046a | |
| E2F3 | 525 | 1.21 (0.85–1.74) | 0.292 | 483 | 1.05 (0.77–1.43) | 0.756 | |
| E2F4 | 525 | 1.24 (0.86–1.78) | 0.243 | 483 | 1.57 (1.15–2.14) | 0.004a | |
| E2F5 | 525 | 1.20 (0.83–1.71) | 0.329 | 483 | 1.09 (0.80–1.48) | 0.581 | |
| E2F6 | 525 | 1.05 (0.73–1.51) | 0.777 | 483 | 0.79 (0.58–1.08) | 0.142 | |
| E2F7 | 489 | 1.66 (1.13–2.44) | 0.010a | 372 | 1.02 (0.71–1.45) | 0.930 | |
| E2F8 | 525 | 2.04 (1.40–2.96) | <0.001a | 483 | 1.12 (0.82–1.52) | 0.482 | |
| HER-2 | E2F1 | 168 | 1.09 (0.65–1.84) | 0.737 | 756 | 1.61 (1.23–2.10) | <0.001a |
| E2F2 | 150 | 0.57 (0.33–0.99) | 0.045a | 635 | 1.80 (1.33–2.44) | <0.001a | |
| E2F3 | 168 | 1.03 (0.61–1.72) | 0.925 | 756 | 1.50 (1.15–1.96) | 0.003a | |
| E2F4 | 168 | 1.09 (0.65–1.84) | 0.736 | 756 | 1.25 (0.96–1.63) | 0.099 | |
| E2F5 | 168 | 0.67 (0.40–1.14) | 0.137 | 756 | 1.17 (0.90–1.52) | 0.253 | |
| E2F6 | 168 | 0.82 (0.48–1.38) | 0.453 | 756 | 1.09 (0.84–1.42) | 0.505 | |
| E2F7 | 150 | 0.79 (0.46–1.36) | 0.396 | 635 | 2.02 (1.48–2.74) | <0.001a | |
| E2F8 | 168 | 0.96 (0.57–1.62) | 0.883 | 756 | 1.84 (1.41–2.42) | <0.001a | |
| Lymph node | E2F1 | 945 | 1.44 (1.16–1.80) | 0.001a | 1813 | 1.60 (1.34–1.91) | <0.001a |
| E2F2 | 665 | 1.52 (1.16–2.00) | 0.003a | 451 | 1.40 (0.93–2.10) | 0.107 | |
| E2F3 | 945 | 1.33 (1.07–1.66) | 0.011a | 1813 | 1.47 (1.23–1.75) | <0.001a | |
| E2F4 | 945 | 1.23 (0.99–1.54) | 0.061 | 1813 | 1.11 (0.93–1.32) | 0.253 | |
| E2F5 | 945 | 1.11 (0.89–1.38) | 0.349 | 1813 | 1.09 (0.91–1.29) | 0.343 | |
| E2F6 | 945 | 1.06 (0.85–1.32) | 0.600 | 1813 | 0.95 (0.80–1.13) | 0.558 | |
| E2F7 | 665 | 1.33 (1.01–1.74) | 0.041a | 451 | 1.89 (1.25–2.86) | 0.002a | |
| E2F8 | 945 | 1.53 (1.22–1.90) | <0.001a | 1813 | 1.73 (1.45–2.07) | <0.001a | |
P<0.05. HR, hazard radio; CI, confidence interval; N/A, not available; HER-2, human epidermal growth factor 2; ER, estrogen receptor; PR, progesterone receptor.
Association between E2F expression and DMFS rates in patients with breast cancer
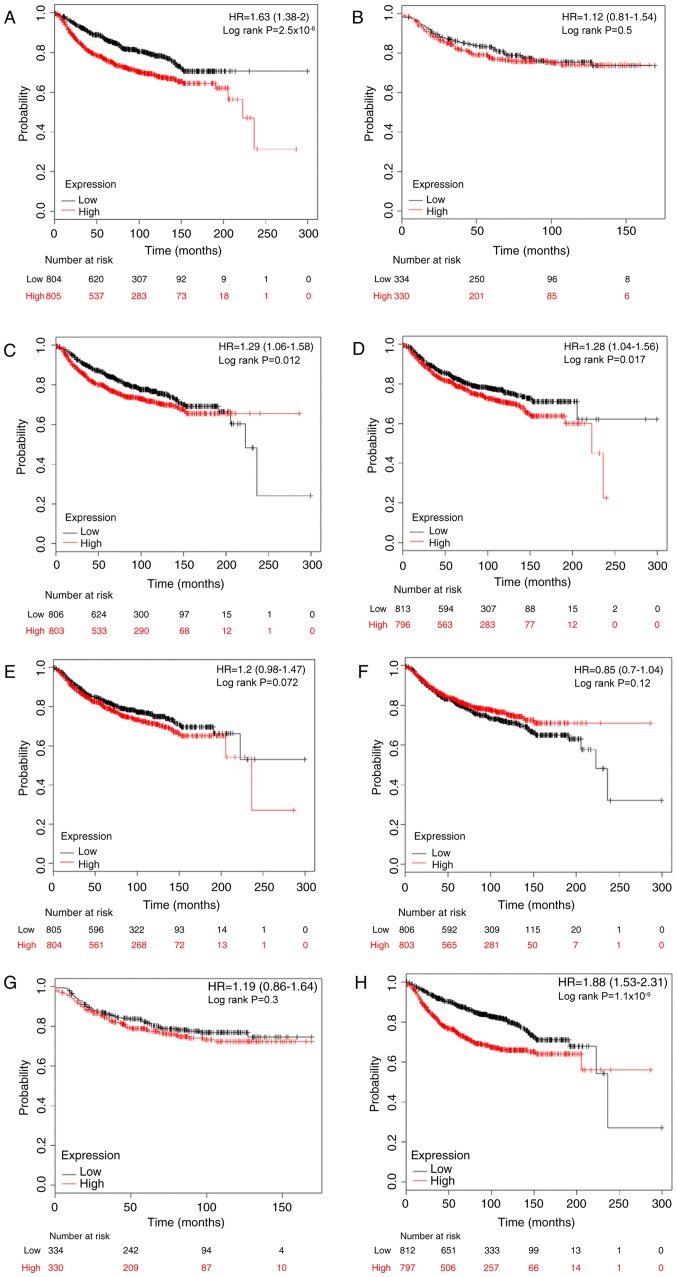
Metastasis is the most common cause of mortality in breast cancer, and 20–30% individuals initially diagnosed with early breast cancer would exhibit distant metastasis (30). Following this, the prognostic significance of E2Fs to DMFS was investigated. High expression levels of E2F1, E2F3, E2F4 and E2F8 were significantly associated with worse DMFS in patients with breast cancer, with HR=1.63; 95% CI, 1.33–2.00 and P<0.001 (Fig. 4A); HR=1.29; 95% CI, 1.06–1.58 and P=0.012 (Fig. 4C); HR=1.28; 95% CI, 1.04–1.56 and P=0.017 (Fig. 4D); and HR=1.88; 95% CI, 1.53–2.31 and P<0.001 (Fig. 4H), respectively. However, there was no difference in DMFS between high and low expression groups for the other four E2Fs (Fig. 4B, E-G).
Figure 4.
The prognostic effects of E2Fs on distant metastasis-free survival. Kaplan-Meier survival curves are presented: (A) E2F1 (204947_at t, n=1609); (B) E2F2 (228361_at, n=664); (C) E2F3 (203693_s_at, n=1609); (D) E2F4 (202248_at, n=1609); (E) E2F5 (221586_s_at, n=1609); (F) (203957_at, n=1609); (G) E2R7 (228033_at, n=664); and (H) E2F8 (219990_at, n=1609). HR, hazard ratio.
The prognostic values of E2Fs were investigated by subgroup analysis. High mRNA expression of E2F1 was associated with reduced DMFS rates in ER-positive patients (HR, 1.89; 95% CI, 1.29–2.75; P<0.001) and lymph node-negative patients (HR, 1.76; 95% CI, 1.32–2.35; P<0.001). E2F2 and E2F4 were not associated with any subgroups. Upregulated E2F3 predicted reduced DMFS rates in lymph node-negative breast cancer (HR, 1.49; 95% CI, 1.12–1.99; P=0.006). In the ER-negative subgroup, a high level of E2F5 was significantly associated with an improved DMFS rate (HR, 0.59; 95% CI, 0.35–0.99; P=0.044). Elevated E2F6 was significantly associated with improved DMFS rates in ER-negative (HR, 0.51; 95% CI, 0.29–0.81; P=0.012), PR-negative (HR, 0.36; 95% CI, 0.16–0.82; P=0.012), HER-2-positive (HR, 0.35; 95% CI, 0.12–0.98; P=0.037), lymph node-positive (HR, 0.65; 95% CI, 0.43–1.00; P=0.046) and lymph node-negative patients (HR, 0.68; 95% CI, 0.51–0.91; P=0.009). However, in contrast to the results in the overall cohort, high expression of E2F7 demonstrated an improved DMFS rate for HER-2-positive patients (HR, 0.25; 95% CI, 0.08–0.75; P=0.007). Finally, increased E2F8 was significantly associated with reduced DMFS rates in ER-positive (HR, 2.74; 95% CI, 1.68–4.04; P<0.001) and lymph node-negative (HR, 2.01; 95% CI, 1.50–2.69; P<0.001) patients. All KM analysis results are summarized in Table IV.
Table IV.
The association between E2Fs and distant metastasis-free survival for patients with breast cancer based on clinicopathological features.
| Positive status | Negative status | ||||||
|---|---|---|---|---|---|---|---|
| Clinicopathological factor | Gene symbol | Cases | HR (95% CI) | P-value | Cases | HR (95% CI) | P-value |
| ER | E2F1 | 577 | 1.89 (1.29–2.75) | <0.001a | 170 | 1.01 (0.61–1.69) | 0.958 |
| E2F2 | 161 | 1.79 (0.69–4.64) | 0.221 | 68 | 0.79 (0.34–1.83) | 0.583 | |
| E2F3 | 577 | 1.05 (0.73–1.50) | 0.812 | 170 | 0.96 (0.57–1.60) | 0.867 | |
| E2F4 | 577 | 1.38 (0.96–1.97) | 0.082 | 170 | 0.82 (0.49–1.37) | 0.451 | |
| E2F5 | 577 | 1.42 (0.99–2.04) | 0.057 | 170 | 0.59 (0.35–0.99) | 0.044a | |
| E2F6 | 577 | 0.93 (0.65–1.34) | 0.707 | 170 | 0.51 (0.29–0.87) | 0.012a | |
| E2F7 | 161 | 1.57 (0.61–4.05) | 0.348 | 68 | 0.69 (0.30–1.62 | 0.394 | |
| E2F8 | 577 | 2.74 (1.86–4.04) | <0.001a | 170 | 0.84 (0.50–1.41) | 0.512 | |
| PR | E2F1 | 122 | 1.49 (0.43–5.12) | 0.522 | 95 | 1.35 (0.64–2.83) | 0.433 |
| E2F2 | 122 | 0.92 (0.28–3.01) | 0.887 | 95 | 1.59 (0.75–3.38) | 0.224 | |
| E2F3 | 122 | 2.32 (0.61–8.85) | 0.203 | 95 | 0.97 (0.46–2.04) | 0.932 | |
| E2F4 | 122 | 0.42 (0.11–1.61) | 0.193 | 95 | 1.90 (0.88–4.08) | 0.095 | |
| E2F5 | 122 | 3.27 (0.86–12.42) | 0.065 | 95 | 0.91 (0.43–1.93) | 0.808 | |
| E2F6 | 122 | 0.60 (0.18–2.03) | 0.409 | 95 | 0.36 (0.16–0.82) | 0.012a | |
| E2F7 | 122 | 0.79 (0.24–2.60) | 0.701 | 95 | 1.16 (0.55–2.44) | 0.697 | |
| E2F8 | 122 | 1.87 (0.55–6.39) | 0.311 | 95 | 1.56 (0.74–3.30) | 0.242 | |
| HER-2 | E2F1 | 66 | 1.12 (0.44–2.82) | 0.810 | 82 | 1.63 (0.46–5.76) | 0.447 |
| E2F2 | 66 | 1.00 (0.40–2.53) | 0.996 | 82 | 2.38 (0.61–9.20) | 0.195 | |
| E2F3 | 66 | 1.09 (0.43–2.76) | 0.853 | 82 | 2.39 (0.62–9.24) | 0.193 | |
| E2F4 | 66 | 1.33 (0.53–3.36) | 0.543 | 82 | 0.64 (0.18–2.28) | 0.492 | |
| E2F5 | 66 | 0.44 (0.17–1.18) | 0.092 | 82 | 1.58 (0.44–5.59) | 0.477 | |
| E2F6 | 66 | 0.35 (0.12–0.98) | 0.037a | 82 | 1.55 (0.44–5.50) | 0.492 | |
| E2F7 | 66 | 0.25 (0.08–0.75) | 0.007a | 82 | 2.43 (0.63–9.39) | 0.184 | |
| E2F8 | 66 | 1.46 (0.57–3.71) | 0.425 | 82 | 4.48 (0.95–21.1) | 0.038 | |
| Lymph node | E2F1 | 337 | 1.32 (0.87–2.01) | 0.185 | 896 | 1.76 (1.32–2.35) | <0.001a |
| E2F2 | 172 | 1.52 (0.82–2.84) | 0.181 | 162 | 1.88 (0.78–4.55) | 0.154 | |
| E2F3 | 337 | 1.17 (0.77–1.77) | 0.462 | 896 | 1.49 (1.12–1.99) | 0.006a | |
| E2F4 | 337 | 1.50 (0.99–2.29) | 0.055 | 896 | 1.16 (0.87–1.54) | 0.305 | |
| E2F5 | 337 | 1.00 (0.66–1.52) | 0.985 | 896 | 1.27 (0.96–1.68) | 0.099 | |
| E2F6 | 337 | 0.65 (0.43–1.00) | 0.046a | 896 | 0.68 (0.51–0.91) | 0.009a | |
| E2F7 | 172 | 0.85 (0.46–1.58) | 0.613 | 162 | 1.90 (0.78–4.61) | 0.149 | |
| E2F8 | 337 | 1.31 (0.86–1.98) | 0.207 | 896 | 2.01 (1.50–2.69) | <0.001a | |
P<0.05. HR, hazard radio; CI, confidence interval; N/A, not available; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor 2.
Association between E2F expression and PPS rates in patients with breast cancer
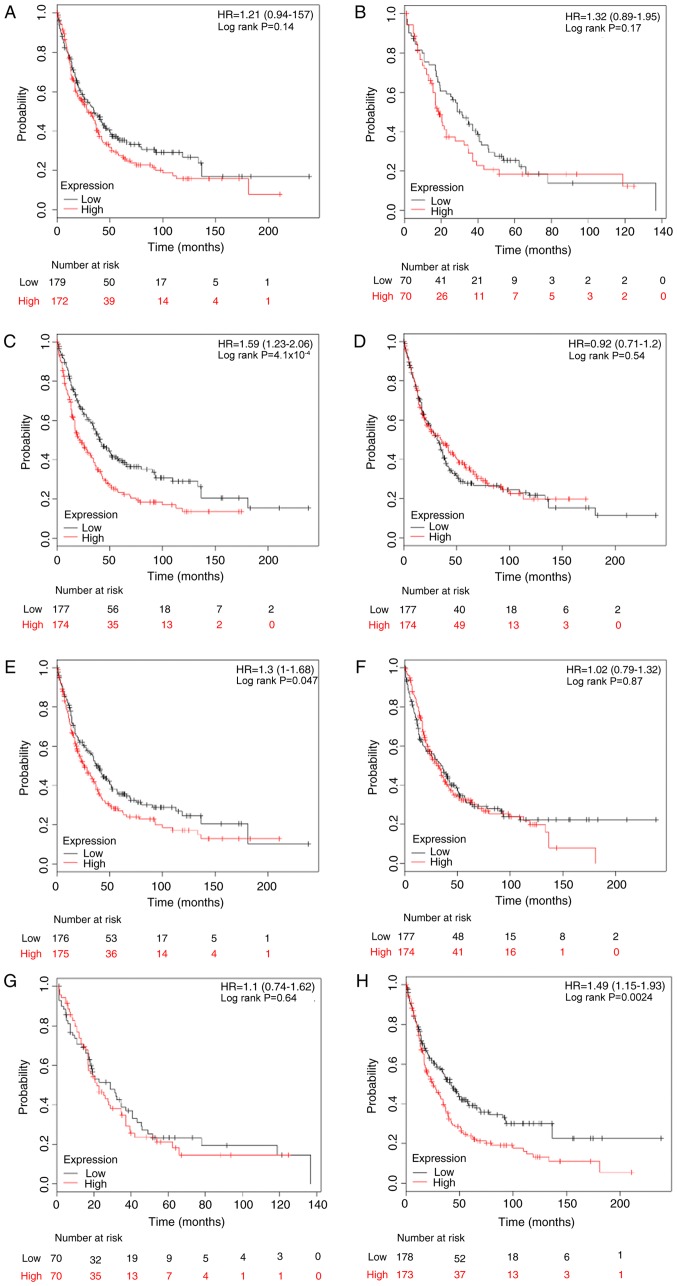
The association between E2F and predictive significance of PPS rates was also determined using the KM plotter database. The results demonstrated that only high expression levels of E2F3, E2F5 and E2F8 were associated with reduced PPS rates in patients with breast cancer, with HR=1.59 (1.23–2.06) and P<0.001; HR=1.30 (1.00–1.68) and P=0.047; and HR=1.49 (1.15–1.93) and P=0.002, respectively (Fig. 5).
Figure 5.
The prognostic effects of E2Fs on post-progression survival. Kaplan-Meier survival curves are presented: (A) E2F1 (204947_at, n=351); (B) E2F2 (228361_at, n=140); (C) E2F3 (203693_s_at, n=351); (D) E2F4 (202248_at, n=351); (E) E2F5 (221586_s_at, n=351); (F) (203957_at, n=351); (G) E2R7 (228033_at, n=140); and (H) E2F8 (219990_at, n=351). HR, hazard ratio.
By stratifying patients into different subgroups by clinicopathological features, it was determined that high expression of E2F3 (HR, 1.73; 95% CI, 1.11–2.71; P=0.015) and E2F8 (HR, 2.22; 95% CI, 1.41–3.49; P<0.001) indicated reduced PPS rates in ER-positive breast cancer (Table V). Furthermore, KM analyses indicated a significant association between PPS rate and patients with lymph node-negative breast cancer with elevated E2F1 (HR, 1.58; 95% CI, 1.01–2.47; P=0.042), E2F4 (HR, 0.60; 95% CI, 0.38–0.93; P=0.022) and E2F8 (HR, 1.75; 95% CI, 1.12–2.74; P=0.015). However, subgroup analysis of the prognostic values for E2Fs in the ER-positive and PR-positive cohort was not conducted for the limited number of patients. No positive result was observed in patients with PR-negative breast cancer (Table V).
Table V.
The association between E2Fs and post-progression survival for patients with breast cancer based on clinicopathological features.
| Positive status | Negative status | ||||||
|---|---|---|---|---|---|---|---|
| Clinicopathological factor | Gene symbol | Cases | HR (95% CI) | P-value | Cases | HR (95% CI) | P-value |
| ER | E2F1 | 144 | 1.18 (0.76–1.84) | 0.452 | 68 | 0.94 (0.52–1.70) | 0.850 |
| E2F2 | N/A | 26 | 0.47 (0.16–1.36) | 0.153 | |||
| E2F3 | 144 | 1.73 (1.11–2.71) | 0.015a | 68 | 1.04 (0.57–1.87) | 0.904 | |
| E2F4 | 144 | 1.05 (0.67–1.64) | 0.836 | 68 | 0.90 (0.50–1.62) | 0.719 | |
| E2F5 | 144 | 1.01 (0.65–1.57) | 0.966 | 68 | 0.86 (0.48–1.55) | 0.614 | |
| E2F6 | 144 | 1.12 (0.72–1.74) | 0.617 | 68 | 0.62 (0.34–1.12 | 0.111 | |
| E2F7 | N/A | 26 | 0.79 (0.30–2.12) | 0.646 | |||
| E2F8 | 144 | 2.22 (1.41–3.49) | <0.001a | 68 | 0.94 (0.52–1.69) | 0.827 | |
| PR | N/A | ||||||
| HER2 | E2F1 | N/A | 27 | 0.88 (0.31–2.52) | 0.808 | ||
| E2F2 | N/A | 27 | 1.06 (0.36–3.06) | 0.920 | |||
| E2F3 | N/A | 27 | 1.61 (0.56–4.62) | 0.372 | |||
| E2F4 | N/A | 27 | 0.75 (0.25–2.20) | 0.594 | |||
| E2F5 | N/A | 27 | 0.89 (0.29–2.71) | 0.837 | |||
| E2F6 | N/A | 27 | 0.40 (0.13–1.20) | 0.090 | |||
| E2F7 | N/A | 27 | 0.92 (0.32–2.64) | 0.887 | |||
| E2F8 | N/A | 27 | 1.35 (0.47–3.89) | 0.576 | |||
| Lymph node | E2F1 | 82 | 0.76 (0.44–1.32) | 0.335 | 148 | 1.58 (1.01–2.47) | 0.042 |
| E2F2 | 44 | 1.61 (0.70–3.73) | 0.257 | N/A | |||
| E2F3 | 82 | 1.60 (0.92–2.77) | 0.091 | 148 | 1.24 (0.80–1.93) | 0.329 | |
| E2F4 | 82 | 1.48 (0.86–2.57) | 0.157 | 148 | 0.60 (0.38–0.93) | 0.022a | |
| E2F5 | 82 | 0.92 (0.53–1.60) | 0.778 | 148 | 1.14 (0.74–1.76) | 0.560 | |
| E2F6 | 82 | 0.93 (0.53–1.61) | 0.795 | 148 | 0.81 (0.52–1.25) | 0.342 | |
| E2F7 | 44 | 1.21 (0.53–2.76) | 0.653 | N/A | |||
| E2F8 | 82 | 0.78 (0.45–1.36) | 0.385 | 148 | 1.75 (1.12–2.74) | 0.013a | |
P<0.05. HR, hazard radio; CI, confidence interval; N/A, not available; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor 2.
Discussion
E2Fs have been implicated in numerous human cancer types (7). Deregulated expression of E2Fs was demonstrated to be a common phenomenon in malignances (31). Depending on the context, E2Fs were regarded as oncogenes or tumor suppressors and exerted exactly opposite functions during tumorigenesis (11); therefore, identifying the underling mechanisms of the E2F-mediated cell cycle, differentiation, apoptosis and numerous other pivotal physiological progressions, and unraveling how they are involved in different types of human cancer may provide novel therapeutic strategies. In addition, a number of studies have confirmed the significant associations between E2Fs, and clinicopathological features and survival outcomes of patients with cancer, which indicated that E2Fs may serve as predictive biomarkers for specific carcinomas (13,18,21). However, inconsistent expression patterns and prognostic significance, even in the same type of carcinoma, have been frequently observed in previous studies (18–20). In the present study, the transcription levels and prognostic significance of all eight E2F genes in breast cancer were systematically investigated using the Oncomine, TCGA and KM plotter databases.
E2F1-3 were classified as activator E2Fs due to their ability to induce the transcription of target genes during the transition from G1 to S phase in cell cycle progression (32). In structure, a nuclear localization signal adjacent to the cyclin-binding domains of E2Fs ensures entrance into the nucleus and modulates their transcriptional activity (33). Two previous studies have indicated that the mRNA expression level of E2F1 was much lower in breast cancer tissues than in normal tissues (34,35); however, it may be contradictory that a high transcription level of E2F1 was positively associated with tumor cell proliferation and indicated a poorer prognosis for patients with breast cancer (21,36). E2F2 was indicated to exhibit oncogenic or tumor suppressive activity depending on the context (37). For example, E2F2 contributed to cell proliferation in primary mouse embryo fibroblasts and downregulation of E2F2 inhibited cell proliferation in breast cancer (38,39). However, Pusapati et al (40) demonstrated that inactivation of E2F2 significantly promoted tumor formation in K5.Myc transgenic mice, which indicated a tumor-suppressor role of E2F2. It has been reported that either E2F3a or E2F3b is sufficient to regulate E2F target gene transcription and cell proliferation in the absence of other E2F activators, E2F1 and E2F2 (41). A recent study demonstrated that E2F3 was upregulated in the majority of breast cancer cell lines and that E2F3 depletion significantly suppressed cell proliferation (42). In the present study, it was determined that E2F1-3 were all upregulated in breast cancer. Notably, high mRNA expression of E2F1 and E2F3 were significantly associated with reduced OS, RFS and DMFS rates. Furthermore, it was determined that E2F1-3 may be associated with survival outcomes in an ER, PR, HER-2 and lymph node status-specific manner. For instance, upregulated E2F1 indicated reduced OS rates in ER-positive but not in patients with ER-negative breast cancer. Although no significant association was observed between E2F2 and clinical outcomes in all breast cancer patients, subgroup analysis determined that E2F2 was associated with reduced RFS rates in patients with PR-negative, HER-2-negative or lymph node-positive breast cancer.
As repressor members of the E2F family, E2F4 and E2F5 were reported to contribute toward cell transformation, proliferation and cell cycle progression in the presence of a dimerization partner and inhibitory pocket proteins (Rbs) (43,44). In a previous study, E2F4 was able to cooperate with any Rbs, while E2F5 was predominantly associated with p130 (45). In breast cancer, the expression level of E2F4 was determined to be lower in primary and metastatic tissues, compared with corresponding normal samples, which indicated a tumor suppressor function for E2F4 (35); however, a more recent study demonstrated that overexpression of E2F4 in the nuclei of breast cancer cells was associated with multiple advanced clinicopathological characteristics and poorer clinical outcomes for patients with breast cancer (22). In the present study, it was determined that there was no mRNA expression difference between tumor and normal tissues; however, a relatively high level of E2F4 was significantly associated with an improved RFS rate, but not with a reduced DMFS rate. Similar to that of E2F4, the present understanding of E2F5 was also limited in breast cancer. A group of microRNAs (miRNAs/miRs), including miR-34a, miR-106, miR-132 and miR-181a, was proven to target E2F5 in a number of cancer types (46). Umemura et al (47) demonstrated that E2F5-positive breast cancer was characterized by a higher Ki-67 index and an aggressive histological pathology. Furthermore, the DFS rate was reduced in lymph node-negative patients with E2F5-positive breast cancer, compared with patients with E2F5-negative breast cancer (47). Consistently, it was determined that E2F5 was upregulated in breast tumors, compared with normal tissues, and a high mRNA expression level of E2F5 predicted reduced RFS and PPS rates. Notably, a high level of E2F5 was significantly associated with improved OS, RFS and DMFS rates in ER-negative patients and with an improved OS rate in HER-2-positive and lymph node-positive patients by subgroup analysis. Accordingly, with these preliminary results, the actual roles of E2F4 and E2F5 require further clarification in breast cancer.
E2F6-8 have similar functions with the repressor group but it is distinct in molecular mechanisms (48,49). Although exhibiting a high level of homology with E2F1-5 in the heterodimerization and DNA binding domains, E2F6-8 lacks a transactivation domain and an Rb-binding domain, thereby acting as pocket protein-independent transcriptional repressor (50). In addition, E2F6 was demonstrated to act as a repressor through interaction with the polycomb complex, whereas E2F7 and E2F8 were able to form homodimers or heterodimers to suppress the transcription of target genes (48,49). Recently, Tang et al (51) reported that the regulation of BRCA1 by miR-185 was mediated by E2F6, which indicated a critical role of E2F6 in breast cancer, though no expression difference of E2F6 was detected between tumors and normal tissue in the present study. In a previous study, E2F7 was overexpressed in tamoxifen-resistant breast cancer cells and silencing E2F7 re-sensitized resistant cells to tamoxifen (52). Furthermore, high expression of E2F7 was significantly associated with reduced RFS rate in patients with ERα-positive breast cancer treated with tamoxifen (52). In the present study, it was determined that high expression of E2F7 was associated with reduced a RFS not only in ER-positive but also in patients with PR-positive and HER-2-negative breast cancer; however, E2F7 was associated with an improved DMFS rate in patients with HER-2-positive breast cancer. Notably, a high expression of E2F8 was significantly associated with reduced OS, RFS, DMFS and PPS rates. This was similar to a recent study reported by Ye et al (53), which indicated that upregulated E2F8 was correlated with a poorer prognosis in breast cancer. Specifically, it was also demonstrated that E2F8 indicated a poorer prognosis in patients with ER-positive, PR-positive and HER-2-negative breast cancer.
In summary, it was concluded that mRNA expression levels of E2F1, E2F2, E2F3, E2F5, E2F7 and E2F8 are notably increased in breast carcinoma, while the expression of E2F4 and E2F6 is not altered in tumors, compared with normal tissues. Furthermore, significant associations between E2Fs and clinical outcomes of patients with breast cancer were also identified. These results indicated that E2Fs may serve as promising biomarkers for breast cancer; however, further studies concerning molecular mechanisms, focusing on individual E2Fs or combining several E2Fs, are required to facilitate the clinical application of E2Fs serving as prognostic indicators or therapeutic targets in breast cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural Science Foundation of China (grant nos. 81472475 and 81102007), Chongqing Science and Technology Commission (grant no. cstc2016jcyjA0313) and Scientific Research Foundation of Chongqing Medical University (grant no. 201408).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Oncomine (http://www.oncomine.org), TCGA (https://cancergenome.nih.gov/), and KM-plotter (http://kmplot.com/analysis/) databases repository.
Authors' contributions
GR and HL conceived and designed the study. YL, JH, SX, DY, and JS performed data analyses. YL, JH, and HL contributed reagents/materials/analysis tools. YL and HL wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chlebowski RT, Manson JE, Anderson GL, Cauley JA, Aragaki AK, Stefanick ML, Lane DS, Johnson KC, Wactawski-Wende J, Chen C, et al. Estrogen plus progestin and breast cancer incidence and mortality in the Women's health initiative observational study. J Natl Cancer Inst. 2013;105:526–535. doi: 10.1093/jnci/djt043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Althuis MD, Dozier JM, Anderson WF, Devesa SS, Brinton LA. Global trends in breast cancer incidence and mortality 1973–1997. Int J Epidemiol. 2005;34:405–412. doi: 10.1093/ije/dyh414. [DOI] [PubMed] [Google Scholar]
- 4.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 5.Redig AJ, McAllister SS. Breast cancer as a systemic disease: A view of metastasis. J Intern Med. 2013;274:113–126. doi: 10.1111/joim.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Assi HA, Khoury KE, Dbouk H, Khalil LE, Mouhieddine TH, El Saghir NS. Epidemiology and prognosis of breast cancer in young women. J Thorac Dis. 2013;5(Suppl 1):S2–S8. doi: 10.3978/j.issn.2072-1439.2013.05.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsantoulis PK, Gorgoulis VG. Involvement of E2F transcription factor family in cancer. Eur J Cancer. 2005;41:2403–2414. doi: 10.1016/j.ejca.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Attwooll C, Lazzerini Denchi E, Helin K. The E2F family: Specific functions and overlapping interests. EMBO J. 2004;23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell LA, Ryan KM. Life and death decisions by E2F-1. Cell Death Differ. 2004;11:137–142. doi: 10.1038/sj.cdd.4401324. [DOI] [PubMed] [Google Scholar]
- 10.Stevens C, La Thangue NB. E2F and cell cycle control: A double-edged sword. Arch Biochem Biophys. 2003;412:157–169. doi: 10.1016/S0003-9861(03)00054-7. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z, Zheng S, Yu Q. The E2F family and the role of E2F1 in apoptosis. Int J Biochem Cell Biol. 2009;41:2389–2397. doi: 10.1016/j.biocel.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Jiang H, Martin V, Gomez-Manzano C, Johnson DG, Alonso M, White E, Xu J, McDonnell TJ, Shinojima N, Fueyo J. The RB-E2F1 pathway regulates autophagy. Cancer Res. 2010;70:7882–7893. doi: 10.1158/0008-5472.CAN-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorgoulis VG, Zacharatos P, Mariatos G, Kotsinas A, Bouda M, Kletsas D, Asimacopoulos PJ, Agnantis N, Kittas C, Papavassiliou AG. Transcription factor E2F-1 acts as a growth-promoting factor and is associated with adverse prognosis in non-small cell lung carcinomas. J Pathol. 2002;198:142–156. doi: 10.1002/path.1121. [DOI] [PubMed] [Google Scholar]
- 14.Huang CL, Liu D, Nakano J, Yokomise H, Ueno M, Kadota K, Wada H. E2F1 overexpression correlates with thymidylate synthase and survivin gene expressions and tumor proliferation in non small-cell lung cancer. Clin Cancer Res. 2007;13:6938–6946. doi: 10.1158/1078-0432.CCR-07-1539. [DOI] [PubMed] [Google Scholar]
- 15.Liu LX, Jiang HC, Liu ZH, Zhu AL, Zhang WH, Wu LF, Zhou J, Wang XQ, Wu M. Expression of cell cycle/growth regulator genes in human hepatocellular carcinoma and adjacent normal liver tissues. Oncol Rep. 2003;10:1771–1775. [PubMed] [Google Scholar]
- 16.Deng Q, Wang Q, Zong WY, Zheng DL, Wen YX, Wang KS, Teng XM, Zhang X, Huang J, Han ZG. E2F8 contributes to human hepatocellular carcinoma via regulating cell proliferation. Cancer Res. 2010;70:782–791. doi: 10.1158/0008-5472.CAN-09-3082. [DOI] [PubMed] [Google Scholar]
- 17.Palaiologou M, Koskinas J, Karanikolas M, Fatourou E, Tiniakos DG. E2F-1 is overexpressed and pro-apoptotic in human hepatocellular carcinoma. Virchows Arch. 2012;460:439–446. doi: 10.1007/s00428-012-1220-4. [DOI] [PubMed] [Google Scholar]
- 18.De Meyer T, Bijsmans IT, Van de Vijver KK, Bekaert S, Oosting J, Van Criekinge W, van Engeland M, Sieben NL. E2Fs mediate a fundamental cell-cycle deregulation in high-grade serous ovarian carcinomas. J Pathol. 2009;217:14–20. doi: 10.1002/path.2452. [DOI] [PubMed] [Google Scholar]
- 19.Reimer D, Sadr S, Wiedemair A, Goebel G, Concin N, Hofstetter G, Marth C, Zeimet AG. Expression of the E2F family of transcription factors and its clinical relevance in ovarian cancer. Ann N Y Acad Sci. 2006;1091:270–281. doi: 10.1196/annals.1378.073. [DOI] [PubMed] [Google Scholar]
- 20.Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- 21.Han S, Park K, Bae BN, Kim KH, Kim HJ, Kim YD, Kim HY. E2F1 expression is related with the poor survival of lymph node-positive breast cancer patients treated with fluorouracil, doxorubicin and cyclophosphamide. Breast Cancer Res Treat. 2003;82:11–16. doi: 10.1023/B:BREA.0000003843.53726.63. [DOI] [PubMed] [Google Scholar]
- 22.Rakha EA, Pinder SE, Paish EC, Robertson JF, Ellis IO. Expression of E2F-4 in invasive breast carcinomas is associated with poor prognosis. J Pathol. 2004;203:754–761. doi: 10.1002/path.1573. [DOI] [PubMed] [Google Scholar]
- 23.Fujiwara K, Yuwanita I, Hollern DP, Andrechek ER. Prediction and genetic demonstration of a role for activator E2Fs in Myc-induced tumors. Cancer Res. 2011;71:1924–1932. doi: 10.1158/0008-5472.CAN-10-2386. [DOI] [PubMed] [Google Scholar]
- 24.Szász AM, Lánczky A, Nagy A, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gluck S, Ross JS, Royce M, McKenna EF, Jr, Perou CM, Avisar E, Wu L. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer Res Treat. 2012;132:781–791. doi: 10.1007/s10549-011-1412-7. [DOI] [PubMed] [Google Scholar]
- 26.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H, Langerød A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Kåresen R, Botstein D, Børresen-Dale AL, Jeffrey SS. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.e03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G, Gish K, Kwok K, Hanna W, Zubovits J, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2005;102:11005–11010. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T, Yasui W, Yokozaki H, Naka K, Ishikawa T, Tahara E. Expression of the E2F family in human gastrointestinal carcinomas. Int J Cancer. 1999;81:535–538. doi: 10.1002/(SICI)1097-0215(19990517)81:4<535::AID-IJC5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 33.Muller H, Moroni MC, Vigo E, Petersen BO, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/MCB.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worku D, Jouhra F, Jiang GW, Patani N, Newbold RF, Mokbel K. Evidence of a tumour suppressive function of E2F1 gene in human breast cancer. Anticancer Res. 2008;28:2135–2139. [PubMed] [Google Scholar]
- 35.Ho GH, Calvano JE, Bisogna M, Van Zee KJ. Expression of E2F-1 and E2F-4 is reduced in primary and metastatic breast carcinomas. Breast Cancer Res Treat. 2001;69:115–122. doi: 10.1023/A:1012230115173. [DOI] [PubMed] [Google Scholar]
- 36.Vuaroqueaux V, Urban P, Labuhn M, Delorenzi M, Wirapati P, Benz CC, Flury R, Dieterich H, Spyratos F, Eppenberger U, Eppenberger-Castori S. Low E2F1 transcript levels are a strong determinant of favorable breast cancer outcome. Breast Cancer Res. 2007;9:R33. doi: 10.1186/bcr1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson DG, Degregori J. Putting the oncogenic and tumor suppressive activities of E2F into context. Curr Mol Med. 2006;6:731–738. doi: 10.2174/1566524010606070731. [DOI] [PubMed] [Google Scholar]
- 38.Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park CH, Giangrande P, Wu L, Saavedra HI, Field SJ, et al. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol Cell. 2001;8:105–113. doi: 10.1016/S1097-2765(01)00275-1. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen-Vu T, Vedin LL, Liu K, Jonsson P, Lin JZ, Candelaria NR, Candelaria LP, Addanki S, Williams C, Gustafsson JÅ, et al. Liver × receptor ligands disrupt breast cancer cell proliferation through an E2F-mediated mechanism. Breast Cancer Res. 2013;15:R51. doi: 10.1186/bcr3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pusapati RV, Weaks RL, Rounbehler RJ, McArthur MJ, Johnson DG. E2F2 suppresses Myc-induced proliferation and tumorigenesis. Mol Carcinog. 2010;49:152–156. doi: 10.1002/mc.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chong JL, Tsai SY, Sharma N, Opavsky R, Price R, Wu L, Fernandez SA, Leone G. E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Mol Cell Biol. 2009;29:414–424. doi: 10.1128/MCB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vimala K, Sundarraj S, Sujitha MV, Kannan S. Curtailing overexpression of E2F3 in breast cancer using siRNA (E2F3)-based gene silencing. Arch Med Res. 2012;43:415–422. doi: 10.1016/j.arcmed.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Dirks PB, Rutka JT, Hubbard SL, Mondal S, Hamel PA. The E2F-family proteins induce distinct cell cycle regulatory factors in p16-arrested, U343 astrocytoma cells. Oncogene. 1998;17:867–876. doi: 10.1038/sj.onc.1202008. [DOI] [PubMed] [Google Scholar]
- 44.Polanowska J, Le Cam L, Orsetti B, Vallés H, Fabbrizio E, Fajas L, Taviaux S, Theillet C, Sardet C. Human E2F5 gene is oncogenic in primary rodent cells and is amplified in human breast tumors. Genes Chromosomes Cancer. 2000;28:126–130. doi: 10.1002/(SICI)1098-2264(200005)28:1<126::AID-GCC15>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 45.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 46.Tian H, Hou L, Xiong YM, Huang JX, Zhang WH, Pan YY, Song XR. miR-132 targeting E2F5 suppresses cell proliferation, invasion, migration in ovarian cancer cells. Am J Transl Res. 2016;8:1492–1501. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Umemura S, Shirane M, Takekoshi S, Kusakabe T, Itoh J, Egashira N, Tokuda Y, Mori K, Osamura YR. Overexpression of E2F-5 correlates with a pathological basal phenotype and a worse clinical outcome. Br J Cancer. 2009;100:764–771. doi: 10.1038/sj.bjc.6604900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trimarchi JM, Fairchild B, Wen J, Lees JA. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc Natl Acad Sci USA. 2001;98:1519–1524. doi: 10.1073/pnas.98.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lammens T, Li J, Leone G, De Veylder L. Atypical E2Fs: New players in the E2F transcription factor family. Trends Cell Biol. 2009;19:111–118. doi: 10.1016/j.tcb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moon NS, Dyson N. E2F7 and E2F8 keep the E2F family in balance. Dev Cell. 2008;14:1–3. doi: 10.1016/j.devcel.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 51.Tang H, Liu P, Yang L, Xie X, Ye F, Wu M, Liu X, Chen B, Zhang L, Xie X. miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol Cancer Ther. 2014;13:3185–3197. doi: 10.1158/1535-7163.MCT-14-0243. [DOI] [PubMed] [Google Scholar]
- 52.Chu J, Zhu Y, Liu Y, Sun L, Lv X, Wu Y, Hu P, Su F, Gong C, Song E, et al. E2F7 overexpression leads to tamoxifen resistance in breast cancer cells by competing with E2F1 at miR-15a/16 promoter. Oncotarget. 2015;6:31944–31957. doi: 10.18632/oncotarget.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye L, Guo L, He Z, Wang X, Lin C, Zhang X, Wu S, Bao Y, Yang Q, Song L, Lin H. Upregulation of E2F8 promotes cell proliferation and tumorigenicity in breast cancer by modulating G1/S phase transition. Oncotarget. 2016;7:23757–23771. doi: 10.18632/oncotarget.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Oncomine (http://www.oncomine.org), TCGA (https://cancergenome.nih.gov/), and KM-plotter (http://kmplot.com/analysis/) databases repository.